Abstract
Contrast-enhanced digital mammography (CEDM) is a promising imaging modality in breast cancer diagnosis. This study aims to investigate how to optimally develop a computer-aided diagnosis (CAD) scheme of CEDM images to classify breast masses. A CEDM dataset of 111 patients was assembled, which includes 33 benign and 78 malignant cases. Each CEDM includes two types of images namely, low energy (LE) and dual-energy subtracted (DES) images. A CAD scheme was applied to segment mass regions depicting on LE and DES images separately. Optimal segmentation results generated from DES images were also mapped to LE images or vice versa. After computing image features, multilayer perceptron based machine learning classifiers that integrate with a correlation-based feature subset evaluator and leave-one-case-out cross-validation method were built to classify mass regions. When applying CAD to DES and LE images with original segmentation, areas under ROC curves (AUC) were 0.759±0.053 and 0.753±0.047, respectively. After mapping the mass regions optimally segmented on DES images to LE images, AUC significantly increased to 0.848±0.038 (p<0.01). Study demonstrated that DES images eliminated overlapping effect of dense breast tissue, which helps improve mass segmentation accuracy. The study demonstrated that applying a novel approach to optimally map mass region segmented from DES images to LE images enabled CAD to yield significantly improved performance.
Keywords: Breast Cancer Diagnosis, Computer-aided diagnosis (CAD), Contrast-enhanced digital mammography (CEDM), Classification of breast masses, Segmentation of breast mass regions, Performance comparison
I. INTRODUCTION
Full-field digital mammography (FFDM) and dynamic contrast-enhanced breast magnetic resonance imaging (DCE-MRI) are two commonly used imaging modalities in breast cancer detection, diagnosis, and prognosis assessment. FFDM has advantages of high image resolution, improved image contrast, low operation cost, faster imaging scan and widely accessibility. However, as a two-dimensional projection imaging modality, FFDM has relatively lower sensitivity and specificity due to the overlap of dense and heterogeneous fibro-glandular tissues (FGT) over the suspicious lesions. For example, mammography has lower sensitivity among women who are younger19, have dense breasts16, use hormone replacement therapy12, and carry certain breast cancer susceptibility genes11. One study reported that mammography sensitivity reduced from 87.0% in women with almost entirely fatty breasts to 62.9% in women with extremely dense breasts or reduced from 83.3% in older women (over 80 years old) to 68.6% in younger women (less than 50 years old)5. Mammography also has lower specificity as reported that during a 10-year screening period, more than 50% women would receive at least one false-positive recall and 7 to 9% have at least one false-positive biopsy21, which can add anxiety with potentially long-term psychosocial consequences to many women4.
On the other hand, DCE-MRI has superior sensitivity in detecting and diagnosing invasive breast cancer comparing to mammography and other existing breast imaging modalities3. However, DCE-MRI has a number of disadvantages including higher cost and longer imaging scanning time. It may also have relatively lower specificity, which may generate unnecessary breast biopsies and/or over-diagnosis2. As a result, both FFDM and DCE-MRI modalities have advantages and disadvantage when they are used in breast cancer imaging.
In order to take advantages of both FFDM and DCE-MRI modalities, while overcome their disadvantages, an alternative imaging modality namely, contrast-enhanced digital mammography (CEDM), emerges and is quickly gaining momentum in recent clinical trials worldwide. When using CEDM modality, contrast agent is injected into breast and two series of scans are conducted at two different X-ray energy levels. Since malignant lesions are often accompanied by increased blood vessels that have unique permeability as compared to benign or normal tissues, use of contrast agent allows analyzing morphology and vascular enhancement of the suspicious lesions. Additionally, when logarithmic subtraction is performed between two scans taken at different instances after contrast agent injection, difference in permeability is further enhanced and overlapping effect of FGT is removed. In general, CEDM generates both low energy (LE) images (similar to FFDM) and contrast enhanced dual-energy subtraction (DES) images (similar to MRI, but it is ~4 times faster than MRI exam). Therefore, the novelty or unique characteristics of using CEDM is that it can overcome effect of tissue overlapping in FFDM and enable detection of tumor’s neovascularity related functional information similar to MRI, while maintaining high image resolution as FFDM8.
In current breast imaging, accurate classification between malignant and benign lesions is still a major challenge. Studies have shown that performance of breast lesion diagnosis varied due to the intra- and inter-reader variability13, and only approximate one in four biopsies are malignant27. Thus, in order to help improve accuracy in classification between malignant and benign breast lesions, developing computer-aided diagnosis (CAD) schemes aiming to assist radiologists in their decision-making for better assessing risk of lesion malignancy has been attracting extensive research interest in medical imaging field for the last two decades15,20. Although CEDM is an emerging imaging modality, our recent pilot study demonstrated that classification results based on a machine learning classifier that fuses the computed quantitative image features extracted from CEDM images might provide complementary information to radiologists in particular to help reduce false-positive recalls18. Thus, based on the well-developed CAD concept, objective of this study is to investigate a novel approach to develop a fully-automated CAD scheme of CEDM images and assess CAD performance in classifying between the malignant and benign mass-type lesions.
II. MATERIALS AND METHODS
2.1. CEDM Dataset
CEDM images were retrospectively collected from the existing clinical database of Mayo Clinic Arizona, USA. All CEDM imaging examinations were performed using the following imaging acquisition protocol. In brief, the patient with mammography suite is seated to minimize vasovagal episodes and the intravenous line is first flushed with 10mL of saline. Next, an iodinated contrast agent of 1.5 mL/Kg of OMNIPAQUE 350 (GE Healthcare, Princeton, NJ, USA) is injected using a single lumen power injector at a rate of 3 mL/second. Last, the intravenous line is flushed again with an additional 10 mL of saline. If possible, the injected arm is raised above patient’s head to facilitate contrast drainage from the arm, which enables maximum contrast circulation. After 2 minutes of contrast agent injection, breast is compressed and image acquisition starts.
