Abstract
Genomic and transcriptome sequencing of bladder cancer (BLCA) has identified multiple molecular alterations during cancer progression. Many of these identified genetic and epigenetic changes play a role in the progression of this disease. Studies have identified molecular subtypes in muscle-invasive bladder cancer (MIBC) with different sensitivities to frontline therapy suggesting the heterogeneity in these tumors and the importance of molecular characterization of MIBC to provide effective treatment. Specifically, it has become increasingly evident, as demonstrated by The Cancer Genome Atlas project, that metabolic enzymes are commonly dysregulated in BLCA. Elevated expression of multiple metabolic enzymes is due to the increased demand from rapidly proliferating BLCA cells requiring extensive nucleotide synthesis. Cancer cells utilize the de novo purine and pyrimidine biosynthetic pathway as a source of their nucleotide needs. In this study, we show that phosphoribosyl aminoimidazole succinocarboxamide synthetase (PAICS), an enzyme involved in de novo purine biosynthetic pathway, is significantly overexpressed in BLCA. Immunohistochemical staining of paraffin-embedded tissue sections showed that PAICS is overexpressed in MIBC. Furthermore, we found that tumor suppressor miR-128 negatively regulated PAICS expression by binding to its 3′-untranslated region. We also found that PAICS induces EMT by positively regulating SNAI1 and by a reduction in E-cadherin expression. Additionally, our in vitro functional studies and in vivo chicken chorioallantoic membrane assay show that PAICS plays a critical role in BLCA cell proliferation, invasion, and tumor growth. Collectively, our data suggest that targeting PAICS may provide a therapeutic option in BLCA.
Introduction
Bladder cancer (BLCA) is a common disease, with an estimated 81,190 new cases and 17,240 deaths in 2018 in the United States [1]. Metastatic urothelial carcinoma of the bladder is generally incurable by current platinum-based first-line chemotherapy and leads to early mortality with a median survival of 12-15 months [2]. T-cell checkpoint inhibitors (e.g., atezolizumab, nivolumab, pembrolizumab, durvalumab, avelumab) have recently provided durable benefits following prior platinum therapy to a minority (~20%) of patients, but the median survival is still only 8-10 months [3], [4], [5], [6]. Multiple molecular alterations play a role in the progression of this disease. Recent studies have identified molecular subtypes of muscle-invasive bladder cancer (MIBC) with different sensitivities to chemotherapy, suggesting that the heterogeneity in these tumors and their molecular characterization have an impact in effectiveness of treatment [7], [8], [9], [10]. Specifically, it has become increasingly evident, as demonstrated by The Cancer Genome Atlas (TCGA) project, that epigenetic and metabolic enzyme changes play a pivotal role in regulating gene expression, cancer metabolism, and the eventual development of BLCA [11].
Alterations in cellular metabolism are now recognized as an emerging hallmark of cancer [12]. The common feature of altered mechanism in tumor cells is the increased glucose uptake and glycolytic rates compared to resting cells under aerobic conditions, which is known as “Warburg Effect” (reviewed in [13]). Several studies have demonstrated the mechanism by which this and other metabolic changes allow cancer cells to accumulate building blocks for the biosynthesis of macromolecules (reviewed in [14]). Apart from serving as building blocks for nucleic acids, purine metabolites provide cofactors and necessary energy for cell survival and proliferation [15]. The purine levels are maintained by a coordinated action of the salvage and de novo biosynthetic pathways. Generally, most of the cellular purine levels are maintained by recycling of degraded bases via the salvage pathway [16], [17], [18]. Cancer cells, with their higher demand for the purines, utilize the de novo biosynthetic pathway [16], [18], [19], [20]. The de novo biosynthetic pathway utilizes phosphoribosyl pyrophosphate (PRPP) to generate inosine 5′-monophosphate and is carried out in 10 steps by 6 sequential enzymes. The first reaction in the de novo purine biosynthetic pathway is the conversion of PRPP to 5-phosphoribosylamine by PRPP amidotransferase and is presumed to be a rate-limiting step. One of the bifunctional enzymes in this cascade, phosphoribosyl aminoimidazole carboxylase/phosphoribosyl aminoimidazole succinocarboxamide synthetase (PAICS), utilizes AIR to generate N-succinocarboxyamide-5-aminoimidazole ribonucleotide and aminoimidazole-4-carboxamide ribonucleotide by adenylosuccinate lyase [15].
Earlier studies have shown that de novo purine biosynthetic pathway enzymes are dysregulated in prostate cancer [21], [22] and glioma [23]. During oncogenic transformation, alternations in cellular metabolism have been shown to be involved in cancer cell proliferation. However, the metabolic changes in promoting cancer cell aggressiveness and epithelial-mesenchymal transition (EMT) are poorly understood.
In the present study, we show that PAICS, which catalyzes a critical step in purine biosynthetic pathway, is overexpressed and plays a role in BLCA cell proliferation, colony formation, and 3D spheroid invasion, suggesting a role in oncogenesis. Furthermore, we found that tumor suppressor miR-128 negatively regulates PAICS expression. In addition, we found that PAICS induces EMT by positively regulating SNAI1 and by a reduction in E-cadherin expression. Our observations support the hypothesis that PAICS plays a crucial role in maintenance of the mesenchymal phenotype by downregulating the expression of E-cadherin. Thus, these results indicate that PAICS is a key player in EMT induction and that PAICS expression is associated with invasion and metastasis of BLCA cells.
Material and Methods
Cell Lines
BLCA cell line VM-CUB1 was grown in MEM (Corning cellgro, ThermoFisher Scientific, Atlanta, GA), while RT112 and 5637 were grown in RPMI; all media were supplemented with penicillin-streptomycin (100 U/ml) and 10% FBS (Invitrogen) in 5% CO2 cell culture incubator. BLCA cells were infected with lentiviruses expressing PAICS shRNA or nontargeting shRNA controls, and stable cell lines were generated by selection with 1 μg/ml puromycin (Life Technologies).
Benign and Tumor Tissues
In this study, we utilized formalin-fixed paraffin-embedded tissues from both normal and clinically localized BLCA patients. The bladder tissues were collected with an Institutional Review Board–approved retrospective study at the University of Alabama at Birmingham (UAB), which allowed the investigation of deidentified sample obtained from human subjects.
Gene Expression from The Cancer Genome Atlas (TCGA)
Gene expression level of PAICS in BLCA and adjacent normal samples was obtained from UALCAN web portal which provides boxplots depicting each gene's expression based on level 3 RNA-seq data from TCGA transcriptome sequencing datasets [24].
