Abstract
Objectives
We investigated whether HLA-B27-mediated experimental spondyloarthritis (SpA) is associated with a common gut microbial signature to identify potential drivers of pathogenesis.
Methods
Effects of HLA-B27 on three genetic backgrounds, Dark Agouti (DA), Lewis, and Fischer were compared, using wild-type littermates and HLA-B7 transgenic Lewis rats as controls. At 2, 3-4, and 6-8 months of age, cecal and colonic tissue or contents were analyzed by histology for inflammation, RNA-Seq for gene expression differences, and 16S rRNA gene sequencing for microbiota differences.
Results
HLA-B27 transgenic Lewis and Fischer rats develop gut inflammation, while DA rats are resistant to effects of HLA-B27, and HLA-B7 transgenic rats remain unaffected. Immune dysregulation in affected Lewis and Fischer rats is similar and dominated by activation of IL-23/IL-17, IFN, TNF, and IL-1 cytokines and pathways in colon and cecum, while DA rats exhibit low-level cytokine dysregulation without inflammation. Gut microbial changes in HLA-B27 transgenic rats are strikingly divergent on the three different backgrounds, including different patterns of dysbiosis in HLA-B27 transgenic Lewis and Fischer strains with some overlap. Interestingly, DA rats lack segmented filamentous bacteria (SFB) that promote CD4+ Th17 T-cell development, which may explain their resistance to disease.
Conclusion
Effects of HLA-B27 on gut microbiota and dysbiosis in SpA are highly dependent on host genetic background and/or environment despite convergence of dysregulated immune pathways. These results indicate an ecological model of dysbiosis where the effects of multiple microbes contribute to the aberrant immune response, rather than a single or small number of microbes driving pathogenesis.
INTRODUCTION
Spondyloarthritis (SpA) is an immune-mediated inflammatory disease encompassing several conditions that exhibit overlapping clinical features and genetic predisposition (1,2). Considerable evidence supports a link between gastrointestinal tract inflammation and the development of SpA (3). In ankylosing spondylitis (AS), the prototype of SpA, 7% of individuals have co-existing inflammatory bowel disease (IBD), and another 60% harbor sub-clinical gut inflammation (4). Similarly, in IBD arthritis occurs in 10-50% of patients, and nearly 10% develop AS (5).
Direct evidence that gut microbiota is critical in the development of SpA derives from an animal model. Rats transgenic for HLA-B27, a major genetic risk factor for SpA, and human β2-microglobulin (hβ2m) (HLA-B27 transgenic), develop key features of human SpA when housed in a conventional or specific-pathogen free (SPF) animal facility. However, in a germ-free environment gut and joint inflammation are prevented (6,7). Re-colonizing the gut with bacteria is sufficient to induce inflammatory gut and joint disease, implicating commensals in the development of HLA-B27-induced SpA (7). More recently, we have shown that expression of HLA-B27 and hβ2m, as well as the non-disease associated HLA-B7 allele, alter gut microbial communities in this animal model, providing compelling evidence that major histocompatibility complex (MHC) class I proteins can shape the microbiome (8). Alterations in gut microbiota in association with human SpA have also been reported in juvenile onset disease (9), psoriatic arthritis (10), and AS (11).
The mechanism (or mechanisms) by which HLA-B27 promotes the development of SpA remains unclear. Despite its central role in presenting peptides to cytotoxic CD8+ T lymphocytes, evidence that these cells drive disease in rats is lacking, and the SpA phenotype develops normally in Cd8a-deficient HLA-B27 transgenic animals (12). In contrast, compelling evidence has implicated CD4+ T-cells in pathogenicity (13), with activation of the IL-23/IL-17 axis in both joint and gastrointestinal inflammation in rats (14) as well as in human SpA (15). Aberrant features of HLA-B27 such as its tendency to misfold and dimerize, which can lead to endoplasmic reticulum stress and expression of aberrant cell surface complexes have been implicated, and both are linked to activation of the IL-23/IL-17 axis (16). In addition, a growing appreciation for the contribution of endogenous microbiota to many disease states led Rosenbaum and Davey to hypothesize that HLA-B27-induced changes in gut microbiota may be a key intermediary in the development of SpA (17). Recent evidence suggests that innate immune activation and Th17 expansion may precede the development of dysbiosis and gut inflammation in HLA-B27 transgenic rats (18). Thus, it is important to understand how HLA-B27 shapes the gut microbiome.
Here, we show that quite unexpectedly three different rat strains exhibit considerable differences in HLA-B27-mediated dysbiosis. On the disease-permissive Lewis and Fischer backgrounds, different patterns of gut microbial dysbiosis are associated with common immune dysregulation reflecting activation of IL-23/IL-17, IFN, TNF, and IL-1 pathways in the cecum and colon. Our results indicate that effects of HLA-B27 on gut microbiota are strongly dependent on host genetic background and/or environment, while converging on similar mechanisms of immune dysregulation and inflammation. These results have important implications for understanding how HLA-B27 may shape the intestinal microbiome and promote disease in the diverse human population.
Materials and Methods
Animals
All HLA-B27/hβ2m transgenic (HLA-B27 transgenic) rats used in this study carry the same transgene locus (33-3), which contains genomic HLA-B27 and hβ2m DNA (19). The 33-3 locus was backcrossed from the original Fischer (F344) strain to the DA background (13), and then subsequently from DA to Lewis for more than 10 generations (20). HLA-B7/hβ2m transgenic (HLA-B7 transgenic) rats homozygous for the 120-4 transgene locus on the Lewis background were used (13) to ensure comparable expression of the HLA class I heavy chains with HLA-B27 transgenic rats (20). While the HLA-B27 TG DA animals are resistant to disease, HLA-B27 TG Lewis animals develop gut inflammation but no arthritis during the study period. HLA-B27 TG Fischer rats are affected with gut inflammation and also develop more frequent arthritis (~30%) beginning at about 3 months of age. For this study, both male and female Lewis, Fischer, and DA transgenic rats were used with transgene negative littermates (wild-type) as controls. All rats were bred and housed at the NIH except for HLA-B27 transgenic and wild-type Fischer rats, which were bred and housed at OHSU. Animals were weaned at 21 days and cohorts were selected randomly for euthanasia at 2, 3-4 or 6-8 months of age. Fischer animals were housed according to their genotype after weaning, whereas the DA animals and the Lewis animals were co-housed for a short period of time before separation (according to weight limit requirements per cage). DA and Lewis cohorts selected for euthanasia at 2 months of age were housed singly after weaning. All animals were maintained under SPF conditions, and fed Purina 5008 rat chow. Experiments were performed following approval by the Institutional Animal Care and Use Committees at NIAMS or OHSU.
Histology
Hematoxylin and eosin stained tissue sections from paraffin-embedded ileum, cecum and colon samples were scored in a blinded fashion by two independent observers using an established scoring system (21). Samples with initial discordant scores (scores differing by >1) were re-randomized and re-scored to eliminate discrepancies.
