Abstract
From an individual bacterium to the cells that compose the human immune system, cellular chemotaxis plays a fundamental role in allowing cells to navigate, interpret, and respond to their environments. While many features of cellular chemotaxis are shared among systems as diverse as bacteria and human immune cells, the machinery that guides the migration of these model organisms varies widely. In this article, we review current literature on the diversity of chemoattractant ligands, the cell surface receptors that detect and process chemotactic gradients, and the link between signal recognition and the regulation of cellular machinery that allow for efficient directed cellular movement. These facets of cellular chemotaxis are compared among E. coli, Dictyostelium discoideum, and mammalian neutrophils to derive organizational principles by which diverse cell systems sense and respond to chemotactic gradients to initiate cellular migration.
Keywords: chemotaxis, communication theory, G protein‐coupled receptor, methyl‐accepting chemotaxis protein receptor
Short abstract
We review the signal recognition process in mammalian neutrophils in order to derive general principles by which diverse cell systems sense and respond to chemotactic gradients to initiate cellular migration.
1. INTRODUCTION
1.1. Cell communication and movement
Biological organisms are open systems, exchanging energy and matter with the environment during replication, growth, reproduction, and death. These exchanges of energy and matter play out on the organismal level as well as the cellular level. Just as ants perceive and respond to their environments by interpreting and depositing pheromones, individual cells similarly communicate with their environments through the bidirectional exchange of biological information across the cell membrane.1, 2, 3 To ensure survival, a cell must be able to not only sense the state of the surrounding extracellular space (e.g., the presence or absence of nutrients), but to also enact an appropriate response (e.g., modification of cellular metabolism). Both prokaryotic and eukaryotic cells have evolved remarkably elegant communication systems for such purposes, and these sophisticated systems directly allow for the existence of complex cellular communities and multicellular organisms.
Importantly, cells are not static entities. When Robert Hooke peered through his microscope at a thin piece of cork, and, for the first time, described the biologic ‘cell’,4 he failed to appreciate the dynamic capabilities of living cells (the piece of cork he was observing was no longer living). The discovery that cells are motile entities was made contemporaneously by Antoni van Leeuwenhoek, who aptly described the animate behavior of the ‘little animalcules’ (microorganisms) he observed.5 Cellular movement is essential for nearly all cell types and enables diverse biologic phenomena ranging from the sourcing of nutrients by bacteria to leukocyte recruitment during inflammation.6
1.2. The diversity and complexity of directed migration
Directed migration occurs on multiple scales. The migration of alphaproteobacteria in response to the earth's magnetic field,7 the migration of neurons in the developing brain,8 and even the remarkable seasonal migration of bar‐tailed godwits,9 follow a common process: in each case, the organism interprets and responds to an environmental cue to guide its migratory route. In the context of individual cells, migration can be stimulated by incredibly diverse environmental cues, including light (i.e., phototaxis 10), chemicals (i.e., chemotaxis 11), temperature (i.e., thermotaxis 12), stiffness (i.e., durotaxis 13), electric fields (i.e., electrotaxis 14), or even gravity (i.e., gravitaxis 15). Importantly, cells do not typically have tactic responses when exposed to constant levels of environmental stimuli but migrate only in response to temporal or spatial gradients. Though gradient‐sensing capabilities exist in nearly all cells, the respective mechanisms by which prokaryotic and eukaryotic cells sense external gradients differ. Due to their small size, prokaryotes sense gradients temporally by randomly tumbling until they stochastically arrive at a region of higher stimulus concentration (i.e., moving “up” the gradient), at which point they decrease their tumbling rate to prolong movement toward the stimulus.16, 17, 18 In contrast, eukaryotic cells integrate both temporal and spatial information to sense gradients, utilizing cell surface receptor occupancy and complex intracellular signaling pathways to measure the stimulant concentration difference between the ends of the cell.19, 20, 21
Chemotaxis, the directed migration of cells in response to a chemical stimulus, is critical for the survival of cells and organisms alike. Prokaryotic chemotactic mechanisms have been most extensively studied in Escherichia coli, where a relatively simple signaling cascade promotes the counterclockwise or clockwise rotation of flagella to produce either forward motion or tumbles, respectively.18, 22, 23 Eukaryotic chemotactic mechanisms have been most extensively studied in the amoeboid Dictyostelium discoideum and mammalian neutrophils, where chemoattractants trigger intricate signaling cascades contributing to diverse cellular processes including the establishment of cellular polarity and extension of the cell membrane.17, 19, 24, 25 Both prokaryotic and eukaryotic cells exhibit unique morphologic modes of migration (e.g., swarming for prokaryotes; collective cell migration for eukaryotes) depending on cell type and environment,26, 27, 28 increasing the complexity surrounding the study of chemotaxis.
1.3. Chemotaxis as a cellular communication system
One approach to simplify the complex phenomenon of chemotaxis is to visualize it as a cellular communication system29, 30, 31, 32, 33 (Fig. 1). Claude Shannon, the father of communication theory, remarked in his landmark paper A Mathematical Theory of Communication, “The fundamental problem of communication is that of reproducing at one point either exactly or approximately a message selected at another point”.34 In an effective communication system, a message must be faithfully encoded by a transmitter, sent over a channel (where it encounters noise), delivered to a receiver, and then reconstructed into the same (or approximately the same) message (Fig. 1A). For the cellular communication system resulting in chemotaxis, the message to be transmitted is the chemotactic signal, or more specifically the gradient of chemoattractant molecules along which a cell should migrate (Fig. 1B and C). The fundamental problem then, as Shannon described, is the decoding and processing of that message by the cell in such a way that it can be reproduced (i.e., the cell can move along the chemotactic gradient).
Figure 1.
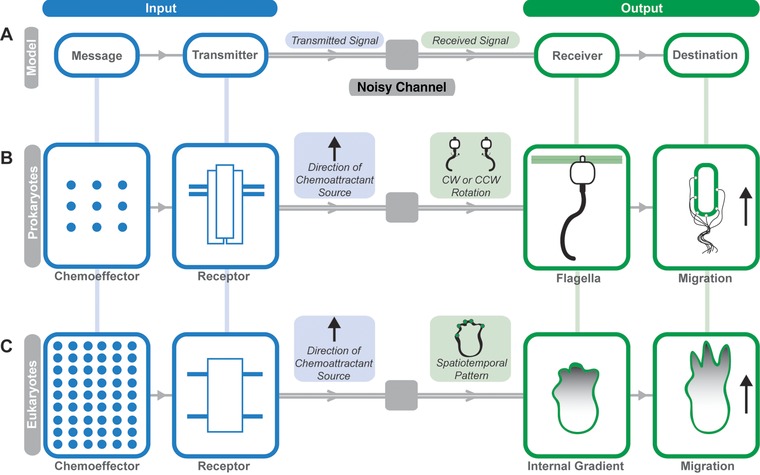
A communication model for cellular chemotaxis in prokaryotic and eukaryotic systems. (A) The Shannon‐Weaver model of communication. The input of the prokaryotic (B) and eukaryotic (C) chemotactic communication system can be considered the interaction of the message (i.e., chemoeffector) and the transmitter (i.e., chemoattractant receptor). The input is transmitted over a noisy channel (i.e., intracellular signaling cascades) to a receiver which decodes and reproduces the message in such a way to cause directed cellular movement. Prokaryotic and eukaryotic chemotactic communication systems use different messages and transmitters to achieve the same outcome (i.e., cellular migration). The objective of this review is to highlight the similarities and differences of the chemotactic input for prokaryotic and eukaryotic systems
The focus of this review is on utilizing an information theoretical framework to help decode the chemotactic signal. Specifically, we will consider how prokaryotic (Fig. 1B) and eukaryotic (Fig. 1C) chemotaxis systems have remarkably diverse input messages yet are able to convey the same meaning, namely cellular movement along the chemoattractant gradient. Additionally, we will explore how chemoattractants and receptor systems work in tandem to generate a signal for transmission over the channel, and the implications of using a common transmitter architecture (i.e., methyl‐accepting chemotaxis proteins (MCPs) in bacteria and G protein‐coupled receptors (GPCRs) in eukaryotes). By understanding chemotaxis by analogy to communication theory, we hope to apply principles such as message size, complexity, and redundancy, as well as transmitter capacity and combinatorial complexity to extract fundamental principles and paradoxes of diverse chemotaxis systems.
2. OVERVIEW OF THE MESSAGE AND TRANSMITTER
2.1. The input for chemotaxis
We begin by discussing general features of the input for chemotaxis, namely the chemoattractant and chemoattractant receptor systems. We draw upon Shannon's model of communication by considering both the message that is being conveyed and the transmitter that “operates on the message in some way to produce a signal suitable for transmission”.34 For our purposes, we designate the chemoattractant ligand as the message and the chemoattractant receptor as the transmitter, and the result of their interaction as the input (Fig. 1). The transmitter (chemoattractant receptor) can be said to “operate” on the message (chemoeffector ligand) to initiate complex signal transduction pathways (through the noisy channel) that ultimately lead to the output of chemotaxis. Here, we will compare and contrast the chemotactic input for prokaryotes (E. coli) and eukaryotes (Dictyostelium discoideum and mammalian neutrophils) in an attempt to derive general design principles for the architecture of effective chemotactic signal transmission.
2.2. The chemotactic message and transmitter for prokaryotes
First, we consider the message driving prokaryotic chemotaxis. As the most extensively studied prokaryotic chemotaxis systems are bacterial, we restrict our considerations here to bacterial species (specifically, to E. coli), though the archaeal systems that have been studied are remarkably similar.35 Bacteria are broadly capable of recognizing diverse chemoattractant and chemorepellant messages including amino acids, sugars, peptides, toxins, alcohols, and oxygen.23 Bacterial chemotactic messages can also be grouped according to the “type” of message they represent, for instance nutrient (e.g., amino acids, sugars), environmental (e.g., pH, temperature), danger (e.g., phenol, lipids, other repellants), or communication (e.g., AI‐2, which choreographs quorum sensing behaviors)36, 37 (Fig. 2A). Finally, bacterial chemotactic messages can also be grouped according to their receptor‐activation mechanism. Some chemotactic ligands freely diffuse into the periplasmic ligand binding domain (LBD) of the chemoreceptor, whereas others (e.g., dipeptides, sugars, AI‐2) bind first to a periplasmic binding protein (BP), and the ligand‐BP complex then binds and activates the receptors.38, 39, 40, 41
Figure 2.