Immunohistochemistry (IHC)
IHC was carried out to evaluate PAICS expression using mouse monoclonal antibody against PAICS (GeneTex, CA, catalog #GTX83950, 1:10,000). Antigen retrieval was performed by boiling the slides (paraffin-embedded tissue sections) for 10 minutes in citrate buffer (Sigma Aldrich, MO, catalog #C9999-1000ML). Immunostaining was performed using Vector Laboratories staining kit following the manufacturer's protocol.
RNA Extraction and Quantitative Real-Time PCR Analysis
Total RNA was isolated from BLCA cell lines using the RNeasy mini kit (Qiagen, Valencia, CA). RNA was reverse transcribed into complementary DNA using Superscript III Reverse Transcriptase (Invitrogen). qPCR was performed as described [25]. SYBR green was used to determine the mRNA expression level of a gene of interest. All primers for SYBR green were synthesized by Integrated DNA Technologies (Coralville, IA). Primer sequences used for SYBR green qPCR are PAICS: forward primer, GTGGCAGGCAGAAGTAATGG, and reverse primer, CACATCCTGAACTCCCCAGT; ACTB: forward primer, GCACAGAGCCTCGCCTT, and reverse primer, GTTGTCGACGACGAGCG; SNAI1: forward primer, TGCCCTCAAGATGCACATCCGA, and reverse primer, GGGACAGGAGAAGGGCTTCTC; SLUG: forward primer, ATCTGCGGCAAGGCGTTTTCCA, and reverse primer, GAGCCCTCAGATTTGACCTGTC; VIM: CCCTCACCTGTGAAGTGGAT, and reverse primer, GCTTCAACGGCAAAGTTCTC. All PCRs were performed in triplicates.
Immunoblot Analyses
Protein samples were harvested with NP-40 lysis buffer (Boston Bioproducts, Ashland, MA). Then, the protein quantification was performed using BioRad DC protein assay (Bio-Rad Laboratories, Hercules, CA). For immunoblot analysis, 10-μg protein samples were separated on a NuPAGE 4%-12% Bis-Tris protein gel and transferred onto Immobilon-P PVDF membrane (EMD Millipore, Billerica, MA). The membrane was incubated for 1 hour in blocking buffer (Tris-buffered saline with 0.1% Tween and 5% nonfat dry milk) followed by incubation overnight at 4°C with the primary antibody. After two washes for 5 minutes each with Tris-buffered saline with 0.1% Tween, the blot was incubated with horseradish peroxidase–conjugated secondary antibody (1:5000) for 1 hour at room temperature, and signals were visualized by Luminata Forte chemiluminescence Western blotting substrate as per manufacturer's protocol (EMD Millipore). Antibodies used in the study are anti-PAICS (Mo): #GTX83950 (IB, 1:5000, IHC, 1:10000; GeneTex, Irvine, CA), anti–E-cadherin (Mo): #610181 (IB, 1: 20,000; BD Pharmingen, San Diego, CA), anti–HRP-β-actin: # HRP-60008 (IB, 1:200000; PTG Labs, Rosemont, IL), and anti-mouse IgG HRP: # SA00001-1 (IB, 1:5000; PTG Labs, Rosemont, IL).
RNA Interference and Transfection
The PAICS and nontargeting small interfering RNA (siRNA) duplexes were purchased from Dharmacon, Lafayette, CO (GE Healthcare). Transfections were performed with Lipofectamine RNAiMAX (Life Technologies) reagent. PAICS shRNAs [22] were purchased from SBI (System Biosciences, Mountain View, CA). Lentiviruses for these stable knockdowns were generated by the UAB Neuroscience NINDS Protein Core (P30 NS47466). For RNA interference or miRNAs transfection, the reverse transfection was performed by co-incubation of BLCA cells in a six-well plate with siRNA duplexes or miRNA as per manufacturer's instructions. Seventy-two hours after the transfection, cells were harvested for RNA isolation or immunoblot analysis.
Cell Proliferation Assays
Cell proliferation was measured by cell counting. For this, transient and stable PAICS knockdowns, and miR-128–treated cells were used. After 72 hours of transfection using specific siRNA or miRNA, the cells were trypsinized and seeded at a density of 10,000 cells/well in 24-well plates (n = 3). Nontargeting si/shRNA/miRNA-treated cells were served as controls. Then, the cells were trypsinized and counted at specified time points by Z2 Coulter particle counter (Beckman Coulter, Brea, CA). Each experiment has been performed with three replicates per sample.
3D Spheroid Cell Invasion Assays
The 3D spheroid cell invasion kit (Trevigen, Gaithersburg, MD) was used according to the manufacturer's protocol. Briefly, the test cells (10,000) were seeded in a basement membrane extract and cultured for 72 hours. Once spheroids formed, the invasion matrix was added and then supplemented with 10%FBS-RPMI after 1 hour. Images were taken after 3 days.
Matrigel Invasion Assay
Matrigel invasion assays were performed as described earlier [22], [25], [26], [27]. Various test cells were seeded onto Corning BioCoat Matrigel matrix (Corning, New York, NY) in the upper chamber of a 24-well culture plate. The lower chamber containing respective medium was supplemented with 10% serum as a chemoattractant. After 48 hours, the noninvading cells and Matrigel matrix were gently removed with a cotton swab. Invasive cells located on the lower side of the chamber were stained with 0.2% crystal violet in methanol, air-dried, and photographed using an inverted microscope (4×).
Colony Formation Assay
After 72 hours of transfection, the various test cells were counted and seeded 800 cells per 1 well of 6-well plates (triplicates) and incubated at 37°C, 5% CO2 for 10 days. Colonies were fixed with 10% (v/v) glutaraldehyde for 30 minutes and stained with crystal violet (Sigma-Aldrich, St. Louis, MO) for 20 minutes. Then, the photographs of the colonies were taken using Amersham Imager 600RGB (GE Healthcare Life Sciences, Pittsburgh, PA).