Host Transcriptome Analysis
Tissue samples from cecum and colon were homogenized in Trizol reagent (Invitrogen, Waltham, MA) and RNA was isolated using standard phenol-chloroform extraction method. All samples used for RNA-Seq had RNA integrity number (RIN) >8. Library preparation from RNA samples was performed according to the Illumina protocol (Illumina Inc., San Diego, CA). Single end sequencing of 50 bases was performed using the Illumina HiSeq 2000 and raw reads were mapped to the rat rn5 genome using TopHat (2.0.8). Transcript expression levels in reads per kilobase million (RPKM) and ANOVA comparisons were performed using Partek Genomics Suite (6.6). Differentially expressed genes were defined as those with a minimum 2-fold change (p <0.05 and q <0.2), with RPKM >1 for various comparisons. Principal Component Analysis (PCA) was performed using Partek Genomics Suite (6.6) and Euler diagrams were calculated with eulerr (https://cran.r-project.org/package=eulerr) using RStudio (1.0.136).
Immune Cell Prediction
The Immunological Genome Project (ImmGen) function in ToppGene (22) was used to predict immune cell types based on functional annotation of differentially expressed genes. Genes with increased expression (>2 fold up, p <0.05, q <0.2) in HLA-B27 or HLA-B7 transgenic rats compared to wild-type controls on various backgrounds were used. Over-representation analysis in ToppGene maps differentially expressed genes to multiple immune cell subtypes from ImmGen, and then groups them into broader immune cell types. These are represented by multiple lines distinguished by color, with each line representing a p-value (y-axis) based on the quality of the match to a specific immune cell subtype.
Microbial Community Analysis
DNA was isolated from ileum (mucosa), cecum (mucosa and lumen), and colon (lumen) using DNeasy Kit (Qiagen, Valencia, CA). 16S rRNA genes were amplified using the 515–806 primers as specified by the EMP (http://www.earthmicrobiome.org/), sequenced on Illumina MiSeq (23) and processed through Qiita (www.qiita.ucsd.edu). Data were quality-filtered using quantitative insights into microbial ecology (QIIME 1.9.1) and taxa were summarized to the species level (Phylogenetic Level 7). We compared relative frequency of the gut microbes at the species level (max >0.1%; p <0.05; q <0.1) with their wild-type controls in the ileal mucosa, cecal (mucosa and lumen) and colon lumen. Operational Taxonomic Units (OTUs) classified to the species level were used. For area-proportional Euler diagrams, eulerr (https://cran.r-project.org/package=eulerr) was used in RStudio (1.0.136).
Statistical Analysis
For RNA-Seq data, analysis of variance (ANOVA) was used to compare differences between group means for log2 transformed RPKM values (offset by 0.1). Using Partek GS, false discovery rate (FDR) analysis was performed to account for multiple tests (q <0.2). For the microbiome analysis, nonparametric (Wilcoxon Signed-Rank Test with FDR corrections for multiple testing) approaches were used for comparing relative frequencies (p <0.05; q<0.1). Individual Wilcoxon tests were performed between groups to calculate statistical significance using JMP (12.2.0). RPKM results for individual genes were assessed with two-tailed t-tests using GraphPad Prism 6.
RESULTS
Variable Penetrance and Severity of HLA-B27-Induced Gut Inflammation
To determine the role of genetic background on HLA-B27-induced gut inflammation and dysbiosis, we compared effects of the same transgene locus (33-3) that encodes HLA-B27 and hβ2m, in DA, Lewis, and Fischer rats (13). HLA-B7/hβ2m transgenic rats (120-4 transgene locus) were used as an HLA class I control for the Lewis background. Animals were weaned at 3 weeks of age, and then randomized for euthanasia at 2, 3-4 or 6-8 months of age. Samples were collected from the terminal ileum, cecum, and distal colon for histology, microbiome and gene expression analysis (Figure 1A and Supplementary Table 1).
Figure 1. Experimental design and histological assessment of HLA-B27-associated gut inflammation.
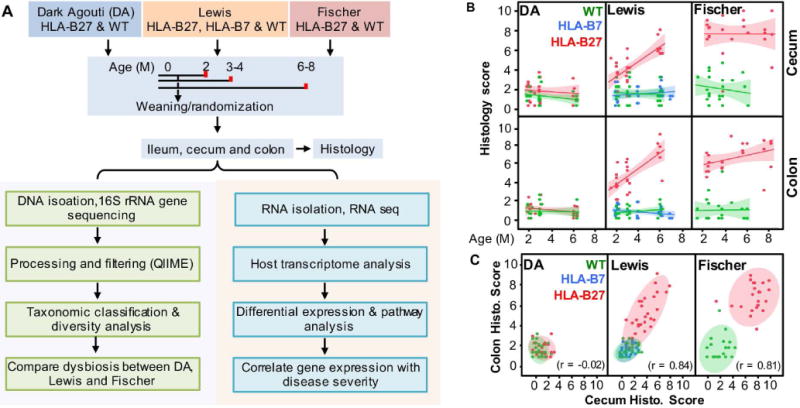
A, Experimental design showing animal cohorts, sample acquisition and processing, and bioinformatic analyses. B, Histological scores for cecum and colon. Each data point represents the average score of 4-8 tissue sections from a single rat at that location. The scoring system is described in Experimental Procedures. The lines represent regression analysis with shadows indicating a 95% confidence interval. Animals with scores > 2.5 are considered abnormal. C, Correlation between cecal and colonic histology scores. Each data point represents the cecal and colonic histology score for an individual rat, with shaded areas depicting a 95% confidence interval. Correlation coefficients (r) for each background are shown.
Histology scores revealed that the presence and severity of gut inflammation is strongly background dependent. DA rats are resistant to the effects of HLA-B27, while Lewis and Fischer rats develop inflammation in the cecum and colon that is already apparent at 2 months of age (Figure 1B), while the ileum is spared (not shown). Two to three month old Fischer animals are affected more severely although by 6 months of age cecum and colon histology scores are comparable between HLA-B27 transgenic Lewis and Fischer rats. HLA-B7 does not cause gastrointestinal inflammation in Lewis rats, consistent with previous observations (13). We did not observe sex differences in gut inflammation in HLA-B27 TG Lewis or Fischer rats (data not shown). In affected rats, there is a strong correlation between cecum and colon histology scores (Figure 1C), indicating consistent involvement of both sites. Representative images of cecum and colon histology are provided in Supplementary Figure 1.
Similar Immune and Inflammatory Response in Lewis and Fischer Animals
To characterize immune dysregulation during gut inflammation, we analyzed differentially expressed genes in cecal and colon tissue. PCA revealed strong clustering of samples from HLA-B27 transgenic Lewis and Fischer rats that exhibit inflammation, away from Lewis and Fischer wild-type controls, respectively (Supplementary Figure 2A). In contrast, samples from HLA-B7 transgenic Lewis and HLA-B27 transgenic DA rats that have normal histology scores clustered with their respective background controls. There was no clustering of samples based on sex (data not shown). Next, we identified differentially expressed genes (HLA-B27 transgenic vs. wild-type) in the cecum and colon of DA, Lewis and Fischer rats (fold change >2 or <−2, p <0.05, q <0.2). Significantly overexpressed (Figure 2A) or underexpressed genes (Supplementary Figure 2B) in cecum and colon are more numerous in Fischer followed by Lewis animals. Disease-resistant DA rats had relatively few differentially expressed genes, consistent with the normal histology scores. The vast majority of up-regulated genes on the Lewis (81%) and DA (93%) backgrounds overlap with Fischer as shown in area-proportionate Euler diagrams for cecum and colon (Figure 2A). In HLA-B7 transgenic rats there are 38 and 45 differentially expressed genes in cecum and colon, respectively, but there is minimal overlap with effects of HLA-B27 on DA, Lewis or Fischer backgrounds (not shown). Pathway analysis of HLA-B27-associated upregulated genes reveals inflammatory pathways such as IFN, TNF, IL-23, IL-17, IL-12, TLR, TCR and BCR-mediated signaling as well as others, while the pathways identified by downregulated genes revealed a loss of metabolism including lipids and proteins, biological oxidation, and digestion and absorption, with a huge overlap between HLA-B27 transgenic Lewis and Fischer animals (Figure 2B and Supplementary Figure 2C, respectively). In addition, the same pathways are identified in cecum and colon.