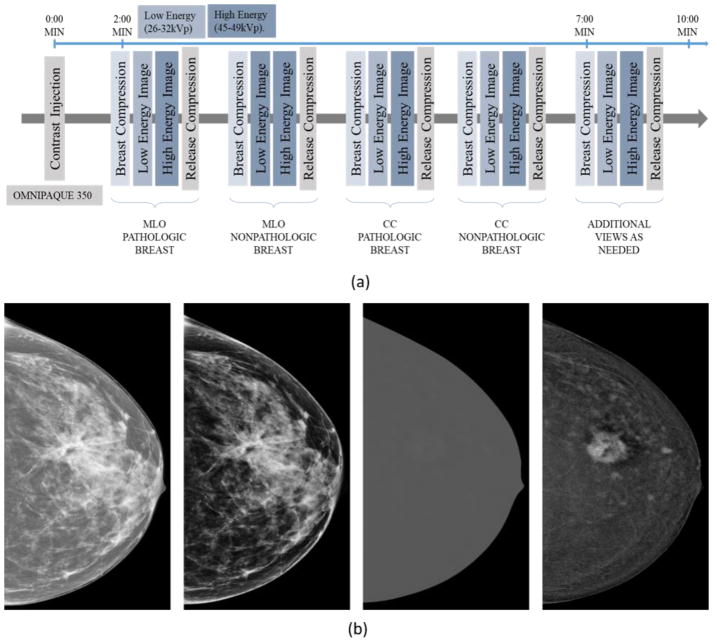
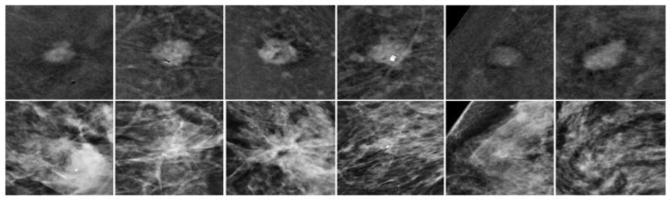
In one CEDM imaging procedure, two sequential images on mediolateral oblique (MLO) and craniocaudal (CC) view are taken at both low and high X-ray energy levels. The low-energy (LE) image is acquired at (26–32kVp), which is less than the K-edge of iodine (33.2keV) to yield higher image contrast of soft tissue and calcifications similar to the regular FFDM. The high-energy (HE) image is acquired at an energy significantly higher than K-edge of iodine at (45–49kVp). Figure 1(a) shows the workflow for the CEDM imaging acquisition with approximate timestamps at each instance (view and energy). Finally, a difference (third) image is obtained by taking subtraction between HE and LE image, which is named as dual-energy subtracted (DES) image as shown in Figure 1(b). DES image is a single contrast medium-enhanced image that improves visual enhancement of neovascularity information in and around the tumors while suppresses the normal breast parenchymal tissues in the background. Figure 2 shows several examples in our dataset where the lesions are almost invisible or undetectable in LE (or regular FFDM) images, but they are clearly visible in DES images with the highly distinguishable lesion boundary contour.
Figure 1.
(a) Illustration of the workflow of a CEDM imaging acquisition procedure and (b) an example of 4 images from left to right: High energy (HE) image, Low energy (LE) image, dual energy subtraction (DES) image displayed at same window and level as HE image, and the DES image displayed at an adjusted window and level for improving visibility, respectively.
Figure 2.
A few samples in which mass-type lesions are clearly visible in DES images (the 1st row), but almost invisible in LE (or regular FFDM) images (the 2nd row).
In summary, we retrieved and assembled a fully anonymous CEDM image test dataset involving 111 women underwent breast cancer diagnosis at Mayo Clinic Arizona. Each case depicts one detected suspicious breast mass. Based on the histopathologic test results of the biopsy samples, 78 masses were confirmed to be malignant and 33 were benign. In this dataset, each mass was considered visible in both CC and MLO views of LE images.
Similar to regular FFDM images, size of the original images acquired from CEDM is either 3328 × 2560 or 4096 × 3328 pixels depending on breast size. Then, based on the standardized approach to develop a CAD scheme for detecting and/or classifying breast masses10, the original images were subsampled using bilinear interpolation method in which output pixel value is a weighted average pixel value from a 5-by-5 neighborhood kernel. The subsampled image size was reduced to corresponding 666 × 512 or 820 × 666 pixels, respectively. Similar image subsampling process has been commonly used in developing previous CAD schemes of FFDM images29.
2.2. Breast Mass Segmentation
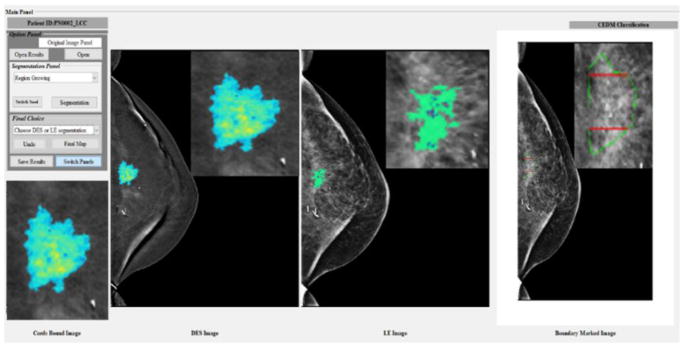
CAD scheme is first applied to automatically segment suspicious mass region. Since CEDM is a diagnostic imaging modality that applies to the recalled patients who have suspicious lesions detected in screening mammograms, locations of each suspicious mass in CC and MLO view images are known and can be easily mapped to CEDM images. Figure 3 shows the graphical user interface (GUI) of our new interactive CAD scheme. After loading an image (either CC or MLO view) of interest, the user can observe and place an initial seed point around the mass center to segment mass region. In this study, all region growth seeds namely, the mass region center pixels, were automatically placed based on the retrieved clinical truth file. In a batched CAD processing, no human intervention is involved. Although a large number of mass segmentation algorithms have been reported in the literature17, we applied and implemented a multi-layer topographic (MLT) region growing algorithm, which has been well-developed and applied in previous CAD schemes9,23.
Figure 3.
Illustration of graphical user interface (GUI) of the CAD scheme.