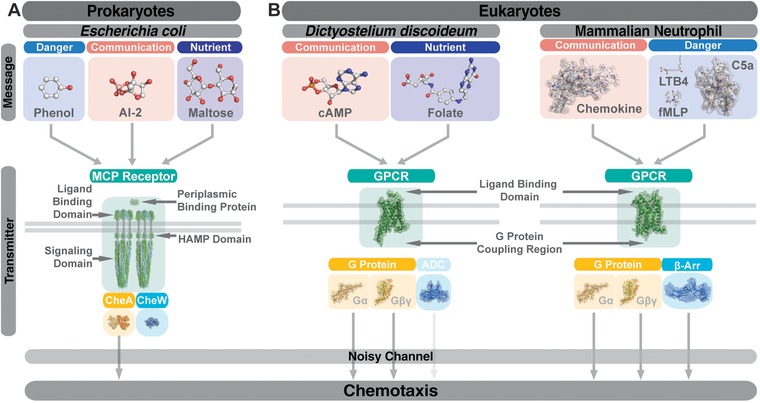
Messages and transmitters in prokaryotic and eukraryotic chemotactic systems. Chemotactic messages of prokaryotes and eukaryotes alike can be grouped by “type” (i.e., danger, communication, or nutrient signal) and each message is transmitted through an integral membrane protein shown here. Comparing the prokaryotic (A) messages shown (phenol, PDB ID: 5KBE; maltose, PDB ID: 1MPD; AI‐2, PDB ID: 2HJ9) to their eukaryotic (B) counterparts (cAMP, PDB ID:5KJY; folate, PDB ID 4QLE; chemokine IL‐8, PDB ID: 1IL8; LTB4, PDB ID 3ZUO; fMLP, PDB ID: 1Q7O; C5a, PDB ID: 5B4P), the increased molecular size and complexity of eukaryotic messages can be appreciated. All messages for Escherichia coli are transmitted through the common MCP Receptor architecture shown (from top to bottom; ligand binding domain, PDB ID:4Z9H; HAMP domain, PDB ID: 3ZX6; signaling domain, PDB ID: 3JA6; CheA and CheW, PDB ID: 3JA6) which is considerably larger that of the eukaryotic transmitter (i.e., GPCR). No structures are available for Dictyostelium discoideum GPCRs, G proteins, or arrestin domain‐containing proteins (ADC), so shown are mammalian examples (GPCR and G Protein, PDB ID: 3SN6; ADC, PDB ID: 4R7X). For mammalian neutrophils, shown is an example of a chemokine receptor (CCR5, PDB ID: 4MBS), the heterotrimeric G protein (PDB ID: 3SN6), and β‐arrestin 1 (PDB ID: 4JQI). Arrows shown indicate which proteins are thought to begin transmission through the noisy channel (i.e., begin the intracellular signaling cascades). The arrow below ADC is in light gray as it is unclear whether or not arrestin domain‐containing proteins play a role in Dictyostelium discoideum cAMP‐mediated chemotaxis
Next, we consider the transmitter that interprets the messages encoded in bacterial chemoeffectors to modulate chemotaxis. Bacteria possess highly conserved machinery for recognizing and responding to environmental chemoeffectors.42 This machinery includes plasma membrane‐spanning chemoreceptors, known as MCP receptors, and a cohort of cytosolic kinases and adaptor proteins that relay MCP chemoattractant recognition signals to the flagellar motor complex to alter chemotactic direction (Fig. 2A). An MCP receptor core‐signaling unit is composed of a trimer of dimers, and the LBD is commonly located at the periplasmic dimer interface. Ligand binding induces conformational and/or dynamic changes that are transmitted through the plasma membrane to a cytosolic “signal conversion module” known as a HAMP domain, which in turn transmits these changes to the signaling domain (SD), the most conserved domain of MCP receptors.36, 43 These changes in the SD inhibit autophosphorylation of the histidine kinase CheA through a cytosolic adaptor, CheW. Decreased CheA phosphorylation results in decreased phosphorylation of the cytosolic effector CheY, which directly regulates the flagellar motor complex to direct chemotaxis. The MCP receptor core‐signaling unit complexes with CheA and CheW to form supramolecular hexagonal arrays (chemosensory arrays) consisting of thousands of proteins, and these chemosensory arrays allow for significant amplification (at least 50‐fold) of the chemoeffector stimulus.44, 45, 46, 47, 48 While the MCP architecture described is the best studied and most common architecture, MCP receptors can adopt alternative architectures, although all contain the conserved SD.43
The model system E. coli encodes five MCP receptors, each of which recognizes on the order of 1–2 ligands: Tsr (taxis to serine and repellants), Tar (taxis to aspartate and repellants), Tap (taxis to dipeptides), Trg (taxis to ribose and galactose), and Aer (taxis to oxygen).36 The amino acid‐binding MCP receptors (Tsr and Tar) are approximately 10‐fold more abundant than the low‐abundance dipeptide and sugar‐biding MCP receptors (Tap and Trg).49 Aer is an unorthodox MCP receptor50 due to the presence of a cytoplasmic N‐terminal Per‐Arnt‐Sim (PAS) sensing domain rather than a periplasmic LBD.36 Regardless, all five E. coli MCP receptors share identical trimer contact residues,22 which enable participation in mixed trimers of dimers or “receptor squads” and allow for complex, amplified signaling.51
2.3. The chemotactic message for eukaryotes
For D. discoideum, the chemoattractant message is well described for each stage of its life cycle,52 and is limited to a few players. During its vegetative growth stage, D. discoideum cells migrate toward food sources by responding selectively to folate.52, 53 Upon starvation, D. discoideum cells aggregate to form a multicellular structure by selectively responding to cAMP secreted by other D. discoideum cells.54, 55 In contrast, the chemoattractant message for mammalian neutrophils (and even more broadly, for leukocytes) is considerably more complex. Neutrophils respond to a wide variety of chemoattractants including lipids (e.g., leukotriene B4 (LTB4),56 platelet‐activating factor (PAF)57), peptides (e.g., fMLP,58 LL‐3759), protein fragments (e.g., C5a60), and proteins (e.g., chemokines61, 62, 63), each of which orchestrates a unique migration response critical for efficient neutrophil function.61 Similar to their prokaryotic counterparts, eukaryotic chemoattractant messages can be grouped according to the “type” of message they represent, whether nutrient (e.g., folate), “danger” (e.g., fMLP, LL‐37, LTB4, PAF, C5a), or communication (e.g., cAMP, chemokines) (Fig. 2B). Alternatively, eukaryotic chemoattractant messages can be grouped according to size (Fig. 3), whether small molecule (e.g., folate, cAMP), lipid (e.g., LTB4, PAF), peptide (e.g., formylated peptides), or protein (e.g., chemokines, complement products).
Figure 3.
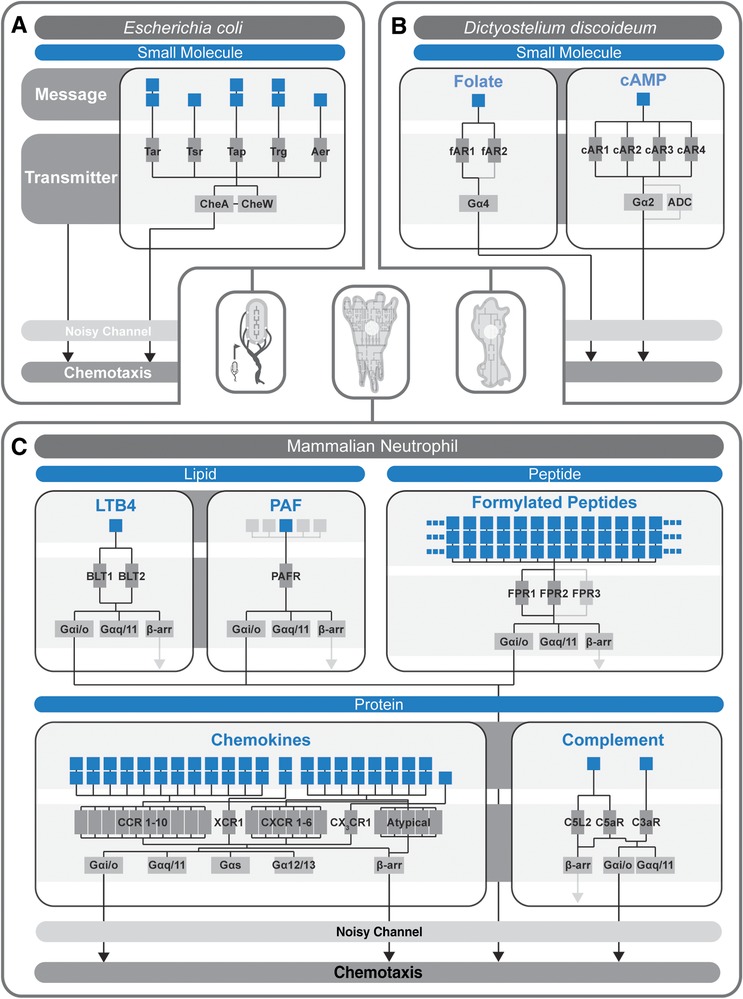
Comparison of communication circuitry among different organisms by message complexity. Chemotactic messages of different chemical complexity (i.e., small molecule, lipid, peptide, and protein) have different repertoire sizes (i.e., the number of ligands) and utilize unique circuitry to transmit their message. (A) Escherichia coli is used as an example to show the message, transmitter, and noisy channel architecture of each circuit. Messages (i.e., ligands) are shown as blue boxes. The transmitters (i.e., MCP receptors, Che proteins) are shown as gray boxes. Messages are connected to their corresponding transmitters via solid black lines, with uncertain or debated connections shown in gray. (B) Dictyostelium discoideum utilizes soley small molecule chemoattractant ligands, while (C) mammalian neutrophils utilize lipid, peptide, and protein messages. The G protein specificity of each message‐transmitter system is shown. Regardless of the specific message chemical complexity and receptor subtype, all systems seem to require Gαi/o for chemotactic signaling, though most also signal through other G protein subtypes (specifically, through the Gαq/11 family). The role of β‐arrestins in chemotactic signaling for many systems is still being elucidated (shown as gray arrows), but β‐arrestin 1 and β‐arrestin 2 have been shown to be required for chemokine‐mediated chemotactic signaling
Eukaryotic chemoattractant messages are primarily transmitted through members of the GPCR superfamily. GPCRs share a common seven transmembrane (7TM) helical architecture with an extracellular N‐terminus, alternating intracellular and extracellular loops, and an intracellular C‐terminus.64, 65 Agonist binding to GPCRs leads to structural and dynamic changes that are transmitted to the intracellular side of the receptor, classically resulting in the activation of heterotrimeric G proteins.64, 65 Heterotrimeric G proteins consist of 3 subunits, Gα, Gβ, and Gγ. Upon activation, the Gα subunit exchanges GDP for GTP and the heterotrimer dissociates into 2 functional units, Gα‐GTP and Gβγ, which then activate a number of distinct downstream signaling pathways.66, 67 In humans, there are 16 genes that express 21 Gα subunits of 4 main classes (Gαi/o, Gαs, Gαq/11, and Gα12/13), grouped according to sequence similarity and the downstream signaling pathways they trigger.67 GPCRs are known to exhibit specificity for one or more Gα classes,68, 69 and while mammalian chemoattractant responses are largely mediated through Gαi/o, most chemoattractant GPCRs also act through Gαq/11 and Gα12/1370 (see Fig. 3C). In contrast, Dictyostelium discoideum cells express 14 distinct Gα subunits, all of which share 40–50% identity and are most similar to the mammalian Gαi/o class.19, 53, 71 D. discoideum chemoattractant GPCRs are associated with 2 specific Gα subunits, Gα2 (for cAMP‐mediated chemotaxis) and Gα4 (for folate‐mediated chemotaxis) (Fig. 3B).53 Compared to the Gα subunit, the Gβγ subunit shows less genetic diversity, with 5 Gβ and 12 Gγ subunit genes in humans and only a single Gβ and Gγ subunit gene in D. discoideum.19, 67 Though it was classically thought that chemotactic responses were mediated through the Gβγ functional unit (with the Gα acting as a “timer”, controlling the release and consequent activation of Gβγ), both Gα‐GTP and Gβγ have been shown to play important roles in directing eukaryotic chemotaxis, each triggering unique signaling cascades that are necessary for both the establishment of cellular polarity and migration.72, 73, 74, 75, 76
Following activation, GPCRs are phosphorylated at their intracellular loops and C‐terminal tail by GPCR kinases (GRKs), facilitating interactions with β‐arrestins.77 β‐arrestins have important roles in receptor desensitization and internalization, but also act as G protein‐independent signal transducers for GPCRs.77 In the context of mammalian cell chemotaxis, β‐arrestins not only modulate receptor G protein signaling and surface expression, but to also contribute to the spatial regulation of actin assembly proteins.78 Mammals express 2 nonvisual arrestins (β‐arrestin1 and β‐arrestin2), both of which play important roles in mammalian cell chemotaxis.78, 79, 80 β‐arrestin1 and β‐arrestin2 are required for chemotaxis in some mammalian cell systems, though their precise role is undefined in many contexts.78 In contrast, D. discoideum is not known to have true arrestins, but rather contains six distinct arrestin‐domain containing proteins (AdcA, AdcB, AdcC, AdcD, AdcE, AdcF).81, 82 Two of these arrestin‐domain containing proteins (AdcB and AdcC) may play a role in cAMP signaling in D. discoideum, but their role in mediating chemotaxis is still unclear (Fig. 2B).81, 83
2.4. Transmitting unique chemotactic messages in eukaryotes
To address message (i.e., chemoattractant) diversity present in eukaryotic systems, we will now consider the chemoattractant messages for eukaryotic chemotaxis systems in more detail. We will consider these messages according to their relative size and chemical complexity (small molecule, lipid, peptide, protein fragment, and protein), and explore their interactions with their respective transmitters (i.e., GPCRs, heterotrimeric G proteins, and β‐arrestins) (Fig. 3).
