Abstract
Bacterial computing is a known candidate in natural computing, the aim being to construct “bacterial computers” for solving complex problems. In this paper, a new kind of bacterial computing system, named the bacteria and plasmid computing system (BP system), is proposed. We investigate the computational power of BP systems with finite numbers of bacteria and plasmids. Specifically, it is obtained in a constructive way that a BP system with 2 bacteria and 34 plasmids is Turing universal. The results provide a theoretical cornerstone to construct powerful bacterial computers and demonstrate a concept of paradigms using a “reasonable” number of bacteria and plasmids for such devices.
Keywords: bacterial computing, bacteria and plasmid system, Turing universality, recursively enumerable function
1. Introduction
In cell biology, bacteria, despite their simplicity, contain a well-developed cell structure that is responsible for some of their unique biological structures and pathogenicity. The bacterial DNA resides inside the bacterial cytoplasm, for which transfer of cellular information, transcription, and DNA replication occurs within the same compartment [1,2]. Along with chromosomal DNA, most bacteria also contain small independent pieces of DNA called plasmids, which can be conveniently obtained and released by a bacterium to act as a gene delivery vehicle between bacteria in the form of horizontal gene transfer [3].
Bacterial computing was coined with the purpose of building biological machines, which are developed to solve real-life engineering and science problems [4]. Practically, bacterial computing proves mechanisms and the possibility of using bacteria for solving problems in vivo. If an individual bacterium can perform computation work as a computer, this envisions a way to build millions of computers in vivo. These “computers”, combined together, can perform complicated computing tasks with efficient communication via plasmids. Using such conjugation, DNA molecules, acting as information carriers, can be transmitted from one cell to another. On the basis of the communication, information in one bacteria can be moved to another and can be used for further information processing [5,6].
Bacterial computing models belong to the field of bio-computing models, such as DNA computing models [7,8,9] and membrane computing models [10,11,12]. Because of the computational intelligence and parallel information processing strategy in biological systems, most of the bio-computing models have been proven to have the desired computational power. Most of these can do what a Turing machine can do (see, e.g., [13,14,15,16,17,18,19]). The proposed bacterial computing models can provide powerful computing models at the theoretical level but a lack of practical results. Current bacterial computing models are designed for solving certain specific biological applications, such as bacteria signal pathway detecting, but give no result for computing power analysis.
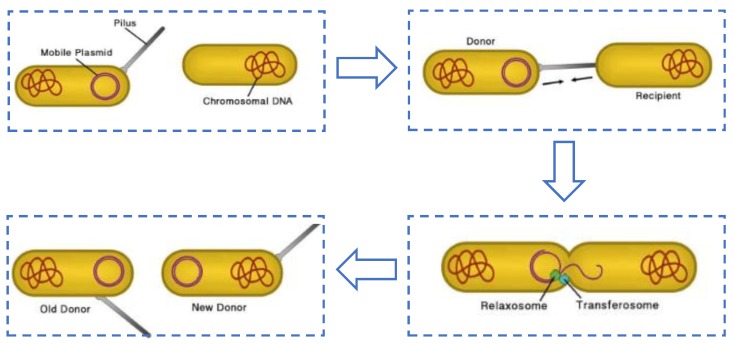
In general bacterial computing models, information to be processed is encoded by DNA sequences, and conjugation is the tool for communicating among bacteria. The biological process is shown in Figure 1.
Figure 1.
Bacteria conjugation from biological point of view.
Looking for small universal computing devices, such as small universal Turing machines [20,21], small universal register machines [22], small universal cellular automata [23], small universal circular Post machines [24], and so on, is a natural and well-investigated topic in computer science. Recently, this topic started to be considered also in the framework of bio-computing models [25,26,27,28,29,30,31].
In this work, we focus on designing small universal bacteria and plasmid computing systems (BP systems); that is, we construct Turing universal BP systems with finite numbers of bacteria and plasmids. Specifically, we demonstrate that a BP system with 2 bacteria and 34 plasmids is universal for computing recursively enumerable functions and families of sets of natural numbers. In the universality proofs, 2 bacteria are sufficient, as in [32], but the numbers of plasmids needed are reduced to about 10 from a possible infinite number. The results provide a theoretical cornerstone to construct powerful “bacterial computers” and demonstrate a concept of paradigms using a “reasonable” number of bacteria and plasmids for these devices.
