Abstract
Mitochondrial accumulation of intracellular β-amyloid (Aβ) peptides is present in the brains of individuals with Alzheimer's disease (AD) as well as in related mouse models of AD. This accumulation is extremely toxic because Aβ disrupts the normal functions of many mitochondrial proteins, resulting in significant mitochondrial dysfunction. Therefore, understanding the mitochondrial accumulation of Aβ is useful for future pharmaceutical design of drugs to address mitochondrial dysfunction in AD. However, the detailed molecular mechanism of this accumulation process remains elusive. Here, using yeast mitochondria, we present direct experimental evidence suggesting that Aβ is specifically recognized by translocase of outer mitochondrial membrane subunit 22 (Tom22 in yeast; TOMM22 in human), a noncanonical receptor within the mitochondrial protein import machinery, and that this recognition is critical for Aβ accumulation in mitochondria. Furthermore, we found that residues 25–42 in the Aβ peptide mediate the specific interaction with TOMM22. On the basis of our findings, we propose that cytosolic Aβ is recognized by TOMM22; transferred to another translocase subunit, TOMM40; and transported through the TOMM channel into the mitochondria. Our results not only confirm that yeast mitochondria can be used as a model to study mitochondrial dysfunction caused by Aβ peptides in AD but also pave the way for future studies of the molecular mechanism of mitochondrial Aβ accumulation.
Keywords: yeast, mitochondria, amyloid-β (Aβ), electron microscopy (EM), chromatography, protein-protein interaction, Ni-NTA conjugated nanogold, protein import, Tom22
Introduction
One of the apparent hallmarks of Alzheimer's disease is the extracellular aggregation of β-amyloid (Aβ)2 peptides in patients' brains (1). However, there is now growing evidence suggesting that the intracellular Aβ contributes to the neurodegeneration and Alzheimer's pathology as well (2–4). It is also well documented that the cytosolic Aβ is produced at different subcellular sites, such as the Golgi compartments (5, 6), endoplasmic reticulum (7), and endosomal/lysosomal system (8). Hence, intracellular Aβ production and removal are carefully balanced under physiological conditions, and this balance can be ruined by genetic and environmental conditions (9, 10). It has been hypothesized that the increased level of intracellular Aβ maybe one of the first steps in the fatal cascade leading to neurodegeneration in Alzheimer's disease (11). In addition, intracellular Aβ peptides, as well as their precursor protein, amyloid precursor protein (APP), are found accumulated in the subcellular mitochondria in the brains of Alzheimer's patients as well as in related mouse models (12–16). This mitochondrial accumulation event is extremely toxic because Aβ interacts with many mitochondrial proteins, such as components of the protein import machinery (translocase of the outer membrane–translocase of the inner membrane (TOM-TIM)) (17), cytochrome c oxidase (18), subunits of the electron transfer chain (19, 20), F1α of ATP synthase (21), and many more. These interactions cause various forms of mitochondrial dysfunction, including but not limited to deficits in morphology or mitochondrial dynamics (22), mitochondrial bioenergetics (23), and protein transport (24). In line with all these findings, mitochondrial dysfunction is now considered as a general pathological feature of Alzheimer's disease (25, 26). Thus, a better understanding of the mitochondrial Aβ accumulation process will be useful to provide new avenues to treat mitochondrial dysfunction in Alzheimer's disease.
At least two distinct pathways of mitochondrial Aβ accumulation have been proposed: 1) nonspecific coaggregation with other mitochondrial preproteins (synthesized protein peptide chains in cytosol that are not yet folded) (27) and 2) specific translocation through the mitochondrial protein import machinery TOM-TIM (28), a similar process to most other mitochondrial targeting proteins. Here, we focused on the second proposed Aβ accumulation pathway and studied the potential specific interactions between Aβ and TOM. The TOM complex consists of at least seven known protein components: Tom5, Tom6, Tom7, Tom20, Tom22, Tom40, and Tom70 (29, 30). Among them, Tom20 and Tom70 are known receptors responsible for the recognition of various mitochondrial targeting signals (MTSs) within the preproteins. Generally, after MTS recognition, the preprotein substrates are transported through the TOM core complex, which consists of Tom22, Tom40, and three other small Tom proteins. It is believed that Tom22 is capable of binding to the positively charged face of the substrates, thus mediating the intermediate step between substrate–receptor binding and substrate translocation through the TOM central pore protein Tom40 (31, 32).
In the case of mitochondrial Aβ accumulation, here, we asked the important question: how does the TOM complex recognize cytosolic Aβ peptides given that those peptides do not have canonical MTSs? In this study, we examined only Aβ(1–42) because it is more fibrillogenic than other forms of Aβ peptides. Because it has been reported that Tom20 may be responsible for the Aβ recognition (28), we carefully examined this potential Tom20/Aβ interaction in our system. To our surprise, we found that Aβ is not able to bind to either purified Tom20 in solution or Tom20 within the context of isolated functional mitochondria. Instead, we discovered that the Aβ recognition process is mainly mediated through another essential Tom component, Tom22. In addition, Tom22 specifically recognizes the C terminus of the Aβ peptides, residues 25–42. These results suggest a noncanonical way of substrate recognition mediated by the mitochondrial TOM machinery.