2.4.1. Small molecule chemoattractants
While prokaryotic chemotactic messages are nearly exclusively small molecules (See Section 2.2, Fig. 3A), eukaryotic chemotactic responses to small molecule messages are largely limited to the soil‐dwelling amoeba D. discoideum (Fig. 3B). As noted above, D. discoideum cells primarily respond to two distinct small molecule chemoattractants, folate and cAMP, and modulate their responsiveness to each molecule with respect to their current life cycle stage.84 Four cAMP receptors have been identified in D. discoideum (cAR1, cAR2, cAR3, and cAR4), each with unique expression patterns and affinities for cAMP.85 D. discoideum cARs signal through Gα2‐associated heterotrimeric G proteins,85, 86 and may interact with arrestin‐domain containing proteins to modulate cAMP responses.83 To date, only 2 folate receptors have been identified in D. discoideum (fAR1 and fAR2), and only fAR1 has been shown to be required for folate‐induced signaling.87 Further, recent evidence suggests that the folate receptor fAR1 couples to Gα4‐associated heterotrimeric G proteins, rather than Gα2.87
2.4.2. Lipid chemoattractants
Eukaryotic chemotactic responses to lipids (e.g., LTB4, PAF) are typically associated with neutrophils (and other mammalian leukocytes) rather than with D. discoideum (Fig. 3C), though there are some reports of D. discoideum responding chemotactically to LPA.86, 88 Leukotriene B4 (LTB4) is an arachidonic acid derivative produced in response to inflammation, and is a potent chemoattractant for neutrophils (as well as for other leukocytes including eosinophils, monocytes, and T cells).89, 90 LTB4 binds 2 highly homologous (45.2% sequence identity) G protein‐coupled receptors, BLT1 and BLT2.90, 91 BLT1, expressed primarily on leukocytes, binds LTB4 with higher affinity than BLT2, which is expressed ubiquitously.91 BLT1 and BLT2 are promiscuous in their affinities for different heterotrimeric G proteins, signaling through members of both the Gαi/o and Gαq/11 classes, though both require Gαi signaling to mediate chemotactic responses.91 Platelet activating factor (1‐O‐alkyl‐2‐acetyl‐sn‐glyceryl‐3‐phosphorylcholine; PAF),92 is a general term for a group of phospholipid molecules whose chemical identity is dependent on both the cell type and the stimulus that lead to its production.57, 93 PAF mediates chemotaxis by interacting with a single GPCR, platelet‐activating factor receptor (PAFR).94 PAFR signals through both pertussis toxin (PTX)‐sensitive Gαi/o and PTX‐insensitive Gαq/11 heterotrimeric G protein classes,95, 96 though, similar to BLT1 and BLT2, PAFR‐driven chemotactic events are mediated exclusively through Gαi/o signaling.97
2.4.3. Peptide chemoattractants
Forymylated peptides are produced and secreted by bacteria, which initiate protein synthesis with a formylated methionine. These peptides act as pathogen‐associated molecular patterns (PAMPs), alerting the immune system to bacterial pathogens by triggering chemotaxis of neutrophils and other immune cells following peptide interactions with formyl peptide receptors 1–3 (FPRs 1–3) (Fig. 3C).98 FPR1 and FPR2 respond to formylated peptides of different lengths, with FPR1 preferring shorter peptides and FPR2 preferring longer peptides.98, 99 FPR3 shares ligands with FPR2, but binds fewer peptides than FPR1 and FPR2, and all 3 receptors are associated with chemotactic responses of mammalian immune cells.99 FPRs 1 and 2 are unusually promiscuous, jointly estimated to recognize over 100,000 different formylated peptides, which, if true, would constitute the largest ligand sets of any GPCRs.100 Like other mammalian chemoattractant receptors, FPRs primarily couple to Gαi/o proteins, but coupling to other G proteins has been reported.101
2.4.4. Protein chemoattractants
With approximately 45 chemokine ligands and 20 receptors, the chemokine‐receptor system represents the most extensive system of ligands and receptors for mediating chemotaxis (Fig. 3C). Chemokine ligands can be subdivided structurally, by the arrangement of a conserved set of cysteines (i.e., CC, CXC, CX3C, and XC) or by their functional role (e.g., inflammatory versus homeostatic).102 Receptors are subdivided based upon the type of ligand they bind (i.e., CC receptors, CCRs, or CXC receptors, or CXCRs). While there are no apparent structural features distinguishing CCRs and CXCRs, instances in which CC chemokines bind CXC receptors and vice versa are rare.103 Members of a third class of chemokine receptors known as atypical chemokine receptors (ACKRs) do not couple to G proteins (although G protein coupling has been reported to the atypical receptor CCRL1104) and are incapable of eliciting chemotaxis. Instead, ACKRs modulate extracellular chemokine gradients by internalizing chemokine ligands and targeting them for degradation.104, 105 Despite widespread promiscuity, some chemokines and receptors are monogamous, binding only one respective partner.106 Receptor promiscuity is regulated at the systems level by the selective expression of different chemokine receptors on different cell types, and the selective expression of chemokines in different physiologic and anatomical contexts. Broadly speaking, chemokine receptor subsets are expressed on all major immune cells.107 Like other chemoattractant GPCRs, chemokine receptors primarily couple to Gαi/o proteins, but have been reported to couple to Gαs, Gαq/11, and Gα12/13.108
Other protein chemoattractants include cleavage products of the complement system. The complement system consists of a set of cytosolic proteins that are activated in innate immune responses to trigger killing, phagocytosis, and clearance of pathogens and infected cells, as well as recruit immune cells to sites of infection.109, 110The principle complement pathways (i.e., classical, alternative, and lectin pathways) are activated by different stimuli to initiate generation of a set of secreted protein fragments. These fragments, in turn, can assemble a lethal pore complex in the pathogen's membrane (i.e., membrane attack complex, MAC), signal to immune cells, or opsonize pathogens for subsequent phagocytosis.109 Two of these secreted complement protein fragments, C3a (∼9 kDa) and C5a (∼11 kDa), interact with cell surface GPCRs to trigger chemotaxis111, 112 (Fig. 3C). C5a binds 2 complement receptors, C5aR and C5L2 which are typically coexpressed with one another on immune and nonimmune cells.112, 113 Whereas C5aR activation results in the activation of G proteins and β‐arrestins, C5L2 activation does not activate G protein, functioning exclusively through β‐arrestin pathways, analogously to ACKRs.113, 114 Like other eukaryotic chemoattractant receptors, C5aR signals through both the PTX‐sensitive Gαi/o and the PTX‐insensitive Gαq/11 heterotrimeric G protein classes.95, 115 C3a binds C3aR on the surface of many immune and nonimmune cells, and stimulates chemotaxis in both a direct and indirect fashion.116, 117, 118, 119 Though C3aR has also been shown to signal through Gαi/o, Gαq/11, and potentially Gα12/13 heterotrimeric G protein classes, its intracellular signaling pathways are cell type‐specific and distinct from those stimulated by C5aR.120, 121, 122
3. HOW MUCH INFORMATION?
3.1. The information in chemotactic communication systems
In communication theory, information is the “measure of one's freedom of choice when one selects a message”.34 For example, each character in the Latin alphabet (26 characters) possesses much more information than each character of Morse code (3 characters: dot, dash, and space), because one has significantly more choice in how to express oneself when choosing from the larger set of Latin characters.123 Indeed, it takes a sequence of dots and dashes to specify a single letter of the alphabet, and thus more dots and dashes than letters to specify the same word, showing how symbols vary in the amount of information they encode.
Like Morse code and the Latin alphabet, chemoattractant ligands encode different amounts of information to coordinate chemotaxis in bacteria, D. discoideum, and immune cells. In this section, we ask the following questions: How diverse are the sets of chemoattractant symbols used by these systems, and how might that change the amount of information each system is able to convey in the context of chemotaxis? How diverse are the sets of cell‐surface receptors that receive and transmit the messages encoded in chemoattractants, and how might their diversity and type alter ability to transmit the full meaning of the encoded messages to the inside of the cell? How are the chemoattractant alphabets used by these systems strung together into sentences that convey more complex instructions? Finally, why do bacteria, D. discoideum, and immune cells use such different alphabets to accomplish the same task?
In this section, we address these questions with the help of 4 principles: (1) message alphabet size, (2) message complexity, (3) transmitter capacity, and (4) combinatorial complexity (Fig. 4). These principles will help inform the way we understand how different organisms encode information to direct complex chemotactic processes. In each subsection, we will highlight illustrative examples of how these principles shed light on cellular chemotaxis in different systems.
Figure 4.