2. The Bacteria and Plasmid System
In this work, as for automata in automata theory, the BP system is formally designed and defined. In general, the system is composed of three main components:
-
–
a set of bacteria;
-
–
a set of plasmids;
-
–
a set of evolution rules in each bacterium, including conjugation rules and gene-editing (inserting/deleting) rules.
The evolution rules are in the form of productions in formal language theory, which are used to process and communicate information among bacteria. Such a system is proven to be powerful for a number of computing devices; that is, they can compute the sets of natural numbers that are Turing computable. However, in the universality proof, the number of plasmids involved is not limited. It is possible to use an infinite number of plasmids for information processing and exchanging. Such a feature is acceptable (as for the infinite tape in Turing machines) in mathematic theory but is not feasible with the biological facts.
A BP system of degree m is a construct of the following form:
is a set of genes in the chromosomal DNA of bacteria.
- is a set of plasmids.
-
–Plasmids in are of the form with , which is used for cutting specific genes.
-
–Plasmids in are of the form , which takes templates of genes to be inserted.
-
–Plasmid is of the form for bacteria conjugation.
-
–
- Variables are m bacteria of the form , where
-
–is a set of genes over O initially placed in bacterium ;
-
–is a set of rules in bacterium of the following forms:
-
(1)Conjugation rule is of the form , by which ATP in bacterium is consumed and a set of plasmids associated with ATP is transmitted into bacterium .
-
(2)CRISPR/Cas9 gene inserting rule is of the form , where , , , and and are two neighboring genes. The insertion is operated if and only if and are neighboring genes and plasmids are present in the bacterium.
-
(3)CRISPR/Cas9 gene deleting rule is of the form with , , and and being two neighboring genes. The rule can be used if and only there exists gene placed between the two neighboring genes.
-
(1)
-
–
Variable is the output bacterium.
It is possible to have more than one enabled conjugation rule at a certain moment in a bacterium, but only one is non-deterministically chosen for use. This is due to the biological fact that ATP can support the transmission of one plasmid but not all of the plasmids. If a bacterium has more than one CRISPR/Cas9 operating rule associated with a certain common plasmid, only one of the rules is non-deterministically chosen for use; if the enabled CRISPR/Cas9 operating rules are associated with different plasmids, all of them will be used to edit the related genes.
The configuration of the system is described by chromosomal DNA encoding the information in each bacterium. Thus, the initial configuration is . Using the conjugation and CRISPR/Cas9 rules defined above, we can define the transitions among configurations. Any sequence of transitions starting from the initial configuration is called a computation. A computation is called successful if it reaches a halting configuration, that is, no rule can be used in any bacterium. The computational result is encoded by the chromosomal DNA in bacterium when the system halts, where denotes the output bacterium. There are several ways to encode numbers by the chromosomal DNA. We use the number of genes in the chromosomal DNA to encode different numbers computed by the system.
The set of numbers computed by system is denoted by . We denote by the family of sets of numbers computed/generated by BP systems with m bacteria and k plasmids (if no limit is imposed on the values of parameters m and k, then the notation is replaced by *).
We need an input bacterium to receive genetic signals in the form of short DNA segments from the environment or certain bacteria, as well as an output bacteria, with which the system can compute functions. The input bacterium is denoted by with . Input bacterium can read/receive information from the environment, where information is encoded by DNA segments or a string of genes. When a BP system has both input and output bacteria, it starts by reading/receiving information from the environment through input bacterium . After reading the input information, the system starts its computation by using the conjugation and CRISPR/Cas9 gene inserting/deleting rules; it then finally halts. The computational result is stored in the output bacterium encoded by a number of certain genes.
Mathematically, if the input information is x, which is encoded by DNA segments composed of x genes, when the system halts, bacterium holds y genes. It is said that the BP system can compute the function . In general, if the inputs are in the form of DNA strands containing copies of gene with , when the system halts, we obtain the computational result y in the output bacterium in the form of y copies of genes. The system is said to compute the function .