Results
Aβ is specifically accumulated on yeast mitochondria
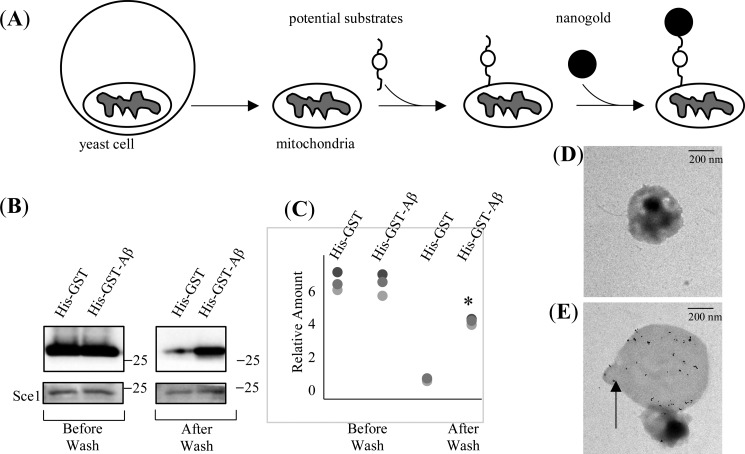
Once again, a recent report has shown that Aβ peptides directly interfere with the mitochondrial preprotein import process in human cells (27). Considering that the mitochondrial protein import machinery is highly conserved among eukaryotes from yeast to mammals, we asked whether yeast mitochondria are also capable of accumulating human Aβ peptides. To answer that question, we designed an “import and stop” experiment using isolated yeast mitochondria and purified recombinant proteins (Fig. 1A). Briefly, by following established protocols (33), we isolated the active mitochondria from WT Saccharomyces cerevisiae strain S288C and used them for the following protein import assays. Originally, to be consistent with the notion that most MTSs are located on the N terminus of the preproteins, we designed a recombinant protein that consists of N-terminal Aβ peptide and C-terminal GST/His tags. However, this recombinant protein was not stable in our experiments because of fast degradation during expression and purification processes (Fig. S1). Thus, we designed a new construct with N-terminal His/GST tag and C-terminal Aβ peptide. The His-GST-Aβ recombinant protein behaved well during expression and purification (Fig. S1). Thus, we used it as a substrate for the following mitochondrial import experiments. The rationale for the import and stop experiment was that mitochondria could only import unfolded preproteins but not folded proteins. Within the purified His-GST-Aβ, the His tag and the Aβ fragment were expected to be as flexible as unfolded peptides, whereas the GST part was expected to be a fully folded structural domain. Thus, if the isolated yeast mitochondria were able to accumulate the human Aβ peptides, we expected that the Aβ fragment in the His-GST-Aβ would be recognized, and the mitochondrial import process would start. Because the GST domain was folded, we then expected that the import reaction would soon stop automatically, and the His-GST-Aβ would be stuck in the TOM channel. To eliminate the possibility that the mitochondrial import was initiated by the His-GST part rather than the Aβ part of His-GST-Aβ, we expressed and purified the His-GST recombinant protein without Aβ. We used it in the import and stop experiments as a negative control. The treated mitochondria were than analyzed by Western blotting and electron microscopy (EM) imaging. The results of Western blot analysis of the treated yeast mitochondria are shown in Fig. 1B and quantified in Fig. 1C. We started with the same amount of His-GST and His-GST-Aβ (same band intensity) as substrates for import. By comparing the control bands of His-GST before and after wash, it is clear that His-GST itself is not mitochondrially accumulated under our experimental settings. However, the high-intensity band of His-GST-Aβ after wash suggests that His-GST-Aβ was specifically recognized by the yeast mitochondria, and the recognition was Aβ-dependent. Furthermore, to confirm that the His-GST-Aβ was trapped in the middle of the import event on the mitochondrial outer membrane, we used the commercially available Ni-NTA–conjugated nanogold solution to label the protein's N-terminal His tag and imaged the mitochondria by EM. It is worth noting that we did not use any stains for the EM analysis here because 1) the large size of the yeast mitochondria created enough visual contrast in the images, and 2) the nanogold particles were easier to visualize without potential interference from heavy metal stains. The rationale of the experiment is that if the substrates were trapped on the mitochondrial surface we expected to see clear nanogold particles in the EM images. In Fig. 1D, the mitochondria treated with the His-GST negative control were imaged, and no nanogold particles were found. In Fig. 1E, the mitochondria treated with His-GST-Aβ were imaged, and we could clearly identify labeled nanogold particles. Altogether, the results suggest that the human Aβ peptides can specifically target the yeast mitochondria, similar to previous observations in the brains of Alzheimer's patients.
Figure 1.
Yeast mitochondria specifically recognize and accumulate human Aβ peptides. A, illustration of the import and stop experiment. The experiment was started by adding the recombinant protein His-GST-Aβ or His-GST into the isolated mitochondrial solution and stopped in 20 min by centrifugation. The treated mitochondria were then washed thoroughly before Western blot analysis and nanogold labeling. B, Western blot analysis of the treated mitochondria from A probed by the anti-GST antibody. The left panel shows the total input proteins His-GST-Aβ and His-GST (as a control) before the wash, and the right panel shows significant accumulation of His-GST-Aβ but not His-GST after the wash. Yeast Sce1 is the loading control for all Western blot analysis in this work. C, quantification of each band in B. Data of three independent experiments are shown. *, p < 0.01 (two-tailed Student's t test, compared with His-GST after wash). D and E, representative EM images of the nanogold-labeled mitochondria treated with His-GST (D) and His-GST-Aβ (E). The arrow indicates labeled nanogold. The scale bar in both micrographs is 200 nm.
To verify that the mitochondrial accumulation of Aβ also happens under conditions closer to physiological, we cloned the His-GST-Aβ into the pRS414 vector, which allows the expression of the recombinant protein in yeast. We then isolated the yeast mitochondria and found that His-GST-Aβ accumulated on the yeast surface as shown in Fig. S2. This result further confirms that human Aβ targets the yeast mitochondria in vivo.
It is also worth noting that the mitochondria we isolated were of high purity and were adequate for this study because 1) in the EM images we could only observe large subcellular structures ranging from ∼0.5 to ∼1.5 μm in diameter and 2) the observed large subcellular structures were mitochondria as confirmed by the following experiment. We derived a genetically modified yeast strain called 20His. It had a His tag inserted at the N terminus of the Tom20 gene, which is exposed to the cytosolic side. Thus, the His-tagged Tom20 could serve as a mitochondrial marker. We isolated mitochondria from the 20His strain, labeled them with Ni-NTA–conjugated nanogold particles, and imaged them by EM. We found that all large subcellular structures are clearly labeled with gold particles and thus are indeed purified mitochondria (Fig. S3).
In the literature, there are confusing reports regarding the manner of the mitochondrial Aβ accumulation process. For example, a recent report suggests that Aβ peptides coaggregate on the mitochondrial surface in a nonspecific manner (27). In contrast, previous studies suggest a more specific Aβ uptake process mediated by the mitochondrial TOM complex (28). Based on our results, we argue that the Aβ mitochondrial targeting process is specific rather than nonspecific. If it was nonspecific, with the excessive amount of His-GST-Aβ we used in the experiments, the mitochondrial surface should be overwhelmingly covered by the protein and thus heavily labeled with nanogold particles in the EM images. However, we found that the nanogold particles were well spread over the mitochondria in all EM micrographs as in the example shown in Fig. 1D, Thus, we propose that the interactions between Aβ peptides and mitochondria are specific.