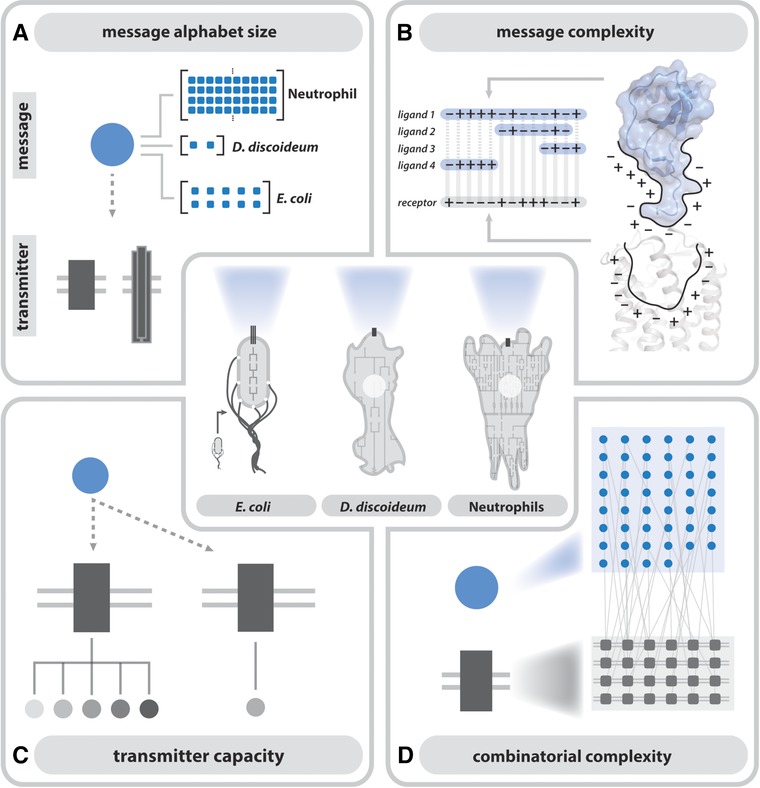
Four principles to decode the chemotactic signal. Comparisons of chemoattractant‐receptor systems used by E. coli, D. discoideum, and neutrophils by message alphabet size (A), message complexity (B), transmitter capacity (C), and combinatorial complexity (D) demonstrate that chemoattractant receptor systems differentially encode complex chemotactic outcomes
3.2. Understanding chemotactic information by message alphabet size
The idea of information as a function of ‘message alphabet size’ was developed by Ralph Hartley. Hartley developed an equation that defines the amount of information in a message as the base 2 logarithm of the number of symbols from which it was selected (i.e., the alphabet size).34, 124 For instance, a symbol chosen at random from a set of 32 symbols carries 5 bits of information per symbol (number of possible symbols = 2amount of information per symbol, or 32 = 25). With this equation, we see that larger alphabets contain more information per symbol (Fig. 4A). If we compare chemoattractant message alphabet sizes of E. coli, D. discoideum, and neutrophils, we find widespread variation in the information content among chemoattractant ligands or messages. For E. coli, whose alphabet contains ∼10 messages (see Section 2.2), each individual message is estimated to contain only ∼3 bits of information per ligand. Similarly, the 2 chemoattractant messages for D. discoideum (i.e., cAMP and folate) each carry only 1 bit of information. The chemoattractant message alphabet for neutrophils is by far the largest, with more than 100,000 estimated messages.100 Each neutrophil chemoattractant message thus contains ∼17 bits of information, significantly more information than either E. coli or D. discoideum chemoattractant messages.
Chemoattractant message alphabet size can be defined in two other ways, the number of ligands belonging to a particular class (e.g., chemokines vs. formylated peptides, see Fig. 3) or the number of ligands binding a single chemoattractant receptor. Considering these definitions of message alphabet size, we find that E. coli and D. discoideum possess restrictive alphabets by all definitions. For example, E. coli MCP receptors recognize exclusively small molecule chemoeffectors and each MCP receptor typically interacts with only 1–2 ligands (though some MCP receptors have been reported to bind upward of 5).36 D. discoideum principally respond to a single nutrient (i.e., folate) and a single communication signal (i.e., cAMP), both of which are small molecule chemoeffectors. The alphabet size of ligands shared by a single receptor is similarly limited, with cAMP and folate constituting the only canonical ligands binding the respective receptors.52
Comparatively, mammalian neutrophils encode significantly larger chemoattractant alphabets. Formylated peptides, which act as bacterial pathogen signals, and chemokines, which act as intercellular communication signals, have alphabet sizes of >100,000100 and ∼45 ligands,103 respectively. In addition to these two large alphabets, mammalian neutrophils respond to two small alphabets: LTB4 and PAF, which encode general‐use inflammation signals, and complement pathway products (e.g., C5a), which encode pathogen danger signals.90 Considering the Hartley principle as above, in which symbols in large alphabets encode more information than those in small alphabets, formylated peptides are the most “information‐rich” chemoattractant class for neutrophils, followed by chemokines, then LTB4, PAF, and complement, which are on par with E. coli and D. discoideum chemoattractant systems. This theoretical information hierarchy agrees with what is known biologically, as diverse bacterial pathogens encode highly specific, “information‐rich” chemical signals as compared to broad‐utility chemoattractant cues like leukotrienes and complement products. Chemokines, which have an intermediate information capacity according to Hartley's principle, are used broadly as a family, although individual ligand‐receptor systems can be employed in highly specific contexts.
Using communication theory to compare chemoattractant systems among E. coli, D. discoideum, and neutrophils highlights the following question: Why do neutrophils need larger, more diverse alphabets than their eukaryotic D. discoideum counterparts, despite sharing much of the same receptor and intracellular machinery? The answer to this question may be understood by comparing the context‐specificity afforded by small versus large chemoattractant alphabets. Large chemoattractant alphabets allow neutrophils to undergo chemotaxis to serve functionally diverse roles as a major arm of the innate immune system, whereas D. discoideum undergo chemotaxis in only two contexts. Thus, while both organisms utilize similar machinery, differences in message alphabet size between neutrophils and D. discoideum emphasize the difference in complexity between the two eukaryotic organisms, illustrating the need for a more complete understanding of organism‐ or cell type‐specific (e.g., mammalian rather than broadly eukaryotic, neutrophil rather than macrophage) chemotaxis.
3.3. Understanding chemotactic information by message complexity
The amount of information encoded in a chemoattractant ligand can also be evaluated in terms of chemical complexity. To demonstrate this principle, we borrow a concept developed by Hann and colleagues, in which ligands and receptors are depicted by strings of “+” and “–“ signs (e.g., receptor: – – + – + – – + –; ligand: + + –) (Fig. 4B).125 In this model, binding events only occur when ligand and receptor contain strings of complementary signs, which represent the combination of interactions (e.g., shape, electrostatics, van der Waals, H bonding, etc.) that create binding specificity. Intuitively, the larger the ligand, the more requirements there are for it to make complementary interactions across a more distributed receptor surface. While the authors use this analysis to derive strategies for optimizing drug discovery efforts, it is useful to consider the effect of increasing ligand complexity in instances where nature has already provided specific ligand–receptor pairs.
The chemical complexity of chemoattractant messages for the mammalian immune system vary drastically by both composition (small molecules, modified peptides, lipids, and proteins) and size (∼330 Da for cAMP vs. 9,000–12,000 Da for chemokines). Comparatively, the composition and size of bacterial chemoattractant (or chemorepellant) ligands is more limited, with the best studied bacterial ligands (e.g., serine, aspartate, dipeptides, O2) < 500 Da (although it should be noted that some chemoattractants bind periplasmic binding proteins which then bind MCP receptors, see section 2.2). Though D. discoideum is a eukaryotic system, the canonical D. discoideum chemoattractants are much closer in size to prokaryotic (i.e., E. coli) chemoattractants than to those of mammalian neutrophils, with cAMP at ∼330 Da and folate at ∼440 Da. Thus, based on chemical complexity, mammalian neutrophil chemoattractants have the potential to encode much more information and conceivably more complex outcomes than either E.coli or D. discoideum. In effect, whereas E.coli and D. discoideum ligands would be represented by short strings of +’s and –‘s, neutrophil ligands would be represented, on average, by much longer strings of +’s and –‘s.
What are the functional implications of utilizing “information‐rich,” complex ligands to organize chemotaxis? The comparatively complex structure of chemokines, for one, allow many of them to encode signaling specificity (i.e., biased signaling) at a given receptor, demonstrating how a more chemically complex message enables complex cellular outcomes.106 Interestingly, despite their structural complexity, chemokines possess a highly conserved tertiary structure, which constrains their chemical diversity to some extent. Moreover, the high degree of chemokine structural similarity suggests that among chemokines, complexity is encoded by substituting residue identities on a structurally conserved scaffold. It should be mentioned that beyond receptor‐interactions, the ligand complexity of chemokines allows them to encode numerous other functions, some of which are associated with chemotaxis, like GAG binding and homo‐/hetero‐dimerization, as well as non‐chemotactic functions, such as antimicrobial activity.126, 127, 128
Formylated peptides, which have lengths between 3 and 10 amino acids on average, demonstrate much less individual complexity than chemokines, but with >100,000 approximated members, they have the potential to encode significantly more information by way of combinatorial complexity.100 Nevertheless, molecular modeling studies suggest that formylated peptides, like chemokines, share a conserved structure, despite divergent amino acid sequences. Bufe and colleagues demonstrated the formation of a common “clawlike” fold, stabilized by residues at only 3 peptide positions, which allows for considerable sequence diversity at other peptide residues.100 Interestingly, substitution of peptide residues outside of 3 conserved positions had little effect on chemotaxis.100 In the context of the Hann model, formylated peptide structural conservation and sequence constraints may functionally limit their complexity such that instead of being represented by >100,000 unique strings of +/– symbols, apparently different formylated peptides may in fact be redundant, such that a common string of +/– symbols may more appropriately represent their chemical complexity.
The chemical complexity framework highlights the following paradox: how do small, low‐complexity chemoattractants, such as LTB4 maintain receptor specificity, while larger, high‐complexity ligands, such as chemokines, demonstrate widespread promiscuity? Regarding the chemokine system, we speculate that chemokines sharing common receptors are more evolutionarily related to one another than those binding different receptors. Consequently, even though all chemokines would be represented by long strings of +/– symbols that should, in principle, be highly specific for an individual receptor, chemokines sharing a common receptor would have closely related +/‐ strings. While numerous structural studies of unique chemokine‐receptor interactions have highlighted how chemokines recognize their receptors for specific ligand–receptor pairs, structures of different chemokines bound to the same receptor (and vice versa) and new bioinformatics approaches will be needed to understand how chemokines are able to be so promiscuous despite highly complex interactions with their receptor counterparts.
3.4. Understanding chemotactic information by transmitter capacity
In our discussion of information content of chemotactic systems, we must also consider how the chemoattractant message is encoded by the transmitter (i.e., receptor) (Fig. 4C). Transmitters may vary (1) in the ways in which they encode different messages, and (2) the extent to which they can encode the full complexity of the message. In other words, while complex (i.e., “information‐rich”) ligands can, in principle, relay more information to the cytosol than can simple ligands (i.e., “information poor”), ligand complexity may be lost when the signal is passed through the receptor such that the output signal contains less information than the original ligand message. Thus, while message alphabet size and message complexity are significant when considering the information content of chemotactic systems, the transmitter of the chemotactic message (i.e., the chemoattractant receptor) plays a significant role as well. In this section, we summarize this significance using the concept of “transmitter capacity.”