3. Universality Results
In this section, we construct two small universal BP systems. Specifically, we construct a Turing universal BP system with 2 bacteria and 34 plasmids to compute recursively enumerable functions. As a natural-number computing device, a universal BP system with 2 bacteria and 34 plasmids is achieved.
In the following universality proofs, the notion of a register machine is used. A register machine is a construct of the form , where m is the number of registers, H is the set of instruction labels, is the start label, is the halt label (assigned to instruction HALT), and R is the set of instructions; each label from H labels only one instruction from R, thus precisely identifying it. The instructions are of the following forms:
ADD (add 1 to register r and then go to one of the instructions with labels and );
SUB (if register r is non-zero, then subtract 1 from it, and go to the instruction with label ; otherwise, go to the instruction with label );
HALT (the halt instruction).
A register machine M generates a set of numbers in the following way: it starts with all registers being empty (i.e., storing the number zero) and then applies the instruction with label ; it continues to apply instructions as indicated by the labels (and made possible by the contents of registers). If the register machine finally reaches the halt instruction, then the number n present in specified register 0 at that time is said to be generated by M. If the computation does not halt, then no number is generated. It is known (e.g., see [33]) that register machines generate all sets of numbers that are Turing computable.
A register machine can also compute functions. In [22], register machines are proposed for computing functions, with the universality defined as follows: Let be a fixed admissible enumeration of the unary partial recursive functions. A register machine M is said to be universal if there is a recursive function g such that for all natural numbers x and y, it holds ; that is, with input and y introduced in registers 1 and 2, the result is obtained in register 0 when M halts.
A specific universal register machine shown in Figure 2 is used here, which was modified by a universal register machine from [22]. Specifically, the universal register machine from [22] contains a separate check for zero of register 6 of the form (SUB(6), ); this instruction was replaced in by (SUB(6), , (ADD(6), (see Figure 2). Therefore, in the modified universal register machine, there are 8 registers (numbered from 0 to 7) and 23 instructions (hence 23 labels), the last instruction being the halting instruction. The input numbers are introduced in registers 1 and 2, and the result is obtained in register 0.
Figure 2.
The universal register machine for computing Turing-computable functions [22].
3.1. A Small Universal BP System as Function Computing Device
Theorem 1.
There exists a Turing universal BP system with 2 bacteria and 34 plasmids that can compute Turing-computable recursively enumerable functions.
Proof.
To this aim, we construct a BP system with 2 bacteria and 34 plasmids to simulate the register machine shown in Figure 2. The system is of the following form:
is set of genes in chromosomal DNA of bacteria.
- –
, whose elements associated with the labels of instructions are used for gene cutting;- –
are plasmids taking templates of genes to be inserted, which are used for simulating ADD instructions;- –
plasmid for bacteria conjugation is used for simulating SUB instructions., where , meaning no initial chromosomal DNA is placed in bacteria ; the set of rules is shown in Table 2.
, where , indicating the initially placed chromosomal DNA in bacterium ; the set of rules is shown in Table 2.
, which means bacterium can read signals from the environment, and when the system halts, the computational result is stored in bacterium .
In general, for each add instruction acting on register , plasmids and are associated; for any SUB instruction acting on register , a plasmid is associated in system . The numbers stored in register r are encoded by the number of copies of gene with in chromosomal DNA of bacterium . Specifically, if the number stored in register r is , then bacterium contains copies of gene .
During the simulation of register machine by system , when bacterium holds a pair of plasmids (respectively ) and ATP, the system starts to simulate an ADD instruction (respectively a SUB instruction) of : plasmids (respectively ) are transmitted to bacterium by the conjugation rule; then one copy of gene between neighboring genes and is inserted (respectively deleted) to simulate increasing (respectively decreasing) the number in register r by 1; after this, bacterium sends ATP and plasmids to bacterium if the proceeding instruction is a SUB instruction or plasmids if the proceeding is an ADD instruction.