In brief, the MLT region growing algorithm first applies a conventional region growing process using a pre-selected small threshold to segment lesion central region. Second, the threshold value is adaptively adjusted based on the pixel intensity difference between the initially segmented region and the surrounding region. The next layer of segmentation is performed with the adjusted threshold. Two parameters namely, growth rate (an increase of size) and center shift (the displacement of centroid) between the prior and current region growth layer, are computed. If the current growth layer passes two boundary conditions in which the growth rate is less than 100% (double the size), and the shift of the region center is less than 10 pixels, this current growth layer is accepted to replace its prior growth layer. Third, this region growing process continues to define the new growth layer until it fails to pass one of the above two boundary conditions. Then, the growing iteration ends and the last “prior” growth layer is selected as the final segmentation output. Figure 3 shows examples of the mass segmentation results on both DES and LE images (from the left to right). For a comparison, image with radiologist’s marking on the mass region is also displayed in the first image from the right.
2.3. Feature Computation
After segmentation of each mass region, the second step of CAD is applied to compute image features. In the development phase, CAD initially computes a set of 109 image features, which can be divided into 4 groups as listed in Table 1. The first group includes 4 mass size and shape related image features, which include mass size, the maximum radius or convexity (smoothness) of mass boundary. The second group includes 13 statistical features related to heterogeneity of mass density (pixel values). The third group includes 8 features to detect variation of density (pixel values) between the mass and its surrounding boundary. These features have been defined and used in our previous CAD schemes of different types of medical images (including FFDM images and lung CT images) to represent the segmented lesions7,25.
Table 1.
Summary of 109 features separated by feature class.
| Feature class | Feature number | Feature description |
|---|---|---|
| Shape | 4 (F1–F4) | Mass size, convexity, maximum radius, standard deviation (STD) of all radii. |
| Tumor Density related | 13 (F5–F17) | Mean, STD, energy, entropy, maximum intensity, mean absolute deviation, median, minimum, range, rms, uniformity, skewness, and kurtosis of a segmented mass region. |
| Tumor background Density related | 8 (F18–F25) | gradient mean, gradient STD, ISO-intensity, fluctuation mean, fluctuation STD, mean contrast, contrast, STD ratio of mass to boundary. |
| Wavelet | 84 (F26–F109) | Apply the density features on the four wavelet components |
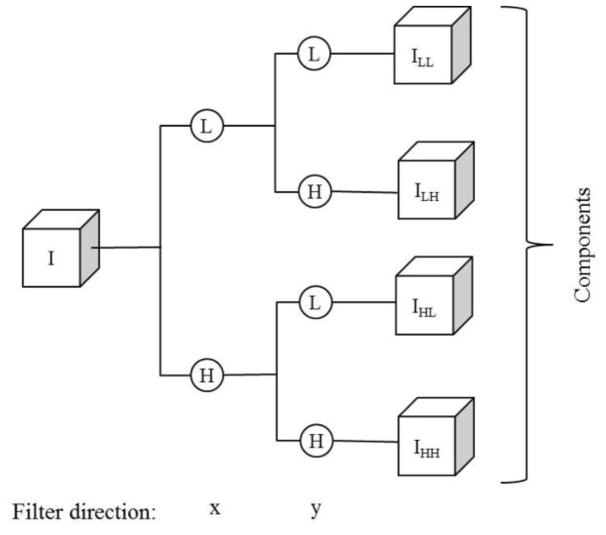
Last, the fourth group includes 84 wavelet transform generated image features. Specifically, a two-dimensional wavelet transform (using a “Coiflet 1” filter) was applied, which decomposes each image into four decompositions. During the decomposition, two-dimensional filters (low pass and high pass) are applied in both x- and y-direction to compute ILL, ILH, IHL, and IHH as represented in Figure 4. For instance, IHL is obtained by applying a high pass filter along the x-direction followed by a low pass filter in the y-direction as described in Equation 1, where L and H indicate low and high pass filters, respectively. NH and NL are the length of filters for high and low pass filter, respectively. In our study both NH and NL have length of 6. All features in the second and third groups are applied individually on each of the four wavelet components to detect density variations in the filtered wavelet decompositions.
Figure 4.
Illustration of the image decomposition using a wavelet transformation (one-level, un-decimated two-dimensional wavelet transforms using "Coiflet 1" filter), where L is a low pass filter and H is a high pass filter.
| (1) |
For non-solid or diffused breast lesions, since there are multiple suspicious masses spread in the images without any connectivity between them, the segmented primary (the largest) mass region is used for computing shape, morphology, and background related features, whereas all the pixels in the diffused suspicious masses are used to compute density related image features, which are independent of its corresponding background information.
In addition, we took two considerations in CAD feature computation. First, each mass is segmented separately from CC and MLO view images. Two segmented mass regions from two view images often do not have the exactly same computed feature values due to the different tissue overlapping in two 2D projection images. Thus, we used average value of two feature values separately computed from CC or MLO view image to represent the final feature value of a mass of interest. Second, due to the possible difference of mass region segmentation results on LE and DES images, GUI of our CAD scheme has a function that allows user to select an optimal segmentation result from either LE or DES image, and then automatically map the selected segmentation result to the matched DES or LE images if necessary in the future clinical applications. Using this mapping method, we are able to compute optimal image features from both LE and DES images.
2.4. Machine Learning Classifier and Performance Assessment
The third step of CAD uses a multi-feature fusion-based machine learning classifier to produce a classification score for each suspicious mass under test, which ranges from 0 to 1. The higher score represents a higher likelihood of the region being malignant. Although many machine learning classifiers have been used in developing CAD schemes, we in this study selected a simple and popular classifier namely, a multilayer perceptron (MLP) based artificial neural network to classify suspicious breast mass. Specifically, we used Weka data mining and machine learning software platform28 to train and test the MLP classifier. In order to build a highly performed and robust machine learning classifier, we needed to address following 3 issues: (1) a relatively small CEDM image dataset of 111 cases, (2) a relatively large pool of initially computed 109 features, and (3) case imbalance in dataset, which includes 29.7% (33/111) of benign masses and 70.3% (78/111) of malignant masses.