Figure 2. HLA-B27-associated gene expression in DA, Lewis and Fischer cecum and colon.
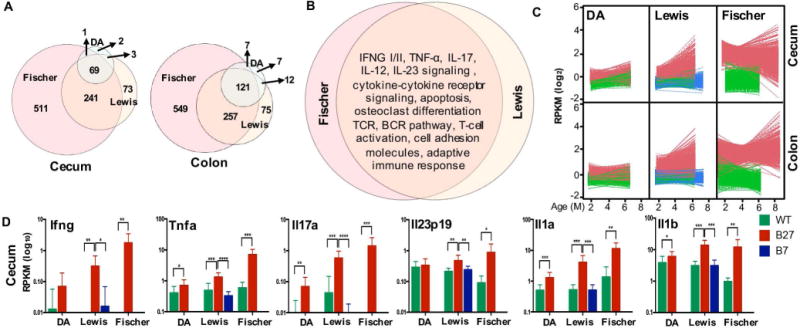
A, The number of differentially expressed genes (HLA-B27 transgenic vs. wild-type) on DA, Lewis and Fischer backgrounds in the cecum and colon are represented in Euler diagrams depicting overlaps and non-overlaps between backgrounds. Genes were selected based on a fold increase of >2, with p <0.05 and q <0.2. B, Pathway analysis (ToppGene) of differentially expressed cecum and colon genes was performed for HLA-B27 transgenic Lewis and Fischer rats. Identical results were obtained for cecum and colon, and Euler diagram depicts complete overlap of inflammatory pathways between Lewis and Fischer (p <0.05). C, Expression patterns for transcripts positively correlating (r >0.6) with disease scores in cecum and colon in DA, Lewis and Fischer rats. D, Expression of selected individual genes in cecum. Individual genes represent key cytokines from pathways identified in Figure 2B are shown. Log10 RPKM is plotted for each gene with each bar representing the average of 9-16 animals. Asterisks represent statistically significant differences for the comparisons shown (*p <0.05, **p <0.01, ***p <0.001 and ****p <0.0001).
To ask whether HLA-B27 associated pathways (Figure 2B) correlate with disease severity, we independently identified genes whose expression is increased and correlates most strongly (r >0.6) with histology scores in the cecum and colon across all genotypes and backgrounds (Figure 1B). The temporal pattern of expression of positively correlating genes is depicted in Figure 2C, while the genes correlating negatively are shown in Supplementary Figure 2D. As expected, there is an age-dependent increase in HLA-B27 transgenic Lewis rats, and consistently high expression in HLA-B27 transgenic Fischer rats, whereas expression levels in HLA-B7 transgenic Lewis animals were no different from control. Pathway analysis of the genes correlating positively and negatively with severity in cecum as well as colon reveals considerable overlap with pathways shown in Figure 2B and Supplementary Figure 2C, respectively. Interestingly, there is low-level up-regulation of a small subset of these genes in HLA-B27 transgenic DA animals (Figure 2C) despite normal histology scores. These differences are most pronounced at 2-3 months of age and are lost in older HLA-B27 DA animals. Expression of several cytokines driving these inflammatory pathways in the cecum (Figure 2D) and colon (Supplementary Figure 2E) is shown. The relative expression of these genes in HLA-B27 transgenic rats compared to their wild-type controls is consistent with differences in disease penetrance and severity between backgrounds (Figure 1B).
Using differentially expressed genes to predict increases in immune cell types of transgenic compared to wild-type rats reveals strikingly similar profiles in cecum and colon and also between HLA-B27 transgenic Lewis and Fischer rats (Supplementary Figure 3). These include myeloid cells, αβ- and γδ-T-cells, NK cells and lymphoid stromal cells. There are no immune cell differences identified in HLA-B7 transgenic rats. Interestingly, despite their lack of clinical phenotype, the DA HLA-B27 transgenic animals demonstrate signals for several cell types, although they are weaker particularly for B-cells and myeloid cells (Supplementary Figure 3).
Shared HLA-B27-Induced Inflammatory Pathways Contrast Background-Dependent Dysbiosis
The striking overlap in dysregulated immune pathways associated with HLA-B27 in the Lewis and Fischer backgrounds, suggested that there might be a core group of dysbiotic microbes. To explore this possibility, we determined relative abundance of microbes at the species level in all tissue sites and strains. This revealed 90 microbes that were differentially abundant (HLA class I transgenic vs. wild-type) in at least one tissue location on at least one genetic background. Hierarchical clustering of these 90 microbes results in distinct groups based on tissue location and genetic background (Supplementary Figure 4). We did not observe sex based differences in dysbiosis (data not shown). The first hierarchal division in the dendrogram is driven by microbial differences in the large intestine (cecum and colon) from the ileum (Supplementary Figure 4). The cecum and colon portion of Supplementary Figure 4 is shown in Figure 3. In contrast to expectations, we found several background-specific differences in effects of HLA-B27 in the cecum and colon (Figure 3). There are HLA-B27-associated increases in Akkermansia and Bacteroides uniformis in Fischer rats, while these microbes were not significantly different in HLA-B27 transgenic Lewis or DA animals compared to their respective wild-type controls. There are also HLA-B27-associated increases in Roseburia and Anaerotruncus, and a decrease in Coprococcus that are limited to the Fischer background. On the other hand, f_Christenellaceae is decreased in the cecum mucosa of HLA-B27 transgenic Lewis animals but not in DA or Fischer rats. Further, there are HLA-B27-associated decrease in Mucispirillum schaedleri and an unknown species of genus Mucispirillum on the Lewis background, not seen in Fischer or DA rats. Lewis rats show an HLA-B27-associated increase in Prevotella in multiple tissue sites (e.g. cecum and colon lumen), but is unchanged in HLA-B27 transgenic Fischer rats in the cecum lumen, and is unchanged in DA animals. Also, there is an HLA-B27-associated increase in an unclassified genus of f_[Barnesiellaceae] in cecal and colon fractions in Lewis rats and in cecal mucosa in DA rats, but not Fischer animals. In the ileum of Lewis rats there is an HLA-B27-associated increase in Sutterella that is also observed in other tissue sites in Lewis animals and in the cecum fractions of DA, while it is not seen in Fischer rats (Supplementary Figure 4). Statistically significant differences for these 90 microbes (q <0.1) and an additional 20 microbes (with q <0.2 but >0.1), and their phylogeny, is shown in Supplementary Table 2 and 3 respectively. These additional microbes are included because many exploratory analyses such as ours use q <0.2 as a selection criteria.
Figure 3. Relative abundance of species level microbes in cecal and colon samples.