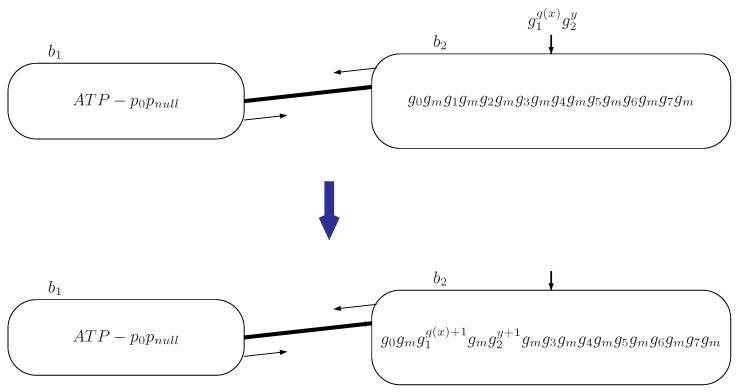
Initially, there is no chromosomal DNA initially placed in bacterium , but bacterium has genes . At the beginning, the system receives copies of gene and y copies of gene from the environment through input bacterium , which simulates the numbers and y being introduced in registers 1 and 2 for register machine . In this way, the chromosomal DNA of bacterium becomes
Once completing the reading of information from the environment, a pair of plasmids and one unit of ATP is placed in bacterium to trigger the computation; meanwhile no plasmid or ATP is initially contained in bacterium . The transition of system by reading input signals encoded by copies of genes and y copies of gene through input bacterium is shown in Figure 3.
In what follows, we explain how system simulates ADD instructions and SUB instructions and outputs the computational result.
Simulating the ADD instruction: (DD(r), .
We assume at a certain moment that system starts to simulate an ADD instruction of , acting on register . At that moment, bacterium holds two plasmids and ATP, such that the conjugation rule is used. By using the conjugation rule, plasmids and ATP are transmitted to bacterium . In system , plasmids and are associated with the ADD instruction , where plasmid is of the form for cutting a certain site of chromosomal DNA, and is of the form carrying the gene to be inserted.
In bacterium , the CRISPR/Cas9 inserting rule is used to insert gene between neighboring genes and . In this way, the number of gene of bacterium is increased by 1, which simulates the number in register r being increased by 1. We note that there is a unique position at which gene can be inserted with the context of neighboring and .
By using the CRISPR/Cas9 inserting rule, plasmid is consumed, and plasmid and ATP remain in bacterium . The conjugation rule in bacterium is designed by the operation of the proceeding instruction . One of the following two cases occurs in bacterium .
If instruction is an ADD instruction, then bacterium has the conjugation rule . By using the rule, plasmids and ATP are conjugated to bacterium . In this case, system starts to simulate the proceeding ADD instruction .
If instruction is a SUB instruction, then bacterium has the conjugation rule , by which plasmids and ATP are transmitted to bacterium . In this case, system starts to simulate the proceeding SUB instruction .
Therefore, system can correctly simulate the ADD instruction of . The system starts from bacterium having plasmid and ATP, which are transmitted to bacterium by the conjugation rule. In bacterium , the number of gene in chromosomal DNA is increased by 1 using the CRISPR/Cas9 gene inserting rule, and plasmids (if the proceeding instruction is an ADD instruction) or (if the proceeding instruction is a SUB instruction) are transmitted to bacterium , which means that system starts to simulate instruction .
Simulating the SUB instruction: (SUB(r), .
We suppose at a certain computation step that system has to simulate a SUB instruction (SUB(r), . For any SUB instruction , plasmid of the form is associated in system . In bacterium , there are plasmids and ATP such that the conjugation rule can be used. In bacterium , it has the following two cases.
- –
If there is at least one gene existing between neighboring genes and in chromosomal DNA of bacterium (corresponding to the case that the number stored in register r is ), then the CRISPR/Cas9 deleting rule is used to delete one copy of gene from chromosomal DNA. This simulates the number stored in register r being decreased by 1. By consuming plasmid , bacterium retains plasmid and ATP such that a conjugation rule or is used, which depends on whether the proceeding instruction would be an ADD or a SUB instruction. In this way, plasmids or and ATP are transmitted to bacterium . The system starts to simulate instruction .
- –
If there is no gene existing between neighboring genes and in chromosomal DNA of bacterium (corresponding to the case that the number stored in register r is 0), then the CRISPR/Cas9 deleting rule cannot be used, but a conjugation rule or is able to be used. Plasmids ( or ) and ATP are conjugated to bacterium , which means the system starts to simulate instruction .