To minimize the potentially biased impact of above 3 issues, we adopted following 3 methods. First, we applied a leave-one-case-out (LOCO) cross-validation method to maximize learning power while minimizing the case partition and testing bias14. Second, we used a correlation-based feature subset (CFS) evaluator to reduce dimensionality of feature space by dropping highly correlated, redundant, irrelevant and noisy features, and thus produce a subset of optimal features from the initial feature pool22. Specifically, a CFS evaluator integrating a BestFirst search method was used with a search termination setting of 5, which means if the number of non-improving nodes in the forward search is greater than 5, CFS stops feature selection process. Features selected before the termination were used to build an optimal feature set to train classifier. Third, we applied a Synthetic Minority Oversampling Technique (SMOTE)6 method to generate synthetic data of benign mass regions to produce a more balanced training dataset to minimize the potential classification bias towards majority (malignant) cases. By applying SMOTE to double “benign cases” from 33 to 66, the dataset becomes more balanced with 45.9% (66/144) benign and 54.1% (78/144) malignant cases. The effectiveness of applying similar SMOTE method has been reported in previous studies1,24.
After taking these considerations and protection steps, we built 4 MLP classifiers. The first 2 MLPs used image features computed from the segmented mass regions depicting on either DES or LE images, respectively. Since mass segmentation results on DES and LE images may vary significantly. Using the GUI tool of CAD scheme (as shown in Figure 3), we also mapped the optimal segmentation results from one image to another (i.e., from DES to LE or vice versa). Then, after optimal mapping, CAD recomputed image features from the mapped mass regions depicting on either LE or DES images.
In training and testing each MLP classifier, we embedded both the CFS evaluator and SMOTE algorithm into the LOCO cross-validation process. Thus, in each LOCO training and testing iteration, one mass was removed from the training dataset. SMOTE algorithm was applied to generate synthetic data to double the number of benign cases. A CFS feature selection evaluator was applied to select a set of optimal features. A MLP classifier was trained using the training dataset and selected optimal features. After training process, the classifier was applied to test one independent testing mass, which was not involved in the training process. The LOCO process repeated 111 times. As a result, each mass in our dataset was independently tested and all classification scores were recorded.
Finally, classification performance of each MLP classifier was evaluated using the following two steps. First, a receiver operating characteristic (ROC) method was used. Each ROC curve and the area under ROC curve (AUC) were computed using a maximum likelihood based ROC curve fitting program (ROCKIT, http://www-radiology.uchicago.edu/krl/, University of Chicago). Second, we applied an operating threshold (T=0.5) on the classification scores to divide masses into two classes (or groups) of malignant and benign cases. From the results, we generated a confusion matrix and computed overall classification accuracy, as well as the positive and negative predictive values (PPV and NPV). The evaluation results of 4 MLP classifiers were tabulated and compared.
III. RESULTS
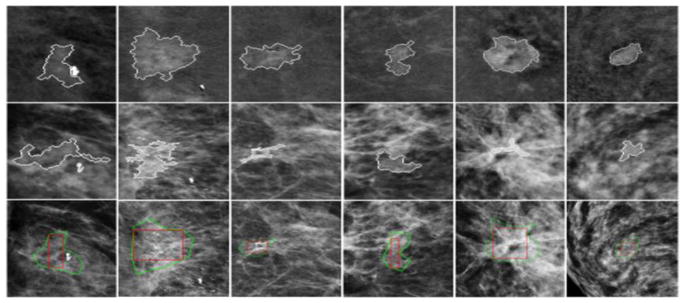
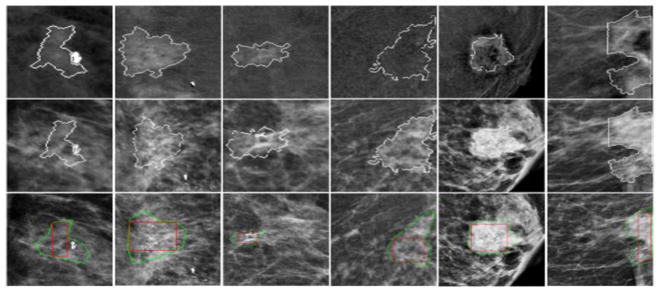
Figure 5 to 7 show examples of comparing the results of applying CAD scheme to segment regions of the same breast masses depicting on both DES (the 1st row) and LE (the 2nd row) images, respectively. Results show that due to the large heterogeneity of breast masses and surrounding parenchymal tissue background, mass segmentation results vary between using LE and DES images as compared to the regions of interest (ROIs) marked by the radiologists (as shown in the third row of Figures 5 to 7). In general, for masses that are partially occulted under the surrounding dense fibro-glandular tissues, it is often difficult for CAD to generate satisfactory segmentation results using LE images due to the mass boundary fuzziness.
Figure 5.
Sample cases illustrating failed segmentation in LE images (2nd row) as compared to DES images (1st row). The 3rd-row shows the lesion bounding boxes placed by the radiologists.
Figure 7.
Sample cases showing optimal segmentation mapping on both DES (1st row) and LE (2nd row) images. The 3rd-row shows the lesion bounding boxes placed by the radiologists.
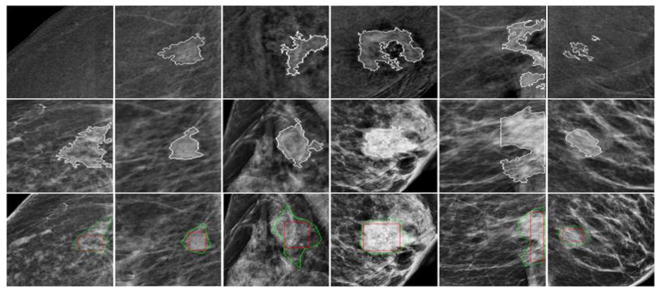
For illustration purpose, Figure 5 shows 6 examples in which segmentation failed in LE images (the middle row) as compared to the better segmentation results yielded using DES images (the top row). On the other hand, some masses may be invisible or only partially visible on DES images due to the lack of enhancement or large necrosis. In these cases, CAD segmentation results on LE images may more accurately represent real mass regions (see Figure 6). Figure 7 shows examples of the mapped “optimal” segmentation results on both LE and DES images. The 3rd row of Figures 5 to 7 also shows the lesion bounding boxes placed by radiologists. By comparing with CAD-generated segmentation results (as shown in the 1st and 2nd rows of these figures), we can observe that CAD-segmented lesion boundary are often more accurate than the results of manually drawing.
Figure 6.
Sample cases illustrating failed segmentation in DES images (1st row) as compared to LE images (2nd row). The 3rd-row shows the lesion bounding boxes placed by the radiologists.
