Abstract
MicroRNAs are found throughout the genome and are processed by the microprocessor complex (MPC) from longer precursors. Some precursor miRNAs overlap intron:exon junctions. These splice site overlapping microRNAs (SO-miRNAs) are mostly located in coding genes. It has been intimated, in the rarer examples of SO-miRNAs in noncoding RNAs, that the competition between the spliceosome and the MPC modulates alternative splicing. However, the effect of this overlap on coding transcripts is unknown. Unexpectedly, we show that neither Drosha silencing nor SF3b1 silencing changed the inclusion ratio of SO-miRNA exons. Two SO-miRNAs, located in genes that code for basal membrane proteins, are known to inhibit proliferation in primary keratinocytes. These SO-miRNAs were up-regulated during differentiation and the host mRNAs were down-regulated, but again there was no change in inclusion ratio of the SO-miRNA exons. Interestingly, Drosha silencing increased nascent RNA density, on chromatin, downstream from SO-miRNA exons. Overall our data suggest a novel mechanism for regulating gene expression in which MPC-dependent cleavage of SO-miRNA exons could cause premature transcriptional termination of coding genes rather than affecting alternative splicing.
Keywords: splice site overlapping miRNA; microprocessor complex; splicing, keratinocytes; small RNA-seq
INTRODUCTION
The nuclear microprocessor complex (MPC) is a heterotrimeric complex that includes one Drosha protein and two DGCR8 proteins (Nguyen et al. 2015). It recognizes and cleaves primary microRNA transcripts (pri-miRNAs), which are stem–loop structures of several kilobases in length transcribed by RNA Pol II that are usually capped and polyadenylated (Cai et al. 2004; Lee et al. 2004). Cleavage by the MPC generates ∼70 nt precursor miRNAs (pre-miRNAs), which are exported to the cytoplasm (Yi et al. 2003) and cleaved by Dicer into 20–25 nt double-stranded mature miRNAs. The guide miRNA strands are then incorporated into the RNA-induced silencing complex (RISC), which contains the Argonaute 2 (AGO2) protein (Gregory et al. 2005). As part of RISC complex, miRNA bases pair to target mRNAs and AGO2 functions as the “slicer” enzyme inducing degradation or translational repression of target mRNAs (Song et al. 2004). Human miRNA genes can be found in intergenic regions as independent transcriptional units. However they can also be transcribed as part of coding or noncoding genes host genes. Processing of these hosted miRNAs may affect the splicing process (Morlando et al. 2008; Macias et al. 2012).
RNA splicing is the mRNA maturation reaction that takes place in the nucleus in which introns are removed from precursor mRNA (pre-mRNA) and the exons are ligated together. Splicing is catalyzed in two consecutive reactions by spliceosome (Moore and Sharp 1993), a highly dynamic macromolecular complex composed of five different small nuclear ribonucleoproteins (snRNPs) and many additional proteins. Recognition of the exons is guaranteed by a combinatorial regulatory mechanism that involved consensus sequences at the 5′ and 3′ splice sites, branch point sequence and polypyrimidine tract and the presence of additional cis-acting elements including exonic and intronic splicing enhancers or silencers bounded by trans-acting factors that either stimulate or repress the assembly of the spliceosome (Smith and Valcarcel 2000).
Most of pri-miRNA hairpins are located in introns of coding transcripts and even if there are cases in which their cleavage can inhibit (Kim and Kim 2007) or improve (Morlando et al. 2008; Janas et al. 2011) splicing, there is a general consensus that their cotranscriptional cleavage, mediated by the MPC, does not affect splicing nor cause a transcriptional termination (Baskerville and Bartel 2005; Kim and Kim 2007). As a result, intronic miRNAs and their hosting mRNAs are coexpressed from the same nascent transcript with no significant interference. A different situation occurs for splice site overlapping miRNAs (SO-miRNAs), a peculiar class of miRNAs whose hairpins overlap with the splice sites (Mattioli et al. 2014). Most human SO-miRNAs are embedded in protein coding transcripts (46 out of 52 SO-miRNAs) and their position on the nascent mRNA implies a competition between the spliceosome and the MPC. As a result, the nascent mRNA is processed to generate either the miRNA or the spliced transcript (Mattioli et al. 2013; Melamed et al. 2013) and changes in splicing efficiency can affect miRNA biosynthesis and, conversely, MPC-dependent cleavage can affect splicing (Mattioli et al. 2013; Melamed et al. 2013). However, this analysis was limited to three human SO-miRNAs (hsa-miR-34b [Mattioli et al. 2013], hsa-miR-202, hsa-miR-365-2 [miR-365b new name]) and to the murine mmu-miR-412 (Melamed et al. 2013), all embedded in noncoding genes, a minor class of SO-miRNAs (six out of 52 in humans). Hsa-miR-34b is also located on the last exon of a noncoding gene and cannot result in the generation of exon skipping (ES) isoforms (Mattioli et al. 2013). For hsa-miR-34b and mmu-miR-412 there was an inverse correlation between the splicing efficiency of corresponding SO-miRNA exons and miRNA biosynthesis in different tissues (Mattioli et al. 2013) or during development (Melamed et al. 2013), respectively. Furthermore, most experimental evidence was obtained using minigene assays (Mattioli et al. 2013; Melamed et al. 2013) that do not take into account the complexity of gene regulation in chromatin. A clear inverse correlation between the miRNA biosynthesis and selection of an alternative exon during development was only seen in, the unconserved and noncoding murine SO-miRNA, mmu-miR-412 (Melamed et al. 2013).
Cleavage of pri-miRNA hairpins can also affect processing of hosting mRNAs (Han et al. 2009; Triboulet et al. 2009; Sundaram et al. 2013; Dhir et al. 2015). For example, during keratinocyte migration Drosha-dependent processing of miR-198, located in the last exon of follistatin-like 1 (FSTL1) gene, inhibits the production of its mRNA. This results in the production of two alternative products (the miRNA or the coding mRNA) from the same nascent transcript (Sundaram et al. 2013). Furthermore, in long noncoding transcripts, the MPC can mediate transcriptional termination. Indeed, the liver-specific miR-122 hosting transcript, along with other noncoding mRNAs, do not use the canonical cleavage and polyadenylation pathways but instead the cleavage that terminates transcription is mediated by the MPC (Dhir et al. 2015). A genome wide analysis of chromatin RNA-seq data showed that Drosha depletion resulted in transcriptional readthrough downstream from these miRNA hairpins (Dhir et al. 2015). Drosha-mediated cleavage occurs cotranscriptionally and it further recruits exonucleases to the cleavage site (Morlando et al. 2008; Ballarino et al. 2009; Wagschal et al. 2012). Drosha can also have an RNA-independent function in the regulation of the gene expression when it binds to promoter-proximal regions (Gromak et al. 2013).
In this study, we used various cellular models to evaluate the crosstalk between splicing and the MPC on SO-miRNA biosynthesis and pre-mRNA processing. Neither silencing of spliceosomal or MPC components, nor a specific switch between miRNA and mRNA expression during keratinocyte differentiation, have an effect on alternative splicing patterns at SO-miRNA exons. Analysis of published genome wide scale chromatin RNA-seq data and validation showed that Drosha depletion resulted in transcriptional readthrough downstream from SO-miRNAs implying that MPC-dependent cleavage at SO-miRNA hairpins might terminate transcription inside genes rather than affect splicing.