We note that when plasmids are conjugated to bacterium from bacterium , it may happen that both the CRISPR/Cas9 deleting rule and (or ) can be used. In this case, the CRISPR/Cas9 deleting rule will be applied because of the fact that it has priority over the plasmid transferring rule.
The simulation of a SUB instruction is correct: System starts from bacterium having plasmid and ATP and ends with plasmid or and ATP (if the number stored in register r is ) to start the simulation of instruction ; otherwise it ends with plasmid or and ATP (if the number stored in register r is 0) to start the simulation of instruction .
Simulating the halt instruction: HALT.
When register machine reaches the halt instruction HALT, the computation of register machine halts. At that moment, bacterium in system holds plasmids and ATP, and the conjugation rule can be used. By using the rule, plasmids and ATP are transmitted to bacterium ; no gene can be edited by plasmid , and no rule can be used. Hence, the computation of system finally halts.
The number of gene in chromosomal DNA of bacterium encodes the number stored in register 0 of . If the number stored in register 0 is , then there are copies of gene in chromosomal DNA of bacterium . The computational result can be obtained by counting the number of gene in chromosomal DNA of bacterium .
From the above description of system and its work, it is clear that system can simulate each computation of . We can check that the constructed system has
2 bacterium for conjugation with each other;
22 plasmids for the 22 ADD and SUB instructions with ;
9 plasmids for 9 ADD instructions with ;
1 plasmid for the 13 SUB instructions;
2 plasmids and for the HALT instruction;
8 genes for encoding numbers in registers i with ;
1 gene for separating gene in chromosomal DNA.
This gives, in total, 2 bacteria, 34 plasmids, and 9 genes.
This concludes the proof. ☐
Table 1.
Plasmids in system .
| Plasmid | Forms of Plasmids | Plasmid | Forms of Plasmids |
|---|---|---|---|
Table 2.
Rules in each bacterium of system .
| Sim. | Rules | Bac. |
|---|---|---|
| , , | ||
| , | ||
| , | ||
| , , | ||
| , , | ||
| , | ||
| , , | ||
| , | ||
| , , | ||
| , | ||
| , , | ||
| , , | ||
| , , | ||
| , , | ||
| , , | ||
| , , | ||
| , | ||
| , | ||
| , , | ||
| , , | ||
| , | ||
| , | ||
Figure 3.
The transition of system by reading input information encoded by copies of genes and y copies of gene through input bacterium .
3.2. A Small Universal BP System as a Number Generator
In this section, we construct a small universal BP system as a number generator. A BP system is universal if, given a fixed admissible enumeration of the unary partial recursive functions , there is a recursive function g such that for each natural number x, whenever we input the number in , the set of numbers generated by the system is equal to { is defined}. In other words, after introducing the “code” of the partial recursive function in the form of copies of certain genes in chromosomal DNA of the input bacterium, the BP system generates all numbers n for which is defined.
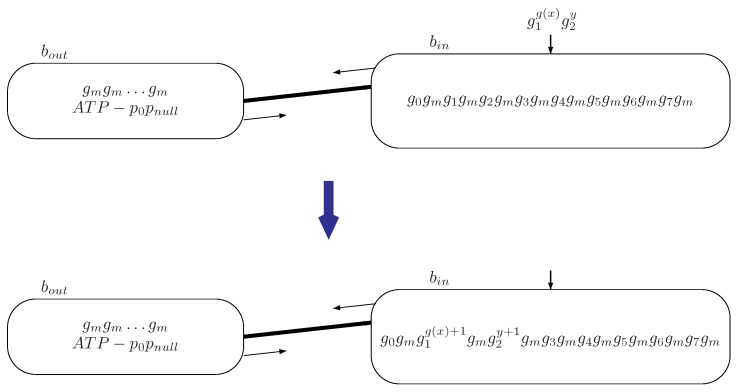
System has the same topological structure, plasmids, and evolution rules as system constructed in Section 3.1, but the input bacterium is and the output bacterium is . Differently from the universal computing devices considered in Section 3.1, the strategy to simulate a universal register machine as a number generator is as follows.