Immunofluorescence
After 72 hours of non–T- or PAICS-siRNA transfection, the RT112 cells were seeded in chamber slides (Lab-Tek II CC2, Nunc, Rochester, NY) and incubated for a day to immunostain with rabbit monoclonal E-cadherin antibody (IF, 1:200; catalog #3195, Cell Signaling Technology, Danvers, MA). The slides were washed with phosphate-buffered saline (PBS) and were fixed using ice-cold methanol. Following three times PBS with 0.05% Tween 20 (PBS-T) wash, the slides were blocked for 2 hours using 5% normal horse serum in PBS-T. A rabbit anti-E-cadherin antibody was added to the slides at 1:200 dilution and incubated for 1 hour at room temperature. Following three times PBS-T wash, the slides were incubated with Alexa 555–conjugated goat, anti-rabbit antibody (Invitrogen) for 1 hour in the dark at room temperature. After three times PBS wash, the slides were mounted using ProLong Gold Antifade Mountant with DAPI (catalog #P36931, Invitrogen, ThermoFisher Scientific, Carlsbad, CA). Confocal images were taken with a 60× lens Nikon A1 High Speed Laser Confocal Spectral Imaging microscope (Nikon Instruments Inc., Melville, NY) from UAB high-resolution imaging facility.
miR Reporter Luciferase Assays
Wild-type or mutant 3′-untranslated regions (UTRs) of PAICS were cloned into the pMIR-REPORT miRNA Expression Reporter Vector (Life Technologies). VM-CUB1 cells were co-transfected with pre-miR-128-3p (#PM11746) or controls (#AM17110) and wild-type or mutant 3′-UTR-luc, as well as pRL-TK vector as an internal control for luciferase activity. Forty-eight hours posttransfection, the cells were lysed, and luciferase assays were conducted using the dual luciferase assay system (Promega, Madison, WI). Each experiment was performed in triplicate.
Chicken Chorioallantoic Membrane (CAM) Assay
The CAM assay for tumor (or xenograft) formation was performed as previously described [22], [25], [26], [27]. After 3 days of implanting the cells in each egg, extraembryonic tumors were isolated and weighed. An average of eight eggs per group was used in each experiment.
Statistical Analysis
To determine significant differences between two groups, Student's two-tailed t test was used. P values < .05 were considered significant.
Results
De Novo Purine Biosynthetic Enzyme PAICS Is Overexpressed in BLCA
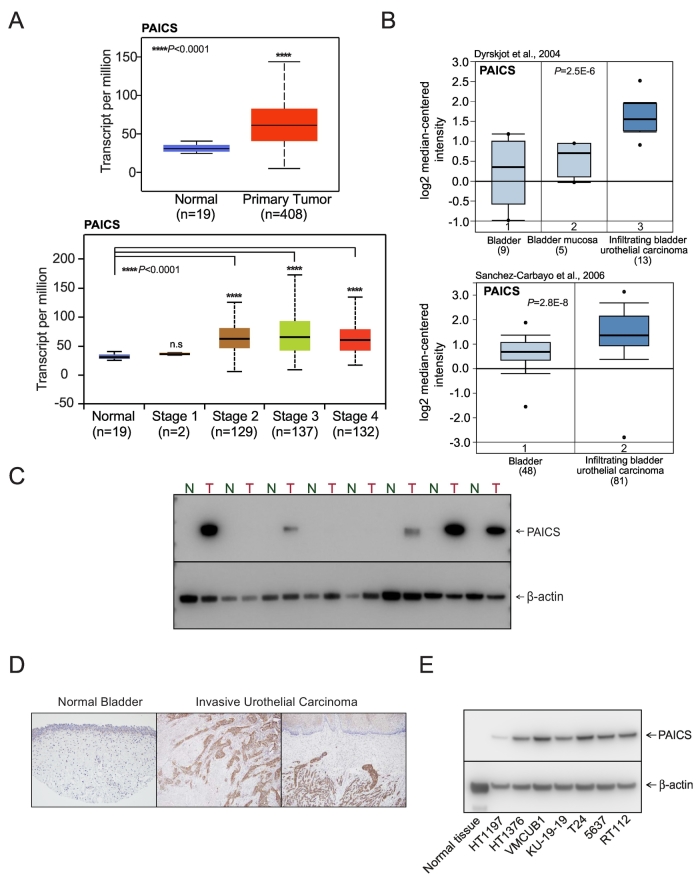
The transcriptome sequence analysis showed upregulation of several oncogenes including PAICS across different stages of bladder urothelial carcinoma (Figure 1A). Additionally, TCGA data also showed the elevated expression of PAICS in primary bladder urothelial carcinoma (Figure 1A). Next, we conducted Oncomine analysis (Life Technologies, Ann Arbor, MI) of previously described expression arrays and found that PAICS is elevated in infiltrating bladder urothelial carcinoma (Figure 1B) when compared with normal tissues. Immunoblot analysis using PAICS-specific antibody indicated PAICS protein overexpression in bladder carcinoma compared to normal tissues (Figure 1C). Additionally, we investigated PAICS protein expression in BLCA samples by immunohistochemical analysis, and invasive urothelial carcinoma showed overexpression of PAICS compared to normal bladder epithelium (Figure 1D). Similarly, elevated levels of PAICS protein were observed in BLCA cell lines (Figure 1E). Together, these findings suggested an elevated expression of PAICS in BLCA.
Figure 1.
PAICS, a de novo purine biosynthetic pathway enzyme, is overexpressed in bladder adenocarcinoma. (A) Expression of PAICS in normal bladder and primary tumor samples from TCGA and a boxplot showing relative expression of PAICS mRNA transcript in normal and stage 1, 2, 3, and 4 BLCA patients. (B) Boxplots represent PAICS expression level across normal bladder, mucosa, and infiltrating bladder urothelial carcinoma (top [49]) and across normal bladder and infiltrating bladder urothelial carcinoma (bottom [50]). The data were retrieved from publicly available microarray datasets of BLCA with log2 median intensity using Oncomine array datasets. (C) Immunoblot analysis showing PAICS expression in matched normal bladder and tumor tissues. Here, b-actin served as a loading control. (D) Immunohistochemical analysis of PAICS in normal bladder tissue and invasive urothelial carcinoma. (E) PAICS protein levels in various BLCA cell lines by immunoblot analysis.