Table 2 lists the highly performed image features, which were selected more than 90% of LOCO training and testing iterations. From the Table, several interesting observations can be made. For example, (1) although lesion shape or boundary margin features (i.e., F1 to F4 as shown in Table 1) are commonly considered as the most important image features in many of previous CAD schemes, this type of features were largely removed or not selected by the classifiers trained using LE images, which indicates that the lesion boundary features can only play important role when the lesions are more accurately segmented. (2) The density heterogeneity features computed from both inside a lesion and its surrounding background can contribute to the CAD scheme to classify between malignant and benign lesions. (3) Extracting optimal density heterogeneity features can also expand to the filtered images (i.e., using wavelet transform as done in this study). From the filtered images, CAD can detect and select optimal features to build the machine leaning classifiers.
Table 2.
List of features selected in ≥90% of LOCO training and testing of 4 MLP classifiers.
| Original segmentation | Optimally mapped segmentation | ||||||
|---|---|---|---|---|---|---|---|
| DES images | LE images | DES images | LE images | ||||
| Feature | % | Feature | % | Feature | % | Feature | % |
| F1 | 97 | F7 | 99 | F2 | 100 | F5 | 100 |
| F2 | 99 | F10 | 99 | F5 | 98 | F6 | 100 |
| F3 | 100 | F11 | 98 | F8 | 95 | F10 | 100 |
| F5 | 100 | F16 | 100 | F12 | 100 | F16 | 100 |
| F20 | 100 | F21 | 93 | F20 | 100 | F33 | 100 |
| F41 | 99 | F24 | 100 | F35 | 100 | F41 | 100 |
| F48 | 95 | F39 | 100 | F41 | 100 | F43 | 97 |
| F56 | 91 | F44 | 100 | F50 | 97 | F44 | 100 |
| F65 | 100 | F45 | 100 | F62 | 100 | F45 | 100 |
| F66 | 100 | F65 | 100 | F77 | 99 | F46 | 99 |
| F83 | 100 | F80 | 100 | F81 | 97 | F65 | 92 |
| F88 | 100 | F81 | 94 | F82 | 99 | F75 | 100 |
| F101 | 100 | F83 | 99 | F88 | 100 | ||
| F88 | 100 | F103 | 100 | ||||
| F96 | 100 | ||||||
| F109 | 100 | ||||||
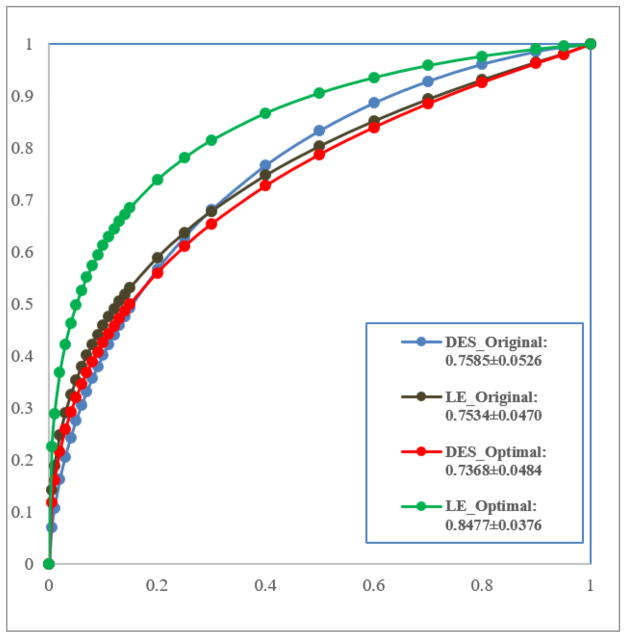
Figure 8 shows 4 ROC curves that are generated using 4 sets of CAD classification scores computed by 4 MLP classifiers. Since in this dataset, 7 masses were not enhanced in CEDM images (i.e., one mass region as shown in the first ROI of the top row of Figure 6) and thus they cannot be segmented, the first MLP classifier was trained and tested using the remaining 104 cases (27 benign vs. 77 malignant masses). Other 3 MLP classifiers were trained and tested using all 111 masses. The computed AUC values are 0.759±0.053 and 0.753±0.047 for the first two MLP classifiers trained and tested using mass regions originally segmented from DES and LE images, respectively. By mapping the optimal segmentation results from LE images to DES images, AUC=0.739±0.048, which did not show classification performance improvement. However, when mapping the optimal segmentation results from DES images to LE images, AUC value of using the new MLP classifier significantly increases to 0.848±0.038 as compared to all other 3 MLP classifiers (p<0.01).
Figure 8.
Comparison of four ROC curves generated using 4 MLP classifiers using the original and optimally mapped mass segmentation results on DES and LE images to distinguish between malignant and benign breast masses.
Two confusion matrices in Table 3 show distribution of the classification scores computed by two MLP classifiers trained using the originally segmented mass regions, while two confusion matrices in Table 4 show distribution of the classification scores computed by two MLP classifiers trained using the optimally mapped mass regions depicting on DES and LE images, respectively. Then, from these 4 confusion matrices, the overall classification accuracy, positive predictive values (PPV) and negative predictive values (NPV) of 4 MLP classifiers were computed and compared as shown in Table 5. Results indicated that using the fourth MLP classifier trained and tested using LE images after mapping the optimal mass region segmentation results from DES images to LE images yielded the highest classification accuracy including both the highest PPV and NPV values. For example, when comparing to the second MLP classifier trained and tested using the originally segmented mass regions depicting on LE images, the overall classification accuracy of the fourth MLP increased 8.7% (from 72.1% to 78.4%).
Table 3.
Two confusion matrices generated when applying MLP classifier to the originally segmented breast mass regions depicting on LE and DES images.
| Actual | LE Images (Total: 111 masses) | DES Images (Total: 104 masses) | ||
|---|---|---|---|---|
|
| ||||
| Prediction | Malignant | Benign | Malignant | Benign |
|
| ||||
| Malignant | 56 (0.5) | 9 (0.08) | 53 (0.51) | 9 (0.09) |
|
|
||||
| Benign | 22 (0.2) | 24 (0.22) | 24 (0.23) | 18 (0.17) |
Table 4.