The Aβ mitochondrial targeting is TOM-dependent
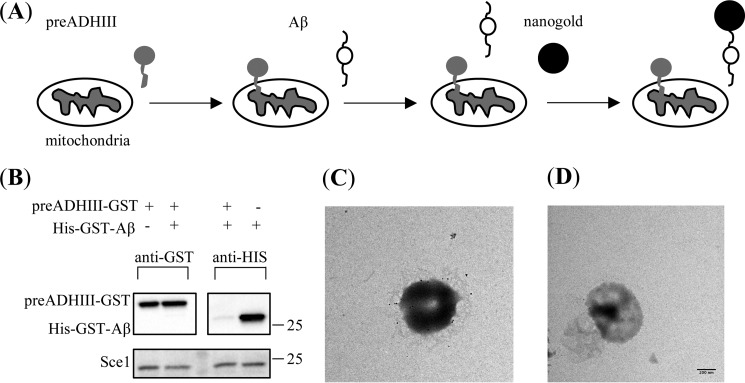
Next, we asked which proteins mediate the specific mitochondrial Aβ accumulation process. Previous studies have shown that a majority of the mitochondrial protein import events are mediated by the TOM-TIM machinery. In addition, it has been proposed that Aβ is imported into the rat liver mitochondria via the mitochondrial TOM machinery (28). Thus, our hypothesis here was that the Aβ mitochondrial accumulation in yeast could also be TOM-dependent. To test this hypothesis, we investigated whether or not Aβ could still accumulate on the mitochondria when the TOM channel is physically blocked by other factors (Fig. 2A). To block the TOM channel, we designed a construct with the N-terminal presequence of the yeast alcohol dehydrogenase III (preADHIII) and C-terminal GST tag, preADHIII-GST. preADHIII is a known efficient MTS with only 27 residues to initiate the mitochondrial import event when attached to a cytosolic enzyme, mouse dihydrofolate reductase (34). Thus, in the import and stop experiment illustrated in Fig. 1A, the preADHIII recombinant proteins would be recognized and imported by the TOM machinery and then would physically block the transport pathway by the structured GST domain. The results verified that the preADHIII recombinant proteins were imported by the yeast mitochondria (Fig. 2B, lane 1) and then stuck on the mitochondrial surface as labeled by the nanogold particles (Fig. 2C). Next, we incubated the preADHIII-GST–blocked mitochondria with His-GST-Aβ to investigate whether Aβ still accumulates on mitochondria by performing the import and stop experiment. The rationale here was that if the Aβ mitochondrial accumulation was TOM-dependent, with the TOM channel already blocked by preADHIII-GST, we expected no His-GST-Aβ proteins to accumulate on the mitochondria surface. The Western blotting results of this experiment, presented in Fig. 2B, show that when mitochondria were blocked by preADHIII-GST mitochondrial accumulation of His-GST-Aβ was insignificant using anti-GST antibodies (compare lanes 1 and 2). In addition, the accumulated amount of His-GST-Aβ on the surface of preADHIII-GST–blocked mitochondria is significantly less than the amount on the surface of treated mitochondria (compare lanes 3 and 4). Furthermore, we labeled the mitochondria (treated with preADHIII-GST and then His-GST-Aβ) with the Ni-NTA–conjugated nanogold and found almost no gold particles on the mitochondrial surface in the EM micrographs (Fig. 2D). Taken together, the results suggest that His-GST-Aβ does not efficiently accumulate on the yeast mitochondrial surface when the TOM complex is occupied by preADHIII-GST, indicating that the Aβ mitochondrial accumulation is also TOM-dependent, similar to the mitochondrial import of preADHIII.
Figure 2.
Mitochondrial Aβ accumulation is TOM-dependent. A, illustration of the experiment. Yeast mitochondria were preloaded with preADHIII substrates and then incubated with His-GST-Aβ for the import and stop assay. B, Western blot analysis of the treated mitochondria. The mitochondria were pretreated with or without preADHIII-GST and then treated with or without His-GST-Aβ. The left two lanes used anti-GST probe, and the right two lanes used anti-His probe. C and D, representative EM micrographs of the nanogold-labeled mitochondria that were either pretreated with preADHIII-GST-His (C) or treated with both preADHIII-GST and His-GST-Aβ. The scale bar is 200 nm.
Tom22, rather than Tom20 or Tom70, is the main receptor for Aβ binding
Tom receptors, namely Tom20 and Tom70, are responsible for recognizing a variety of MTSs. As a result of extensive biochemical and structural studies, the detailed molecular mechanism of the substrate recognition mediated by the Tom receptors has been partially revealed. It has been suggested that Tom20 recognizes the hydrophobic part of the substrates' amphipathic helices (35, 36), whereas Tom70 recognizes both hydrophobic and hydrophilic faces of the MTS (37, 38). It is also known that Tom22 is capable of binding to the preprotein substrates, but it seems to happen only after the initial recognition via Tom20 or Tom70 (31). Here, we sought to understand how Aβ was recognized by the TOM machinery, and we asked exactly which Tom component was responsible for the Aβ recognition. To our surprise, we discovered that Tom22, rather than Tom20 or Tom70, was the major receptor for the Aβ binding.
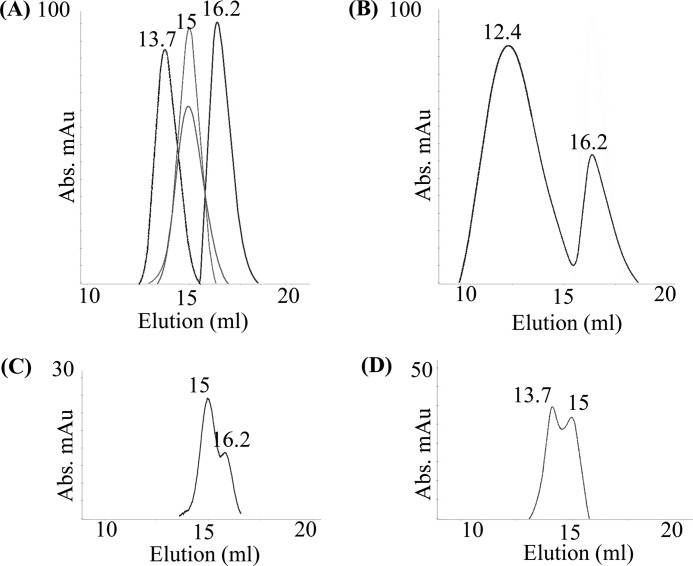
We started the study by trying to capture the Tom20/Aβ interaction in vitro because it was previously suggested that Tom20 may be the receptor responsible for binding to Aβ (28). We know that Tom20 is a membrane protein with one transmembrane helix, and its receptor function is solely mediated through the cytosolic domain (35, 39). Thus, we expressed and purified the cytosolic domain of Tom20 (cytoTom20) to homogeneity as shown in gel filtration chromatography (Fig. 3A). We then mixed the purified homogenous cytoTom20 and His-GST-Aβ at a molar ratio of 3:1 and subjected the sample to analytical gel filtration analysis using a Superdex 200 column. The rationale for this analysis was that if the two proteins interacted with each other we would observe a shifted peak in the chromatography representing the newly formed protein complexes. To demonstrate this, we did a control experiment using a mixture of cytoTom20 and preADHIII-GST at a 3:1 molar ratio. The result (Fig. 3) showed that the original peak of cytoTom20 (∼16.2 ml) remained unchanged. However, the original peak of preADHIII-GST (∼15 ml; Fig. 3A) disappeared completely, and the new peak (∼12.4 ml; Fig. 3B) represented the preADHIII-GST–cytoTom20 complex. However, in the case of cytoTom20 and His-GST-Aβ, we found that both original peaks of cytoTom20 and His-GST-Aβ remained unchanged as shown in Fig. 3C, indicating no apparent interaction between the two.
Figure 3.
Neither Tom20 nor Tom70 binds to Aβ peptides in vitro. A, cytoTom20, cytoTom70, His-GST-Aβ, and preADHIII-GST were purified by gel filtration on a Superdex 200 column, respectively, in buffer containing 20 mm HEPES, pH 7, 150 mm NaCl. The peaks, from left to right, are cytoTom70 (∼13.7 ml), preADHIII-GST (tall, ∼15 ml), His-GST-Aβ (short, ∼15 ml), and cytoTom20 (∼16.2 ml). B, C, and D, mixtures of preADHIII-GST–cytoTom20 (1:3 molar ratio), His-GST-Aβ–cytoTom20 (1:3), and His-GST-Aβ–cytoTom70 (2:1) were examined by analytical gel filtration, respectively. Abs, absorbance; mAU, milli-absorbance units.