Knockdown of PAICS Inhibits Cell Proliferation, Colony Formation, and 3D Spheroid Formation
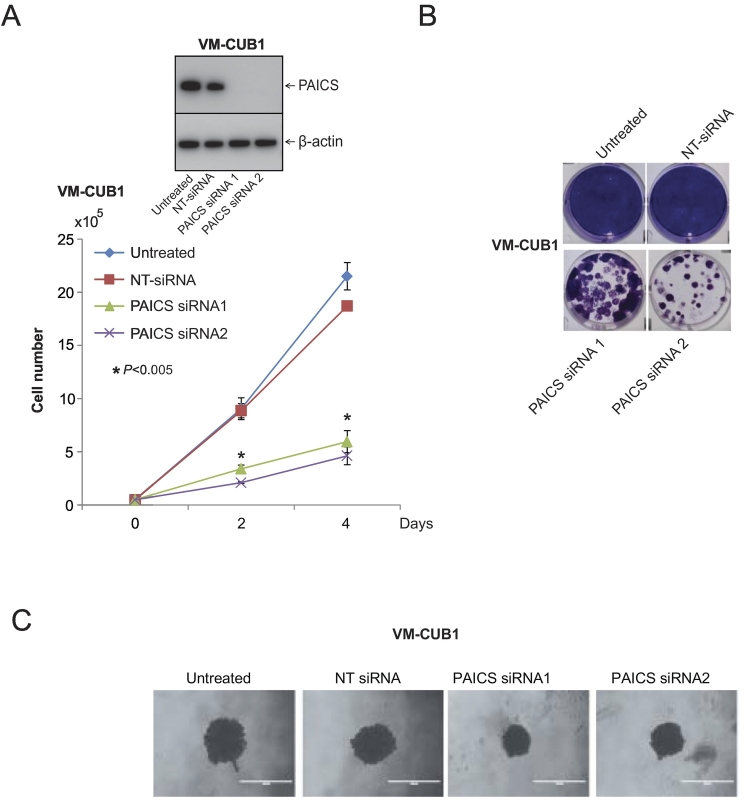
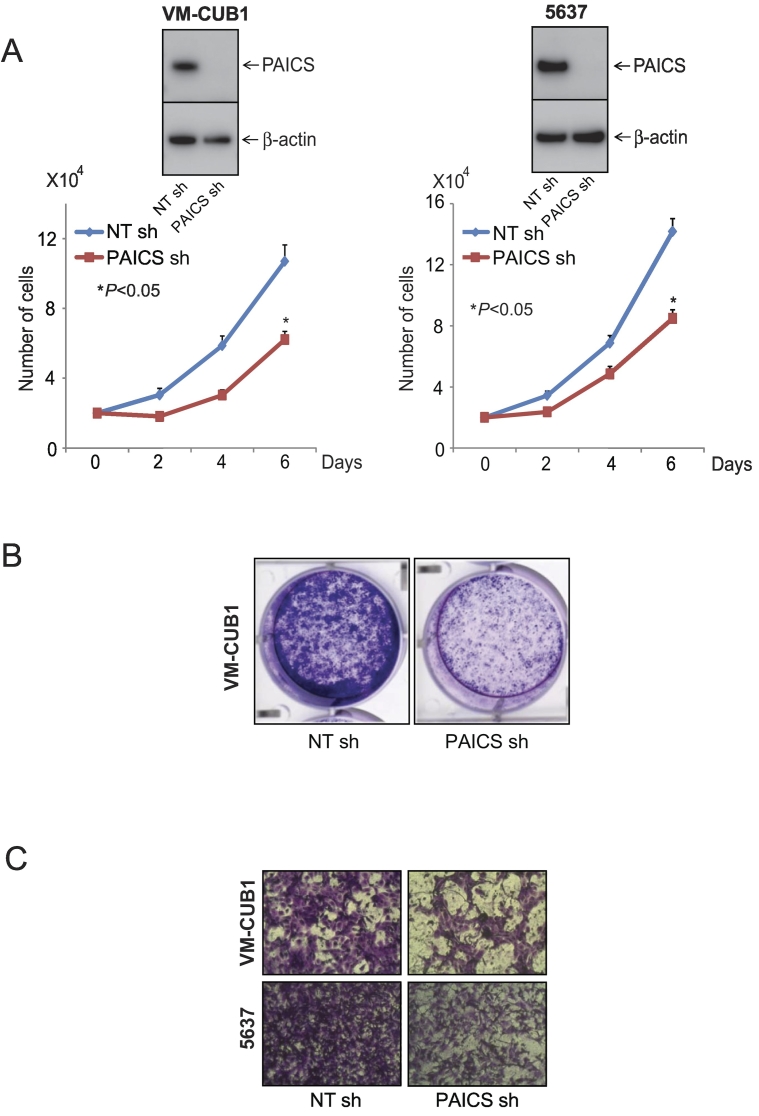
To investigate the role of PAICS in BLCA, we performed transient knockdown of PAICS using two independent and specific siRNAs in BLCA cell line VM-CUB1 and tested cell proliferation, colony formation, and 3D spheroid formation. Similarly, we also performed stable knockdown of PAICS in VM-CUB1 and 5637 BLCA cells using PAICS shRNA. To confirm the knockdown efficiency, we performed immunoblot analysis using the protein lysates prepared after 72 hours of transient transfection (Figure 2A, inset) or after puromycin selection (Figure 3, A and B, insets). The cell proliferation assay using these test cells indicated a significant decrease in cell number upon knockdown of PAICS (Figures 2A and 3A). Next, we performed colony formation assay using both transiently and stably knocked down PAICS cells (Figure 2B and 3B, respectively). Similarly, the knockdown cells showed reduced spheroid formation when compared with untreated and nontargeting siRNA treatments in 3D assay (Figure 2C). Next, we tested the invasive potential of these cells after stable knockdown. PAICS knockdown in VM-CUB1 and 5637 reduced the invasive potential of these cells as assessed by Boyden chamber Matrigel invasion assay (Figure 3C). The above results suggest that PAICS is involved in BLCA cell proliferation, colony formation, 3D spheroid formation, and invasion in vitro.
Figure 2.
PAICS is essential for BLCA cell growth. (A) Transient knockdown of PAICS in BLCA cell line VM-CUB1 reduces cell proliferation. Transient knockdown was achieved by two specific and independent PAICS siRNA duplexes. Nontargeting siRNA served as a negative control. PAICS knockdown was confirmed by immunoblot analysis using specific monoclonal antibody. Here, we used b-actin as a loading control. (B) Colony formation assay of VM-CUB1 cells after treating with either PAICS siRNA or NT-siRNA. The photographic images of colony formation assay are shown. (C) Photographic images of 3D Spheroid cell invasion assay. VM-CUB1 cells were treated with PAICS siRNA or NT-siRNA for 72 hours followed by seeding in a basement membrane extract to allow spheroids to form. Images were taken after 3 days of the invasion matrix addition.
Figure 3.