Shannon discusses encoding as a process of “operating on a message” to change its format in a way that is “suitable for transmission over the channel,” for instance encoding words into Morse code or encoding speech into a digital signal to send over a telephone wire.34 In chemotactic signal transduction, the receptor (e.g., MCP receptor or GPCR) encodes the message of the chemoattractant by undergoing conformational and dynamic changes in the ligand‐bound state, and these changes are interpreted by cytosolic signaling molecules (e.g., CheA/W proteins in bacteria or G proteins/β‐arrestins in eukaryotes) which relay the converted message to the interior of the cell. Transmitter capacity, or the ability of chemoattractant receptors to adequately decode the complexity of chemoattractant messages for transmission to the intracellular environment, is functionally constrained (1) at the structural level, in the extent to which a given receptor can interpret the differences of its numerous chemoattractant binding partners (i.e., receptor promiscuity), and (2) at the signaling level, in the extent to which the same receptor can initiate complex outcomes through any number of cytosolic effectors (i.e., biased signaling).
Considering first transmitter capacity at the structural level, bacteria and eukaryotes have evolved vastly different receptor systems to transmit information encoded in chemoattractant ligands to cytosolic effectors that can regulate chemotaxis. Comparing the structures used by prokaryotic and eukaryotic chemoattractant receptors, the E. coli MCP receptors are >3000 kDa (i.e., as trimer‐of‐dimers) and span ∼350 Å, whereas GPCRs are typically 40–50 kDa and span ∼ 50 Å.129, 130 It is interesting to consider the size discrepancy of the two systems, relative to the sizes of their respective ligands. MCP receptors are on the order of 104 times larger than their ligands by mass, whereas GPCRs range from ∼1–10 times the mass of their ligands. Despite this size difference, receptor promiscuity, or the ability of a chemoattractant receptor to respond to more than one chemoattractant ligand, is common to both bacterial MCP receptors and eukaryotic GPCRs. However, the number of chemoattractant messages that a single receptor can transmit (i.e., the degree of receptor promiscuity) differs for each organism. The ∼10 chemotactic messages for E. coli are transmitted through only 5 MCP receptors, with each MCP receptor transmitting 1–2 messages (or 10–20% of the total), on average. In contrast, the 6 chemoattractant GPCRs (4 cAMP, 2 folate) for D. discoideum transmit only 2 chemotactic messages, with a given GPCR only transmitting 1 of the 2 messages (or 50% of the total). Mammalian neutrophils are considerably more complex than either E. coli or D. discoideum, with ∼30 chemoattractant GPCRs to interact with >100,000 potential messages (see Section 3.2). For mammalian neutrophils, however, it is perhaps more valuable to consider the transmitter fidelity for each chemoattractant class (e.g., chemokines vs formylated peptides) rather than the group together. For example, we say that mammalian neutrophils have >100,000 potential chemoattractant messages, but most of these are of the formylated peptide class which are transmitted through only 2 (or possibly 3) receptors (Fig. 3).
Further, the activation mechanisms of GPCRs and MCP receptors share both “piston” type movements (e.g., at TM2 in MCP receptors and at TM5 in GPCRs) as well as large scale dynamic alterations, although the structural differences between the two receptors make the dynamic changes very different.129, 131, 132 In both cases, long clusters of alpha helices allow the receptor systems to amplify the small changes occurring at the ligand binding pocket into much larger changes intracellularly, although despite their increased length, conformational changes in MCP receptors are subtle and localized to the distal CheA and CheW contact sites.133 Despite their distinctive size advantage, bacterial MCP receptors are much more limited than D. discoideum or mammalian neutrophil GPCRs in the amount of information they can transmit to the cytosol, and eventually, to the chemotaxis machinery. As discussed above, bacterial MCP receptors alter chemotaxis by modulating the rate of autophosphorlation of the cytosolic effector CheA, which directly interacts with the flagellar motor complex. While the rate of CheA autophosphorylation can be modulated by additional proteins (e.g., methyltransferase CheR and methylesterase CheB), the information encoded in individual chemoattractant ligands is limited to altering the function of a single effector in CheA.
Contrastingly, GPCRs are capable of eliciting activation of numerous cytosolic effectors, including but not limited to G proteins, G protein‐coupled receptor kinases (GRKs), and β‐arrestins. Moreover, GPCRs exhibit widespread biased signaling, in which different ligands preferentially activate a subset of the available effectors to elicit ligand‐specific outcomes.134 Biased signaling is widespread in the chemokine system, with different chemokine ligands eliciting unique signaling profiles at the same receptor.106 While much less established, biased signaling may also play a role in the formylated peptide receptor system.98, 99, 135, 136, 137 Although biased signaling has not been explicitly described in D. discoideum, D. discoideum nevertheless has numerous cytosolic effectors available to initiate complex signaling outcomes that are adaptable in different environmental circumstances.138 In effect, eukaryotic chemoattractant receptor systems are better equipped to initiate diverse outcomes. In other words, the large, diverse, and complex chemoattractant messages used by eukaryotes are in many cases transformed into complex downstream outcomes. Comparatively, the diverse E. coli chemoattrantant messages are funneled into activation of a single cytosolic effector, thus limiting the extent to which diverse messages can encode diverse processes.
A close inspection of GPCR‐chemoattractant interactions sheds some light on the ways in which eukaryotic signals are capable of eliciting such complex intracellular pathways. Recent structural studies of chemokine–receptor interactions demonstrate an enormous ligand–receptor interface, spanning as much as 1700 Å2.139 Importantly, other recent studies suggest that differences in chemokine‐receptor interactions far from the traditional receptor binding pocket influence how a single receptor can mediate alternative functional responses.140, 141 In one example, CXCL12 provokes and arrests chemotaxis in a monomeric and dimeric form, respectively, and structures of CXCL12 bound to its receptor CXCR4 in both forms demonstrate mutually exclusive interactions that may influence CXCL12's opposing effects on chemotaxis.141 Comparison of three chemokine‐receptor crystal structures demonstrates diverse binding orientations, suggesting that in addition to making unique ligand–receptor contacts, ligand orientation may also contribute to signal transmission.139, 142, 143 These and other examples demonstrate the ways in which eukaryotic chemoattractant receptors, which transmit messages encoded in generally large and diverse chemoattractants, are high “capacity” transmitters insofar as they convey the full extent of the encoded messages by activating diverse functional responses.
3.5. Understanding chemotactic information by combinatorial complexity
Bacterial and eukaryotic cells navigate through complex environments in which they integrate multiple chemoattractant messages to devise a resultant chemotactic route. In an analogous way, Shannon demonstrates how one can construct complex sentences resembling English by semi‐randomly choosing letters from the alphabet with the help of transition probability tables, which define the frequency at which each letter follows all other letters.34 In both Shannon's stochastic English sentence algorithm and in chemotactic organisms, individual symbols or messages are “strung together” to create more complex outcomes. We will discuss this principle in the context of chemoattractant‐receptor systems as “combinatorial complexity” (Fig. 4D).
Prokaryotes and eukaryotes are both capable of “stringing together” numerous chemoattractant messages to “build up” more complex chemotactic routes. In the case of prokaryotes, specifically E. coli, cocktails of environmental stimuli must be continually synthesized into a single, binary response (i.e., clockwise or counterclockwise rotation of flagella). As previously discussed (see Section 2.2), bacteria use the same machinery to sense and transmit diverse chemoeffector messages. Expansive chemoreceptor arrays, composed of mixed or uniform trimers of MCP receptor homodimers, work cooperativity to both amplify the chemotactic signal and to integrate diverse chemoattractant and chemorepellant messages.144, 145, 146, 147 Consequently, the net chemotactic response of a bacterium in a complex environment has been shown to be dependent on the relative abundance of the MCP receptor types,148 the sum of agonistic or antagonistic responses elicited by environmental stimuli at each MCP receptor,149, 150 as well as the methylation status or adaptation of each MCP receptor.32, 151 MCPs adapt to (or retain “memory” of) previous signaling events through the alteration of methylated glutamate residues by the methyltransferase CheR and the methylesterase CheB.151 The slow methylation–demethylation adaptation process allows for the temporal gradient sensing necessary for both efficient bacterial chemotaxis and for maintaining MCP receptor sensitivity over a wide range of concentrations (from nanomolar to millimolar, in some cases).144, 152 Furthermore, although MCP receptors display cooperativity in the chemoreceptor arrays, methylation “memory” at MCP receptors is specific and thus allows for complex signal integration and signal prioritization.153
Eukaryotic cells similarly navigate through remarkably complex chemotactic environments. For example, mammalian neutrophils must navigate through the bloodstream, exit the vasculature via transendothelial migration, and subsequently migrate to the precise site of infection to perform their phagocytic function. In order to accomplish such a complex task, neutrophils (and other mammalian immune cells) utilize remarkably elegant systems to integrate and prioritize various chemotactic signals. As discussed in Section 3.2, eukaryotic immune cells recognize at least 4 different alphabets that evoke chemotaxis at different stages of an immune response (i.e., chemokines, complement fragments, formylated peptides, leukotrienes). Previous studies have demonstrated that neutrophils prioritize these different alphabets, utilizing “regulatory” (non‐dominant) chemoattractants (i.e., chemokines and leukotrienes) to migrate to the vicinity of an infection and prioritizing “end target” (dominant) chemoattractants (i.e., at the site of the pathogen, complement fragments and formylated peptides) to home to the pathogen site, likely through a process of heterologous receptor desensitization.154, 155 Furthermore, studies have shown that neutrophils can integrate signals from competing non‐dominant chemoattractants (e.g., the chemokine IL‐8 and lipid LTB4) and migrate along their vector sum (e.g., between the 2 agonist sources).156 Similar to bacteria, neutrophils navigate through complex chemotactic environments utilizing “memory” of their environment, though neutrophils do not encode their “memories” via methylation–demethylation adaptation but rather through a proposed mechanism of long‐lived intracellular asymmetry of cytoskeletal elements.157 To this end, a recent study by Prentice‐Mott and colleagues demonstrated moesin, actomyosin, and microtubules to be among the key cytoskeletal elements mediating memory in neutrophil‐like cells.157
4. CONCLUSION
In utilizing an information theoretical framework to compare the chemotactic signal processing of prokaryotic and eukaryotic cells, the unifying significance of directed cellular movement can be readily appreciated. Though prokaryotes (i.e., E. coli) and eukaryotes (i.e., D. discoideum and mammalian neutrophils) use their respective chemotactic machinery to navigate through unique and diverse environments, common principles including message transmission, signal amplification and capacity, and memory are shared.