At this point, we suspected that maybe another known TOM receptor, Tom70, was responsible for the Aβ binding. Tom70 is also a membrane protein with one transmembrane helix and a cytosolic domain. Similar to Tom20, the receptor function of Tom70 is carried out by its cytosolic domain, cytoTom70. We expressed and purified cytoTom70 to homogeneity as a peak at ∼13.7 ml as shown in Fig. 3A. The analytical gel filtration experiment on the mixture of cytoTom70 and His-GST-Aβ showed unchanged peaks of the two proteins, again suggesting no apparent interactions between them (Fig. 3D).
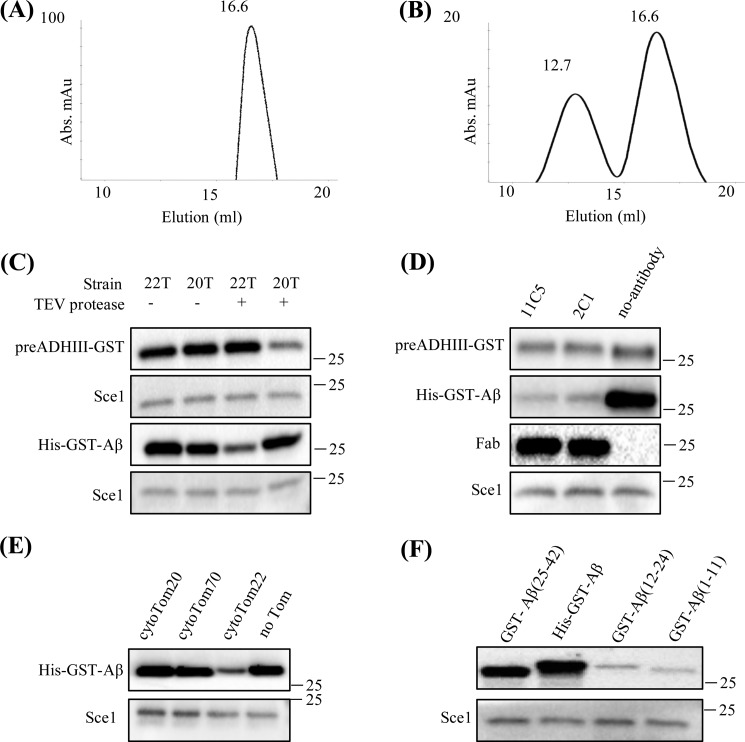
These results led us to hypothesize that Tom22 might be the receptor we were looking for. To test the hypothesis in vitro, we purified the His-tagged cytosolic domain of Tom22, cytoTom22, and performed the analytical gel filtration experiment. The homogenous single peak of cytoTom22 appears at the elution volume of ∼16.6 ml (Fig. 4A). When His-GST-Aβ was mixed with an excessive amount of cytoTom22, its elution peak shifted to ∼12.7 ml (Fig. 4B and Fig. S4), behavior similar to the positive control cytoTom20–preADHIII-GST presented in Fig. 3B. As a negative control, we incubated preADHIII-GST with cytoTom22 and performed the same gel filtration analysis. We found that there was no apparent peak shift. This indicates no strong binding between cytoTom22 and preADHIII (Fig. S5). Thus, we conclude that Tom22 is able to recognize and bind to Aβ in vitro.
Figure 4.
Tom22 is responsible for the Aβ peptides binding. A, gel filtration chromatography of the purified cytoTom22. B, the mixture of His-GST-Aβ–cytoTom22 (1:3) was examined by analytical gel filtration. C, Western blot analysis of the substrate accumulation by isolated mitochondria from 22T and 20T strains with and without TEV protease digestion. D, Western blot analysis of the mitochondrial Aβ accumulation using antibody-shielded mitochondria. E, mitochondrial Aβ accumulation under competitive experiments with purified cytosolic domains of Tom20, Tom70, or Tom22. F, mitochondrial accumulation of various GST-tagged Aβ fragments tested in the import and stop experiments. Abs, absorbance; mAU, milli-absorbance units.
Next, we asked whether or not the cytoTom22–His-GST-Aβ binding exists in the context of isolated functional mitochondria. To answer that, we tested whether His-GST-Aβ could still accumulate on the mitochondrial surface without Tom22. We used two distinct methods to abolish the function of Tom22. The first method was to remove the Tom22 cytosolic domain using a genetically modified yeast strain, 22T (40). Tom22 in this strain contains a tobacco etch virus (TEV) protease cleavage site in between its cytosolic domain and transmembrane helix. Meanwhile, 20T strain (40), which has a similar genetic modification in Tom20, was used as a control. We used isolated mitochondria from both 22T and 20T strains, treated them with an excessive amount of TEV protease, and then performed the import and stop experiments using two substrates, preADHIII-GST and His-GST-Aβ. We found that preADHIII-GST accumulated on mitochondria from both 22T and 20T as expected (Fig. 4C, lanes 1 and 2, and Fig. S6A). Upon TEV protease digestion, 20T mitochondria accumulated much less preADHIII-GST (only ∼30% compared with nondigested 20T; Fig. 4C, lane 4, and Fig. S6A), whereas 22T mitochondria accumulated as much preADHIII-GST as the controls. The results agreed well with the fact that Tom20, not Tom22, is the major receptor for preADHIII substrate. In the case of His-GST-Aβ, the non-TEV protease–treated 22T and 20T mitochondria were able to accumulate the substrate as expected (Fig. 4C, lanes 1 and 2, and Fig. S6B). However, upon TEV protease digestion, 22T mitochondria recognized much less Aβ substrate (∼35% compared with nondigested 22T; Fig. 4C, lane 3, and Fig. S6B), whereas 20T mitochondria remained similar to the controls (Fig. 4C, right panel, lanes 2 and 4). Thus, together the results strongly suggest that Tom22, instead of Tom20, acts as the major receptor for the Aβ substrate.
It is worth noting that the modified TEV protease cleavage site was in close approximation to the mitochondrial outer membrane and could be partially buried within the TOM complex, leading to incomplete digestion (∼65–70% efficiency as estimated by Western blotting quantifications) by the TEV protease. To confirm the obtained results, we used a second method to abolish Tom22's function, which is to shield cytoTom22 by specific conformational antibodies. Here, we generated monoclonal antibodies that bound to the folded Tom22 by recognizing its structured portion. These antibodies were different from the antibodies used in general Western blot analysis that bind to a specific peptide sequence on denatured Tom22. We obtained two such conformational antibodies, 11C5 and 2C1. When binding to cytoTom22 on the mitochondrial surface, the ∼150-kDa antibody molecules might be too large and could shield other parts of the TOM complex, leading to nonconclusive results. To minimize the potential interference, we decided to use only the Fab fragments (∼50 kDa) generated from such antibodies. We incubated the Fab fragments of 11C5 and 2C1 with the isolated yeast mitochondria to perform the import and stop experiments. The results showed that the amount of His-GST-Aβ proteins accumulated on the antibody-shielded mitochondrial surface (Fig. 4D, lanes 1 and 2, and Fig. S6C) was reduced dramatically compared with the control unshielded mitochondria (Fig. 4D, lane 3, and Fig. S6C). In addition, we labeled the antibody-shielded mitochondria with the Ni-NTA–conjugated nanogold, and found very few, if any, gold particles in EM micrographs (data not shown).