PAICS regulates BLCA cell proliferation, colony formation and invasion. (A) Stable knockdown of PAICS reduces cell proliferation of BLCA cell lines VM-CUB1 and 5637. Immunoblot analysis showing PAICS protein in VM-CUB1 and 5637 cells stably transduced with a shRNA specific to PAICS or control nontargeting shRNA. (B) Photographic images of colony formation assay using above test cells. (C) PAICS shRNA-treated BLCA cells showed reduced invasion in Boyden chamber Matrigel invasion assay. Invaded cells were stained with crystal violet.
miR-128 Regulates PAICS in BLCA
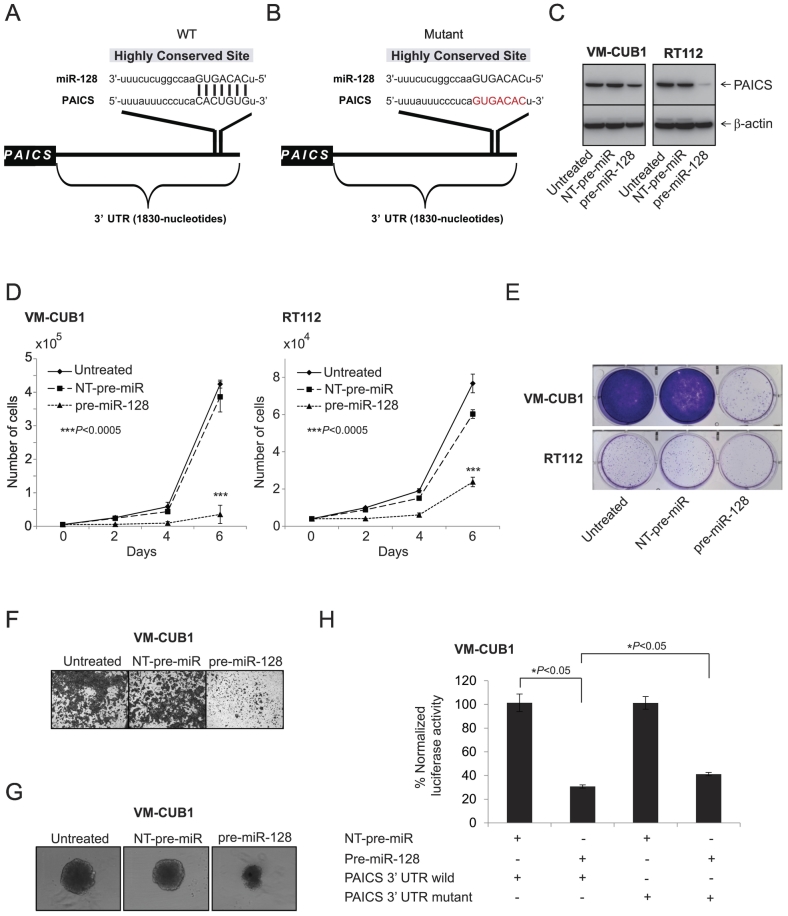
To investigate the potential miRNAs that target PAICS, we used freely available web-based miR target prediction resources: miRanda and TargetScan. We identified that miR-128 could potentially target PAICS. The binding site for miR-128 at 3′-UTR of PAICS is indicated (Figure 4, A and B). MiR-128 is known to have tumor suppressor function by targeting several oncogenes including ZEB1 [28] and RPS6KB1 [29] in prostate cancer, VEGF-C in BLCA [30], CYP2C9 in hepatocellular carcinoma [31], and p70S6K1 in glioma [32]. Therefore, we sought to determine its role in PAICS regulation. We treated BLCA cell lines VM-CUB1 and RT112 with a precursor miR-128 and tested PAICS. We observed downregulation of PAICS protein levels significantly by mir-128 (Figure 4C). Further, miR-128–treated cells showed a decrease in cell proliferation (Figure 4D), colony formation (Figure 4E), Matrigel invasion (Figure 4F), and 3D spheroid formation (Figure 4G). Next, to determine whether miR-128 directly binds PAICS 3′-UTR and regulates it, VM-CUB1 cells were co-transfected with miR-128 and pMir-REPORT-PAICS 3′-UTR plasmids. MiR-128 showed substantial reduction in luciferase reporter activity compared with nontargeting control miR (Figure 4H). This reduction in luciferase activity is curtailed when miR-128 target site is mutated (Figure 4H). These results indicate that PAICS is a direct target of miR-128.
Figure 4.
miR-128 targets and downregulates PAICS in BLCA. (A, B) The predicted miR-128 binding sites in the 3′-UTRs of PAICS. Schematic representation of wild-type and mutant PAICS 3′-UTRs is given, respectively. Red nucleotides represent mutations that are predicted to disrupt potential miR-128 binding. BLCA cells VM-CUB1 and RT112 were treated with pre-miR-128, and (C) immunoblot analysis was performed showing PAICS protein expression, (D) cell proliferation, (E) colony formation, (F) Matrigel invasion, and (G) 3D spheroid formation. (H) Luciferase reporter assay of PAICS-3′-UTR. 293T cells were transfected either with pre-miR-128 or nontargeting pre-miR (NT-pre-miR) along with either PAICS-3′-UTR wild-type or mutant luciferase constructs. pRL-TK vector was used as an internal control.
PAICS Is Involved in EMT
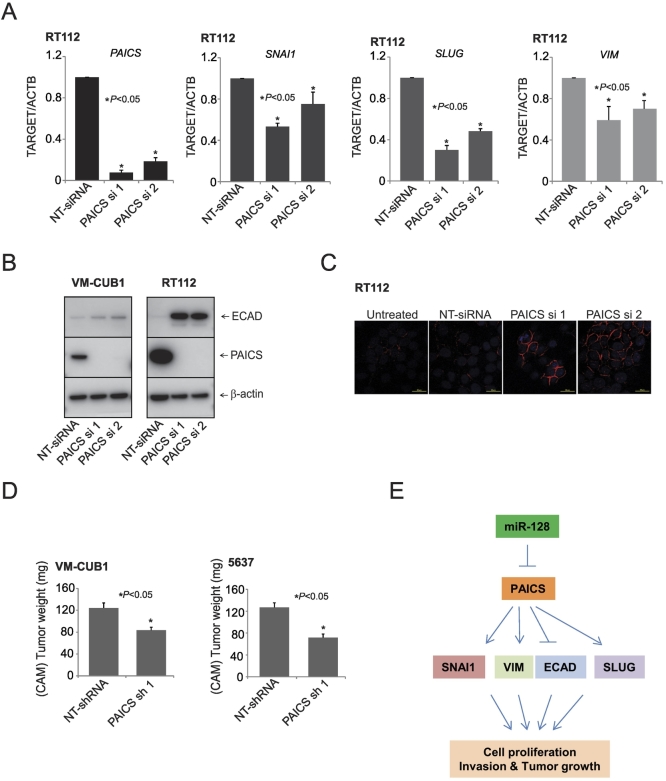
The invasion and metastasis of the tumor cells are known to be regulated by EMT [12]. Therefore, to understand the molecular mechanism underlying the PAICS-dependent growth and invasion of BLCA cells, we examined the levels of E-cadherin protein after PAICS depletion in BLCA cells. For this, we used VM-CUB1 and RT112 cells which express various EMT markers. Our qPCR analysis showed a decrease in SNAI1, a key transcription factor involved in EMT [33], [34]. SNAI1 is known to repress E-cadherin expression transcriptionally by binding to three E-boxes present in the human E-cadherin promoter [35], [36]. Additionally, we also observed decreased levels of SLUG and VIM transcript upon PAICS knockdown (Figure 5A). Among these, PAICS depletion increased E-cadherin expression in both VM-CUB1 and RT112 cells (Figure 5B). Concurrently, the E-cadherin protein levels were elevated in situ in RT112 cells upon PAICS knockdown (Figure 5C). Taken together, these results demonstrated the role of PAICS in EMT of BLCA.