As discussed in this review, the chemotactic inputs for E. coli and D. discoideum are arguably simpler than that of mammalian neutrophils. Mammalian neutrophils (as well as other leukocytes) need to be able to migrate through considerably more complex environments than either E. coli or D. discoideum to mediate effective biologic responses. To do this, neutrophils utilize a large (>100,000 chemoattractants, see Section 3.2) and chemically diverse (i.e., lipids, peptides, and proteins, see Section 3.3) message alphabet. Fascinatingly, the entire alphabet for stimulating neutrophil chemotaxis (i.e., every chemoattractant ligand) is encoded or transmitted through the same receptor architecture (i.e., GPCRs, see Section 3.4), which is also shared with D. discoideum. Furthermore, the common transmitter architecture extends beyond the cell membrane, with nearly all chemotactic responses being at least partially mediated through Gαi/o or Gαi/o‐like heterotrimeric G proteins (see Section 2.4). This remarkable “funneling” phenomenon allows mammalian neutrophils to chemotactically respond to an incredibly diverse repertoire of ligands, but complicates our understanding of how neutrophils are able to integrate a plethora of signals to orchestrate movement through complex environments (see Section 3.5). Moreover, as aberrant cell migration mediates a variety of pathologies, the common transmitter architecture is significant when considering therapeutic targeting of the chemotactic input. Indeed, GPCRs are among the most popular proteins for pharmacologic modulation, with ∼40% of approved drugs targeting GPCRs.158
It is interesting to compare the inherent information capacity of prokaryotic and eukaryotic chemotactic outputs (i.e., chemotactic mechanism). The output of prokaryotic chemotactic systems (i.e., clockwise or counterclockwise flagellar rotation) can be thought of as a simple binary response corresponding to one bit of information,33 while the output of eukaryotic chemotactic systems is considerably more complex. As eukaryotic cell migration requires the successful integration of a plethora of cellular events that are still being elucidated including directional sensing, establishment of cellular polarity, and movement by pseudopod extension,159 the corresponding information capacity of eukaryotic chemotactic systems is considerably greater. Though here we focused primarily on the input of chemotactic systems (i.e., the chemoattractant and chemoattractant receptor), we refer an interested reader to a number of recent reviews detailing the current understanding of the complexities of the output of eukaryotic chemotactic systems.21, 138
AUTHORSHIP
M. A. T. and A. B. K. contributed equally to this work.
ACKNOWLEDGMENTS
This work was supported in part by National Institutes of Health Grants F30 HL134253 (to M.A.T.), F30 CA196040 (to A.B.K.), and R01 AI058072 and R01 AI120655 (to B.F.V.). A.B.K. and M.A.T. are members of the NIH supported (T32 GM080202) Medical Scientist Training Program at MCW.
DISCLOSURE
B.F.V. has ownership interests in Protein Foundry, LLC. The other authors declare no conflict of interest.
Thomas MA, Kleist AB, Volkman BF. Decoding the chemotactic signal. J Leukoc Biol. 2018;104:359–374. 10.1002/JLB.1MR0218-044
REFERENCES
- 1. Nicholson DJ. Biological atomism and cell theory. Stud Hist Philos Biol Biomed Sci. 2010;41:202–211. [DOI] [PubMed] [Google Scholar]
- 2. Bloemendal S, Kück U. Cell‐to‐cell communication in plants, animals, and fungi: a comparative review. Naturwissenschaften. 2013;100:3–19. [DOI] [PubMed] [Google Scholar]
- 3. Steck K. Just follow your nose: homing by olfactory cues in ants. Curr Opin Neurobiol. 2012;22:231–235. [DOI] [PubMed] [Google Scholar]
- 4. Hooke R. Micrographia. London: Royal Society; 1665. [Google Scholar]
- 5. Lane N, The unseen world: reflections on Leeuwenhoek. ‘Concerning little animals’. Philos Trans R Soc Lond B Biol Sci. 1677;370:20140344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ananthakrishnan R, Ehrlicher A. The forces behind cell movement. Int J Biol Sci. 2007;3:303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lefèvre CT, Bazylinski DA. Ecology, diversity, and evolution of magnetotactic bacteria. Microbiol Mol Biol Rev. 2013;77:497–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nadarajah B, Parnavelas JG. Modes of neuronal migration in the developing cerebral cortex. Nat Rev Neurosci. 2002;3:423–432. [DOI] [PubMed] [Google Scholar]
- 9. Battley PF, Warnock N, Tibbitts TL, et al. Contrasting extreme long‐distance migration patterns in bar‐tailed godwits Limosa lapponica. J. Avian Biol. 2012;43:21–32. 10.1111/j.1600-048X.2011.05473.x. [DOI] [Google Scholar]
- 10. Schuergers N, Lenn T, Kampmann R, Meissner MV. Cyanobacteria use micro‐optics to sense light direction. ELife. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Parent CA, Devreotes PN. A cell's sense of direction. Science. 1999;284:765–770. [DOI] [PubMed] [Google Scholar]
- 12. Bahat A, Eisenbach M. Sperm thermotaxis. Mol Cell Endocrinol. 2006;252:115–119. [DOI] [PubMed] [Google Scholar]
- 13. Plotnikov SV, Waterman CM. Guiding cell migration by tugging. Curr Opin Cell Biol. 2013;25:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tai G, Reid B, Cao L, Zhao M. Electrotaxis and wound healing: experimental methods to study electric fields as a directional signal for cell migration Chemotaxis. Totowa, NJ: Humana Press; 2009:77–97. [DOI] [PubMed] [Google Scholar]
- 15. Häder DP. Gravitaxis in unicellular microorganisms. Adv Space Res. 1999;24:843–850. [DOI] [PubMed] [Google Scholar]
- 16. Macnab RM, Koshland DE. The gradient‐sensing mechanism in bacterial chemotaxis. Proc Natl Acad Sci USA. 1972;69:2509–2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Haastert PJM, Devreotes PN. Chemotaxis: signalling the way forward. Nat Rev Mol Cell Biol. 2004;5:626–634. [DOI] [PubMed] [Google Scholar]
- 18. Sourjik V, Wingreen NS. Responding to chemical gradients: bacterial chemotaxis. Curr Opin Cell Biol. 2012;24:262–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Swaney KF, Huang C‐H, Devreotes PN. Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity. Annu Rev Biophys. 2010;39:265–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Devreotes P, Janetopoulos C. Eukaryotic chemotaxis: distinctions between directional sensing and polarization. J Biol Chem. 2003;278:20445–20448. [DOI] [PubMed] [Google Scholar]
- 21. Devreotes PN, Bhattacharya S, Edwards M, Iglesias PA, Lampert T, Miao Y. Excitable signal transduction networks in directed cell migration. Annu Rev Cell Dev Biol. 2017;33:103–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parkinson JS, Hazelbauer GL, Falke JJ. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015;23:257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wadhams GH, Armitage JP. Making sense of it all: bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. [DOI] [PubMed] [Google Scholar]
- 24. Rappel W‐J, Levine H. Receptor noise and directional sensing in eukaryotic chemotaxis. Phys Rev Lett. 2008;100:228101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Insall RH. Understanding eukaryotic chemotaxis: a pseudopod‐centred view. Nat Rev Mol Cell Biol. 2010;11:453–458. [DOI] [PubMed] [Google Scholar]
- 26. Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harshey RM. Bacterial motility on a surface: many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. [DOI] [PubMed] [Google Scholar]
- 28. Henrichsen J. Bacterial surface translocation: a survey and a classification. Bacteriol Rev. 1972;36:478–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kimmel JM, Salter RM, Thomas PJ. An information theoretic framework for eukaryotic gradient sensing In: Schölkopf B, Platt JC, Hoffman T, eds. Advances in Neural Information Processing Systems 19, Cambridge, MA: MIT Press; 2007:705–712. [Google Scholar]
- 30. Hu B, Chen W, Levine H, Rappel W‐J. Quantifying information transmission in eukaryotic gradient sensing and chemotactic response. J Stat Phys. 2011;142:1167–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. [DOI] [PubMed] [Google Scholar]
- 32. Lan G, Tu Y. Information processing in bacteria: memory, computation, and statistical physics: a key issues review. Rep Prog Phys. 2016;79:052601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Micali G, Endres RG. Bacterial chemotaxis: information processing, thermodynamics, and behavior. Curr Opin Microbiol. 2016;30:8–15. [DOI] [PubMed] [Google Scholar]
- 34. Shannon CE, Weaver W, The Mathematical Theory of Communication, Urbana, IL: University of Illinois Press; 1964. [Google Scholar]
- 35. Szurmant H, Ordal GW. Diversity in chemotaxis mechanisms among the bacteria and archaea. Microbiol Mol Biol Rev. 2004;68:301–319. 10.1128/MMBR.68.2.301-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bi S, Lai L. Bacterial chemoreceptors and chemoeffectors. Cell Mol Life Sci. 2015;72:691–708. 10.1007/s00018-014-1770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Laganenka L, Colin R, Sourjik V. Chemotaxis towards autoinducer 2 mediates autoaggregation in Escherichia coli. Nat Commun. 2016;7:12984 10.1038/ncomms12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hegde M, Englert DL, Schrock S, et al. Chemotaxis to the quorum‐sensing signal AI‐2 requires the Tsr chemoreceptor and the periplasmic LsrB AI‐2‐binding protein. J Bacteriol. 2011;193:768–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Manson MD, Blank V, Brade G, Higgins CF. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature. 1986;321:253–256. [DOI] [PubMed] [Google Scholar]
- 40. Vyas NK, Vyas MN, Quiocho FA. Sugar and signal‐transducer binding sites of the Escherichia coli galactose chemoreceptor protein. Science. 1988;242:1290–1295. [DOI] [PubMed] [Google Scholar]
- 41. Zhang Y, Conway C, Rosato M, Suh Y, Manson MD. Maltose chemotaxis involves residues in the N‐terminal and C‐terminal domains on the same face of maltose‐binding protein. J Biol Chem. 1992;267:22813–22820. [PubMed] [Google Scholar]
- 42. Wuichet K, Zhulin IB. Origins and diversification of a complex signal transduction system in prokaryotes. Sci Signal. 2010;3:ra50–ra50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Salah Ud‐Din AIM, Roujeinikova A. Methyl‐accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell Mol Life Sci. 2017;74:3293–3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jones CW, Armitage JP. Positioning of bacterial chemoreceptors. Trends Microbiol. 2015;23:247–256. [DOI] [PubMed] [Google Scholar]
- 45. Briegel A, Li X, Bilwes AM, Hughes KT, Jensen GJ, Crane BR. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109:3766–3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cassidy CK, Himes BA, Alvarez FJ, et al. CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling. ELife. 2015;4:e08419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: high‐performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dahlquist FW. Amplification of signaling events in bacteria. Sci STKE. 2002;2002:pe24–pe24. [DOI] [PubMed] [Google Scholar]