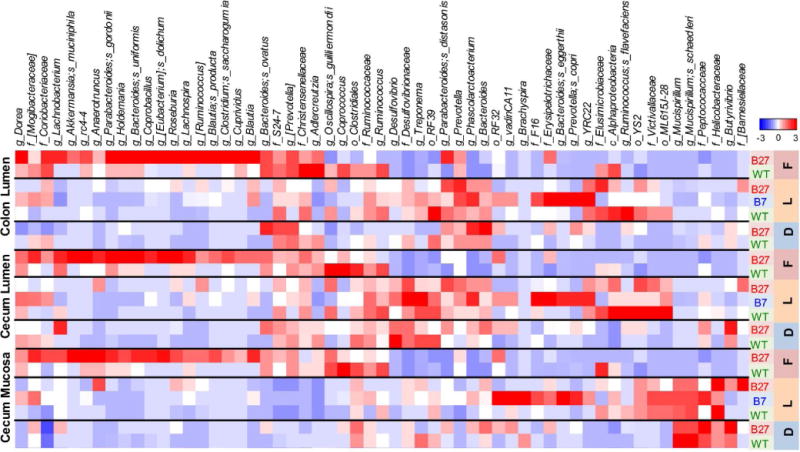
Heat map showing unsupervised hierarchical clustering of species level microbes in HLA-B27/HLA-B7 transgenics compared with wild-type controls on DA, Lewis and Fischer backgrounds. Samples from ileal mucosa, cecal (mucosa and lumen) and colon lumen were used. Microbes with differential relative abundance (non-parametric Wilcoxon each pair analysis, p <0.05, q <0.1) in at least one tissue type on at least one background were used for constructing the heat map. The groups denote major hierarchical clusters observed and these genera have high relative abundance in the cecum and colon fraction. The genera with high relative abundance in ileal fraction are shown in Supplementary Figure 4.
A comparison of differentially abundant (HLA-B27 vs. wild-type) microbes at the species level for each background and each tissue site is shown using area-proportional Euler diagrams (Figure 4). This demonstrates the lack of overlap in HLA-B27-associated differences between Lewis and Fischer backgrounds in ileal and cecal mucosa, and small overlap in the cecum and colon lumen. Together, these results show that there is a substantial effect of genetic background and/or environment on HLA-B27-associated differences in gut microbiota, although shared microbes are also apparent.
Figure 4. Microbial dysbiosis is background dependent.
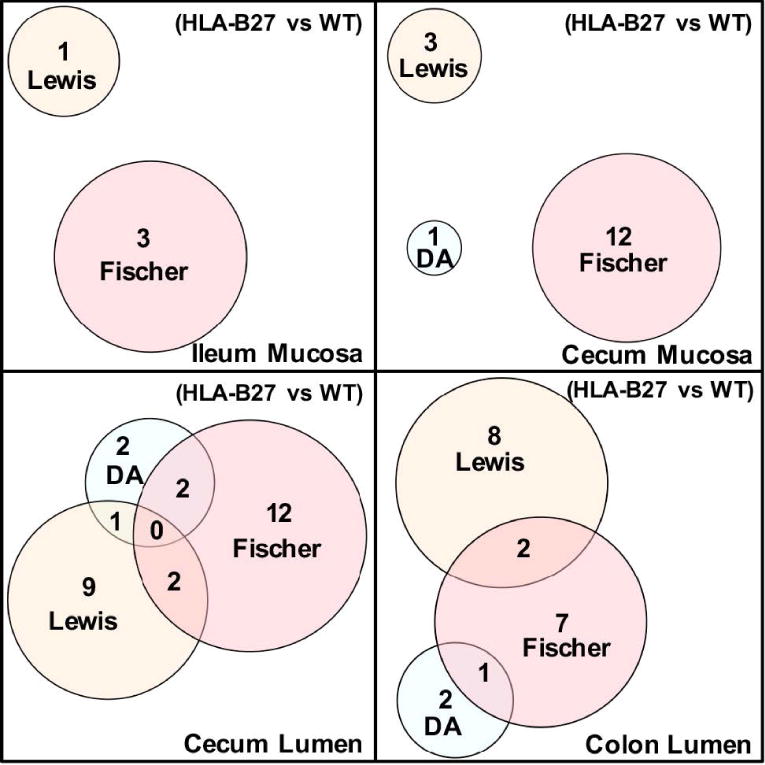
Area proportional Euler graphs representing the overlap and non-overlap between HLA-B27-associated microbes. Species level microbes which are significantly different in HLA-B27 vs wild-type (WT) controls (p <0.05, q<0.1) in DA (blue), Lewis (yellow) and Fischer (pink) backgrounds. Data from ileum mucosa, cecum mucosa, cecum lumen and colon lumen is shown.
Absence of Segmented Filamentous Bacteria in Disease-Resistant DA Rats
A potentially important difference between the backgrounds is the absence of Candidatus arthromitus in the DA animals, independent of HLA-B27. Candidatus arthromitus, more commonly known as segmented filamentous bacteria (SFB) is a major constituent (although variable) in the ileum of both HLA-B27 transgenic and wild-type Lewis and Fischer animals (Figure 5A). SFB are known to attach to intestinal epithelial cells and potentiate induction of Th17 T-cells that play a key role in several inflammatory diseases (24) including colitis (25,26), and thus their absence in DA rats could be responsible for resistance to HLA-B27-mediated disease in this background. It is also worth noting that SFB are very low in HLA-B7 transgenic Lewis rats. In DA and HLA-B7 transgenic Lewis rats lacking SFB, two major contributors to this niche appear to be f_Clostridiaceae (unknown genera) and Lactobacillus, respectively (Figure 5B).
Figure 5. Absence of segmented filamentous bacteria in DA rats.
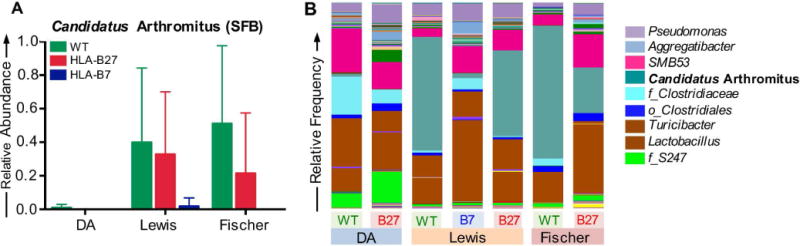
A, Bars represent means for relative frequency of segmented filamentous bacteria (Candidatus arthromitus). Each bar represents the mean of 9-31 animals per genotype per background and the SE is plotted as error bars. B, Relative frequency of species level microbes in ileum mucosa of DA, Lewis and Fischer transgenic and wild-type rats. Each bar represents the mean of 9-31 animals per genotype per background.
HLA-B27-Induced Microbial Dysbiosis is Background Dependent
To further assess effects of HLA-B27 on microbiota in disease-permissive backgrounds, we first compared global relative abundance of 16s rRNA gene of wild-type and HLA-B27 transgenic in both Lewis and Fischer animals using PCA. This revealed major differences between the Lewis and Fischer backgrounds (Figure 6A, ellipsoids). However, within each background there is a clear distinction between HLA-B27 transgenic rats and wild-type controls. The lack of convergence of HLA-B27 transgenic Lewis and HLA-B27 transgenic Fischer samples in the PCA suggests that HLA-B27 drives dysbiosis in a background-specific fashion. Indeed, when we compare effects of HLA-B27 on the two backgrounds, there is a paucity of core dysbiotic organisms (Figure 6B). To ensure that background-specific effects of HLA-B27 were not due to differences in disease severity, we selected samples from HLA-B27 transgenic rats (Lewis and Fischer) with similar histology scores (range 5-8) and re-analyzed them in comparison to their age-matched controls. This revealed slightly altered profiles for each background, but there was no increase in overlap (data not shown), confirming background-dependence of HLA-B27-associated dysbiosis.