Figure 5.
PAICS is involved in tumor growth and epithelial to mesenchymal transition. (A) qPCR analysis of PAICS, SNAI1, SLUG, and Vimentin mRNA in PAICS knockdown or NT-siRNA–treated RT112 cells. (B) Immunoblot analysis showing the levels of PAICS and E-cadherin proteins after PAICS knockdown. (C) Immunostaining of the BLCA cell line RT112 transfected with PAICS and nontargeting siRNA. Red staining represents E-cadherin, and blue represents nuclear staining with DAPI. (D) Tumor growth of VMCUB1 and 5637 PAICS stable knockdown cells or nontargeting control shRNA cells in the CAM tumor assay. Extraembryonic tumors were harvested and weights were measured 72 hours postimplantation of cells. (E) Proposed model of miR-128, PAICS, and EMT regulatory axis in BLCA progression. miR-128 downregulates PAICS expression, and further, PAICS regulates EMT markers SNAI1, VIM, ECAD, and SLUG. PAICS induces cell proliferation, invasion, and tumor growth via these EMT markers.
Role of PAICS in BLCA Growth In Vivo
To investigate the role of PAICS in tumorigenesis, we generated VM-CUB1and 5637 cells stably expressing PAICS or nontargeting shRNA through the lentivirus expression system. Next, we confirmed the knockdown of PAICS by Western blot before proceeding for in vivo CAM assay. The fertilized eggs injected with the PAICS-depleted cells produced small tumors than the eggs that were injected with the control cells (Figure 5D). Tumor weights were also significantly smaller in eggs that were injected with PAICS-depleted cells. These results suggest that PAICS promotes tumor initiation and progression in vivo.
Discussion
In this study, we evaluated the expression, role, and regulation of the de novo purine metabolic enzyme PAICS in BLCA. We identified and validated the overexpression of the de novo purine biosynthetic enzyme PAICS in BLCA using RNA and protein expression analyses. Cancer cells need increased nucleotides and amino acids to cater to the needs of the enhanced cell division. For purine and pyrimidine biosynthesis in cancer cells which produce nucleotides, the salvage pathway will not be sufficient. Hence, cancer cells resort to de novo biosynthesis using the intermediate 5′-phosph-D-rybosyl-1 pyrophosphate (PRPP), a glucose metabolic intermediate. Through a series of enzymatic conversions, PRPP is converted to inosine monophosphate. Gene expression profiling as well as transcriptome sequencing of BLCA suggested upregulation of de novo biosynthetic pathway enzymes. PAICS is a critical enzyme which produces N-succinocarboxyamide-5-aminoimidazole ribonucleotide (SAICAR), which is shown to be involved in PKM2 activation under limiting glucose conditions. We confirmed overexpression of PAICS in BLCA by qPCR and protein expression by immunoblot analyses. IHC analyses using BLCA tissues further showed increased PAICS expression in invasive tumors. In order to test if PAICS in required for BLCA cell growth, we performed multiple functional assays, all of which suggested a critical role for PAICS in BLCA cell proliferation and invasion. These findings are similar to our earlier studies that demonstrated the essential role of PAICS in prostate cancer growth [22]. We further demonstrated using BLCA lines that PAICS knockdown resulted in significant reduction of cell viability, colony formation, 3D spheroid formation, cell migration, and cell invasion. In addition, we found that PAICS knockdown resulted in the induction of E-cadherin, a mesenchymal to epithelial transition marker, as well as the suppression of EMT markers SNAI1, SLUG, and vimentin. In order to investigate the role of PAICS tumor growth, we performed in vivo tumor growth assay using CAM. We found that PAICS knockdown considerably inhibited tumor formation and subsequent tumor growth. Thus, our studies showed that the de novo purine biosynthetic enzyme PAICS promotes bladder tumorigenesis and tumor progression.
EMT represents a series of major cellular events and signals for a change in cellular demand from rapid proliferation and metastasis. The changes in the cellular metabolism have been shown to trigger cancer cell proliferation, but the link between cell metabolism and cancer cell growth remains unclear. Indeed, several studies have reported the alterations of multiple metabolic enzymes upon EMT induction in various cancer types [37], [38], [39], [40], [41]. The function of E-cadherin is essential for the integrity of epithelial cells. Thereby, the downregulation of E-cadherin by EMT modulators is commonly observed [42]. During EMT, several pathways merge into a common end point of E-cadherin repression by activation of a series of transcription factors. Among these transcription factors, SNAI1 has been identified as a powerful E-cadherin repressor [35], [36]. In the present study, we observed the downregulation of SNAI1 in PAICS-depleted cells, thereby increasing the levels of E-cadherin.
Differential microRNAs expression has been linked to tumor growth, invasion, angiogenesis, and immune evasion [43], [44]. In prostate cancer, miR-128 was reported to be downregulated, functioning as a tumor suppressor to influence EMT and cancer stem cell growth [45], [46]. It has also been demonstrated that miR-128-3p accelerated cell cycle arrest and chromosomal instability in mitomycin-c–treated lung cancer cells [47]. Conversely, miR-128 mediates genomic stability by suppressing the mobility of endogenous retrotransposons [48]. However, further investigation needs to be performed to detect the upstream regulators of miR-128 in cancer.
Conclusion
In summary, our study identifies a role for tumor suppressor miR-128 in regulating PAICS, which in turn regulates EMT markers SNAI1, VIM, SLUG, and E-cadherin, thereby increasing cell proliferation, invasion, and bladder tumor growth in vivo (Figure 5E). Present studies also show that PAICS, a purine metabolic enzyme, is overexpressed in BLCA and promotes tumor growth. Our findings may therefore offer a potential therapeutic strategy for BLCA via targeting PAICS. PAICS inhibition may complement T-cell checkpoint inhibitors or chemotherapy, and these combinations may be worthy of investigation. Further assessment of the functions of PAICS is required to determine its biological significance in BLCA.