- 49. Feng X, Baumgartner JW, Hazelbauer GL. High‐ and low‐abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J Bacteriol. 1997;179:6714–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gosink KK, Burón‐Barral MDC, Parkinson JS. Signaling interactions between the aerotaxis transducer Aer and heterologous chemoreceptors in Escherichia coli . J Bacteriol. 2006;188:3487–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Studdert CA, Parkinson JS. Crosslinking snapshots of bacterial chemoreceptor squads. Proc Natl Acad Sci USA. 2004;101:2117–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Meena NP, Kimmel AR. Biochemical responses to chemically distinct chemoattractants during the growth and development of dictyostelium. Methods Mol Biol. 2016;1407:141–151. [DOI] [PubMed] [Google Scholar]
- 53. Manahan CL, Iglesias PA, Long Y, Devreotes PN. Chemoattractant signaling in dictyostelium discoideum. Annu Rev Cell Dev Biol. 2004;20:223–253. [DOI] [PubMed] [Google Scholar]
- 54. Schneider IC, Haugh JM. Mechanisms of gradient sensing and chemotaxis: conserved pathways, diverse regulation. Cell Cycle. 2006;5:1130–1134. [DOI] [PubMed] [Google Scholar]
- 55. von Philipsborn A, Bastmeyer M. Mechanisms of gradient detection: a comparison of axon pathfinding with eukaryotic cell migration. Int Rev Cytol. 2007;263:1–62. [DOI] [PubMed] [Google Scholar]
- 56. Afonso PV, Janka‐Junttila M, Lee YJ, et al. LTB4 is a signal‐relay molecule during neutrophil chemotaxis. Dev Cell. 2012;22:1079–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Carolan EJ, Casale TB. Degree of platelet activating factor‐induced neutrophil migration is dependent upon the molecular species. J Immunol. 1990;145:2561–2565. [PubMed] [Google Scholar]
- 58. Dorward DA, Lucas CD, Chapman GB, Haslett C, Dhaliwal K, Rossi AG. The role of formylated peptides and formyl peptide receptor 1 in governing neutrophil function during acute inflammation. Am J Pathol. 2015;185:1172–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. De Yang QChe, Schmidt AP, Anderson GM, et al. LL‐37, the neutrophil granule‐ and epithelial cell‐derived cathelicidin, utilizes formyl peptide receptor‐like 1 (FPRL1) as a receptor to chemoattract human peripheral blood neutrophils, monocytes, and T cells. J Exp Med. 2000;192:1069–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ehrengruber MU, Geiser T, Deranleau DA. Activation of human neutrophils by C3a and C5A. Comparison of the effects on shape changes, chemotaxis, secretion, and respiratory burst. FEBS Lett. 1994;346:181–184. [DOI] [PubMed] [Google Scholar]
- 61. McDonald B, Kubes P. Cellular and molecular choreography of neutrophil recruitment to sites of sterile inflammation. J Mol Med. 2011;89:1079–1088. [DOI] [PubMed] [Google Scholar]
- 62. de Oliveira S, Rosowski EE, Huttenlocher A. Neutrophil migration in infection and wound repair: going forward in reverse. Nat Rev Immunol. 2016;16:378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Su Y, Richmond A. Chemokine regulation of neutrophil infiltration of skin wounds. Adv Wound Care (New Rochelle). 2015;4:631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Katritch V, Cherezov V, Stevens RC. Structure‐function of the G protein‐coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 2013;53:531–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rosenbaum DM, Rasmussen SGF, Kobilka BK. The structure and function of G‐protein‐coupled receptors. Nature. 2009;459:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moreira IS. Structural features of the G‐protein/GPCR interactions. Biochim Biophys Acta. 2014;1840:16–33. [DOI] [PubMed] [Google Scholar]
- 67. Oldham WM, Hamm HE. Heterotrimeric G protein activation by G‐protein‐coupled receptors. Nat Rev Mol Cell Biol. 2008;9:60–71. [DOI] [PubMed] [Google Scholar]
- 68. Flock T, Hauser AS, Lund N, Gloriam DE, Balaji S, Babu MM. Selectivity determinants of GPCR‐G‐protein binding. Nature. 2017;545:317–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Offermanns S, Simon MI. Genetic analysis of mammalian G‐protein signalling. Oncogene. 1998;17:1375–1381. [DOI] [PubMed] [Google Scholar]
- 70. Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1159–1204. [DOI] [PubMed] [Google Scholar]
- 71. Brzostowski JA, Sawai S, Rozov O, et al. Phosphorylation of chemoattractant receptors regulates chemotaxis, actin reorganization and signal relay. J Cell Sci. 2013;126:4614–4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Wu L, Valkema R, Van Haastert PJ, Devreotes PN. The G protein beta subunit is essential for multiple responses to chemoattractants in Dictyostelium. J Cell Biol. 1995;129:1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Neptune ER, Bourne HR. Receptors induce chemotaxis by releasing the betagamma subunit of Gi, not by activating Gq or Gs. Proc Natl Acad Sci USA. 1997;94:14489–14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kamakura S, Nomura M, Hayase J, et al. The cell polarity protein mInsc regulates neutrophil chemotaxis via a noncanonical G protein signaling pathway. Dev Cell. 2013;26:292–302. [DOI] [PubMed] [Google Scholar]
- 75. Kamp ME, Liu Y, Kortholt A. Function and regulation of heterotrimeric G proteins during chemotaxis. IJMS. 2016;17:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Liu Y, Lacal J, Firtel RA, Kortholt A. Connecting G protein signaling to chemoattractant‐mediated cell polarity and cytoskeletal reorganization. Small GTPases. 2016;85:0–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Luttrell LM, Lefkowitz RJ. The role of beta‐arrestins in the termination and transduction of G‐protein‐coupled receptor signals. J Cell Sci. 2002;115:455–465. [DOI] [PubMed] [Google Scholar]
- 78. DeFea KA. Stop that cell! Beta‐arrestin‐dependent chemotaxis: a tale of localized actin assembly and receptor desensitization. Annu Rev Physiol. 2007;69:535–560. [DOI] [PubMed] [Google Scholar]
- 79. Min J, Defea K. β‐arrestin‐dependent actin reorganization: bringing the right players together at the leading edge. Mol Pharmacol. 2011;80:760–768. [DOI] [PubMed] [Google Scholar]
- 80. Peterson YK, Luttrell LM. The diverse roles of arrestin scaffolds in G protein‐coupled receptor signaling. Pharmacol Rev. 2017;69:256–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nichols JM, Veltman D, Kay RR. Chemotaxis of a model organism: progress with Dictyostelium . Curr Opin Cell Biol. 2015;36:7–12. [DOI] [PubMed] [Google Scholar]
- 82. Aubry L, Guetta D, Klein G. The arrestin fold: variations on a theme. Curr Genomics. 2009;10:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cao X, Yan J, Shu S, Brzostowski JA, Jin T. Arrestins function in cAR1 GPCR‐mediated signaling and cAR1 internalization in the development of Dictyostelium discoideum . Mol Biol Cell. 2014:3210–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Cai H, Huang C‐H, Devreotes PN, Iijima M. Analysis of chemotaxis in Dictyostelium . Methods Mol Biol. 2012;757:451–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Cai H, Devreotes PN. Moving in the right direction: how eukaryotic cells migrate along chemical gradients. Semin Cell Dev Biol. 2011;22:834–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chen MY, Insall RH, Devreotes PN. Signaling through chemoattractant receptors in Dictyostelium. Trends Genet. 1996;12:52–57. [DOI] [PubMed] [Google Scholar]
- 87. Pan M, Xu X, Chen Y, Jin T. Identification of a chemoattractant G‐protein‐coupled receptor for folic acid that controls both chemotaxis and phagocytosis. Dev Cell. 2016;36:428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jalink K, Moolenaar WH, Van Duijn B. Lysophosphatidic acid is a chemoattractant for Dictyostelium discoideum amoebae. Proc Natl Acad Sci USA. 1993;90:1857–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Luna‐Gomes T, Bozza PT, Bandeira‐Melo C. Eosinophil recruitment and activation: the role of lipid mediators. Front Pharmacol. 2013;4:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sadik CD, Luster AD. Lipid‐cytokine‐chemokine cascades orchestrate leukocyte recruitment in inflammation. J Leukoc Biol. 2012;91:207–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Yokomizo T, Kato K, Terawaki K, Izumi T, Shimizu T. A second leukotriene B(4) receptor, BLT2. A new therapeutic target in inflammation and immunological disorders. J Exp Med. 2000;192:421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Demopoulos CA, Pinckard RN, Hanahan DJ. Platelet‐activating factor. Evidence for 1‐O‐alkyl‐2‐acetyl‐sn‐glyceryl‐3‐phosphorylcholine as the active component (a new class of lipid chemical mediators). J Biol Chem. 1979;254:9355–9358. [PubMed] [Google Scholar]
- 93. Reznichenko A, Korstanje R. The role of platelet‐activating factor in mesangial pathophysiology. Am J Pathol. 2015;185:888–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Honda Z‐I, Ishii S, Shimizu T. Platelet‐activating factor receptor. J Biochem. 2002;131:773–779. [DOI] [PubMed] [Google Scholar]
- 95. Amatruda TT, Gerard NP, Gerard C, Simon MI. Specific interactions of chemoattractant factor receptors with G‐proteins. J Biol Chem. 1993;268:10139–10144. [PubMed] [Google Scholar]
- 96. Haribabu B, Zhelev DV, Pridgen BC, Richardson RM, Ali H, Snyderman R. Chemoattractant receptors activate distinct pathways for chemotaxis and secretion. Role of G‐protein usage. J Biol Chem. 1999;274:37087–37092. [DOI] [PubMed] [Google Scholar]
- 97. Brown SL, Jala VR, Raghuwanshi SK, Nasser MW, Haribabu B, Richardson RM. Activation and regulation of platelet‐activating factor receptor: role of G(i) and G(q) in receptor‐mediated chemotactic, cytotoxic, and cross‐regulatory signals. J Immunol. 2006;177:3242–3249. [DOI] [PubMed] [Google Scholar]
- 98. Dahlgren C, Gabl M, Holdfeldt A, Winther M, Forsman H. Basic characteristics of the neutrophil receptors that recognize formylated peptides, a danger‐associated molecular pattern generated by bacteria and mitochondria. Biochem Pharmacol. 2016;114:22–39. [DOI] [PubMed] [Google Scholar]
- 99. He H‐Q, Ye RD. The formyl peptide receptors: diversity of ligands and mechanism for recognition. Molecules. 2017;22:455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bufe B, Schumann T, Kappl R, et al. Recognition of bacterial signal peptides by mammalian formyl peptide receptors: a new mechanism for sensing pathogens. J Biol Chem. 2015;290:7369–7387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rabiet M‐J, Huet E, Boulay F. The N‐formyl peptide receptors and the anaphylatoxin C5a receptors: an overview. Biochimie. 2007;89:1089–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zlotnik A, Yoshie O. The chemokine superfamily revisited. Immunity. 2012;36:705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Stone MJ, Hayward JA, Huang C, Huma ZE, Sanchez J. Mechanisms of regulation of the chemokine‐receptor network. IJMS. 2017;18:342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Watts AO, Verkaar F, van der Lee MMC, et al. β‐Arrestin recruitment and G protein signaling by the atypical human chemokine decoy receptor CCX‐CKR. J Biol Chem. 2013;288:7169–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Bonecchi R, Graham GJ. Atypical chemokine receptors and their roles in the resolution of the inflammatory response. Front Immunol. 2016;7:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Steen A, Larsen O, Thiele S, Rosenkilde MM. Biased and g protein‐independent signaling of chemokine receptors. Front Immunol. 2014;5:277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Zabel BA, Rott A, Butcher EC. Leukocyte chemoattractant receptors in human disease pathogenesis. Annu Rev Pathol. 2015;10:51–81. [DOI] [PubMed] [Google Scholar]