RESULTS
MPC inhibition does not affect alternative splicing of exons with SO-miRNAs
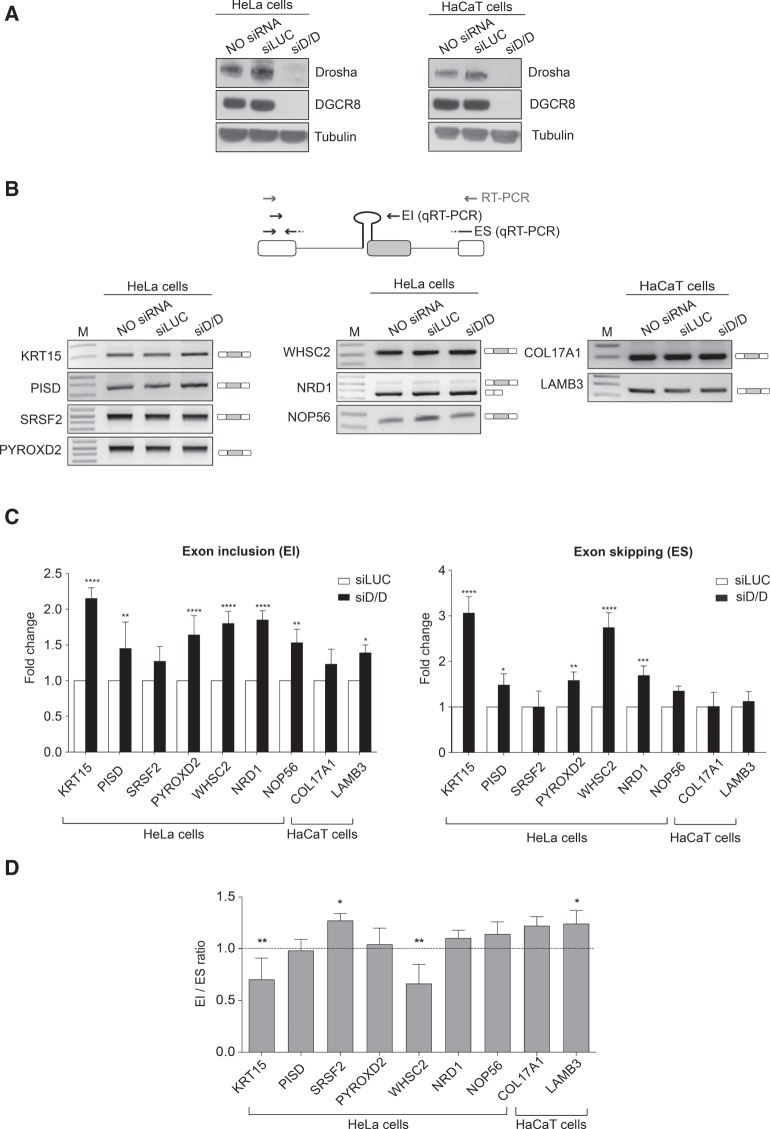
Bioinformatic analysis and manual annotation identify 52 SO-miRNAs in the human genome (Table 1). Most of them (n = 46) are located within protein-coding transcripts (44 in well characterized genes and a further two have an open reading frame) and, among them, 41 are in nonterminal exons. Six SO-miRNAs are in noncoding transcripts and three of them overlap with the last exon. To better understand the relationship between the MPC and pre-mRNA splicing we evaluated the processing of coding SO-miRNA transcripts after Drosha and DGCR8 silencing. Some SO-miRNA transcripts have a cell type-specific expression. Therefore, we used both HeLa and keratinocyte-derived HaCaT cells. Western blot analysis showed significant Drosha and DGCR8 silencing for both cells (Fig. 1A). We used endpoint RT-PCR to evaluate the splicing pattern and in all but one case the SO-miRNA exons were highly included in the final transcripts: The exon inclusion (EI) isoform was present in KRT15, PISD, SRSF2, PYROXD2, WHSC2, NOP56, COL17A1, and LAMB3, carrying the SO-miRNAs miR-6510, miR-7109, miR-636, miR-1287, miR-943, miR-1292, miR-936, and miR-4260, respectively (Fig. 1B). The NRD1 transcript with the embedded SO-miR-761 was the only exception to this. 5′ SO-miR-761 is located at the boundary between intron 3 and exon 4 of NRD1, and the major product had combined exon 3 and exon 4 skipping (Fig. 1B). To detect the absolute levels of alternative splicing isoforms that might eventually originate from MPC-cleaved transcripts we set up a quantitative PCR assay with specific primers to detect the abundance of EI and ES isoforms and the relative splicing ratio. Drosha and DGCR8 depletion increased both the EI and ES isoforms in most of the genes analyzed (Fig. 1C). The general decline in miRNAs biosynthesis after Drosha/DGCR8 silencing likely caused the increase in both isoforms, which might be due to a direct or indirect effect of the miRNAs on the mature transcripts. However, most of the genes did not show a change in the splicing ratio (Fig. 1D). There was just a small reduction of splicing ratio for KRT15 and WHSC2 transcripts and an equally small increase in ratio for SRSF2 and LAMB3 transcripts. These differences might be due to a differential effect exerted by the concomitant miRNAs depletion on the alternative mature products. To exclude the possibility that the SO-miRNA skipped exons could be degraded by nonsense-mediated decay pathway (NMD), we treated the cells with the translation inhibitor puromycin. NMD inhibition as expected increased the amount of the well-known NMD target Down syndrome critical region 1 (DSCR1) mRNA (Supplemental Fig. S1A; Zubović et al. 2012). This treatment did not change the pattern of splicing (Supplemental Fig. S1B) and the splicing ratio (Supplemental Fig. S1C) in most of the SO-miRNA exons, excluding a major effect of NMD on the degradation of SO-miRNA skipped exons. All together this data indicates that inhibition of MPC-dependent cleavage at the SO-miRNA hairpins does not affect alternative selection of the corresponding SO-miRNA exons.
TABLE 1.
Human splice site overlapping miRNAs (SO-miRNAs)
FIGURE 1.
Microprocessor complex depletion does not increase the splicing ratio of the SO-miRNA exons. (A) Western blot analysis of Drosha, DGCR8, and Tubulin proteins from untreated (NO siRNA), control treated (siLUC), and Drosha/DGCR8-depleted (siD/D) HeLa and HaCaT cells. (B) Schematic representation of a SO-miRNA transcript. Boxes represent exons and lines introns. The SO-miRNA exon is highlighted in gray. Gray arrows indicate primers used for endpoint PCR. Black arrows represent the primer sets used for quantitative analysis of exon inclusion (EI) and exon skipping (ES) splice isoforms. RT-PCR analysis of SO-miRNA transcripts in untreated, siLUC-treated, and Drosha/DGCR8-depleted cells. Band identity is depicted on the right. M, marker. (C) qRT-PCR analysis of EI and ES splice isoforms abundance in siD/D-treated HeLa and HaCaT cells, relative to siLUC-treated cells. Two different primer sets are shown in panel B (black arrows). Graphs show fold changes in the EI (left panel) and ES (right panel) splice isoforms levels, relative to siLUC-treated cells set to one. Data were normalized to GAPDH for HeLa cells and for the geometric mean of TBP, RPLPO, and RLFP13A genes for HaCaT cells. Error bars show SD (three independent experiments). P-values were calculated using two-way ANOVA test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001. (D) Graph showing expression ratio of splice isoforms analyzed in panel C. Error bars show SD (three independent experiments). P-values were calculated using two-way ANOVA test. (*) P < 0.05; (**) P < 0.01.
Splicing inhibition mediated by SF3b1 silencing increases SO-miRNA biosynthesis but does not affect inclusion of SO-miRNA exons
To understand the effect of splicing on miRNA biosynthesis, we performed a global screening of miRNAs expression after depleting the splicing factor SF3b1 in human cells (Supplemental Fig. S2A). We compared miRNA-expression profiling from four SF3b1-depleted samples compared to four controls. We used an unsupervised hierarchical clustering based on the variation of expression of each miRNA (2020 miRNAs analyzed) across the samples examined and, as expected, the four controls were clustered together, as well as the four SF3b1 siRNA-treated samples (Supplemental Fig. S2B). We found that depletion of SF3b1 induced changes in 266 miRNAs. We validated changes in miRNA expression using qRT-PCR on four up-regulated miRNAs (miR-5088-3p, miR-3655, miR-483-3p, miR-6510-5p) and four down-regulated miRNAs (miR-155-3p, miR-148a-3p, miR-98-5p, miR-21-3p) in a consistent direction to the RNA-seq (Supplemental Fig. S2C).