Additionally, we designed a third competition experiment to further confirm that Tom22, rather than Tom20 or Tom70, on the mitochondrial surface is responsible for the accumulation of His-GST-Aβ. We incubated the same amount of purified cytoTom20, cytoTom22, and cytoTom70 proteins with an equal amount of isolated yeast mitochondria, respectively. Then we added the substrate His-GST-Aβ into the mixtures and determined which of the purified Tom receptors in solution were able to compete with the mitochondrial accumulation of His-GST-Aβ. We found that, with cytoTom22 in the mixture, the accumulation of His-GST-Aβ on the mitochondrial surface is significantly reduced (Fig. 4E and Fig. S6D), suggesting that cytoTom22 binds to His-GST-Aβ. All together, these results suggest that neither Tom20 nor Tom70, but Tom22, is the major receptor for Aβ peptides.
Aβ(25–42) peptide is responsible for the specific interaction with Tom22
Furthermore, we asked which part of Aβ peptide interacts with Tom22. It has been shown in multiple NMR structures that Aβ could adopt partially folded structures in solution (41, 42). The Aβ(1–40) structure has a central hydrophobic region forming a 310 helix (residues 12–23) with both N and C termini unstructured, whereas the Aβ(1–42) structure appears to adopt two helical regions (residues 8–25 and 28–38) with the rest being mostly flexible. Based on these structures, we divided the Aβ peptide into three segments, Aβ(1–11), Aβ(12–24), and Aβ(25–42), and linked them to an N-terminal GST domain, respectively. We then performed the mitochondrial import and stop assay using these three recombinant proteins. We found that GST-Aβ(25–42) significantly accumulated on the mitochondrial surface, and the accumulated amount is similar to His-GST-Aβ. In contrast, GST-Aβ(1–11) and GST-Aβ(12–24) did not accumulate on the mitochondrial surface (Fig. 4F and Fig. S6E). These results suggest that Aβ(25–42) is the main sequence responsible for the specific interaction between Aβ and Tom22.
Discussion
We demonstrated that yeast mitochondria can specifically accumulate human Aβ peptides through the TOM complex. To our surprise, the recognition process is unconventionally mediated through Tom22 instead of canonical receptors Tom20 and Tom70. Furthermore, Aβ peptide residues 25–42 are responsible for its specific interaction with Tom22. These conclusions provided partial answers to an ongoing question in the field: how do Aβ peptides get into the mitochondria? We propose that the mitochondrial Aβ accumulation process begins when cytosolic Aβ is recognized by Tom22, then transferred to Tom40, and transported through the TOM channel into the mitochondria.
Our proposed model does not rule out other possible origins of the harmful mitochondrial Aβ. For example, in the mitochondrial intermembrane space, the Aβ peptide can still be produced through sequential processing of amyloid precursor protein by several secretases (43). In addition, Aβ can also be directly transported from the endoplasmic reticulum into the mitochondria through the connecting points between the two organelles (44). After all, the relative amount of the mitochondrial Aβ from different origins is not clear and needs to be quantified. Nevertheless, the Aβ/TOM interaction appears to be a significant part of mitochondrial Aβ accumulation because it is in line with previous observations of protein import malfunction caused by mitochondrial Aβ (27, 28).
We know that Aβ accumulation in mitochondria is associated with mitochondrial dysfunction observed in the brains of patients with Alzheimer's disease (45). The mitochondrial Aβ is extremely toxic because of its broad interactions with important proteins in all compartments within the mitochondria (46). Thus, the approach to stop the mitochondrial Aβ accumulation may have great pharmaceutical potential. We have revealed important information regarding the initial step of the mitochondrial Aβ accumulation: the direct interaction between Tom22 and Aβ. The detailed molecular mechanism of this interaction remains to be further studied by structural biology and other biochemical/biophysical methods.
Experimental procedures
Yeast mitochondrial purification
S. cerevisiae strain S228C was grown in yeast extract–peptone–glycerol (YPG) medium at 30 °C. Yeast mitochondrial isolation was performed by following established protocols (33). Specifically, 4 liters of yeast culture were grown to an A600 value of 2 and harvested by centrifugation at 3,000 × g for 20 min. The cells were washed with double distilled H2O; resuspended in buffer containing 20 mm 100 mm Tris-H2SO4, pH 9.4, 10 mm dithiothreitol (DTT); and gently shaken at 30 °C for 20 min before another round of centrifugation and resuspension in the buffer containing 20 mm potassium phosphate, pH 7.4, 1.2 m sorbitol. The cells were then treated with Zymolyase (3 mg of enzyme/g of yeast) at 30 °C for 45 min. The yeast spheroplasts were harvested again and gently homogenized in a glass homogenizer in buffer containing 10 mm Tris-HCl, pH 7.4, 1 mm EDTA, 1 mm phenylmethylsulfonyl fluoride (PMSF), 0.2% bovine serum albumin (BSA), 0.6 m sorbitol. The homogenate was centrifuged at 1,500 × g for 5 min (the pellet was discarded), 4,000 × g for 5 min (the pellet was discarded), and 12,000 × g for 15 min. The pellet containing crude intact mitochondria was resuspended in buffer containing 10 mm MOPS-KOH, pH 7.2, 250 mm sucrose, 1 mm EDTA. Finally, a sucrose step gradient ultracentrifugation at 134,00 × g for 1 h was performed. The highly purified yeast mitochondria were harvested at the 60%/32% sucrose interface, pelleted, and resuspended in buffer containing 20 mm MOPS-KOH, pH 7.2, 250 mm sucrose, 5 mm ATP, 5 mm MgCl2 at a protein concentration of ∼10 mg/ml for later experiments. Protein concentrations reported in this study were all determined using a Pierce Coomassie (Bradford) assay kit.
Mitochondrial import and stop experiments
The import experiment was started by adding corresponding recombinant proteins, such as His-GST-Aβ, preADHIII-GST, and various GST-Aβ fragments, at 0.2 mg/ml into the mitochondrial resuspension at 25 °C and stopped in 20 min by centrifugation at 12,000 × g for 15 min. The treated mitochondria were collected by centrifugation at 12,000 × g for 15 min and then resuspended in buffer containing 20 mm MOPS-KOH, pH 7.2, 250 mm sucrose. The centrifugation/resuspension steps were performed three times to ensure that the nonimported substrates in the buffer were completely washed away. Finally, the collected mitochondria were analyzed by 12% SDS-PAGE followed by Western blotting using either anti-His or anti-GST antibody. The loading control for all Western blot analyses in this study was the yeast mitochondrial intron-encoded endonuclease Sce1 detected by its polyclonal antibody ab216263 (Abcam).