Acknowledgements
S. V. was supported by NIH/NCI R01CA157845 and R01CA154980.
Footnotes
Disclosure of Potential Conflicts of Interest: Guru Sonpavde, MD, was a consultant for BMS, Exelixis, Bayer, Sanofi, Pfizer, Novartis, Eisai, Janssen, Amgen, AstraZeneca, Merck, Genentech, Astellas/Agensys; research support to institution from Bayer, Amgen, Boehringer-Ingelheim, Merck, Sanofi, Pfizer; author for Uptodate; speaker for Clinical Care Options, Physicians Education Resource, Research to Practice, Onclive.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A, Arning M. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602–4608. doi: 10.1200/JCO.2005.07.757. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg JE, Hoffman-Censits J, Powles T, van der Heijden MS, Balar AV, Necchi A, Dawson N, O'Donnell PH, Balmanoukian A, Loriot Y. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma P, Retz M, Siefker-Radtke A, Baron A, Necchi A, Bedke J, Plimack ER, Vaena D, Grimm MO, Bracarda S. Nivolumab in metastatic urothelial carcinoma after platinum therapy (CheckMate 275): a multicentre, single-arm, phase 2 trial. Lancet Oncol. 2017;18:312–322. doi: 10.1016/S1470-2045(17)30065-7. [DOI] [PubMed] [Google Scholar]
- 5.Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, Vogelzang NJ, Climent MA, Petrylak DP, Choueiri TK. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376:1015–1026. doi: 10.1056/NEJMoa1613683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonpavde G. PD-1 and PD-L1 inhibitors as salvage therapy for urothelial carcinoma. N Engl J Med. 2017;376:1073–1074. doi: 10.1056/NEJMe1701182. [DOI] [PubMed] [Google Scholar]
- 7.Choi W, Porten S, Kim S, Willis D, Plimack ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee IL. Identification of distinct basal and luminal subtypes of muscle-invasive bladder cancer with different sensitivities to frontline chemotherapy. Cancer Cell. 2014;25:152–165. doi: 10.1016/j.ccr.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McConkey DJ, Choi W, Ochoa A, Siefker-Radtke A, Czerniak B, Dinney CP. Therapeutic opportunities in the intrinsic subtypes of muscle-invasive bladder cancer. Hematol Oncol Clin North Am. 2015;29:377–394. doi: 10.1016/j.hoc.2014.11.003. [x-xi] [DOI] [PubMed] [Google Scholar]
- 9.Seiler R, Ashab HA, Erho N, van Rhijn BW, Winters B, Douglas J, Van Kessel KE, Fransen van de Putte EE, Sommerlad M, Wang NQ. Impact of molecular subtypes in muscle-invasive bladder cancer on predicting response and survival after neoadjuvant chemotherapy. Eur Urol. 2017;72:544–554. doi: 10.1016/j.eururo.2017.03.030. [DOI] [PubMed] [Google Scholar]
- 10.Damrauer JS, Hoadley KA, Chism DD, Fan C, Tiganelli CJ, Wobker SE, Yeh JJ, Milowsky MI, Iyer G, Parker JS. Intrinsic subtypes of high-grade bladder cancer reflect the hallmarks of breast cancer biology. Proc Natl Acad Sci U S A. 2014;111:3110–3115. doi: 10.1073/pnas.1318376111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Research N Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507:315–322. doi: 10.1038/nature12965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cantor JR, Sabatini DM. Cancer cell metabolism: one hallmark, many faces. Cancer Discov. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pedley AM, Benkovic SJ. A new view into the regulation of purine metabolism: the purinosome. Trends Biochem Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Henderson JF, Khoo KY. On the mechanism of feedback inhibition of purine biosynthesis de novo in Ehrlich ascites tumor cells in vitro. J Biol Chem. 1965;240:3104–3109. [PubMed] [Google Scholar]
- 17.Murray AW. The biological significance of purine salvage. Annu Rev Biochem. 1971;40:811–826. doi: 10.1146/annurev.bi.40.070171.004115. [DOI] [PubMed] [Google Scholar]
- 18.Yamaoka T, Kondo M, Honda S, Iwahana H, Moritani M, Ii S, Yoshimoto K, Itakura M. Amidophosphoribosyltransferase limits the rate of cell growth-linked de novo purine biosynthesis in the presence of constant capacity of salvage purine biosynthesis. J Biol Chem. 1997;272:17719–17725. doi: 10.1074/jbc.272.28.17719. [DOI] [PubMed] [Google Scholar]
- 19.Mayer D, Natsumeda Y, Ikegami T, Faderan M, Lui M, Emrani J, Reardon M, Olah E, Weber G. Expression of key enzymes of purine and pyrimidine metabolism in a hepatocyte-derived cell line at different phases of the growth cycle. J Cancer Res Clin Oncol. 1990;116:251–258. doi: 10.1007/BF01612899. [DOI] [PubMed] [Google Scholar]
- 20.Natsumeda Y, Prajda N, Donohue JP, Glover JL, Weber G. Enzymic capacities of purine de Novo and salvage pathways for nucleotide synthesis in normal and neoplastic tissues. Cancer Res. 1984;44:2475–2479. [PubMed] [Google Scholar]
- 21.Barfeld SJ, Fazli L, Persson M, Marjavaara L, Urbanucci A, Kaukoniemi KM, Rennie PS, Ceder Y, Chabes A, Visakorpi T. Myc-dependent purine biosynthesis affects nucleolar stress and therapy response in prostate cancer. Oncotarget. 2015;6:12587–12602. doi: 10.18632/oncotarget.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chakravarthi BV, Goswami MT, Pathi SS, Dodson M, Chandrashekar DS, Agarwal S, Nepal S, Hodigere Balasubramanya SA, Siddiqui J, Lonigro RJ. Expression and role of PAICS, a de novo purine biosynthetic gene in prostate cancer. Prostate. 2017;77:10–21. doi: 10.1002/pros.23243. [DOI] [PubMed] [Google Scholar]