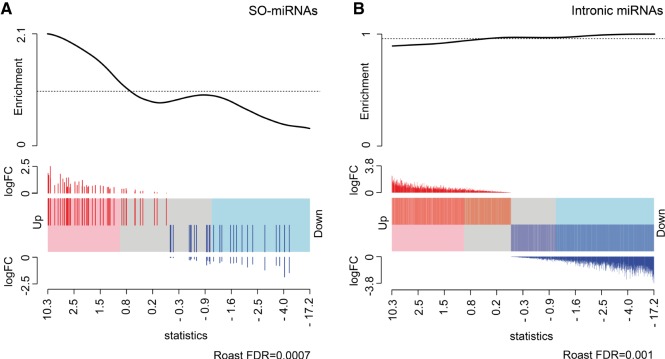
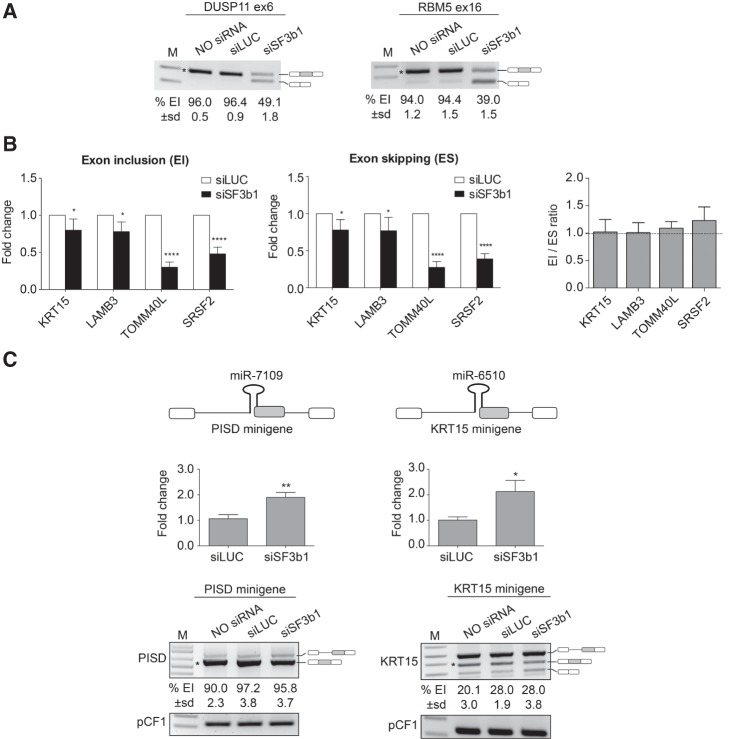
Next, we separately analyzed the differential expression profile of intronic and SO-miRNAs after SF3b1 depletion. Differential expression analysis of the SO-miRNA group by the ROAST gene set test showed a general positive effect of SF3b1 silencing on the production of the mature SO-miRNAs. Overall, there was a statistically significant enrichment of up-regulated SO-miRNAs in SF3b1-depleted samples versus normal samples (FDR < 0.001, Fig. 2A). The expression level of 39% of mature SO-miRNAs was significantly increased compared to control samples, especially miR-205-3p, miR-5187-3p, miR-6510-5p, miR-3614-3p, miR-7109-5p, miR-4260, miR-3655, miR-3656, miR-8069, and miR-636. In contrast, in the largest group of intronic miRNAs, we observed that splicing inhibition has the opposite effect inducing a small but significant reduction in the expression levels of mature miRNAs, with 32% of miRNAs down-regulated in SF3b1-depleted cells (FDR = 0.001, Fig. 2B). We evaluated two previously reported SF3b1-target exons, DUSP11 exon 6 and RBM5 ex 16 (Corrionero et al. 2011), to confirm that SF3b1 silencing had inhibited EI (Fig. 3A). In the four SO-miRNA exons expressed in this cell type (PISD, LAMB3, TOMM40L, and SRSF2), SF3b1 silencing up-regulated the corresponding SO-miRNAs, as confirmed by the RNA-seq data, decreased both EI and ES isoforms, but did not affect the splicing ratio (Fig. 3B).
FIGURE 2.
SF3b1 silencing increases the expression level of SO-miRNAs and slightly decreases the production of intronic miRNAs. (A,B) Gene set analysis computed by the ROAST method compares differential expression of SO-miRNAs (A) and intronic miRNAs (B) in the SF3b1-depleted and control-treated MEC-1 cells. Barcode plot shows each miRNA ranked by the empirical Bayes moderated t-statistics and horizontally represented by bars in a shaded rectangle. miRNAs up-regulated upon SF3b1 depletion are shaded pink and down-regulated ones are blue. The log2FC of each miRNA is indicated above and below the barcode as red and blue lines. (A) Gene set analysis on 52 SO-miRNAs. Barcode plot shows a relevant enrichment of up-regulated SO-miRNAs with a statistically significant FDR (<0.001). (B) Gene set analysis on intronic miRNAs. Barcode plot shows a small enrichment of down-regulated intronic miRNAs with a significant FDR (0.001).
FIGURE 3.
SF3b1 silencing increases the production of SO-miRNAs without affecting the splicing pattern of the corresponding exons. (A) RT-PCR analysis of DUSP11 and RBM5 mRNAs in untreated, siLUC-treated, and SF3b1-depleted MEC-1 cells. Alternative splicing events are indicated on the right. Numbers below each line indicate percentage of EI quantified by ImageJ and expressed as mean ± SD of three independent experiments. (*) indicates the specific quantified isoform. M, marker. (B) qRT-PCR analysis of exon inclusion (EI) and exon skipping (ES) splice isoforms abundance in SF3b1-depleted MEC-1 cells, relative to siLUC-treated cells. Graphs show fold changes in the EI (left panel) and ES (middle panel) splice isoforms levels, relative to siLUC-treated cells set to one. Data were normalized to GAPDH gene. Error bars show SD (three independent experiments). P-values were calculated using two-way ANOVA test. (*) P < 0.05; (****) P < 0.0001. Right panel shows expression ratio of EI and ES isoforms. Error bars show SD (three independent experiments). No significant differences were observed by two-way ANOVA test. (C) Schematic representation of the PISD and KRT15 minigene systems, containing the pri-miR-7109 and pri-miR-6510 (black hairpins) along with their flanking genomic regions. SO-miRNA exons are highlighted in gray. miR-7109 and miR-6510 expression by TaqMan qRT-PCR in siLUC-treated and SF3b1-depleted cells. Expression fold change values are depicted relative to siLUC-treated cells set to one. Data were normalized to RNU6B. Error bars show SD of three independent experiments. P-values were calculated using unpaired t-test. (*) P < 0.05; (**) P < 0.01. Splicing pattern of PISD and KRT15 minigenes transiently transfected in untreated, siLUC-treated, and SF3b1-depleted cells. The identity of the band is depicted on the right. Numbers below each line indicate the percentage of EI quantified by imageJ and expressed as mean ± SD of three independent experiments. (*) indicates the specific quantified isoform. pCF1 plasmid was used as control for normalization of transfection and reverse transcriptase efficiencies. M, marker.
To better clarify the effect of splicing inhibition on the expression of SO-miRNAs and processing of the corresponding transcripts, we prepared two minigenes with the PISD and KRT15 genomic regions surrounding the pri-miR-7109 and pri-miR-6510 hairpins, respectively (Fig. 3C, upper panel). Quantitative TaqMan analysis of mature miRNAs expression in cells that had been control-treated or transfected with SF3b1 siRNA showed that both miR-7109 and miR-6510 were up-regulated approximately twofold (Fig. 3C, middle panel). Interestingly, as found in vivo in cell cultures, SF3b1 silencing did not affect splicing of the corresponding SO-miRNA exons (Fig. 3C, lower panel). Collectively, these experiments indicate that splicing inhibition, mediated by SF3b1 depletion, differentially affects the production of miRNAs depending on the position of the pri-miRNA hairpins in the transcripts: Intronic miRNAs are depleted, whereas SO-miRNAs are up-regulated. However, as for MPC-inhibition, SF3b1 silencing had no effect on alternative splicing of the SO-miRNA exons.