Step 1. The output bacterium initially has n copies of gene .
Step 2. System starts by loading copies of gene and n copies of gene in the input bacterium .
Step 3. The computation of is activated by using plasmid to simulate the register machine from Figure 2, with stored in register 1, and number n stored in register 2.
If the computation in register machine halts, instruction can finally be activated. To simulate register machine reaching the HALT instruction, system holds plasmids and transmits them to bacterium . After this, system halts, as no rule can be used in bacterium . When the system halts, the number of gene in the output bacterium is the computational result, which is exactly the number n. Hence, the number n can be computed/generated by system .
The difference between systems and is the loading input information process. The initial configuration and transition of system by reading input signals encoded by copies of genes and n copies of gene through input bacterium are shown in Figure 4.
Figure 4.
The initial configuration and transition of system by reading input information encoded by copies of genes and n copies of gene through input bacterium .
We can check that the constructed system has
2 bacterium for the conjugation with each other;
22 plasmids for the 22 ADD and SUB instructions with ;
9 plasmids for 9 ADD instructions with ;
1 plasmid for the 13 SUB instructions;
2 plasmids and for the HALT instruction;
8 genes for encoding numbers in registers i with ;
1 gene for separating gene in chromosomal DNA.
This gives, in total, 2 bacteria, 34 plasmids, and 9 genes.
Therefore, we have the following theorem.
Theorem 2.
There is a Turing universal BP system with 2 bacteria and 34 plasmids that can compute a Turing-computable set of natural numbers.
4. Conclusions
In this work, we construct two small universal BP systems. Specifically, it is obtained that a BP system with 2 bacteria, 34 plasmids, and 9 genes is universal for both computing recursively enumerable functions and computing/generating a family of sets of natural numbers. It is obtained that 34 plasmids are sufficient for constructing Turing universal BP systems. This provides theoretical support as well as paradigms using a reasonable number of bacteria and plasmids to construct powerful bacterial computers.
Following the research line, finding smaller universal BP systems deserves further research. A possible way to slightly decrease the number of plasmids used in small universal BP systems is using code optimization, exploiting some particularities of the register machine . For example, as considered in [25], for the sequence of two consecutive ADD instructions : (ADD and : (ADD, without any other instruction addressing the label , the two ADD modules can be combined. However, a challenging problem regards what the minimum size of a universal BP system is—in other words, what the borderline between universality and non-universality is. Characterization of universality by BP systems is expected. A balance between the number of bacteria and plasmids in universal BP systems can be considered, that is, using more bacteria to reduce the number of plasmids.
It is worth developing the applications of BP systems. Bio-inspiring computing models perform well in computations, particularly in solving computational complex problems in feasible time [34,35,36]. It is of interest to use BP systems to solve computationally hard problems. Some specific applications using BP systems would be of interest to researchers from biological fields.
In artificial intelligence, there are many bio-inspired algorithms (see, e.g., [37,38]). It is worth designing bacteria-computing-inspired algorithms or introducing bacteria computing operators in classical algorithms. Additionally, it would be meaningful to construct powerful bacterial computers or computing devices in biological labs.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant Nos. 61502535, 61572522, 61572523, 61672033, and 61672248), PetroChina Innovation Foundation (2016D-5007-0305), Key Research and Development Program of Shandong Province (No. 2017GGX10147), Natural Science Foundation of Shandong Province (No. ZR2017MF004), Talent introduction project of China University of Petroleum (No. 2017010054), Research Project TIN2016-81079-R (AEI/FEDER, Spain-EU) and Grant 2016-T2/TIC-2024 from Talento-Comunidad de Madrid, Project TIN2016-81079-R (MINECO AEI/FEDER, Spain-EU), and the InGEMICS-CM Project (B2017/BMD-3691, FSE/FEDER, Comunidad de Madrid-EU).
Author Contributions
Conceptualization, X.W. and T.S.; Methodology, X.W.; Software, T.M.; Validation, P.Z., T.S. and X.W.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available.