Two confusion matrices generated when applying MLP classifier to the optimally mapped segmented breast mass regions depicting on LE and DES images.
| Actual | LE Images (Total: 111 masses) | DES Images (Total: 111 masses) | ||
|---|---|---|---|---|
|
| ||||
| Prediction | Malignant | Benign | Malignant | Benign |
|
| ||||
| Malignant | 63 (0.57) | 9 (0.08) | 55 (0.49) | 12 (0.11) |
|
|
||||
| Benign | 15 (0.13) | 24 (0.22) | 23 (0.21) | 21 (0.19) |
Table 5.
Summarization and comparison of classification performance using 4 MLP classifiers.
| MLP Classifier | Original segmentation | Optimally mapped segmentation | ||
|---|---|---|---|---|
| DES images | LE images | DES images | LE images | |
| Overall accuracy | 68.3% | 72.1% | 68.5% | 78.4% |
| AUC±STD | 0.769±0.053 | 0.753±0.047 | 0.737±0.048 | 0.848±0.038 |
| PPV | 85.5% | 86.2% | 82.1% | 87.5% |
| NPV | 42.9% | 52.2% | 47.7% | 61.5% |
IV. DISCUSSION
In this study, we proposed and tested several novel approaches aiming to optimally develop a fully-automated CAD scheme of CEDM images to classify between malignant and benign breast masses. The novelty (or difference) of this study as comparing to the previous CAD schemes of FFDM images include to (1) optimally map the segmentation results between the LE and DES images, (2) compute and add more lesion density heterogeneity features to the machine learning classifier, (3) develop a case-based scheme using the average image features computed from both CC and MLO views, and (4) implement an interactive visual aid tool for CAD scheme of CEDM images. Thus, the study has following unique characteristics and/or observations.
First, in breast cancer imaging, accurate classification between malignant and benign breast lesions remains a challenging task to date. Although CAD schemes of FFDM and breast MRI images have been developed aiming to assist radiologists in classifying between malignant and benign breast lesions in previous studies, these CAD schemes have not been accepted and used in the clinical practice. One of the primary difficulties is the lack of capability of accurately segmenting breast lesions depicting on images, in particular, using FFDM images due to the fuzzy lesion boundary caused by tissue overlapping. Segmentation of breast lesion is not only difficult for CAD, but also for radiologists, which generates large intra- and inter-reader variability. Thus, inaccurate lesion segmentation reduces accuracy and robustness of the computed image features used to develop machine learning classifiers. In CEDM imaging modality, DES images enable to enhance breast lesion regions, while removing or suppressing normal parenchymal tissues that overlap or surround the lesions. Thus, segmentation of lesion regions from DES images becomes much more accurate and robust. This is a unique contribution of including DES images in CAD schemes. This study demonstrated that by mapping the optimal lesion segmentation results on DES images to LE images, CAD scheme yielded significantly higher performance in mass classification than using the CAD scheme applying to the originally segmented mass regions depicting on LE images.
Second, although using DES images enhances lesion boundary and makes lesion segmentation easier and more accurate than using LE images, it also has potential disadvantages in developing CAD schemes. For example, we observed that after contrast enhancement, lesions depicting on DES images become more homogeneous, which lose much density heterogeneity information of the lesions depicting on LE images. Thus, when using density heterogeneity and texture related image features computed from the segmented lesions to train and develop machine learning classifiers, CAD classification performance using DES images does not yield significantly higher performance than using LE images. It seems that the advantage of more accurate lesion segmentation using DES images is partially cancelled out by its disadvantage of losing density or texture heterogeneity information. As a result, if we want to improve CAD classification results using the lesion regions segmented from DES images, different strategy or image features need to be explored and used in future studies.
Third, unlike the most of previous CAD schemes of FFDM images, which are region-based schemes to independently classify two suspicious mass regions based on the image features computed from one (i.e., either CC or MLO) view image26, we in this study developed and tested a unique case-based scheme that computes average image features extracted from two corresponding mass regions detected on CC and MLO view images and fuse the average image features to develop or train the machine learning classifier. In order to demonstrate the advantages of this new fusion approach, we also did a comparison experiment. The comparison results shown in Table 6 demonstrate that CAD schemes developed using the averaging features yield the higher performance, which also indicates that using this new case-based CAD approach enables to reduce the impact of image feature difference due to the variation of tissue overlap in the 2D projected CC and MLO view images.
Table 6.
Comparison of CAD performance between the new averaging (CC, MLO) method applied in this study and the conventional independent CC and MLO method.
| Method | Original segmentation | Optimally mapped segmentation | ||||||
|---|---|---|---|---|---|---|---|---|
| DES images | LE images | DES images | LE images | |||||
| AUC | Accuracy | AUC | Accuracy | AUC | Accuracy | AUC | Accuracy | |
| Average (CC, ML) | 0.76±0.05 | 68.3% | 0.75±0.05 | 72.1% | 0.74±0.05 | 68.5% | 0.85±0.04 | 78.4% |
| Independent (CC, ML) | 0.73±0.04 | 63.28% | 0.81±0.03 | 74.77% | 0.72±0.04 | 63.96% | 0.78±0.03 | 72.07% |
Fourth, CAD performance depends on the difficult and diverse levels of testing datasets. For example, our previous study reviewed 8 published CAD studies conducted by different research groups in classifying breast mass-type lesions, which reported AUC values ranging from 0.70 to 0.87 due to use of different datasets26. Thus, although it is not feasible to directly compare lesion classification performance between our new CAD scheme of CEDM images and other previously developed CAD schemes of FFDM images, we conducted a specific comparative analysis. In brief, we compared performance of two CAD schemes applying to LE images only and the complete set of CEDM images, respectively. In order to avoid or minimize the bias in comparison, two CAD schemes used the same lesion segmentation algorithm, image feature computation and selection method, and machine learning classifier training and testing approach. Comparative results showed that CAD scheme of CEDM images yielded the significantly higher performance (AUC=0.848±0.038) than CAD scheme of LE images with AUC=0.753±0.047 based on the same study cases (as shown in Table 5), which supports advantages of developing CAD schemes of CEDM images.