Analysis of SO-miRNAs in keratinocyte differentiation genes
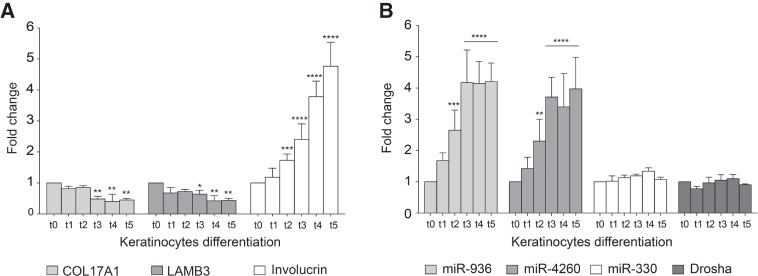
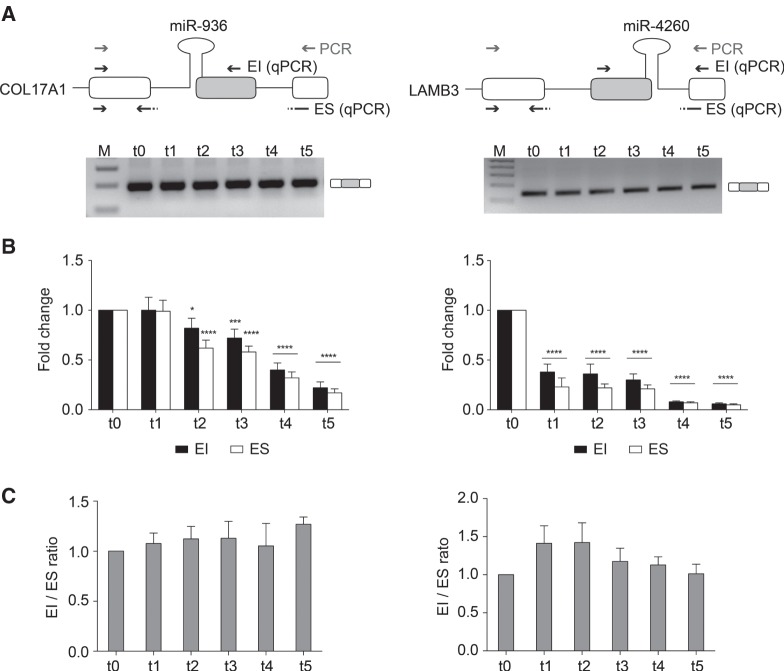
Inspection of the genes that contain SO-miRNAs showed that two of them, COL17A1 and LAMB3, code for adhesion machinery as part of the hemidesmosome, which is involved in keratinocyte differentiation. The renewal of epidermis is accomplished by basal keratinocytes that periodically stop to divide and commit to differentiate by moving toward the skin surface, where they progressively stop proliferating and lastly terminally differentiate in corneocytes (Aberdam et al. 2008). Thus, the keratinocyte differentiation process involves progressive loss of adhesion of basal cells, due to COL17A1 and LAMB3 down-regulation and a concomitant reduced proliferation capacity. COL17A1 and LAMB3 are also involved in maintaining a physiological cell lifespan in culture (Robbins et al. 2001; Turcan and Jonkman 2015). Thus, to study the functional relationship between the spliceosome and the MPC in a more physiological context we focus on keratinocyte differentiation and analyzed the expression of COL17A1 and LAMB3 along with their corresponding miR-936 and miR-4260 SO-miRNAs. As expected, keratinocyte differentiation, verified by the increase in the expression of the differentiation marker involucrin (Fig. 4A) resulted in a gradual decrease in COL17A1 and LAMB3 mRNAs (Fig. 4A). Interestingly, and in contrast to the corresponding host genes, quantitative TaqMan analysis at different time points showed a progressive increase in the expression of miR-936 and miR-4260 from day 0 to day 5 of cell culture (Fig. 4B). To assure that up-regulation of SO-miRNAs was strictly related to changes in the expression levels of corresponding mRNAs and was not a general effect of the differentiation process, we quantified the expression of the intronic miR-330. miR-330 stayed constant throughout differentiation, as did the major MPC component Drosha (Fig. 4B). This strongly suggests that the increase in SO-miRNA levels during differentiation are not due to a general increase in the MPC processivity.
FIGURE 4.
mRNAs’ and SO-miRNAs’ expression level during keratinocytes differentiation. qRT-PCR analysis of (A) COL17A1, LAMB3, and involucrin (used as keratinocyte differentiation marker) transcripts, and (B) miR-936, miR-4260, miR-330 expression by TaqMan assay and Drosha transcript abundance in proliferating keratinocytes (t0) and calcium-treated differentiated cells (t1 to t5). Graphs show expression fold changes in t1–t5 cells, relative to t0 cells set to one. Data were normalized against the geometric mean of TBP, RPLP0, and RPL13A genes. Error bars represent SD between three independent calcium-induced differentiation experiments. P-values were calculated using two-way ANOVA test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001.
As inhibition of proliferation is an important event associated with keratinocyte differentiation, we analyzed the effect of miR-936 and miR-4260 on proliferation of the immortalized human keratinocyte cell line HaCaT. miR-936 and miR-4260 mimics were transfected, and after 72 h the cells were stained for 5-ethynyl-2′-deoxyuridine (EdU), a uridine analog that is incorporated into newly synthesized DNA. miR-936 and miR-4260 significantly decreased the number of EdU-positive, proliferating cells from 60% to <35% (Supplemental Fig. S3). Collectively, the above results demonstrate an inverse correlation between the expression of COL17A1 and LAMB3 transcripts and their corresponding SO-miRNAs, during keratinocyte differentiation and that SO-miRNAs have an antiproliferative effect. The production of SO-miRNAs along with the reduced expression of the host genes might contribute, with antiproliferative and antiadhesion effects, respectively, to exiting basal keratinocytes from the cell cycle, to their upward migration and to their commitment to terminal differentiation. Overall this suggests that the competition between the MPC and the spliceosome on SO-miRNAs in LAMB3 and COL17A1 host genes regulates the proliferation/differentiation switch in keratinocytes.
We next investigated whether increased SO-miRNA biosynthesis in differentiated keratinocytes changed the splicing pattern of COL17A1 and LAMB3 transcripts. As found in the related HaCaT cell line, endpoint RT-PCR showed that both COL17A1 and LAMB3 SO-miRNA exons are included in the final transcripts (Fig. 5A). Furthermore, analysis by qRT-PCR of the relative abundance of the SO-miRNA isoforms showed a decrease in both EI and ES isoforms (Fig. 5B), but did not identify any substantial decrease in the splicing ratio at the different time points of differentiation (Fig. 5C). These results indicate that the splicing pattern is independent of the increase of miRNA biosynthesis and the differentiation process.
FIGURE 5.
Keratinocyte differentiation does not affect alternative splicing of COL17A1 and LAMB3 exons. (A) Schematic representation of the genomic regions surrounding the COL17A1 and LAMB3 SO-miRNA exons. SO-miRNA exons are gray boxes, flanking exons are white boxes and the pri-miRNA hairpins, located across the intron-exon junction, are indicated. Gray arrows indicate primers used for endpoint RT-PCR. Black arrows represent the primer sets used for quantitative analysis of exon inclusion (EI) and exon skipping (ES) splice isoforms. Endpoint RT-PCR of COL17A1 and LAMB3 transcripts in proliferating keratinocytes (t0) and calcium-treated differentiated cells (t1 to t5). Band identity is depicted on the right. M, marker. (B) qRT-PCR analysis of EI and ES splice isoforms abundance in proliferating keratinocytes (t0) and calcium-treated differentiated cells (t1 to t5). Two different primer sets are shown in panel A (black arrows). Graphs show fold changes in the EI (black columns) and ES (white columns) splice isoforms levels, relative to t0 keratinocytes, set to one. Data were normalized to the geometric mean of TBP, RPLPO, and RLFP13A genes. Error bars show SD (three independent experiments). P-values were calculated using two-way ANOVA test. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001. (C) Graph showing expression ratio of splice isoforms analyzed in panel B. Error bars show SD (three independent calcium-induced differentiation experiments). No significant differences across the different time points of differentiation were observed by one-way ANOVA test.
To exclude the possibility that a putative intronic promoter located just upstream the SO-miRNAs might drive an alternative transcription (Monteys et al. 2010), we took advantage of a minigene system. We generated a plasmid carrying the SO-miR-936 genomic region composed of COL17A1 exon 29 along with part of the flanking introns and a plasmid variant (ΔAP) with the deletion of the putative intronic promoter (Supplemental Fig. S4A). As shown in Supplemental Figure S4A, transfection of the ΔAP construct in HeLa cells induced a cryptic 3′ splice site to be used, instead of the canonical site. In addition, the WT COL17A1 minigene was correctly processed, whereas the COL17A1 exon 29 in the mutant ΔAP COL17A1 was only ∼15% included. Deletion of the putative intronic promoter increased the biosynthesis of mature miR-936 ∼1.8 fold (Supplemental Fig. S4B). This evidence precludes the possibility that miR-936 is predominantly transcribed from an intronic promoter and suggests that the selection of a cryptic 3′ splice site induces the conversion of a SO-miRNA into an exonic miRNA that is more easily processed by the MPC.