Figure 6. Microbial dysbiosis in Lewis and Fischer rats.
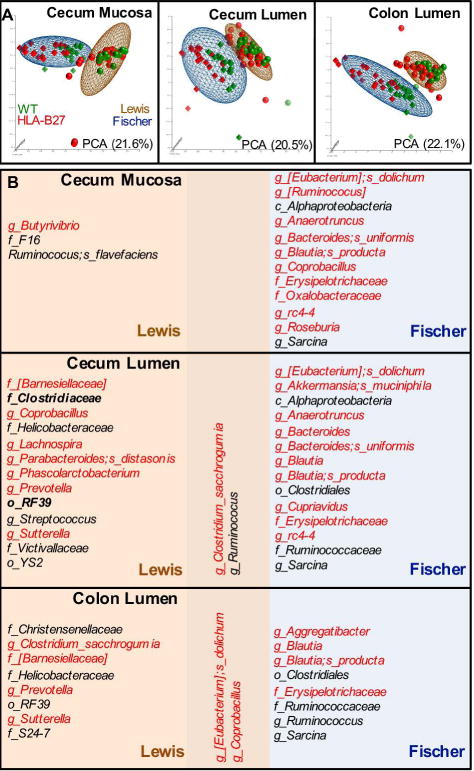
A, PCA of species level data from samples taken from the sites indicated. Each data point represents the first three principal components from the analysis of a single sample from one animal. The distribution of samples from Lewis (sphere) and Fischer (tetrahedron) backgrounds are represented by ellipsoids (brown and blue, respectively), with HLA-B27 and wild-type genotypes indicated by red and green, respectively. B, Overlap between HLA-B27-induced dysbiosis on Lewis and Fischer backgrounds. Microbes with increased (red) or decreased (black) relative frequencies in HLA-B27 transgenic rats compared to wild-type are listed. All microbes shown are differentially represented in HLA-B27 Lewis and Fischer rats (p <0.05, q<0.1).
Discussion
Host-microbe interactions play an important role in the development of many immune-mediated inflammatory diseases (27). In SpA this ranges from the pathogenic Gram-negative intracellular bacteria that trigger reactive arthritis in HLA-B27-positive individuals, to gut commensals that are necessary for development of experimental SpA in HLA-B27 transgenic rats (7) and possibly in humans. While many additional genes contribute to risk for SpA in humans, the prominent role of HLA-B27 has fueled the idea that it might promote SpA by altering gut microbiota (17).
For this study, we hypothesized that comparing gut microbiota in three distinct rat strains that differ in disease penetrance and severity of HLA-B27-mediated disease would facilitate the identification of common microbial signatures associated with experimental SpA. However, we show that gut microbial changes associated with HLA-B27 are strikingly different between DA, Lewis and Fischer strains, that also differ in disease penetrance. Therefore, to better understand microbial shifts in the context of inflammation (dysbiosis), we compared disease susceptible Lewis and Fischer strains. This revealed persistent differences in microbiota between the strains that were not due to differences in disease severity. To ensure that the lack of overlap was not due to the conservative FDR q-value (q <0.1), we performed the same analysis using q <0.2 (Supplementary Tables 2 and 3). While this resulted in an increased number of overlapping and non-overlapping microbes, the percentage of overlapping microbes remained unaltered. It should be noted that while wild-type rats of these three backgrounds exhibit considerable overlap in their gut microbiota, the relative abundance of these microbes differs substantially. These data are provided in Supplementary Figure 5 and Supplementary Table 4 for the interested reader. The paucity of shared dysbiotic microbiota associated with HLA-B27 is in striking contrast to the common dysregulated immune pathways in Lewis and Fischer strains, characterized by prominent increases in IL-1, IL-23, IL-17, IFNγ, and TNF cytokines and pathways. Taken together, our data indicate that background has a prominent effect on HLA-B27-induced microbial dysbiosis despite common immune dysregulation.
While arthritis is an important component of SpA, HLA-B27 TG Lewis animals exhibit infrequent arthritis prior to six months of age in our facility, thus precluding any relevant analysis. On the Fischer background, 32% of the HLA-B27 TG rats developed arthritis (mostly older animals). Comparison of the species level gut microbiota with age matched HLA-B27 TG rats without arthritis did not reveal any significant microbial differences. However, given the relatively small number of rats with arthritis, further studies will be needed before drawing any conclusions.
We have implied that different effects of HLA-B27 on gut microbiota in Lewis and Fischer rats are due primarily to genetic background. However, it is important to note that the Fischer rat colony was maintained at OHSU while all other rats were bred and housed at the NIH. While all animals were maintained on the same commercially available rat chow, environmental differences are likely to alter microbial communities between Lewis and Fischer rats. However, an environmental contribution to differences between Lewis and Fischer strains does not alter the fact that dramatically different patterns of dysbiosis occur in the context of common immune dysregulation caused by HLA-B27. We have previously housed Fischer rats at the NIH and observed increased severity of gut inflammation in HLA-B27 transgenics at disease onset (based on the stool consistency), similar to what we report here for animals housed in two different facilities. This further supports an important role for genetic background in causing differences between Fischer and Lewis HLA-B27 transgenic rats.
While changes in microbiota composition can cause, or be a response to an immune stimulus, in reality they may be a combination of both factors and evolve rapidly, making it difficult to establish a temporal relationship (28). Previous work in HLA-B27 transgenic Fischer rats detected gut inflammation prior to microbial changes (18), but this analysis was limited to select microbiota and thus does not rule out other earlier differences. In the current study, while we initially analyzed cohorts based on age, we eventually grouped all ages in order to simplify the analyses. However, our initial analysis showed immune dysregulation and microbial dysbiosis at the earliest time point in HLA-B27 transgenic Lewis and Fischer rats (data not shown), thus precluding any conlcusions about cause and effect. Nevertheless, the results from DA rats underscore the fact that HLA-B27 can cause immune dysregulation (e.g. low-level increase in Il1a, Il1b, TNF, and Il17a transcripts, with immune cell changes) without progression to a disease phenotype. These changes seen in young HLA-B27 TG DA rats were not sustained in older animals. This could be due to the lack of SFB in DA rats. SFB is required for Th17 development in several rodent strains (24), and thus may be critical for experimental SpA. This bacterium is known to colonize ileal epithelia shortly before weaning and is highly refractory to in vitro culturing (29). It should be noted that HLA-B27 transgenic DA animals were reported to develop severe gut inflammation and cachexia that was not seen in wild-type rats when inadvertently exposed to an unknown infectious agent (13). The episode was transient, but nevertheless suggests they are not completely resistant to the effects of HLA-B27.
It is clear from the present study and previous work (8) that HLA-B7 is associated with major shifts in gut microbial composition (Supplementary Table 2). Here, we document the absence of immune dysregulation in these animals, both at the level of individual cytokines and immune cell signatures, indicating that microbial differences are not sufficient to provoke an inflammatory response (8), and underscoring unique effects of HLA-B27.