Fifth, this study took three measures namely, (1) a leave-one-case-out (LOCO) cross-validation method, (2) a correlation-based feature subset (CFS) evaluator based feature selection method and (3) a synthetic minority oversampling technique (SMOTE) method, to overcome limitation of a relatively small and unbalanced dataset with 111 cases (33 benign vs. 78 malignant cases). Both CFS and SMOTE were embedded into LOCO cross-validation. In order to support advantage of this embedded approach, we also tested CAD performance by removing SMOTE and CFS. Table 7 shows the performance changes and we observed that (1) when SMOTE was not applied to balance the dataset (33 benign, 78 malignant), the performance reduced as comparing to the embedded method used in this study, and (2) when the CFS feature selection step was also removed, the performance further decreased.
Table 7.
Comparison of classification performance changes between three methods.
| Method | Original segmentation | Optimally mapped segmentation | ||||||
|---|---|---|---|---|---|---|---|---|
| DES images | LE images | DES images | LE images | |||||
| AUC | Accuracy | AUC | Accuracy | AUC | Accuracy | AUC | Accuracy | |
| Proposed MLP | 0.76±0.05 | 68.3% | 0.75±0.05 | 72.1% | 0.74±0.05 | 68.5% | 0.85±0.04 | 78.4% |
| Remove SMOTE | 0.63±0.06 | 70.19% | 0.70±0.05 | 68.46% | 0.75±0.05 | 62.16% | 0.69±0.05 | 65.76% |
| Remove CFS and SMOTE | 0.56±0.07 | 65.38% | 0.71±0.05 | 66.67% | 0.61±0.06 | 64.86% | 0.59±0.06 | 63.06% |
Sixth, besides a MLP classifier, we also applied the same CFS evaluator and SMOTE algorithm embedded with LOCO training and testing iteration method to build several other popular machine learning classifiers including logistic regression (LR), Bayesian belief network (BNN), k-nearest neighbor (KNN), RandomForest (RF) and RandomCommittee (RC) algorithms, which are available in Weka data mining software platform28, to classify between malignant and benign masses using DES and LE images. Although performance levels of different classifiers vary (i.e., ranging from AUC=0.735±0.047 for logistic regression to AUC=0.895±0.030 for BNN when using LE images after mapping the optimal lesion segmentation results from DES images), the performance change trend in each classifier maintains consistent. This supports the results produced using the MLP classifier as reported in the Results section of this paper. The additional testing results using different machine learning classifiers clearly indicate when using the original lesion segmentation, classification performance levels on DES and LE images are quite comparable. However, when mapping the optimal lesion segmentation results generated on DES images to LE images, all classifiers using different machine learning models yielded the highest classification performance.
Last, this study also has a number of limitations. For example, the size of dataset remains small. Thus, the performance and robustness of our CAD scheme of CEDM images need to be further optimized and validated using new large and diverse image dataset in the future studies. In addition, we used a well-developed CAD pipeline with new lesion segmentation mapping methods and the computed image features mainly focusing on density heterogeneity of lesion and its surrounding background. Thus, more studies in developing new CAD approaches also need in future studies.
In summary, we investigated and tested a new approach to develop the first fully-automated CAD scheme of breast lesion classification using CEDM images. Study results demonstrated that LE and DES images generated from CEDM contain complementarily valuable information. Using DES images helps more accurately segment suspicious lesions if the lesions are enhanced. Then, by mapping the optimal lesion segmentation results (lesion boundary contour) from DES images onto LE images, the density heterogeneity and texture based image features can be more accurately computed from LE images. Thus, the lesion classification performance of using this new CAD scheme that combines these two types of images can be significantly improved. As a result, new knowledge learned from this proof-of-concept study helps establish a new foundation for us and/or other researchers in CAD related medical imaging informatics field to continue develop and optimize novel CAD schemes of CEDM images with improved performance in future studies.
Acknowledgments
This work is supported in part by Grant R01 CA197150 from the National Cancer Institute, National Institutes of Health, USA.
VI. REFERENCES
- 1.Aghaei F, Tan M, Hollingsworth AB, Zheng B. Applying a new quantitative global breast MRI feature analysis scheme to assess tumor response to chemotherapy. J Magn Reson Imaging. 2016;44:1099–1106. doi: 10.1002/jmri.25276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baltzer PAT, Benndorf M, Dietzel M, Gajda M, Runnebaum IB, Kaiser WA. False-positive findings at contrast-enhanced breast MRI: A BI-RADS descriptor study. Am J Roentgenol. 2010;194:1658–1663. doi: 10.2214/AJR.09.3486. [DOI] [PubMed] [Google Scholar]
- 3.Berg WA, Zhang Z, Lehrer D, Jong RA. Detection of Breast Cancer With Addition of Annual Screening Ultrasound or a Single Screening MRI to Mammography in Women With Elevated Breast Cancer Risk. J Am Med Assoc. 2012;307:1394–1404. doi: 10.1001/jama.2012.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodersen J, Siersma V. Long-Term Psychosocial Consequences ofFalse-Positive Mammography Screening. Ann Fam Med. 2013;11:106–115. doi: 10.1370/afm.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carney PA, Miglioretti DL, Yankaskas BC, Kerlikowske K, Rosenberg R, Rutter CM, Geller BM, Abraham LA, Taplin SH, Dignan MM, Gary Cutter R. Individual and combined effects of age, breast density, and hormone replacement therapy use on the accuracy of screening mammography. Ann Intern Med. 2003;138:168–175. doi: 10.7326/0003-4819-138-3-200302040-00008. [DOI] [PubMed] [Google Scholar]
- 6.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: Synthetic minority over-sampling technique. J Artif Intell Res. 2002;16:321–357. [Google Scholar]