The MPC may terminate transcription at SO-miRNA exons
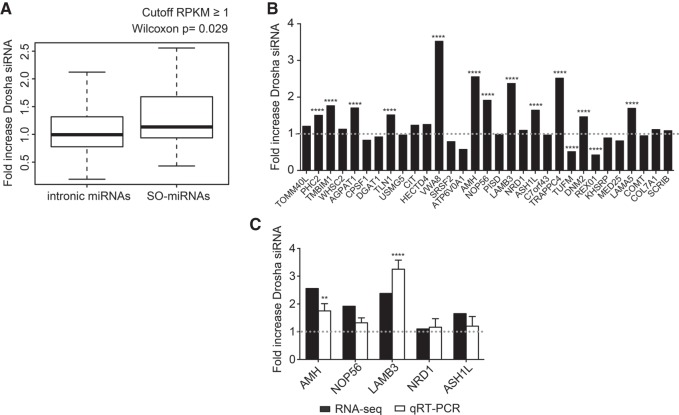
The absence of changes in the alternative splicing pattern at SO-miRNA exons after Drosha/DGCR8 silencing (Fig. 1), splicing inhibition by SF3b1 depletion (Fig. 3), and during keratinocyte differentiation (Fig. 5) suggests that MPC-dependent cleavage of RNA might result in premature transcriptional termination. To investigate this possibility we took advantage of the previously published data of chromatin RNA sequencing (Gene Expression Omnibus, accession number GSE58838) that evaluated the effect of MPC silencing by either Drosha or DGCR8 depletion on nascent pre-mRNA (Dhir et al. 2015). This analysis performed in HeLa cells showed that the MPC-dependent cleavage at pri-miRNA hairpin sites in some long noncoding pri-miRNA transcripts could mediate transcriptional termination. We reasoned that the SO-miRNA hairpins that are mostly in central exons of coding transcripts, might also mediate a MPC-dependent transcriptional termination inside genes, due to their peculiar position on the splice site. We compared intronic miRNAs and SO-miRNAs and analyzed the downstream/upstream ratio between the normalized read counts of Drosha siRNA (mean Y1/mean X1) and control siRNA (mean Y0/mean X0) treated cells (Supplemental Fig. S5). Consistent with a MPC-dependent transcriptional termination at SO-miRNA exons, there were significantly more reads downstream from the cleavage site in Drosha-depleted cells in the SO-miRNA group, than in intronic miRNAs (P = 0.029, Mann-Whitney-Wilcoxon test Fig. 6A). All the expressed 31 SO-miRNA-hosting transcripts were coding and twelve increased very significantly (P < 0.0001, two-way ANOVA test, Fig. 6B). The decrease or the absence of changes in the downstream/upstream ratio after Drosha depletion seen in some cases might be due to a lower MPC cleavage efficiency of the pri-miRNA hairpins at basal conditions.
FIGURE 6.
Effect of Drosha siRNA effect on intronic and SO-miRNA-containing transcripts. Chromatin RNA-seq analysis. (A) Box plot showing fold increase of Drosha distribution in the intronic miRNAs and SO-miRNAs expressed in HeLa cells with a gene body RPKM ≥ 1. The P-value calculated with a two-sided Mann-Whitney-Wilcoxon test is indicated above. (B) List of the 31 SO-miRNA-hosting transcripts expressed in HeLa cells with a gene body RPKM ≥ 1. Graph showing fold increase of normalized read counts of Drosha siRNA-treated cells relative to control siRNA-treated cells were plotted in the graph. P-values were calculated using two-way ANOVA test. (****) P < 0.0001. Dotted line is set to one. (C) Fold increase of normalized reads counts of Drosha siRNA relative to control siRNA-treated HeLa cells detected by chromatin RNA-seq of five selected SO-miRNA transcripts from panel B were plotted in the graph (black columns) and compared to qRT-PCR data obtained from the analysis of the nascent transcripts in the RNA isolated from nuclear chromatin of Drosha siRNA and control siRNA-treated HeLa cells (white columns). Dotted line is set to one. qRT-PCR data are normalized to GAPDH. Error bars represent standard deviations between three independent validation experiments. P-values were calculated using two-way ANOVA test. (**) P < 0.01; (****) P < 0.0001.
To validate the results obtained from the chromatin RNA-seq analysis, we depleted Drosha in HeLa cells by siRNA treatment and then performed cell fractionation and chromatin RNA isolation as this is enriched in nascent transcripts. There were significantly more reads downstream from two out of five selected pri-miRNA hairpins (AMH and LAMB3) after Drosha depletion and a smaller increase for the other three selected SO-miRNA-hosting transcripts (Fig. 6C).
DISCUSSION
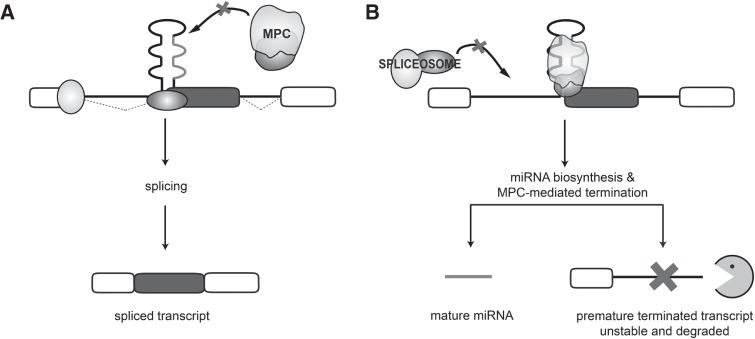
SO-miRNAs originate from MPC-dependent cleavage of pri-miRNA hairpins juxtaposed to intron-exon splice junctions and they are mostly located in central exons of protein-coding genes (Table 1). In this study, we have evaluated how the competition between the MPC and the spliceosome on SO-miRNA exons affects processing of the nascent transcripts. Using different systems in which either the MPC or the spliceosome was inhibited or in the more physiologic keratinocyte differentiation model, our results suggest that the MPC-dependent cleavage might regulates transcriptional termination at the SO-miRNA exons. This mechanism is confirmed by the unexpected observation that alternative splicing of SO-miRNA exons was unchanged in any of the cellular models we investigated including silencing of Drosha/DGCR8, SF3b1, or keratinocyte differentiation (Figs. 1, 3, and 5, respectively). We found evidence of a SO-miRNA-dependent transcriptional termination mechanism by analyzing nascent RNAs abundance upstream and downstream from SO-miRNA hairpins. In fact, the inhibition of the MPC activity increases the amount of chromatin-bound RNA downstream from the SO-miRNA hairpins, both on a global basis, as detected through re-assessment of published data (Fig. 6B; Dhir et al. 2015) and through experimental validation in selected cases (Fig. 6C). As depicted in the model (Fig. 7A), the reduced MPC activity, obtained experimentally with Drosha/DGCR8 silencing, generates more substrate for the spliceosome that, maintaining its unaltered activity, will normally process the pre-mRNA. On the other hand, splicing inhibition mediated by SF3b1 silencing, might increase the amount of unspliced transcript: The subsequent cleavage mediated by the MPC will increase the SO-miRNA but the resulting truncated nascent RNA is rapidly degraded probably by Drosha-mediated recruitment of exonucleases to the cleavage site (Fig. 7B), as already reported (Morlando et al. 2008; Ballarino et al. 2009; Wagschal et al. 2012). Altering transcriptional termination at the canonical polyadenylation site has been shown to cause defects in gene expression and RNA processing (West and Proudfoot 2009). It is possible that the transcriptional termination induced by the MPC-dependent cleavage inside SO-miRNA-hosting genes might act as a signal to reduce the expression of the host gene at the expense of the SO-miRNA biosynthesis. Thus, the nascent transcript cleaved by the MPC at the SO-miRNA exon generates the miRNA but the cleaved transcript is not further processed for splicing because of a concomitant termination defect (Fig. 7).
FIGURE 7.
Model of MPC-dependent processing of SO-miRNA-hosting transcripts. The panel depicts the two processing events that can affect a nascent SO-miRNA-hosting transcript. (A) A reduced cropping activity at the SO-pri-miRNA hairpin and/or a faster assembly of the spliceosome induces normal splicing with no mature SO-miRNA production. (B) Efficient cleavage of the SO-pri-miRNA hairpin by the MPC, occurring for example after splicing inhibition or during keratinocyte differentiation, induces the premature termination of the nascent transcript at the site of cropping and produces the mature SO-miRNA. The resulting unstable transcript is not alternatively spliced, but it is rapidly degraded by Drosha-mediated recruitment of exonucleases to the cleavage site. The SO-miRNA exon and flanking exons are represented as gray and white boxes, respectively. Black thin lines are introns. The SO-miRNA hairpin overlaps the intron:exon junction and the mature miRNA is highlighted in light gray.