Since the majority of microbes associated with HLA-B27-induced disease differ between the Lewis and Fischer backgrounds, we considered that there might be important functional overlaps. Two prominent examples are HLA-B27-associated increases in Akkermansia muciniphila (p_Verrucomicrobia) on Fischer and Prevotella (p_Bacteroidetes) on Lewis strains. While they are phylogenetically diverse, both have been linked to gut inflammation previously, by disrupting mucosal homeostasis (9,30). Akkermansia exacerbates inflammation by degrading the mucous layer overlying gut epithelial cells, thereby weakening the protective barrier (31). A. muciniphila has been reported to be increased in a subset of patients with juvenile SpA (9), and exacerbates Salmonella-induced gut inflammation (31). In contrast, a decrease in the abundance of A. muciniphila has been linked to obesity in mouse models (32,33). Our results suggest that mucus degradation by A. muciniphila, along with dysregulated goblet cell production during inflammation, may be sufficient to bring lumenal bacteria closer to the gut epithelium and promote inflammation. Similarly, Prevotella has been implicated in dysbiosis due to NLRP6 deficiency (34), colitis (30), ankylosing spondylitis (35) and psoriatic arthritis (10). Prevotella encodes enzymes (e.g. superoxide reductase and phosphoadenosine phosphosulphate reductase) enabling it to resist host reactive oxygen species and invade epithelial crypts, most likely by outcompeting commensals that normally maintain mucosal homeostasis (30). Lipopolysaccharide (LPS) from A. muciniphila has been shown to be more immunostimulatory than Prevotella LPS (36), which may also explain the differences in disease severity between Fischer and Lewis backgrounds.
While we have highlighted a surprising lack of common dysbiotic microbes between different backgrounds, there are core similarities between HLA-B27 transgenic Lewis and Fischer rats, particularly in the lumen of the cecum and colon that may be important for disease. This group includes short chain fatty acid (SCFA) producers such as Clostridium and Coprobacillus that are increased, and Ruminococcus, which is decreased (37). SCFA such as butyrate are important as they influence regulatory T-cell homeostasis (38). In addition, colonic epithelial cells from germ-free mice exhibit a mitochondrial respiration deficit and enhanced autophagy as a consequence of reduced SCFA (39). Recent studies have shown an increase in SCFA producer Ruminococcus gnavus (40) and medium chain fatty acid (MCFA) producer Dialister (41) in SpA. Since some SCFA producers in HLA-B27 transgenic rats are increased while others are decreased, additional studies will be needed to determine the overall impact of these changes. Despite the phylogenetic differences in the HLA-B27-associated Lewis and Fischer microbiota, these non-overlapping microbes may perturb common metabolic pathways, thus explaining shared immune dysregulation in these animals. Future studies using metagenomic sequencing along with metabolomic analysis will enable further characterizion and provide functional relevance to these microbial changes.
The gut microbiota play an important role in the development of the host immune system which in turn shapes the composition of the gut microbiota (42,43). It is clear from animal models that gut microbial communities co-evolve with their host and exhibit strong ecological interactions, varying with host genotype, diet and colonization history (43). Our results demonstrate that HLA-B27 effect on the gut microbiota in experimental SpA is largely dependent on host genetics and environment. Also, dysbiotic microbes contributing to HLA-B27-associated inflammation need not be linked taxonomically, but rather similarities in their metabolic functions and/or gut microenvironment are key drivers of pathogenesis. In addition, HLA-B27-mediated disease in rats might depend on SFB, although this needs to be determined experimentally.
In summary, gene expression analysis and microbial community profiling to characterize the effect of HLA-B27 on three different genetic backgrounds provides an unprecedented and comprehensive view of the complexity of microbial dysbiosis associated with common immune dysregulation and gut inflammation in SpA. While we found a common immune-signature associated with gut inflammation, microbial dysbiosis was surprisingly different with fewer overlapping microbes on different backgrounds. The absolute dependence of this model on gut microbiota, together with the striking differences between host backgrounds, suggests an ecological model for dysbiosis in HLA-B27-induced experimental SpA rather than a single or small number of microbial genera driving pathogenesis. Given that recombinant inbred animal strains are analogous to unrelated individuals in the human population, our results suggest that characterizing functionally similar dysbiotic microbial communities may be critical to reveal underlying mechanisms of host-microbe interactions contributing to spondyloarthritis.
Supplementary Material
Supplementary Figure 1. Cecum and colon histology Representative images of H&E stained cecum (20X) and colon (10X) tissue from wild-type and HLA-B27 rats from DA, Lewis and Fischer backgrounds and Lewis HLA-B7 with wild-type controls at 6 months of age. The score for that section is given in the left corner of each image. Cecum and colon sections of HLA-B27 rats in the Lewis and Fischer background show loss of goblet cells, erosion of epithelial layer and inflammatory infiltrates. Each bar represents 100mm.
Supplemental Figure 2. Differential gene expression in DA, Lewis and Fischer A, PCA of cecum and colon transcriptomes in HLA class I TG and wild-type DA, Lewis and Fischer rats. Each data point represents the first three principal components from the analysis of a single animal HLA-B27 TG (red), HLA-B7 TG (blue) and wild-type (green). B, The number of differentially downregulated genes (HLA-B27 transgenic vs. wild-type) on DA, Lewis and Fischer backgrounds in the cecum and colon are represented in Euler diagrams depicting overlaps and non-overlaps between backgrounds. Downregulated genes were selected based on a fold difference of <−2, with p <0.05 and q <0.2. C, Pathway analysis (ToppGene) of differentially expressed cecum and colon genes was performed for HLA-B27 transgenic Lewis and Fischer rats. Identical results were obtained for cecum and colon, and Euler diagram depicts complete overlap of metabolic pathways between Lewis and Fischer (p <0.05). D, Expression patterns for transcripts positively correlating (r >0.6) with disease scores in cecum and colon in DA, Lewis and Fischer rats. E, Raw reads in RPKM (Reads Per Kilobase Million) for IFN-γ, TNF, IL17a, IL23a, IL1a and IL1b in colon are plotted (Log10) in the bar graphs, each bar representing the average between 9-16 animals. The x-axis represents the HLA-B27/HLA-B7genotype with their wild-type controls from DA, Lewis and Fischer backgrounds. The stars represent the degree of significance (*p <0.05, ** p <0.01, *** p <0.001 and ****p <0.0001).
Supplementary Figure 3. Immune cell enrichment profiles Differentially expressed genes in HLA-B27 and HLA-B7 transgenic rats on the DA, Lewis and Fischer background were employed for immune cell enrichment analysis using ToppGene (see Materials and Methods). On the x axis, each color represents one immune cell type comprised of multiple lines depicting individual immune cell subtypes (up to 450 hits for each x-axis). The y axis shows significance for individual cell subtypes as –log10 (p-value) descending from highest (left) to lowest (right) for each cell type.
Supplementary Figure 4. Relative abundance of species level microbes in ileum, cecum and colon samples Species level bacteria demonstrating differential relative abundance in HLA-B27/HLA-B7 transgenic rats compared to wild-type in at least one the fraction (ileal, cecal or colon) on at least DA, Lewis or Fischer background significantly made this list. Significance was calculated using non-parametric Wilcoxon each pair analysis and 90 species level microbes with differential relative abundance (p <0.05, q <0.1) are shown.
Supplementary Figure 5. Microbial community structure comparison across wild-type rats of all backgrounds Area proportional Euler graphs representing the overlap and non-overlap between species level microbes (max percentage >0.1(relative abundance 0.001)) present in wild-type (WT) DA, Lewis and Fischer rats. Data from ileum mucosa, cecum mucosa, cecum lumen and colon lumen samples is shown.