20His yeast strain modification
A His tag was inserted into the C terminus of the Tom20 gene in the genome of yeast strain S228C by homologous recombination. Specifically, a DNA fragment containing a His tag and a hygromycin B resistance gene was amplified from the vector pTF271(pFA6a-TEV-6xGly-12xHis-HphMX) (Addgene 44081) with a 40-bp homologous sequence on both ends. The PCR product was then transformed into yeast competent cells by lithium acetate/single-stranded carrier DNA/PEG method as described previously (47). Transformed cells were plated on yeast extract–peptone–dextrose (YPD) plates with 200 μg/ml hygromycin B. Positive colonies with the inserted gene fragment were grown in appropriate culture and confirmed by Western blotting against His tag.
TEV protease digestion
22T and 20T yeast strains were grown in conditions reported previously (40). Their mitochondria were also isolated using the same protocol and resuspended in buffer containing 20 mm MOPS-KOH, pH 7.2, 250 mm sucrose at a final protein concentration of ∼10 mg/ml. The digestion experiments were performed at 25 °C for 2 h by adding a final TEV protease concentration of ∼2 mg/ml into the respective 22T and 20T mitochondrial suspensions. The reactions were stopped by centrifugation at 12,000 × g for 15 min. Again, the treated mitochondria were washed three times with buffer containing 20 mm MOPS-KOH, pH 7.2, 250 mm sucrose and subjected to Western blot analysis using the Bio-Rad ChemiDoc imaging system. The protease digestion efficiency of 22T was estimated to be ∼65–70% by Western blotting using our own antibody, 13D12, against denatured Tom22, which was consistent with published reports (40).
Cloning, expression, and purification of recombinant proteins
Molecular cloning of all constructs used in this study was performed with either pET15b or pGEX4T1 vector using an In-Fusion HD cloning kit. Specifically, cytoTom20 (UniProtKB accession number P35180, residues 29–183), cytoTom70 (UniProtKB accession number P07213, residues 31–617), and cytoTom22 (UniProtKB accession number P49334, residues 1–97) were inserted in the pET15b vector with an N-terminal His tag. The human Aβ sequence used for cloning was derived from UniProtKB accession number P05067 (DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA). All GST-tagged constructs, His-GST-Aβ (266 residues), His-GST (243 residues), preADHIII-GST (304 residues), GST-Aβ(1–11) (229 residues), GST-Aβ(12–24) (231 residues), and GST-Aβ(25–42) (236 residues), were made with pGEX4T1 vector. All recombinant proteins were expressed in the Escherichia coli C43 strain at 37 °C with 1 mm isopropyl 1-thio-β-d-galactopyranoside. The expressed proteins were purified by Ni-NTA–affinity chromatography followed by gel filtration using a Superdex 200 column in buffer containing 20 mm HEPES, pH 7.0, 150 mm NaCl. The purified recombinant proteins were used for all mitochondrial import and analytical gel filtration experiments.
Endogenous expression of Aβ-related constructs in S. cerevisiae
His-GST-Aβ and Aβ alone were cloned into the pRS414 vector with a glyceraldehyde-3-phosphate dehydrogenase (GPD) promoter and then transformed into yeast strain BCY123. The yeast cells were grown in Trp-dropout medium at 30 °C for 24 h before harvesting to allow recombinant protein expression. The yeast mitochondria were isolated the same way as described above and then subjected to Western blot analysis using antibody sc-53822 (Santa Cruz Biotechnology) against Aβ.
EM analysis using nanogold labeling
The 1.8-nm Ni-NTA–conjugated nanogold was purchased from Nanoprobes and prepared following their online instructions. The treated mitochondrial suspension was incubated with the nanogold solution for 20 min on ice and centrifuged at 12,000 × g for 15 min, and the pellet was resuspended in buffer containing 20 mm MOPS-KOH, pH 7.2, 250 mm sucrose. The centrifugation/resuspension steps were performed three times to ensure that unbound nanogold particles in the solution were completely washed away and would not interfere with the following analysis. 3 μl of the samples were then applied onto the EM grids (prepared in a glow discharger using the following settings: 25-mA current, negative charge, and 1-min processing time) and washed with double distilled H2O three times. The grids were imaged using an FEI T12 microscope, which was operated at 80 keV and room temperature. The images were taken with a 2,000 × 2,000 charge-coupled device camera at a nominal magnification of 45,000. The scale bars in all micrographs represent 200 nm.
Competition experiments with cytosolic domains of Tom receptors
Isolated mitochondria at 10 mg/ml were incubated with purified cytoTom20, cytoTom70, and cytoTom22 at 0.1 mg/ml, respectively. 0.2 mg/ml His-GST-Aβ was added into each mixture to perform the import and stop assay. The mitochondria were washed thoroughly and subjected to Western blot analysis.
Analytical gel filtration
The gel filtration experiments were performed using a Superdex 200 column on a GE Healthcare ÄKTA FPLC machine. The samples were prepared by mixing purified recombinant proteins at certain molar ratios as indicated in the main text, concentrated to ∼50 μl, and incubated for 1 h before injection. All gel filtration experiments were performed in buffer containing 20 mm HEPES, pH 7, 150 mm NaCl at 4 °C.
Author contributions
W. H. and H. Z. conceptualization; W. H. and Z. W. data curation; W. H. and H. Z. formal analysis; W. H., Z. W., and H. Z. validation; W. H. and H. Z. investigation; H. Z. resources; H. Z. supervision; H. Z. funding acquisition; H. Z. methodology; H. Z. writing-original draft.
Supplementary Material
Acknowledgments
We thank D. Cawley for helping with the mAb development and Dr. Toshiya Endo for kindly providing the 20T and 22T yeast strains.
This work was supported by the Boettcher Foundation Webb-Waring Biomedical Research Award (to H. Z.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
- Aβ
- β-amyloid
- TOM
- translocase of the outer membrane
- TIM
- translocase of the inner member
- MTS
- mitochondrial targeting signal
- Ni-NTA
- nickel-nitrilotriacetic acid
- preADHIII
- presequence of the yeast alcohol dehydrogenase III
- cytoTom
- cytosolic domain of Tom
- TEV
- tobacco etch virus.