The transcriptional block we observed at SO-miRNAs is reminiscent of the recently identified Drosha-mediated transcriptional termination event occurring on human long noncoding RNA transcripts, although SO-miRNA-mediated termination is likely less efficient (Dhir et al. 2015). In these noncoding RNAs, the cleavage, that occurs within either intronic or exonic miRNAs, might prevent the potentially deleterious accumulation of transcripts after miRNA biosynthesis and/or avoid transcriptional interference with downstream genes (Dhir et al. 2015). In the case of SO-miRNAs, our results suggest that Drosha cleavage, mainly acting inside coding transcripts, might regulate the production of two alternative products from the same gene. This might be used in physiological conditions for expressing alternatively a SO-miRNA or a coding transcript with antagonistic functions. In keratinocytes, the activation of the differentiation program shifts the balance toward a preferential MPC cleavage of the COL17A1 and LAMB3 nascent transcripts increasing the production of hosted mature miR-936 and miR-4260, while at the same time down-regulating the corresponding mRNAs (Fig. 4). COL17A1 and LAMB3 are involved in cell-extracellular matrix interactions whereas their embedded SO-miRNAs have an antiproliferative effect (Supplemental Fig. S3). Therefore, the shift from coding transcripts to miRNA production might help keratinocyte transition from the basal layer to the cornified stratum. A similar shift from mRNA to miRNA production has been reported in follistatin-like 1 (FSTL1). Here the mRNA and miRNA respectively promote or inhibit keratinocyte migration. In this example, the MPC-dependent cleavage occurs at the 3′UTR of FSTL1, promoted by the KHSRP splicing factor (Sundaram et al. 2013). The presence of a common MPC-dependent processing event for SO-miRNA-hosting transcripts and FSTL1 suggests that keratinocytes can specifically use this mechanism to produce two alternative products from several genes to orchestrate proliferation and differentiation, migration, re-epithelialization and wound healing.
The determinants that promote transcriptional termination at the sites where the MPC cleaves are unknown. One possibility is that different Pol II phospho-CTD isoforms might be involved. There is evidence for this from long intergenic noncoding RNAs that have been recently shown to have a peculiar transcriptional Pol II CTD code that causes inefficient splicing and polyadenylation (Schlackow et al. 2017). It is tempting to speculate that the presence of specific transcriptional Pol II CTD code might be particularly relevant for processing coding SO-miRNAs and in particular for promoting transcriptional termination. Interestingly, previous analysis on three noncoding SO-miRNAs showed that, in contrast to coding counterparts, the competition between the MPC and the spliceosome modulates alternative splicing (Mattioli et al. 2013; Melamed et al. 2013). Since noncoding SO-miRNAs represent just a small number of human SO-miRNAs (six out of 52) and two of them are expressed in only one cellular system analyzed here, we could not perform a systematic comparison between them in our study. However, the coding or noncoding nature of the transcript is probably not the only determinant involved. The promoter architecture or the local context influenced by the histone modifications might also affect the Pol II CTD code, inducing the transcriptional termination at SO-miRNAs. Alternatively, the Pol II, engaged concomitantly on SO-miRNAs in splicing and the MPC's cleavage activities might lose its previously reported exon tethering activity (Dye et al. 2006) detaching from active transcription.
Cell- and tissue-specific factors might regulate the cleavage efficiency of the MPC at the SO-miRNA hairpins promoting transcriptional termination. Splicing factors that are known to regulate Drosha processing efficiency at specific pri-miRNAs (Krol et al. 2010; Finnegan and Pasquinelli 2013) might be also involved in SO-miRNA cleavage efficiency.
In conclusion, our results suggest that the MPC-dependent cleavage of coding SO-miRNAs does not result in alternative splicing of the SO-miRNA exons but rather that it might promote transcriptional termination. This competition provides a system for regulating gene expression that will generate two alternative products from a single shared transcript through a MPC-dependent cleavage activity inside genes. This likely has important functional consequences in regulating various physiological and pathological conditions.
MATERIALS AND METHODS
Cell cultures
HeLa and HaCaT cells were grown in Dulbecco's modified Eagle medium (DMEM) with GlutaMAX-I (Thermo Fisher Scientific) supplemented with 10% Fetal Bovine Serum (FBS) and antibiotic antimycotic solution (Sigma-Aldrich). Human B-chronic lymphocytic leukemia MEC-1 cells (courtesy of D. Efremov), growing in suspension, were cultured in RPMI 1640 Medium (Thermo Fisher Scientific) supplemented with 10% FBS and antibiotic, antimycotic solution. Normal human primary epidermal keratinocytes (NHEK) were cultured as previously described (Zambruno et al. 1995). Keratinocytes were seeded at a concentration of 1–1.5 × 104 cells/cm2 in serum-free Keratinocyte Growth Medium (KGM, Invitrogen) containing 0.15 mM Ca2+, cultured till confluence, and then induced to differentiate by culturing in KGM containing 1.2 mM Ca2+ for up to 5 d (Dal Mas et al. 2015). Cells were maintained at 37°C with 5% CO2.
Puromycin treatment, plasmid, and siRNA transfection
For puromycin experiments, HeLa and HaCaT cells were seeded at 2.5 × 105 cells/well and after 24 h were treated with puromycin (Sigma-Aldrich) at a concentration of 100 µg/mL and were harvested 8 h post-treatment for RNA analysis.
For transfection experiments, HeLa cells grown on six-well plates were transiently transfected with 500 ng of DNA plasmid with Effectene Transfection Reagent (Qiagen) according to manufacturer's instructions. pCF1 plasmid (100 ng) was cotransfected with each minigene and used for normalization of transfection and reverse transcriptase efficiencies (Pastor et al. 2011). Cells were harvested 24 h post-transfection for RNA analysis.
For silencing of Drosha and DGCR8, HeLa and HaCaT cells were seeded at 2.5 × 105 cells/well and 24 h later transfected with 10 µL of 20 µM siRNA duplex (Dharmacon) against Drosha (5′-CGAGUAGGCUUCGUGACUU-3′), DGCR8 (5′-CAUCGGACAAGAGUGUGAU-3′) or negative control Luciferase (5′-UAAGGCUAUGAAGAGAUAC-3′) by using Oligofectamine (Invitrogen) as per manufacturer's instructions. Cells were harvested 72 h post-transfection for RNA and protein analysis. For silencing of SF3b1, 2 × 106 MEC-1 cells were transfected with the Amaxa Cell Line Nucleofector Kit L (Lonza) and 2.5 µg of siRNA duplex against SF3b1 (5′-CGCCAAGACUCACGAAGAU-3′) or the control siRNA against Luciferase. Nucleofection was performed on an Amaxa Nucleofector II device using the C-005 program. Cells were collected 72 h later for RNA and protein analysis.
RNA isolation, splicing analysis, and quantitative real-time PCR
Total RNA was extracted using TRI Reagent (Invitrogen), according to the manufacturer's instructions. One microgram of total RNA was reverse transcribed using random primers and the M-MLV Reverse Transcriptase (Invitrogen).
Endpoint PCR analysis of PISD, KRT15, and COL17A1 minigenes splicing was performed using the PISD ex3 dir and pCI rev, pY7 ex2 dir and KRT15 ex3 rev, alpha 2–3 dir and pFAN 904 rev sets of primers, respectively (Supplemental Table 1). PCR conditions were 94°C for 5′, 30 cycles at 94°C for 45″, 56°C for 45″, 72°C for 45″, and 72°C for 10′. Minigenes were transfected into cells along with 100 ng of pCF1 plasmid, which was used as control for normalization of transfection and reverse transcriptase efficiencies as previously reported (Pastor et al. 2011). PCR amplification of control pCF1 plasmid was performed under the same condition using CF ex1 248 dir and CF ex3 292 rev primers (Supplemental Table 1). PCR products were resolved by 2% agarose gel electrophoresis. The 1kb Plus DNA Ladder (Invitrogen) was used as molecular weight reference marker (M). The intensity of the bands was quantified with the ImageJ software. The percentage of EI isoform (%EI) is expressed as mean ± SD and calculated as the percentage of the isoform divided by the total abundance of the two isoforms.
Transcripts were quantified by using the iQ SYBR Green Supermix (Bio-Rad) in a CFX96 Real-Time PCR system (Bio-Rad). Primers used for cDNA amplification are listed in Supplemental Table 1. Each primer set used was tested by qRT-PCR using a 10-fold serial dilution of cDNA and water as nontemplate control in order to determine amplification efficiency, specificity, and the presence/absence of primer dimers (Supplemental Fig. S6). The amplification of a singular product with a uniform Tm was confirmed by the presence of a single peak in the melt peak curve. The amplification efficiency was determined from the linear slope of the standard curve: Only primers with a standard curve slope between −3.1 and −3.6 and an amplification efficiency comprised between 90% and 110% were considered suitable for further quantification. qRT-PCR cycling protocol was: 95°C for 3′, 40 cycles at 95°C for 15″, and 60°C for 1′. GAPDH and for keratinocytes samples RPL13A, RPLP0, and TBP (expressed as geometric mean) were amplified as internal controls.