Supplementary Table 1. Animal cohorts used for this study.
Supplemetary Table 2. Differentially abundant microbes in the ileum mucosa, cecum mucosa, cecum lumen and colon lumen.
Supplementary Table 3. Phylogeny of the differential species level microbes.
Supplementary Table 4. Average species abundance in wild-type DA, Lewis and Fischer animals in ileum mucosa, cecum mucosa, cecum lumen and colon lumen.
Acknowledgments
We thank Maxime Breban for providing the HLA-B27 transgenic Fischer rats, and Joel Taurog for providing the HLA-B7 transgenic Lewis animals. We thank Gustavo Gutierrez-Cruz, Kristina Zaal and Kalyani Mishra for RNA-Seq, imaging and genotyping core facility at NIAMS, respectively. Thanks are due to Rob Knight and Justine Debelius at UCSD for microbiome sequencing. We also thank Patrick Stauffer and Paul Montgomery at OHSU, and the NIAMS animal facility for their assistance with the necropsies. We utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov) for analysis.
This work was supported by the NIAMS Intramural Research Program, Z01AR041184 to R.A.C. and grants to J.T.R. from the Stan and Madelle Rosenfeld Family Trust, William and Mary Bauman Foundation, the Spondylitis Association of America, and Research to Prevent Blindness. M.A. is a Jane Bruckel Scholar designated by the Spondylitis Association of America. Funding institutes/associations had no influence over experimental design, data collection and analysis, as well as preparation of manuscript.
Footnotes
The authors declare no conflict of interests for the present study.
Author Contributions
T.G., M.A., J.T.R., and R.A.C. designed the experiments and T.G. and M.A. performed the experiments. Data were analyzed by T.G., S.R.B. and R.A.C.; T.G. and R.A.C. wrote the paper, and S.R.B., M.A., and J.T.R. provided editing.
References
- 1.Taurog JD, Chhabra A, Colbert RA. Ankylosing Spondylitis and Axial Spondyloarthritis. N Engl J Med. 2016;374:2563–74. doi: 10.1056/NEJMra1406182. [DOI] [PubMed] [Google Scholar]
- 2.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immune-mediated diseases. Nat Rev Genet. 2013;14:661–73. doi: 10.1038/nrg3502. [DOI] [PubMed] [Google Scholar]
- 3.De Wilde K, Debusschere K, Beeckman S, Jacques P, Elewaut D. Integrating the pathogenesis of spondyloarthritis: gut and joint united? Curr Opin Rheumatol. 2015;27:189–96. doi: 10.1097/BOR.0000000000000144. [DOI] [PubMed] [Google Scholar]
- 4.Jacques P, Elewaut D. Tumor necrosis factor alpha-induced proteins: natural brakes on inflammation. Arthritis Rheum. 2012;64:3831–4. doi: 10.1002/art.34664. [DOI] [PubMed] [Google Scholar]
- 5.Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis Rheum. 2008;58:15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 6.Taurog JD, Richardson JA, Croft JT, Simmons WA, Zhou M, Fernandez-Sueiro JL, et al. The germfree state prevents development of gut and joint inflammatory disease in HLA-B27 transgenic rats. J Exp Med. 1994;180:2359–64. doi: 10.1084/jem.180.6.2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rath HC, Herfarth HH, Ikeda JS, Grenther WB, Hamm TE, Jr, Balish E, et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. J Clin Invest. 1996;98:945–53. doi: 10.1172/JCI118878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin P, Bach M, Asquith M, Lee AY, Akileswaran L, Stauffer P, et al. HLA-B27 and human beta2-microglobulin affect the gut microbiota of transgenic rats. PLoS One. 2014;9:e105684. doi: 10.1371/journal.pone.0105684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoll ML, Kumar R, Morrow CD, Lefkowitz EJ, Cui X, Genin A, et al. Altered microbiota associated with abnormal humoral immune responses to commensal organisms in enthesitis-related arthritis. Arthritis Res Ther. 2014;16:486. doi: 10.1186/s13075-014-0486-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scher JU, Ubeda C, Artacho A, Attur M, Isaac S, Reddy SM, et al. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015;67:128–39. doi: 10.1002/art.38892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Costello ME, Ciccia F, Willner D, Warrington N, Robinson PC, Gardiner B, et al. Intestinal dysbiosis in ankylosing spondylitis. Arthritis Rheumatol. 2014 doi: 10.1002/art.38967. [DOI] [PubMed] [Google Scholar]
- 12.May E, Dorris ML, Satumtira N, Iqbal I, Rehman MI, Lightfoot E, et al. CD8 alpha beta T cells are not essential to the pathogenesis of arthritis or colitis in HLA-B27 transgenic rats. J Immunol. 2003;170:1099–105. doi: 10.4049/jimmunol.170.2.1099. [DOI] [PubMed] [Google Scholar]
- 13.Taurog JD, Maika SD, Satumtira N, Dorris ML, McLean IL, Yanagisawa H, et al. Inflammatory disease in HLA-B27 transgenic rats. Immunol Rev. 1999;169:209–23. doi: 10.1111/j.1600-065x.1999.tb01317.x. [DOI] [PubMed] [Google Scholar]
- 14.DeLay ML, Turner MJ, Klenk EI, Smith JA, Sowders DP, Colbert RA. HLA-B27 misfolding and the unfolded protein response augment interleukin-23 production and are associated with Th17 activation in transgenic rats. Arthritis Rheum. 2009;60:2633–43. doi: 10.1002/art.24763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith JA, Colbert RA. Review: The interleukin-23/interleukin-17 axis in spondyloarthritis pathogenesis: Th17 and beyond. Arthritis Rheumatol. 2014;66:231–41. doi: 10.1002/art.38291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowness P. HLA-B27. Annu Rev Immunol. 2015;33:29–48. doi: 10.1146/annurev-immunol-032414-112110. [DOI] [PubMed] [Google Scholar]
- 17.Rosenbaum JT, Davey MP. Time for a gut check: evidence for the hypothesis that HLA-B27 predisposes to ankylosing spondylitis by altering the microbiome. Arthritis Rheum. 2011;63:3195–8. doi: 10.1002/art.30558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asquith MJ, Stauffer P, Davin S, Mitchell C, Lin P, Rosenbaum JT. Perturbed Mucosal Immunity and Dysbiosis Accompany Clinical Disease in a Rat Model of Spondyloarthritis. Arthritis Rheumatol. 2016;68:2151–62. doi: 10.1002/art.39681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hammer RE, Maika SD, Richardson JA, Tang JP, Taurog JD. Spontaneous inflammatory disease in transgenic rats expressing HLA-B27 and human beta 2m: an animal model of HLA-B27-associated human disorders. Cell. 1990;63:1099–112. doi: 10.1016/0092-8674(90)90512-d. [DOI] [PubMed] [Google Scholar]
- 20.Layh-Schmitt G, Yang EY, Kwon G, Colbert RA. HLA-B27 alters the response to tumor necrosis factor alpha and promotes osteoclastogenesis in bone marrow monocytes from HLA-B27-transgenic rats. Arthritis Rheum. 2013;65:2123–31. doi: 10.1002/art.38001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izcue A, Hue S, Buonocore S, Arancibia-Carcamo CV, Ahern PP, Iwakura Y, et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 2008;28:559–70. doi: 10.1016/j.immuni.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Heng TS, Painter MW. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9:1091–4. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]
- 23.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme j. 2012;6:1621–4. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell. 2015;163:367–80. doi: 10.1016/j.cell.2015.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caselli M, Tosini D, Gafa R, Gasbarrini A, Lanza G. Segmented filamentous bacteria-like organisms in histological slides of ileo-cecal valves in patients with ulcerative colitis. Am J Gastroenterol. 2013;108:860–1. doi: 10.1038/ajg.2013.61. [DOI] [PubMed] [Google Scholar]