References
- 1. Hardy J. A., and Higgins G. A. (1992) Alzheimer's disease: the amyloid cascade hypothesis. Science 256, 184–185 10.1126/science.1566067 [DOI] [PubMed] [Google Scholar]
- 2. Wirths O., and Bayer T. A. (2012) Intraneuronal Aβ accumulation and neurodegeneration: lessons from transgenic models. Life Sci. 91, 1148–1152 10.1016/j.lfs.2012.02.001 [DOI] [PubMed] [Google Scholar]
- 3. LaFerla F. M., Green K. N., and Oddo S. (2007) Intracellular amyloid-β in Alzheimer's disease. Nat. Rev. Neurosci. 8, 499–509 10.1038/nrn2168 [DOI] [PubMed] [Google Scholar]
- 4. Wirths O., Multhaup G., and Bayer T. A. (2004) A modified β-amyloid hypothesis: intraneuronal accumulation of the β-amyloid peptide—the first step of a fatal cascade. J. Neurochem. 91, 513–520 10.1111/j.1471-4159.2004.02737.x [DOI] [PubMed] [Google Scholar]
- 5. Xu H., Sweeney D., Wang R., Thinakaran G., Lo A. C., Sisodia S. S., Greengard P., and Gandy S. (1997) Generation of Alzheimer β-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc. Natl. Acad. Sci. U.S.A. 94, 3748–3752 10.1073/pnas.94.8.3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hartmann T., Bieger S. C., Brühl B., Tienari P. J., Ida N., Allsop D., Roberts G. W., Masters C. L., Dotti C. G., Unsicker K., and Beyreuther K. (1997) Distinct sites of intracellular production for Alzheimer's disease A β40/42 amyloid peptides. Nat. Med. 3, 1016–1020 10.1038/nm0997-1016 [DOI] [PubMed] [Google Scholar]
- 7. Greenfield J. P., Tsai J., Gouras G. K., Hai B., Thinakaran G., Checler F., Sisodia S. S., Greengard P., and Xu H. (1999) Endoplasmic reticulum and trans-Golgi network generate distinct populations of Alzheimer β-amyloid peptides. Proc. Natl. Acad. Sci. U.S.A. 96, 742–747 10.1073/pnas.96.2.742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cam J. A., Zerbinatti C. V., Li Y., and Bu G. (2005) Rapid endocytosis of the low density lipoprotein receptor-related protein modulates cell surface distribution and processing of the β-amyloid precursor protein. J. Biol. Chem. 280, 15464–15470 10.1074/jbc.M500613200 [DOI] [PubMed] [Google Scholar]
- 9. Agostinho P., Pliássova A., Oliveira C. R., and Cunha R. A. (2015) Localization and trafficking of amyloid-β protein precursor and secretases: impact on Alzheimer's disease. J. Alzheimers Dis. 45, 329–347 10.3233/JAD-142730 [DOI] [PubMed] [Google Scholar]
- 10. Mawuenyega K. G., Sigurdson W., Ovod V., Munsell L., Kasten T., Morris J. C., Yarasheski K. E., and Bateman R. J. (2010) Decreased clearance of CNS β-amyloid in Alzheimer's disease. Science 330, 1774 10.1126/science.1197623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Poduslo J. F., Howell K. G., Olson N. C., Ramirez-Alvarado M., and Kandimalla K. K. (2012) Alzheimer's disease amyloid β-protein mutations and deletions that define neuronal binding/internalization as early stage nonfibrillar/fibrillar aggregates and late stage fibrils. Biochemistry 51, 3993–4003 10.1021/bi300275g [DOI] [PubMed] [Google Scholar]
- 12. Manczak M., Anekonda T. S., Henson E., Park B. S., Quinn J., and Reddy P. H. (2006) Mitochondria are a direct site of Aβ accumulation in Alzheimer's disease neurons: implications for free radical generation and oxidative damage in disease progression. Hum. Mol. Genet. 15, 1437–1449 10.1093/hmg/ddl066 [DOI] [PubMed] [Google Scholar]
- 13. Devi L., Prabhu B. M., Galati D. F., Avadhani N. G., and Anandatheerthavarada H. K. (2006) Accumulation of amyloid precursor protein in the mitochondrial import channels of human Alzheimer's disease brain is associated with mitochondrial dysfunction. J. Neurosci. 26, 9057–9068 10.1523/JNEUROSCI.1469-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Caspersen C., Wang N., Yao J., Sosunov A., Chen X., Lustbader J. W., Xu H. W., Stern D., McKhann G., and Yan S. D. (2005) Mitochondrial Aβ: a potential focal point for neuronal metabolic dysfunction in Alzheimer's disease. FASEB J. 19, 2040–2041 10.1096/fj.05-3735fje [DOI] [PubMed] [Google Scholar]
- 15. Yamaguchi H., Yamazaki T., Ishiguro K., Shoji M., Nakazato Y., and Hirai S. (1992) Ultrastructural localization of Alzheimer amyloid β/A4 protein precursor in the cytoplasm of neurons and senile plaque-associated astrocytes. Acta Neuropathol. 85, 15–22 10.1007/BF00304629 [DOI] [PubMed] [Google Scholar]
- 16. Brooksbank B. W., and Balazs R. (1984) Superoxide dismutase, glutathione peroxidase and lipoperoxidation in Down's syndrome fetal brain. Brain Res. 318, 37–44 [DOI] [PubMed] [Google Scholar]
- 17. Sirk D., Zhu Z., Wadia J. S., Shulyakova N., Phan N., Fong J., and Mills L. R. (2007) Chronic exposure to sub-lethal β-amyloid (Aβ) inhibits the import of nuclear-encoded proteins to mitochondria in differentiated PC12 cells. J. Neurochem. 103, 1989–2003 10.1111/j.1471-4159.2007.04907.x [DOI] [PubMed] [Google Scholar]
- 18. Hernandez-Zimbron L. F., Luna-Muñoz J., Mena R., Vazquez-Ramirez R., Kubli-Garfias C., Cribbs D. H., Manoutcharian K., and Gevorkian G. (2012) Amyloid-β peptide binds to cytochrome C oxidase subunit 1. PLoS One 7, e42344 10.1371/journal.pone.0042344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hauptmann S., Scherping I., Dröse S., Brandt U., Schulz K. L., Jendrach M., Leuner K., Eckert A., and Müller W. E. (2009) Mitochondrial dysfunction: an early event in Alzheimer pathology accumulates with age in AD transgenic mice. Neurobiol. Aging 30, 1574–1586 10.1016/j.neurobiolaging.2007.12.005 [DOI] [PubMed] [Google Scholar]
- 20. Keil U., Bonert A., Marques C. A., Scherping I., Weyermann J., Strosznajder J. B., Müller-Spahn F., Haass C., Czech C., Pradier L., Müller W. E., and Eckert A. (2004) Amyloid β-induced changes in nitric oxide production and mitochondrial activity lead to apoptosis. J. Biol. Chem. 279, 50310–50320 10.1074/jbc.M405600200 [DOI] [PubMed] [Google Scholar]
- 21. Schmidt C., Lepsverdize E., Chi S. L., Das A. M., Pizzo S. V., Dityatev A., and Schachner M. (2008) Amyloid precursor protein and amyloid β-peptide bind to ATP synthase and regulate its activity at the surface of neural cells. Mol. Psychiatry 13, 953–969 10.1038/sj.mp.4002077 [DOI] [PubMed] [Google Scholar]
- 22. DuBoff B., Feany M., and Götz J. (2013) Why size matters—balancing mitochondrial dynamics in Alzheimer's disease. Trends Neurosci. 36, 325–335 10.1016/j.tins.2013.03.002 [DOI] [PubMed] [Google Scholar]