- 108. Kleist AB, Getschman AE, Ziarek JJ, et al. New paradigms in chemokine receptor signal transduction: moving beyond the two‐site model. Biochemical Pharmacology. 2016;114:53–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Holers VM. Complement and its receptors: new insights into human disease. Annu Rev Immunol. 2014;32:433–459. [DOI] [PubMed] [Google Scholar]
- 110. Gasque P. Complement: a unique innate immune sensor for danger signals. Mol Immunol. 2004;41:1089–1098. [DOI] [PubMed] [Google Scholar]
- 111. Hartmann K, Henz BM, Krüger‐Krasagakes S, et al. C3a and C5a stimulate chemotaxis of human mast cells. Blood. 1997;89:2863–2870. [PubMed] [Google Scholar]
- 112. Lee H, Whitfeld PL, Mackay CR. Receptors for complement C5a. The importance of C5aR and the enigmatic role of C5L2. Immunol Cell Biol. 2008;86:153–160. [DOI] [PubMed] [Google Scholar]
- 113. Li R, Coulthard LG, Wu MCL, Taylor SM, Woodruff TM. C5L2: a controversial receptor of complement anaphylatoxin, C5a. FASEB J. 2013;27:855–864. [DOI] [PubMed] [Google Scholar]
- 114. Braun L, Christophe T, Boulay F. Phosphorylation of key serine residues is required for internalization of the complement 5a (C5a) anaphylatoxin receptor via a beta‐arrestin, dynamin, and clathrin‐dependent pathway. J Biol Chem. 2003;278:4277–4285. [DOI] [PubMed] [Google Scholar]
- 115. Vanek M, Hawkins LD, Gusovsky F. Coupling of the C5a receptor to Gi in U‐937 cells and in cells transfected with C5a receptor cDNA. Mol Pharmacol. 1994;46:832–839. [PubMed] [Google Scholar]
- 116. Reca R, Mastellos D, Majka M, et al. Functional receptor for C3a anaphylatoxin is expressed by normal hematopoietic stem/progenitor cells, and C3a enhances their homing‐related responses to SDF‐1. Blood. 2003;101:3784–3793. [DOI] [PubMed] [Google Scholar]
- 117. Gutzmer R, Lisewski M, Zwirner J, et al. Human monocyte‐derived dendritic cells are chemoattracted to C3a after up‐regulation of the C3a receptor with interferons. Immunology. 2004;111:435–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Nilsson G, Johnell M, Hammer CH, et al. C3a and C5a are chemotaxins for human mast cells and act through distinct receptors via a pertussis toxin‐sensitive signal transduction pathway. J Immunol. 1996;157:1693–1698. [PubMed] [Google Scholar]
- 119. Bénard M, Raoult E, Vaudry D, et al. Role of complement anaphylatoxin receptors (C3aR, C5aR) in the development of the rat cerebellum. Mol Immunol. 2008;45:3767–3774. [DOI] [PubMed] [Google Scholar]
- 120. Norgauer J, Dobos G, Kownatzki E, et al. Complement fragment C3a stimulates Ca2+ influx in neutrophils via a pertussis‐toxin‐sensitive G protein. Eur J Biochem. 1993;217:289–294. [DOI] [PubMed] [Google Scholar]
- 121. Schraufstatter IU, Trieu K, Sikora L, Sriramarao P, DiScipio R. Complement c3a and c5a induce different signal transduction cascades in endothelial cells. J Immunol. 2002;169:2102–2110. [DOI] [PubMed] [Google Scholar]
- 122. Yang M, Sang H, Rahman A, Wu D, Malik AB, Ye RD. G alpha 16 couples chemoattractant receptors to NF‐kappa B activation. J Immunol. 2001;166:6885–6892. [DOI] [PubMed] [Google Scholar]
- 123. Gleick J. The Information. New York, NY: Pantheon Books; 2011. [Google Scholar]
- 124. Hartley R. Transmission of information. Bell Labs Tech J. 1928;7:535–563. [Google Scholar]
- 125. Hann MM, Leach AR, Harper G. Molecular complexity and its impact on the probability of finding leads for drug discovery. J Chem Inf Comput Sci. 2001;41:856–864. [DOI] [PubMed] [Google Scholar]
- 126. Wang X, Sharp JS, Handel TM, Prestegard JH. Chemokine oligomerization in cell signaling and migration. Prog Mol Biol Transl Sci. 2013;117:531–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Yang D, Chen Q, Hoover DM, et al. Many chemokines including CCL20/MIP‐3alpha display antimicrobial activity. J Leukoc Biol. 2003;74:448–455. [DOI] [PubMed] [Google Scholar]
- 128. Proudfoot AEI, Johnson Z, Bonvin P, Handel TM. Glycosaminoglycan interactions with chemokines add complexity to a complex system. Pharmaceuticals (Basel). 2017;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Falke JJ, Piasta KN. Architecture and signal transduction mechanism of the bacterial chemosensory array: progress, controversies, and challenges. Curr Opin Struct Biol. 2014;29:85–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Cherezov V, Rosenbaum DM, Hanson MA, et al. High‐Resolution crystal structure of an engineered human β2‐adrenergic G protein‐coupled receptor. Science. 2007;318:1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Rasmussen SGF, Choi H‐J, Fung J‐J, et al. Structure of a nanobody‐stabilized active state of the β(2) adrenoceptor. Nature. 2011;469:175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Latorraca NR, Venkatakrishnan AJ, Dror RO. GPCR dynamics: structures in motion. Chem Rev. 2016;117:139–155. [DOI] [PubMed] [Google Scholar]
- 133. Briegel A, Ames P, Gumbart JC, Oikonomou CM, Parkinson JS, Jensen GJ. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Mol Microbiol. 2013;89:831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven‐transmembrane receptors. Nat Rev Drug Discov. 2010;9:373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Cooray SN, Gobbetti T, Montero‐Melendez T, et al. Ligand‐specific conformational change of the G‐protein‐coupled receptor ALX/FPR2 determines proresolving functional responses. Proc Natl Acad Sci USA. 2013;110:18232–18237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Gabl M, Holdfeldt A, Sundqvist M, Lomei J, Dahlgren C, Forsman H. FPR2 signaling without β‐arrestin recruitment alters the functional repertoire of neutrophils. Biochem Pharmacol. 2017;145:114–122. [DOI] [PubMed] [Google Scholar]
- 137. Qin CX, May LT, Li R, Cao N, Rosli S, Deo M, et al. Small‐molecule‐biased formyl peptide receptor agonist compound 17b protects against myocardial ischaemia‐reperfusion injury in mice. Nat Commun. 2017;8:14232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Artemenko Y, Lampert TJ, Devreotes PN. Moving towards a paradigm: common mechanisms of chemotactic signaling in Dictyostelium and mammalian leukocytes. Cell Mol Life Sci. 2014;71:3711–3747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Zheng Y, Han GW, Abagyan R, et al. Structure of CC chemokine receptor 5 with a potent chemokine antagonist reveals mechanisms of chemokine recognition and molecular mimicry by HIV. Immunity. 2017;46:1005–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Smith JS, Alagesan P, Desai NK, et al. C‐X‐C motif chemokine receptor 3 splice variants differentially activate beta‐arrestins to regulate downstream signaling pathways. Mol Pharmacol. 2017;92:136–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Ziarek JJ, Kleist AB, London N, et al. Structural basis for chemokine recognition by a G protein‐coupled receptor and implications for receptor activation. Sci Signal. 2017;10:eaah5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Burg JS, Ingram JR, Venkatakrishnan AJ, et al. Structural biology. Structural basis for chemokine recognition and activation of a viral G protein‐coupled receptor. Science. 2015;347:1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Qin L, Kufareva I, Holden LG, et al. Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science. 2015:1261064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Colin R, Sourjik V. Emergent properties of bacterial chemotaxis pathway. Curr Opin Microbiol. 2017;39:24–33. [DOI] [PubMed] [Google Scholar]
- 145. Briegel A, Wong ML, Hodges HL, et al. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry. 2014;53:1575–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Parkinson JS, Ames P, Studdert CA. Collaborative signaling by bacterial chemoreceptors. Curr Opin Microbiol. 2005;8:116–121. [DOI] [PubMed] [Google Scholar]
- 147. Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. [DOI] [PubMed] [Google Scholar]
- 148. Kalinin Y, Neumann S, Sourjik V, Wu M. Responses of Escherichia coli bacteria to two opposing chemoattractant gradients depend on the chemoreceptor ratio. J Bacteriol. 2010;192:1796–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Khan S, Spudich JL, McCray JA, Trentham DR. Chemotactic signal integration in bacteria. Proc Natl Acad Sci USA. 1995;92:9757–9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Yang Y, Sourjik V. Opposite responses by different chemoreceptors set a tunable preference point in Escherichia coli pH taxis. Mol. Microbiol. 2012;86:1482–1489. [DOI] [PubMed] [Google Scholar]
- 151. Krembel AK, Neumann S, Sourjik V. Universal response‐adaptation relation in bacterial chemotaxis. J Bacteriol. 2015;197:307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Vladimirov N, Sourjik V. Chemotaxis: how bacteria use memory. Biol Chem. 2009;390:1097–1104. [DOI] [PubMed] [Google Scholar]
- 153. Lan G, Schulmeister S, Sourjik V, Tu Y. Adapt locally and act globally: strategy to maintain high chemoreceptor sensitivity in complex environments. Mol Syst Biol. 2011;7:475–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Kitayama J, Carr MW, Roth SJ, Buccola J, Springer TA. Contrasting responses to multiple chemotactic stimuli in transendothelial migration: heterologous desensitization in neutrophils and augmentation of migration in eosinophils. J Immunol. 1997;158:2340–2349. [PubMed] [Google Scholar]
- 155. Foxman EF, Campbell JJ, Butcher EC. Multistep navigation and the combinatorial control of leukocyte chemotaxis. J Cell Biol. 1997;139:1349–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Foxman EF, Kunkel EJ, Butcher EC. Integrating conflicting chemotactic signals. The role of memory in leukocyte navigation. J Cell Biol. 1999;147:577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Prentice‐Mott HV, Meroz Y, Carlson A, et al. Directional memory arises from long‐lived cytoskeletal asymmetries in polarized chemotactic cells. Proc Natl Acad Sci USA. 2016;113:1267–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Hutchings CJ, Koglin M, Olson WC, Marshall FH. Opportunities for therapeutic antibodies directed at G‐protein‐coupled receptors. Nat Rev Drug Discov. 2017;16:1–24. [DOI] [PubMed] [Google Scholar]
- 159. Wang M‐J, Artemenko Y, Cai W‐J, Iglesias PA, Devreotes PN. The directional response of chemotactic cells depends on a balance between cytoskeletal architecture and the external gradient. Cell Rep. 2014;9:1110–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]