For mature miRNA quantification, 1 µg of total RNA was reverse transcribed using the M-MLV Reverse Transcriptase in a reaction mixture containing a miRNA-specific stem–loop reverse transcription primer (Applied Biosystems). Quantification of mature miRNAs was performed using the TaqMan Universal Master Mix II and predesigned TaqMan probes (Applied Biosystems, Supplemental Table 1). qRT-PCR was conducted at 95°C for 10′, followed by 40 cycles of 95°C for 15″, and 60°C for 1′. RNU6B small nuclear RNA was amplified as an internal control. Results of SYBR Green and TaqMan assays were normalized to the internal control and relative quantification of each mRNA and miRNA was determined using the 2−ΔΔCt method (Livak and Schmittgen 2001).
Protein isolation and western blot analysis
For protein analysis, cells were lysed in 1× lysis buffer (100 mM KCl, 200 µM EDTA pH 8.0, 20 mM Hepes pH 7.5, 0.5 mM DTT, 1% NP-40, 6% glycerol) with Proteinase Inhibitor Cocktail (Roche). Thirty micrograms of total proteins were separated on a NuPAGE 4%–12% Bis-Tris precast gel (ThermoFisher) and transferred onto a PVDF membrane (GE Healthcare). After blocking with 5% nonfat milk in PBS, membranes were probed with primary antibodies (Abcam) as follows: anti-Drosha (rabbit polyclonal, 1:625), anti-DGCR8 (goat polyclonal, 1:500), anti-SF3b1 (rabbit monoclonal, 1:1000), and anti-Tubulin (mouse monoclonal, 1:5000) as loading control, all overnight at +4°C. Proteins of interest were detected with HRP-conjugated secondary antibodies (1:2000, Agilent) and visualized with the Pierce ECL Western blotting substrate (Thermo Scientific), according to the provided protocol, followed by exposure to X-ray film (Kodak). Uncropped blots are shown in Supplemental Figure S7.
miRNA transfection
miRNA mimics (miRIDIAN miRNA mimics) corresponding to the human mature miR-936 (hsa-miR-936, MIMAT0004979, 5′-ACAGUAGAGGGAGGAAUCGCAG-3′) and miR-4260 (hsa-miR-4260, MIMAT0016881, 5′-CUUGGGGCAUGGAGUCCCA-3′) were obtained from GE Dharmacon. miRNAs were transfected into HaCaT cells using a standard reverse transfection protocol, at a final miRNA concentration of 50 nM. Briefly, the transfection reagent (Lipofectamine RNAiMAX, Life Technologies) was diluted in Opti-MEM (Invitrogen) and added to the miRNAs arrayed on 96-well plates; 30′ later 1.0 × 103 cells were seeded per well. Nontargeting siRNA #1 (NT siRNA #1, 5′-UGGUUUACAUGUCGACUAA-3′, GE Dharmacon) was used as negative control. Screening was performed in triplicate. Sixty hours after plating, the culture medium was replaced with medium containing 5 µM 5-ethynyl-2′-deoxyuridine (EdU, Thermo Fisher Scientific) for 12 h. Cells were fixed 72 h after plating and processed for immunofluorescence.
Immunofluorescence
Cells were fixed with 4% paraformaldehyde for 20′, permeabilized with 0.5% Triton X-100 in PBS solution for 20′. Cells were further processed using the Click-iT EdU Alexa Fluor 488 Imaging Kit (Thermo Fisher Scientific) to reveal EdU incorporation, according to the manufacturer's instructions. Nuclei were counterstained with Hoechst 33342 (Life Technologies). The screening was performed at the ICGEB High-Throughput Screening Facility (http://www.icgeb.org/high-throughput-screening.html).
Small RNA-seq sample preparation and sequencing
Small RNA-sequencing was performed by IGA Technology Services. Briefly, RNA purity, integrity, and concentration were determined using an Agilent 2100 Bioanalyzer (Agilent Technologies). Only RNA with a RIN value >8.0 and an rRNA 28S/18S ratio >1.8 was taken forward for sample preparation. Two micrograms of total RNA (minimum concentration of 200 ng/µL) from four control (siRNA against Luciferase) and four treated (siRNA against SF3b1) MEC-1 cells were sequenced on an Illumina HiSeq2000. One lane in 12-plex was run, obtaining 10 million of single-reads per sample, 50-bp long. Raw sequencing data were deposited in NCBI's Sequence Read Archive (SRA) and are accessible through SRA STUDY accession SRP099431.
Bioinformatic analysis
Real time image analysis and base calling were performed on a HiSeq2000 instrument using the HiSeq Sequencing Control Software. CASAVA software (version 1.8.2) was used for demultiplexing and production of FASTQ sequence files. FASTQ raw sequence files were subsequently quality checked using FASTQC software (version 0.11.3). RNA-seq reads were mapped to the human reference genome (GRCh38/hg38) and to known mature miRNAs (miRBase release 21) with Bowtie 2 alignment software (Langmead and Salzberg 2012). Rounded Gene counts were used as input to perform differential gene expression analysis using the Bioconductor package DESeq2, version 1.4.5 (Love et al. 2014), estimating the per-gene negative binomial distribution dispersion parameter. To detect outlier data after normalization, we used the R package arrayQualityMetrics (Kauffmann et al. 2009), and before testing differential gene expression, we removed all genes with low normalized mean counts to improve testing power while maintaining type I error rates. Estimated FDR values for each gene were adjusted using the Benjamini-Hochberg method. Features with adjusted FDR < 0.01 and absolute log2FC > 1 were considered having a significant altered expression. Raw count data were transformed to log2CPM before computing appropriate self-contained gene set test, in the sense defined by Goeman and Bühlmann (Goeman and Bühlmann 2007), using the ROAST (Wu et al. 2010) gene set test. For assessing differential expression, the empirical Bayes moderated t-statistics test was used to moderate the standard errors of the estimated log2FC.
Chromatin RNA-seq analysis
Online available chromatin RNA-seq data from HeLa cells treated with a control siRNA and Drosha siRNA deposited in the Gene Expression Omnibus (GEO) data set under GEO Series reference number GSE58838 (Dhir et al. 2015) were analyzed. miRNA-containing genes with length ≥2 kb and gene-body RPKM ≥ 1 were considered for this study as suggested by the original authors (Dhir et al. 2015). This resulted in 475 intronic miRNA-containing genes and 31 SO-miRNA-containing genes. Read counts, normalized to total sequencing depth were calculated across the region extending from the TSS of the miRNA-containing gene to the start of the pri-miRNA hairpin and the region from the end of the pri-miRNA hairpin to the TES. The average number of reads mapping upstream (mean X0 and X1) and downstream (mean Y0 and Y1) of the pri-miRNA hairpin in the control siRNA- and Drosha siRNA-treated cells was estimated and the ratio between reads mapping downstream and upstream of the pri-miRNA hairpin was calculated (meanY0/meanX0 and meanY1/meanX1). The fold increase of Drosha-depleted cells was then calculated as the average number of Drosha siRNA reads over the average number of control siRNA reads [Drosha siRNA FI = (meanY1/meanX1)/(meanY0/meanX0)] (Supplemental Fig. S5).
Nuclear fractionation and chromatin RNA isolation
Nuclear fractionation was performed similar to Wuarin and Schibler (Wuarin and Schibler 1994). A total of 20 × 106 cells were collected by scraping and centrifugation. Cell pellets were resuspended in 300 µL of buffer A (10 mM HEPES pH 7.5, 10 mM KCl, 10% glycerol, 340 mM sucrose, 4 mM MgCl2, 1 mM DTT, 1× Protease Inhibitor Cocktail) and then 300 µL of buffer A with 0.2% (v/v) Triton X-100 was added, incubated on ice for 12′ to lyse cells and centrifuged. The crude nuclear pellet was then resuspended in 250 µL of NRB buffer (20 mM HEPES pH 7.5, 50% glycerol, 75 mM NaCl, 1 mM DTT, 1× Protease Inhibitor Cocktail) and centrifuged again to wash. The pellet was resuspended in 250 µL of NRB buffer and then 250 µL of NUN buffer (20 mM HEPES pH 7.5, 300 mM NaCl, 1 M Urea, 1% NP-40, 10 mM MgCl2, 1 mM DTT) was added, incubated on ice for 5′ and centrifuged. A total of 125 µL of the soluble nuclear extract supernatant were collected and the depleted nuclear pellet was resuspended in 1 mL of buffer A and centrifuged. The resulting chromatin pellet was resuspended in 50 µL of buffer A. Nuclear-soluble RNA and chromatin RNA were then isolated by using TRI Reagent, following manufacturer's instructions.