- 26.Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, et al. Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis. 2007;13:1202–11. doi: 10.1002/ibd.20221. [DOI] [PubMed] [Google Scholar]
- 27.Haberman Y, Tickle TL, Dexheimer PJ, Kim MO, Tang D, Karns R, et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest. 2014;124:3617–33. doi: 10.1172/JCI75436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huttenhower C, Kostic AD, Xavier RJ. Inflammatory bowel disease as a model for translating the microbiome. Immunity. 2014;40:843–54. doi: 10.1016/j.immuni.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ericsson AC, Hagan CE, Davis DJ, Franklin CL. Segmented filamentous bacteria: commensal microbes with potential effects on research. Comp Med. 2014;64:90–8. [PMC free article] [PubMed] [Google Scholar]
- 30.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One. 2013;8:e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110:9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schneeberger M, Everard A, Gomez-Valades AG, Matamoros S, Ramirez S, Delzenne NM, et al. Akkermansia muciniphila inversely correlates with the onset of inflammation, altered adipose tissue metabolism and metabolic disorders during obesity in mice. Sci Rep. 2015;5:16643. doi: 10.1038/srep16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Elinav E, Strowig T, Kau AL, Henao-Mejia J, Thaiss CA, Booth CJ, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–57. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wen C, Zheng Z, Shao T, Liu L, Xie Z, Le Chatelier E, et al. Quantitative metagenomics reveals unique gut microbiome biomarkers in ankylosing spondylitis. Genome Biol. 2017;18:142. doi: 10.1186/s13059-017-1271-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vatanen T, Kostic AD, d’Hennezel E, Siljander H, Franzosa EA, Yassour M, et al. Variation in Microbiome LPS Immunogenicity Contributes to Autoimmunity in Humans. Cell. 2016;165:1551. doi: 10.1016/j.cell.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 37.Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. 2016;7:189–200. doi: 10.1080/19490976.2015.1134082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015;6:7320. doi: 10.1038/ncomms8320. [DOI] [PubMed] [Google Scholar]
- 39.Donohoe DR, Garge N, Zhang X, Sun W, O’Connell TM, Bunger MK, et al. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011;13:517–26. doi: 10.1016/j.cmet.2011.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breban M, Tap J, Leboime A, Said-Nahal R, Langella P, Chiocchia G, et al. Faecal microbiota study reveals specific dysbiosis in spondyloarthritis. Ann Rheum Dis. 2017;76:1614–22. doi: 10.1136/annrheumdis-2016-211064. [DOI] [PubMed] [Google Scholar]
- 41.Tito RY, Cypers H, Joossens M, Varkas G, Van Praet L, Glorieus E, et al. Brief Report: Dialister as a Microbial Marker of Disease Activity in Spondyloarthritis. Arthritis Rheumatol. 2017;69:114–21. doi: 10.1002/art.39802. [DOI] [PubMed] [Google Scholar]
- 42.Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–7. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 43.Dethlefsen L, McFall-Ngai M, Relman DA. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature. 2007;449:811–8. doi: 10.1038/nature06245. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Cecum and colon histology Representative images of H&E stained cecum (20X) and colon (10X) tissue from wild-type and HLA-B27 rats from DA, Lewis and Fischer backgrounds and Lewis HLA-B7 with wild-type controls at 6 months of age. The score for that section is given in the left corner of each image. Cecum and colon sections of HLA-B27 rats in the Lewis and Fischer background show loss of goblet cells, erosion of epithelial layer and inflammatory infiltrates. Each bar represents 100mm.
Supplemental Figure 2. Differential gene expression in DA, Lewis and Fischer A, PCA of cecum and colon transcriptomes in HLA class I TG and wild-type DA, Lewis and Fischer rats. Each data point represents the first three principal components from the analysis of a single animal HLA-B27 TG (red), HLA-B7 TG (blue) and wild-type (green). B, The number of differentially downregulated genes (HLA-B27 transgenic vs. wild-type) on DA, Lewis and Fischer backgrounds in the cecum and colon are represented in Euler diagrams depicting overlaps and non-overlaps between backgrounds. Downregulated genes were selected based on a fold difference of <−2, with p <0.05 and q <0.2. C, Pathway analysis (ToppGene) of differentially expressed cecum and colon genes was performed for HLA-B27 transgenic Lewis and Fischer rats. Identical results were obtained for cecum and colon, and Euler diagram depicts complete overlap of metabolic pathways between Lewis and Fischer (p <0.05). D, Expression patterns for transcripts positively correlating (r >0.6) with disease scores in cecum and colon in DA, Lewis and Fischer rats. E, Raw reads in RPKM (Reads Per Kilobase Million) for IFN-γ, TNF, IL17a, IL23a, IL1a and IL1b in colon are plotted (Log10) in the bar graphs, each bar representing the average between 9-16 animals. The x-axis represents the HLA-B27/HLA-B7genotype with their wild-type controls from DA, Lewis and Fischer backgrounds. The stars represent the degree of significance (*p <0.05, ** p <0.01, *** p <0.001 and ****p <0.0001).
Supplementary Figure 3. Immune cell enrichment profiles Differentially expressed genes in HLA-B27 and HLA-B7 transgenic rats on the DA, Lewis and Fischer background were employed for immune cell enrichment analysis using ToppGene (see Materials and Methods). On the x axis, each color represents one immune cell type comprised of multiple lines depicting individual immune cell subtypes (up to 450 hits for each x-axis). The y axis shows significance for individual cell subtypes as –log10 (p-value) descending from highest (left) to lowest (right) for each cell type.
Supplementary Figure 4. Relative abundance of species level microbes in ileum, cecum and colon samples Species level bacteria demonstrating differential relative abundance in HLA-B27/HLA-B7 transgenic rats compared to wild-type in at least one the fraction (ileal, cecal or colon) on at least DA, Lewis or Fischer background significantly made this list. Significance was calculated using non-parametric Wilcoxon each pair analysis and 90 species level microbes with differential relative abundance (p <0.05, q <0.1) are shown.
Supplementary Figure 5. Microbial community structure comparison across wild-type rats of all backgrounds Area proportional Euler graphs representing the overlap and non-overlap between species level microbes (max percentage >0.1(relative abundance 0.001)) present in wild-type (WT) DA, Lewis and Fischer rats. Data from ileum mucosa, cecum mucosa, cecum lumen and colon lumen samples is shown.
Supplementary Table 1. Animal cohorts used for this study.
Supplemetary Table 2. Differentially abundant microbes in the ileum mucosa, cecum mucosa, cecum lumen and colon lumen.
Supplementary Table 3. Phylogeny of the differential species level microbes.
Supplementary Table 4. Average species abundance in wild-type DA, Lewis and Fischer animals in ileum mucosa, cecum mucosa, cecum lumen and colon lumen.