- 23. Crouch P. J., Harding S. M., White A. R., Camakaris J., Bush A. I., and Masters C. L. (2008) Mechanisms of Aβ mediated neurodegeneration in Alzheimer's disease. Int. J. Biochem. Cell Biol. 40, 181–198 10.1016/j.biocel.2007.07.013 [DOI] [PubMed] [Google Scholar]
- 24. Sheng Z. H. (2014) Mitochondrial trafficking and anchoring in neurons: new insight and implications. J. Cell Biol. 204, 1087–1098 10.1083/jcb.201312123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Selfridge J. E., Lezi E., Lu J., and Swerdlow R. H. (2013) Role of mitochondrial homeostasis and dynamics in Alzheimer's disease. Neurobiol. Dis. 51, 3–12 10.1016/j.nbd.2011.12.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mattson M. P., Gleichmann M., and Cheng A. (2008) Mitochondria in neuroplasticity and neurological disorders. Neuron 60, 748–766 10.1016/j.neuron.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cenini G., Rüb C., Bruderek M., and Voos W. (2016) Amyloid β-peptides interfere with mitochondrial preprotein import competence by a coaggregation process. Mol. Biol. Cell 27, 3257–3272 10.1091/mbc.e16-05-0313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hansson Petersen C. A., Alikhani N., Behbahani H., Wiehager B., Pavlov P. F., Alafuzoff I., Leinonen V., Ito A., Winblad B., Glaser E., and Ankarcrona M. (2008) The amyloid β-peptide is imported into mitochondria via the TOM import machinery and localized to mitochondrial cristae. Proc. Natl. Acad. Sci. U.S.A. 105, 13145–13150 10.1073/pnas.0806192105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schmidt O., Pfanner N., and Meisinger C. (2010) Mitochondrial protein import: from proteomics to functional mechanisms. Nat. Rev. Mol. Cell Biol. 11, 655–667 10.1038/nrm2959 [DOI] [PubMed] [Google Scholar]
- 30. Neupert W. (2015) A perspective on transport of proteins into mitochondria: a myriad of open questions. J. Mol. Biol. 427, 1135–1158 10.1016/j.jmb.2015.02.001 [DOI] [PubMed] [Google Scholar]
- 31. Shiota T., Mabuchi H., Tanaka-Yamano S., Yamano K., and Endo T. (2011) In vivo protein-interaction mapping of a mitochondrial translocator protein Tom22 at work. Proc. Natl. Acad. Sci. U.S.A. 108, 15179–15183 10.1073/pnas.1105921108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Wilpe S., Ryan M. T., Hill K., Maarse A. C., Meisinger C., Brix J., Dekker P. J., Moczko M., Wagner R., Meijer M., Guiard B., Hönlinger A., and Pfanner N. (1999) Tom22 is a multifunctional organizer of the mitochondrial preprotein translocase. Nature 401, 485–489 10.1038/46802 [DOI] [PubMed] [Google Scholar]
- 33. Meisinger C., Pfanner N., and Truscott K. N. (2006) Isolation of yeast mitochondria. Methods Mol. Biol. 313, 33–39 10.1385/1-59259-958-3:033 [DOI] [PubMed] [Google Scholar]
- 34. van Loon A. P., Brändli A. W., and Schatz G. (1986) The presequences of two imported mitochondrial proteins contain information for intracellular and intramitochondrial sorting. Cell 44, 801–812 10.1016/0092-8674(86)90846-9 [DOI] [PubMed] [Google Scholar]
- 35. Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., and Kohda D. (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100, 551–560 10.1016/S0092-8674(00)80691-1 [DOI] [PubMed] [Google Scholar]
- 36. Rimmer K. A., Foo J. H., Ng A., Petrie E. J., Shilling P. J., Perry A. J., Mertens H. D., Lithgow T., Mulhern T. D., and Gooley P. R. (2011) Recognition of mitochondrial targeting sequences by the import receptors Tom20 and Tom22. J. Mol. Biol. 405, 804–818 10.1016/j.jmb.2010.11.017 [DOI] [PubMed] [Google Scholar]
- 37. Brix J., Rüdiger S., Bukau B., Schneider-Mergener J., and Pfanner N. (1999) Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J. Biol. Chem. 274, 16522–16530 10.1074/jbc.274.23.16522 [DOI] [PubMed] [Google Scholar]
- 38. Wu Y., and Sha B. (2006) Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p. Nat. Struct. Mol. Biol. 13, 589–593 10.1038/nsmb1106 [DOI] [PubMed] [Google Scholar]
- 39. Saitoh T., Igura M., Miyazaki Y., Ose T., Maita N., and Kohda D. (2011) Crystallographic snapshots of Tom20-mitochondrial presequence interactions with disulfide-stabilized peptides. Biochemistry 50, 5487–5496 10.1021/bi200470x [DOI] [PubMed] [Google Scholar]
- 40. Yamano K., Yatsukawa Y., Esaki M., Hobbs A. E., Jensen R. E., and Endo T. (2008) Tom20 and Tom22 share the common signal recognition pathway in mitochondrial protein import. J. Biol. Chem. 283, 3799–3807 10.1074/jbc.M708339200 [DOI] [PubMed] [Google Scholar]
- 41. Vivekanandan S., Brender J. R., Lee S. Y., and Ramamoorthy A. (2011) A partially folded structure of amyloid-β(1–40) in an aqueous environment. Biochem. Biophys. Res. Commun. 411, 312–316 10.1016/j.bbrc.2011.06.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Crescenzi O., Tomaselli S., Guerrini R., Salvadori S., D'Ursi A. M., Temussi P. A., and Picone D. (2002) Solution structure of the Alzheimer amyloid β-peptide (1–42) in an apolar microenvironment. Similarity with a virus fusion domain. Eur. J. Biochem. 269, 5642–5648 10.1046/j.1432-1033.2002.03271.x [DOI] [PubMed] [Google Scholar]
- 43. Anandatheerthavarada H. K., Biswas G., Robin M. A., and Avadhani N. G. (2003) Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J. Cell Biol. 161, 41–54 10.1083/jcb.200207030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Takuma K., Fang F., Zhang W., Yan S., Fukuzaki E., Du H., Sosunov A., McKhann G., Funatsu Y., Nakamichi N., Nagai T., Mizoguchi H., Ibi D., Hori O., Ogawa S., et al. (2009) RAGE-mediated signaling contributes to intraneuronal transport of amyloid-β and neuronal dysfunction. Proc. Natl. Acad. Sci. U.S.A. 106, 20021–20026 10.1073/pnas.0905686106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pagani L., and Eckert A. (2011) Amyloid-β interaction with mitochondria. Int. J. Alzheimers Dis. 2011, 925050 10.4061/2011/925050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pinho C. M., Teixeira P. F., and Glaser E. (2014) Mitochondrial import and degradation of amyloid-β peptide. Biochim. Biophys. Acta 1837, 1069–1074 10.1016/j.bbabio.2014.02.007 [DOI] [PubMed] [Google Scholar]
- 47. Gietz R. D., and Schiestl R. H. (2007) High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2, 31–34 10.1038/nprot.2007.13 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.