Fractionation was validated by confirming the chromatin enrichment of a canonically chromatin-associated lncRNA XIST (Kalantry et al. 2009) and soluble nuclear enrichment of the beta-actin (ACTB) mRNA relative to FNBP4 mRNA that had an equivalent level of abundance in both extracts.
Statistical analysis
All data are presented as mean ± standard deviation of at least three independent experiments. Statistical analysis was carried out using Prism Software (GraphPad Software). For statistical comparison the one-way ANOVA and two-way ANOVA followed by Dunnett's multiple comparison test or uncorrected Fisher's LSD test was used. A P-value <0.05 was considered significant.
DATA DEPOSITION
Raw sequencing data for the reported small RNA-sequencing are deposited in NCBI's Sequence Read Archive (SRA) and are accessible through SRA STUDY accession number SRP099431.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dimitar Efremov (ICGEB, Trieste) for providing MEC-1 cell lines. We thank Luca Braga (ICGEB, Trieste) for help with acquisition of fluorescence images. This work was supported by an Italian Ministry of Health grant to D.C. and F.P. (RF-2011-02347596). Funding to pay the open access charges was provided by institutional ICGEB funds.
Author contributions: G.P. designed and performed the experiments, analyzed the data, and wrote and edited the manuscript. P.F. and D.C. performed keratinocyte in vitro differentiation and critically revised the manuscript; I.P. performed qRT-PCR efficiency experiments; D.L. performed bioinformatics analysis; F.P. supervised the study and also wrote and edited the manuscript.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.063438.117.
Freely available online through the RNA Open Access option.
REFERENCES
- Aberdam D, Candi E, Knight RA, Melino G. 2008. miRNAs, ‘stemness’ and skin. Trends Biochem Sci 33: 583–591. [DOI] [PubMed] [Google Scholar]
- Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ, Bozzoni I. 2009. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol 29: 5632–5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S, Bartel DP. 2005. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA 11: 241–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. 2004. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 10: 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrionero A, Minana B, Valcarcel J. 2011. Reduced fidelity of branch point recognition and alternative splicing induced by the anti-tumor drug spliceostatin A. Genes Dev 25: 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Mas A, Fortugno P, Donadon I, Levati L, Castiglia D, Pagani F. 2015. Exon-specific U1s correct SPINK5 exon 11 skipping caused by a synonymous substitution that affects a bifunctional splicing regulatory element. Hum Mutat 36: 504–512. [DOI] [PubMed] [Google Scholar]
- Dhir A, Dhir S, Proudfoot NJ, Jopling CL. 2015. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol 22: 319–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye MJ, Gromak N, Proudfoot NJ. 2006. Exon tethering in transcription by RNA polymerase II. Mol Cell 21: 849–859. [DOI] [PubMed] [Google Scholar]
- Finnegan EF, Pasquinelli AE. 2013. MicroRNA biogenesis: regulating the regulators. Crit Rev Biochem Mol Biol 48: 51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goeman JJ, Bühlmann P. 2007. Analyzing gene expression data in terms of gene sets: methodological issues. Bioinformatics 23: 980–987. [DOI] [PubMed] [Google Scholar]
- Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. 2005. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell 123: 631–640. [DOI] [PubMed] [Google Scholar]
- Gromak N, Dienstbier M, Macias S, Plass M, Eyras E, Caceres JF, Proudfoot NJ. 2013. Drosha regulates gene expression independently of RNA cleavage function. Cell Rep 5: 1499–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. 2009. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136: 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janas MM, Khaled M, Schubert S, Bernstein JG, Golan D, Veguilla RA, Fisher DE, Shomron N, Levy C, Novina CD. 2011. Feed-forward microprocessing and splicing activities at a microRNA-containing intron. PLoS Genet 7: e1002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalantry S, Purushothaman S, Bowen RB, Starmer J, Magnuson T. 2009. Evidence of Xist RNA-independent initiation of mouse imprinted X-chromosome inactivation. Nature 460: 647–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann A, Gentleman R, Huber W. 2009. arrayQualityMetrics—a bioconductor package for quality assessment of microarray data. Bioinformatics 25: 415–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YK, Kim VN. 2007. Processing of intronic microRNAs. EMBO J 26: 775–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krol J, Loedige I, Filipowicz W. 2010. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet 11: 597–610. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9: 357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. 2004. MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23: 4051–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macias S, Plass M, Stajuda A, Michlewski G, Eyras E, Cáceres JF. 2012. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat Struct Mol Biol 19: 760–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli C, Pianigiani G, Pagani F. 2013. A competitive regulatory mechanism discriminates between juxtaposed splice sites and pri-miRNA structures. Nucleic Acids Res 41: 8680–8691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattioli C, Pianigiani G, Pagani F. 2014. Cross talk between spliceosome and microprocessor defines the fate of pre-mRNA. Wiley Interdiscip Rev RNA 5: 647–658. [DOI] [PubMed] [Google Scholar]
- Melamed Z, Levy A, Ashwal-Fluss R, Lev-Maor G, Mekahel K, Atias N, Gilad S, Sharan R, Levy C, Kadener S, et al. 2013. Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions. Mol Cell 50: 869–881. [DOI] [PubMed] [Google Scholar]
- Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. 2010. Structure and activity of putative intronic miRNA promoters. RNA 16: 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. 1993. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature 365: 364–368. [DOI] [PubMed] [Google Scholar]
- Morlando M, Ballarino M, Gromak N, Pagano F, Bozzoni I, Proudfoot NJ. 2008. Primary microRNA transcripts are processed co-transcriptionally. Nat Struct Mol Biol 15: 902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS. 2015. Functional anatomy of the human microprocessor. Cell 161: 1374–1387. [DOI] [PubMed] [Google Scholar]
- Pastor T, Dal Mas A, Talotti G, Bussani E, Pagani F. 2011. Intron cleavage affects processing of alternatively spliced transcripts. RNA 17: 1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins PB, Lin Q, Goodnough JB, Tian H, Chen X, Khavari PA. 2001. In vivo restoration of laminin 5 β3 expression and function in junctional epidermolysis bullosa. Proc Natl Acad Sci 98: 5193–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlackow M, Nojima T, Gomes T, Dhir A, Carmo-Fonseca M, Proudfoot NJ. 2017. Distinctive patterns of transcription and RNA processing for human lincRNAs. Mol Cell 65: 25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CW, Valcarcel J. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem Sci 25: 381–388. [DOI] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. 2004. Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305: 1434–1437. [DOI] [PubMed] [Google Scholar]
- Sundaram GM, Common JE, Gopal FE, Srikanta S, Lakshman K, Lunny DP, Lim TC, Tanavde V, Lane EB, Sampath P. 2013. ‘See-saw’ expression of microRNA-198 and FSTL1 from a single transcript in wound healing. Nature 495: 103–106. [DOI] [PubMed] [Google Scholar]
- Triboulet R, Chang HM, Lapierre RJ, Gregory RI. 2009. Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 15: 1005–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan I, Jonkman MF. 2015. Blistering disease: insight from the hemidesmosome and other components of the dermal-epidermal junction. Cell Tissue Res 360: 545–569. [DOI] [PubMed] [Google Scholar]
- Wagschal A, Rousset E, Basavarajaiah P, Contreras X, Harwig A, Laurent-Chabalier S, Nakamura M, Chen X, Zhang K, Meziane O, et al. 2012. Microprocessor, Setx, Xrn2, and Rrp6 co-operate to induce premature termination of transcription by RNAPII. Cell 150: 1147–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S, Proudfoot NJ. 2009. Transcriptional termination enhances protein expression in human cells. Mol Cell 33: 354–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Lim E, Vaillant F, Asselin-Labat ML, Visvader JE, Smyth GK. 2010. ROAST: rotation gene set tests for complex microarray experiments. Bioinformatics 26: 2176–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuarin J, Schibler U. 1994. Physical isolation of nascent RNA chains transcribed by RNA polymerase II: evidence for cotranscriptional splicing. Mol Cell Biol 14: 7219–7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. 2003. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambruno G, Marchisio PC, Marconi A, Vaschieri C, Melchiori A, Giannetti A, De Luca M. 1995. Transforming growth factor-β1 modulates β1 and β5 integrin receptors and induces the de novo expression of the ανβ6 heterodimer in normal human keratinocytes: implications for wound healing. J Cell Biol 129: 853–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubović L, Baralle M, Baralle FE. 2012. Mutually exclusive splicing regulates the Nav 1.6 sodium channel function through a combinatorial mechanism that involves three distinct splicing regulatory elements and their ligands. Nucleic Acids Res 40: 6255–6269. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.