References
- 1.Gitai Z. The new bacterial cell biology: Moving parts and subcellular architecture. Cell. 2005;120:577–586. doi: 10.1016/j.cell.2005.02.026. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein E., Drlica K. Regulation of bacterial dna supercoiling: Plasmid linking numbers vary with growth temperature. Proc. Natl. Acad. Sci. USA. 1984;81:4046–4050. doi: 10.1073/pnas.81.13.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Summers D. The Biology of Plasmids. John Wiley & Sons; Hoboken, NJ, USA: 2009. [Google Scholar]
- 4.Poet J.L., Campbell A.M., Eckdahl T.T., Heyer L.J. Bacterial computing. XRDS Crossroads ACM Mag. Stud. 2010;17:10–15. doi: 10.1145/1836543.1836550. [DOI] [Google Scholar]
- 5.Gupta V., Irimia J., Pau I., Rodríguez-Patón A. Bioblocks: Programming protocols in biology made easier. ACS Synth. Biol. 2017;6:1230–1232. doi: 10.1021/acssynbio.6b00304. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez M., Gregorio-Godoy P., del Pulgar G.P., Munoz L.E., Sáez S., Rodríguez-Patón A. A new improved and extended version of the multicell bacterial simulator gro. ACS Synth. Biol. 2017;6:1496–1508. doi: 10.1021/acssynbio.7b00003. [DOI] [PubMed] [Google Scholar]
- 7.Adleman L.M. Molecular computation of solutions to combinatorial problems. Sciences. 1994;266:1021–1024. doi: 10.1126/science.7973651. [DOI] [PubMed] [Google Scholar]
- 8.Carell T. Molecular computing: Dna as a logic operator. Nature. 2011;469:45–46. doi: 10.1038/469045a. [DOI] [PubMed] [Google Scholar]
- 9.Xu J. Probe machine. IEEE Trans. Neural Netw. Learn. Syst. 2016;27:1405–1416. doi: 10.1109/TNNLS.2016.2555845. [DOI] [PubMed] [Google Scholar]
- 10.Păun G., Rozenberg G., Salomaa A. The Oxford Handbook of Nembrane Computing. Oxford University Press; Oxford, UK: 2010. [Google Scholar]
- 11.Păun G., Rozenberg G. A guide to membrane computing. Theor. Comput. Sci. 2002;287:73–100. doi: 10.1016/S0304-3975(02)00136-6. [DOI] [Google Scholar]
- 12.Păun G. Computing with membranes. J. Comput. Syst. Sci. 2000;61:108–143. doi: 10.1006/jcss.1999.1693. [DOI] [Google Scholar]
- 13.Freund R., Kari L., Păun G. DNA computing based on splicing: The existence of universal computers. Theory Comput. Syst. 1999;32:69–112. doi: 10.1007/s002240000112. [DOI] [Google Scholar]
- 14.Kari L., Păun G., Rozenberg G., Salomaa A., Yu S. DNA computing, sticker systems, and universality. Acta Inform. 1998;35:401–420. doi: 10.1007/s002360050125. [DOI] [Google Scholar]
- 15.Martın-Vide C., Păun G., Pazos J., Rodrıguez-Patón A. Tissue P systems. Theor. Comput. Sci. 2003;296:295–326. doi: 10.1016/S0304-3975(02)00659-X. [DOI] [Google Scholar]
- 16.Ionescu M., Păun G., Yokomori T. Spiking neural P systems. Fundam. Inform. 2006;71:279–308. [Google Scholar]
- 17.Song T., Pan L., Păun G. Asynchronous spiking neural p systems with local synchronization. Inform. Sci. 2013;219:197–207. doi: 10.1016/j.ins.2012.07.023. [DOI] [Google Scholar]
- 18.Ezziane Z. DNA computing: Applications and challenges. Nanotechnology. 2005;17:R27. doi: 10.1088/0957-4484/17/2/R01. [DOI] [Google Scholar]
- 19.Chen X., Pérez-Jiménez M.J., Valencia-Cabrera L., Wang B., Zeng X. Computing with viruses. Theor. Comput. Sci. 2016;623:146–159. doi: 10.1016/j.tcs.2015.12.006. [DOI] [Google Scholar]
- 20.Rogozhin Y. Small universal turing machines. Theor. Comput. Sci. 1996;168:215–240. doi: 10.1016/S0304-3975(96)00077-1. [DOI] [Google Scholar]