- 23.Wang X, Yang K, Xie Q, Wu Q, Mack SC, Shi Y, Kim LJY, Prager BC, Flavahan WA, Liu X. Purine synthesis promotes maintenance of brain tumor initiating cells in glioma. Nat Neurosci. 2017;20:661–673. doi: 10.1038/nn.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chakravarthi BV, Pathi SS, Goswami MT, Cieslik M, Zheng H, Nallasivam S, Arekapudi SR, Jing X, Siddiqui J, Athanikar J. The miR-124-prolyl hydroxylase P4HA1-MMP1 axis plays a critical role in prostate cancer progression. Oncotarget. 2014;5:6654–6669. doi: 10.18632/oncotarget.2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakravarthi BV, Goswami MT, Pathi SS, Robinson AD, Cieslik M, Chandrashekar DS, Agarwal S, Siddiqui J, Daignault S, Carskadon SL. MicroRNA-101 regulated transcriptional modulator SUB1 plays a role in prostate cancer. Oncogene. 2016;35:6330–6340. doi: 10.1038/onc.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang R, Asangani IA, Chakravarthi BV, Ateeq B, Lonigro RJ, Cao Q, Mani RS, Camacho DF, McGregor N, Schumann TE. Role of transcriptional corepressor CtBP1 in prostate cancer progression. Neoplasia. 2012;14:905–914. doi: 10.1593/neo.121192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun X, Li Y, Yu J, Pei H, Luo P, Zhang J. miR-128 modulates chemosensitivity and invasion of prostate cancer cells through targeting ZEB1. Jpn J Clin Oncol. 2015;45:474–482. doi: 10.1093/jjco/hyv027. [DOI] [PubMed] [Google Scholar]
- 29.Tao T, Li G, Dong Q, Liu D, Liu C, Han D, Huang Y, Chen S, Xu B, Chen M. Loss of SNAIL inhibits cellular growth and metabolism through the miR-128-mediated RPS6KB1/HIF-1alpha/PKM2 signaling pathway in prostate cancer cells. Tumour Biol. 2014;35:8543–8550. doi: 10.1007/s13277-014-2057-z. [DOI] [PubMed] [Google Scholar]
- 30.Zhou XU, Qi L, Tong S, Cui YU, Chen J, Huang T, Chen Z, Zu XB. miR-128 downregulation promotes growth and metastasis of bladder cancer cells and involves VEGF-C upregulation. Oncol Lett. 2015;10:3183–3190. doi: 10.3892/ol.2015.3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu D, Green B, Marrone A, Guo Y, Kadlubar S, Lin D, Fuscoe J, Pogribny I, Ning B. Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma. Sci Rep. 2015;5:8534. doi: 10.1038/srep08534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi ZM, Wang J, Yan Z, You YP, Li CY, Qian X, Yin Y, Zhao P, Wang YY, Wang XF. MiR-128 inhibits tumor growth and angiogenesis by targeting p70S6K1. PLoS One. 2012;7:e32709. doi: 10.1371/journal.pone.0032709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 34.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 35.Batlle E, Sancho E, Franci C, Dominguez D, Monfar M, Baulida J, Garcia De Herreros A. The transcription factor snail is a repressor of E-cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84–89. doi: 10.1038/35000034. [DOI] [PubMed] [Google Scholar]
- 36.Cano A, Perez-Moreno MA, Rodrigo I, Locascio A, Blanco MJ, del Barrio MG, Portillo F, Nieto MA. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2:76–83. doi: 10.1038/35000025. [DOI] [PubMed] [Google Scholar]
- 37.Shaul YD, Freinkman E, Comb WC, Cantor JR, Tam WL, Thiru P, Kim D, Kanarek N, Pacold ME, Chen WW. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell. 2014;158:1094–1109. doi: 10.1016/j.cell.2014.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masin M, Vazquez J, Rossi S, Groeneveld S, Samson N, Schwalie PC, Deplancke B, Frawley LE, Gouttenoire J, Moradpour D. GLUT3 is induced during epithelial-mesenchymal transition and promotes tumor cell proliferation in non-small cell lung cancer. Cancer Metab. 2014;2:11. doi: 10.1186/2049-3002-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun Y, Daemen A, Hatzivassiliou G, Arnott D, Wilson C, Zhuang G, Gao M, Liu P, Boudreau A, Johnson L. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab. 2014;2:20. doi: 10.1186/2049-3002-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aspuria PP, Lunt SY, Varemo L, Vergnes L, Gozo M, Beach JA, Salumbides B, Reue K, Wiedemeyer WR, Nielsen J. Succinate dehydrogenase inhibition leads to epithelial-mesenchymal transition and reprogrammed carbon metabolism. Cancer Metab. 2014;2:21. doi: 10.1186/2049-3002-2-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dong C, Yuan T, Wu Y, Wang Y, Fan TW, Miriyala S, Lin Y, Yao J, Shi J, Kang T. Loss of FBP1 by Snail-mediated repression provides metabolic advantages in basal-like breast cancer. Cancer Cell. 2013;23:316–331. doi: 10.1016/j.ccr.2013.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y, Shi J, Chai K, Ying X, Zhou BP. The role of Snail in EMT and tumorigenesis. Curr Cancer Drug Targets. 2013;13:963–972. doi: 10.2174/15680096113136660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choudhury Y, Tay FC, Lam DH, Sandanaraj E, Tang C, Ang BT, Wang S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest. 2012;122:4059–4076. doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med. 2013;5:111. doi: 10.1186/gm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan AP, Poisson LM, Bhat VB, Fermin D, Zhao R, Kalyana-Sundaram S, Michailidis G, Nesvizhskii AI, Omenn GS, Chinnaiyan AM. Quantitative proteomic profiling of prostate cancer reveals a role for miR-128 in prostate cancer. Mol Cell Proteomics. 2010;9:298–312. doi: 10.1074/mcp.M900159-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nanta R, Kumar D, Meeker D, Rodova M, Van Veldhuizen PJ, Shankar S, Srivastava RK. NVP-LDE-225 (Erismodegib) inhibits epithelial-mesenchymal transition and human prostate cancer stem cell growth in NOD/SCID IL2Rgamma null mice by regulating Bmi-1 and microRNA-128. Oncogenesis. 2013;2:e42. doi: 10.1038/oncsis.2013.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang R, Liu C, Niu Y, Jing Y, Zhang H, Wang J, Yang J, Zen K, Zhang J, Zhang CY. MicroRNA-128-3p regulates mitomycin C-induced DNA damage response in lung cancer cells through repressing SPTAN1. Oncotarget. 2017;8:58098–58107. doi: 10.18632/oncotarget.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, Pedersen IM. miR-128 represses L1 retrotransposition by binding directly to L1 RNA. Nat Struct Mol Biol. 2015;22:824–831. doi: 10.1038/nsmb.3090. [DOI] [PubMed] [Google Scholar]
- 49.Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K, Orntoft TF. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res. 2004;64:4040–4048. doi: 10.1158/0008-5472.CAN-03-3620. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-Carbayo M, Socci ND, Lozano J, Saint F, Cordon-Cardo C. Defining molecular profiles of poor outcome in patients with invasive bladder cancer using oligonucleotide microarrays. J Clin Oncol. 2006;24:778–789. doi: 10.1200/JCO.2005.03.2375. [DOI] [PubMed] [Google Scholar]