- 7.Danala G, Thai T, Gunderson CC, Moxley KM, Moore K, Mannel RS, Liu H, Zheng B, Qiu Y. Applying Quantitative CT Image Feature Analysis to Predict Response of Ovarian Cancer Patients to Chemotherapy. Acad Radiol. 2017;24:1233–1239. doi: 10.1016/j.acra.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dromain C, Balleyguier C, Muller S, Mathieu MC, Rochard F, Opolon P, Sigal R. Evaluation of Tumor Angiogenesis of Breast Carcinoma Using Contrast-Enhanced Digital Mammography. Am J Roentgenol. 2006;187:W528–W537. doi: 10.2214/AJR.05.1944. [DOI] [PubMed] [Google Scholar]
- 9.Eltonsy NH, Tourassi GD, Elmaghraby AS, Member S. A Concentric Morphology Model for the Detection of Masses in Mammography. IEEE Trans Med Imaging. 2007;26:880–889. doi: 10.1109/TMI.2007.895460. [DOI] [PubMed] [Google Scholar]
- 10.Gur D, Stalder J, Hardesty La. CAD performance on sequentially ascertained mammographic examinations of masses: an assessment. Radiology. 2004;233:418–423. doi: 10.1148/radiol.2332040277. [DOI] [PubMed] [Google Scholar]
- 11.Kriege M, Brekelmans CTM, Boetes C, Besnard PE. Efficacy of MRI and Mammography for Breast-Cancer Screening in Women with a Familial or Genetic Predisposition. N Engl J Med. 2004;351:427–437. doi: 10.1056/NEJMoa031759. [DOI] [PubMed] [Google Scholar]
- 12.Laya MB, Larson EB, Taplin SH, White E. Effect of estrogen replacement therapy on the specificity and sensitivity of screening mammography. J Natl Cancer Inst. 1996;88:643–649. doi: 10.1093/jnci/88.10.643. [DOI] [PubMed] [Google Scholar]
- 13.Lee AY, Wisner DJ, Aminololama-Shakeri S, Arasu VA, Feig SA, Hargreaves J, Ojeda-Fournier H, Bassett LW, Wells CJ, De Guzman J, Flowers CI, Campbell JE, Elson SL, Retallack H, Joe BN. Inter-reader Variability in the Use of BI-RADS Descriptors for Suspicious Findings on Diagnostic Mammography: A Multi-institution Study of 10 Academic Radiologists. Acad Radiol. 2017;24:60–66. doi: 10.1016/j.acra.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 14.Li Q, Doi K. Reduction of bias and variance for evaluation of computer-aided diagnostic schemes. Med Phys. 2006;33:868–875. doi: 10.1118/1.2179750. [DOI] [PubMed] [Google Scholar]
- 15.Lu W, Li Z, Chu J. A novel computer-aided diagnosis system for breast MRI based on feature selection and ensemble learning. Comput Biol Med. 2017;83:157–165. doi: 10.1016/j.compbiomed.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 16.Mandelson TM. Breast density as a predictor of breast cancer risk. J Natl Cancer Inst. 2000;12:1081–1087. doi: 10.1093/jnci/92.13.1081. [DOI] [PubMed] [Google Scholar]
- 17.Oliver A, Freixenet J, Martí J, Pérez E, Pont J, Denton ERE, Zwiggelaar R. A review of automatic mass detection and segmentation in mammographic images. Med Image Anal. 2010;14:87–110. doi: 10.1016/j.media.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 18.Patel BK, Ranjbar S, Wu T, Pockaj BA, Li J, Zhang N, Lobbes M, Zhang B, Mitchell JR. Computer-aided diagnosis of contrast-enhanced spectral mammography : A feasibility study. Eur J Radiol. 2018;98:207–213. doi: 10.1016/j.ejrad.2017.11.024. [DOI] [PubMed] [Google Scholar]
- 19.Peer PGM, Verbeek ALM, Straatman H, Hendriks JHCL, Holland R. Age-specific sensitivities of mammographic screening for breast cancer. Breast Cancer Res. 1996:153–160. doi: 10.1007/BF01806669. [DOI] [PubMed] [Google Scholar]
- 20.Qiu Y, Yan S, Gundreddy RR, Wang Y, Cheng S, Liu H, Zheng B. A new approach to develop computer-aided diagnosis scheme of breast mass classification using deep learning technology. J Xray Sci Technol. 2017;25:751–763. doi: 10.3233/XST-16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rebecca AH, Kerlikowske K, Flowers CI, Yankaskas BC, Weiwei Z, Miglioretti DL. Cumulative probability of false-positive recall or biopsy recommendation after 10 years of screening mammography. Ann Intern Med. 2011;155:481–492. doi: 10.1059/0003-4819-155-8-201110180-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saeys Y, Inza I, Larranaga P. A review of feature selection techniques in bioinformatics. Bioinformatics. 2007;23:2507–2517. doi: 10.1093/bioinformatics/btm344. [DOI] [PubMed] [Google Scholar]
- 23.Tan M, Aghaei F, Wang Y, Zheng B. Developing a new case based computer-aided detection scheme and an adaptive cueing method to improve performance in detecting mammographic lesions. Phys Med Biol. 2017;62:358–376. doi: 10.1088/1361-6560/aa5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan M, Zheng B, Leader JK, Gur D. Association Between Changes in Mammographic Image Features and Risk for Near-Term Breast Cancer Development. IEEE Trans Med Imaging. 2016;35:1719–1728. doi: 10.1109/TMI.2016.2527619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Lederman D, Tan J, Wang XH, Zheng B. Computerized Prediction of Risk for Developing Breast Cancer Based on Bilateral Mammographic Breast Tissue Asymmetry. Med Eng Phys. 2011;27:934–942. doi: 10.1016/j.medengphy.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, Aghaei F, Zarafshani A, Qiu Y, Qian W, Zheng B. Computer-aided classification of mammographic masses using visually sensitive image features. J Xray Sci Technol. 2017;25:171–186. doi: 10.3233/XST-16212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weaver DL, Rosenberg RD, Barlow WE, Ph D, Ichikawa L, Carney PA, Ph D, Kerlikowske K, Buist DSM, Ph D, Geller BM, Ed D, Key CR, Ballard-barbash R. Pathologic Findings from the Breast Cancer Surveillance Consortium Population-Based Outcomes in Women Undergoing Biopsy after Screening Mammography. Cancer. 2006;106:732–742. doi: 10.1002/cncr.21652. [DOI] [PubMed] [Google Scholar]
- 28.Witten IH, Frank E, Hall MA. Data mining: practical machine learning tools and techniques. 3. Elsevier; 2011. [Google Scholar]
- 29.Zheng B, Sumkin JH, Zuley ML, Lederman D, Wang X, Gur D. Computer-aided detection of breast masses depicted on full-field digital mammograms: a performance assessment. Br J Radiol. 2012;85:e153–61. doi: 10.1259/bjr/51461617. [DOI] [PMC free article] [PubMed] [Google Scholar]