- 21.Baiocchi C. Machines, Computations, and Universality. Springer; Berlin/Heidelberg, Germany: 2001. Three small universal turing machines; pp. 1–10. [Google Scholar]
- 22.Korec I. Small universal register machines. Theor. Comput. Sci. 1996;168:267–301. doi: 10.1016/S0304-3975(96)00080-1. [DOI] [Google Scholar]
- 23.Iirgen Albert J., Culik K., II A simple universal cellular automaton and its one-way and totalistic version. Complex Syst. 1987;1:1–16. [Google Scholar]
- 24.Kudlek M., Yu R. Small universal circular post machines. Comput. Sci. J. Mold. 2001;9:25. [Google Scholar]
- 25.Păun A., Păun G. Small universal spiking neural P systems. BioSystems. 2007;90:48–60. doi: 10.1016/j.biosystems.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhang X., Zeng X., Pan L. Smaller universal spiking neural P systems. Fundam. Inform. 2008;87:117–136. [Google Scholar]
- 27.Pan L., Zeng X. A note on small universal spiking neural P systems. Lect. Notes Comput. Sci. 2010;5957:436–447. [Google Scholar]
- 28.Păun A., Sidoroff M. Membrane Computing. Springer; Berlin/Heidelberg, Germany: 2012. Sequentiality induced by spike number in SNP systems: Small universal machines; pp. 333–345. [Google Scholar]
- 29.Song T., Jiang Y., Shi X., Zeng X. Small universal spiking neural P systems with anti-spikes. J. Comput. Theor. Nanosci. 2013;10:999–1006. doi: 10.1166/jctn.2013.2799. [DOI] [Google Scholar]
- 30.Song T., Xu J., Pan L. On the universality and non-universality of spiking neural P systems with rules on synapses. IEEE Trans. Nanobiosci. 2015;14:960–966. doi: 10.1109/TNB.2015.2503603. [DOI] [PubMed] [Google Scholar]
- 31.Song T., Pan L. Spiking neural P systems with request rules. Neurocomputing. 2016;193:193–200. doi: 10.1016/j.neucom.2016.02.023. [DOI] [Google Scholar]
- 32.Song T., Rodríguez-Patón A., Gutiérrez M., Pan Z. Computing with bacteria conjugation and crispr/cas9 gene editing operations. Sci. Rep. 2018 sumitted. [Google Scholar]
- 33.Minsky M.L. Computation: Finite and Infinite Machines. Prentice-Hall, Inc.; Upper Saddle River, NJ, USA: 1967. [Google Scholar]
- 34.Valencia-Cabrera L., Orellana-Martín D., Martínez-del Amor M.A., Riscos-Nú nez A., Pérez-Jiménez M.J. Computational efficiency of minimal cooperation and distribution in polarizationless p systems with active membranes. Fundam. Inform. 2017;153:147–172. doi: 10.3233/FI-2017-1535. [DOI] [Google Scholar]
- 35.Song B., Pérez-Jiménez M.J., Pan L. An efficient time-free solution to qsat problem using p systems with proteins on membranes. Inf. Comput. 2017;256:287–299. doi: 10.1016/j.ic.2017.06.005. [DOI] [Google Scholar]
- 36.Macías-Ramos L.F., Pérez-Jiménez M.J., Riscos-Nú nez A., Valencia-Cabrera L. Membrane fission versus cell division: When membrane proliferation is not enough. Theor. Comput. Sci. 2015;608:57–65. doi: 10.1016/j.tcs.2015.06.025. [DOI] [Google Scholar]
- 37.Ma X., Sun F., Li H., He B. Neural-network-based sliding-mode control for multiple rigid-body attitude tracking with inertial information completely unknown. Inf. Sci. 2017;400:91–104. doi: 10.1016/j.ins.2017.03.013. [DOI] [Google Scholar]
- 38.Alsaeedan W., Menai M.E.B., Al-Ahmadi S. A hybrid genetic-ant colony optimization algorithm for the word sense disambiguation problem. Inf. Sci. 2017;417:20–38. doi: 10.1016/j.ins.2017.07.002. [DOI] [Google Scholar]