Abstract
Interest toward the human microbiome, particularly gut microbiome has flourished in recent decades owing to the rapidly advancing sequence-based screening and humanized gnotobiotic model in interrogating the dynamic operations of commensal microbiota. Although this field is still at a very preliminary stage, whereby the functional properties of the complex gut microbiome remain less understood, several promising findings have been documented and exhibit great potential toward revolutionizing disease etiology and medical treatments. In this review, the interactions between gut microbiota and the host have been focused on, to provide an overview of the role of gut microbiota and their unique metabolites in conferring host protection against invading pathogen, regulation of diverse host physiological functions including metabolism, development and homeostasis of immunity and the nervous system. We elaborate on how gut microbial imbalance (dysbiosis) may lead to dysfunction of host machineries, thereby contributing to pathogenesis and/or progression toward a broad spectrum of diseases. Some of the most notable diseases namely Clostridium difficile infection (infectious disease), inflammatory bowel disease (intestinal immune-mediated disease), celiac disease (multisystemic autoimmune disorder), obesity (metabolic disease), colorectal cancer, and autism spectrum disorder (neuropsychiatric disorder) have been discussed and delineated along with recent findings. Novel therapies derived from microbiome studies such as fecal microbiota transplantation, probiotic and prebiotics to target associated diseases have been reviewed to introduce the idea of how certain disease symptoms can be ameliorated through dysbiosis correction, thus revealing a new scientific approach toward disease treatment. Toward the end of this review, several research gaps and limitations have been described along with suggested future studies to overcome the current research lacunae. Despite the ongoing debate on whether gut microbiome plays a role in the above-mentioned diseases, we have in this review, gathered evidence showing a potentially far more complex link beyond the unidirectional cause-and-effect relationship between them.
Keywords: microbiome, colorectal cancer, dysbiosis, celiac disease, obesity, autism, inflammatory bowel disease, Clostridium difficile infection
Introduction
The human microbiome comprises of collective genomes of microbiota inhabiting us, namely protozoa, archaea, eukaryotes, viruses and predominantly bacteria that live symbiotically on and within various sites of the human body. Examples of occupied habitats include our oral cavity, genital organs, respiratory tract, skin and gastrointestinal system (Lloyd-Price et al., 2016). The human microbiota is estimated to be ∼1013–1014 microbial cells, with around 1:1 microbial cells to human cells ratio (Sender et al., 2016). These numbers are derived from the total bacterial cells in colon (3.8 × 1013 bacteria), the organ that harbors the densest number of microbes (Sender et al., 2016). The diverse gastrointestinal microbiota is predominantly composed of bacteria from three major phyla, namely Firmicutes, Bacteroidetes, and Actinobacteria (Tap et al., 2009). This diverse and complex microbiome serves as a functional expansion of host genomes, and is estimated to harbor 50- to 100-fold more genes, compared to the host. These extra genes have added various types of enzymatic proteins which were non-encoded by the host, and play a critical role in facilitating host metabolism, thus contributing to the regulation of host physiology (Hooper and Gordon, 2001). Until recent decades, the properties of the human microbiome and the host–microbiota interactions have been largely unknown due to technology limitations especially in examining non-cultivable microbes of interest, and lack of population-scale data depicting the microbiota compositions and functions. However, advances in sequencing technologies and subsequent large-scale sequence-based microbiome projects such as the Human Microbiome Project (HMP) consortium funded by The United States National Institutes of Health (NIH), as well as the MetaHIT (Metagenomics of the Human Intestinal Tract) consortium funded by the European Commission, have served as catalysts in nourishing research on the human microbiome. These large-scale endeavors both share similar missions in characterizing the human microbiome and their roles in health and disease states, with MetaHIT solely focusing on gut microbiome. Several analyses have been incorporated in these meta-omics projects including 16S ribosomal RNA (rRNA) sequencing to taxonomically characterize the microbiota communities; Whole Genome Shotgun (WGS) metagenomic sequencing of body-site specific whole community DNA, followed by reference genome mapping, metagenomic assembly, gene cataloging and metabolic reconstruction, to facilitate maximal capture of organismal and functional data of human microbiota (The Human Microbiome Project Consortium, 2012).
Due to the inherent complexity and heterogeneity of the human microbiome, experiments are required to counteract the limitation of empirical methods in examining the causation or correlation links between microbiota disequilibrium (dysbiosis) and human diseases. Robust experimental modeling enables systematic manipulation of variables to investigate hypotheses deduced from “omics” studies. For this, the application of ‘humanized’ gnotobiotic animal model that harbors defined collection of sequenced microbial communities, has gained momentum in recent years in microbiome research (Faith et al., 2010). This allows proof-of-mechanism study to examine the potential impacts of diet (Turnbaugh et al., 2009b), antibiotic, environmental toxicants (Stedtfeld et al., 2017) and host genotypic variation (Ley et al., 2005) on the microbiota and disease manifestation, due to changes in microbiota composition, transcriptomes, proteomes or metabolomes post-induced variations can be extracted and characterized to understand the operation of the microbiota. Besides, ‘humanized’ gnotobiotic mice can be used in preliminary testing of the therapeutic efficacy in treating dysbiosis-associated diseases, as they allow monitoring of the pharmacokinetic–pharmacodynamic changes in microbial communities, thus facilitating optimization of treatment and dosage regime (Mahe et al., 1987; Silva et al., 1999).
Undoubtedly, these efforts shed light on the clinical significance of the human microbiome which is pretty much a ‘black box.’ Although the human microbiome research is still at its preliminary stage, the findings are deemed intriguing yet promising in terms of filling the knowledge gap in microbiome-host relationships, and their role in disease pathogenesis, as well as therapeutic value, which requires more in-depth investigations to uncover this exciting yet mysterious field of research. Below, we review recent investigations specifically related to the bacterial microbiome in the GIT – the largest microbial reservoir of the human body. Gut microbiota and the microbial-synthesized metabolites are discussed along with their roles in human wellness and normal functioning. Further, we review several studies related to the gut microbiome dysbiosis and its association with specific human diseases. We introduce novel microbiome-based therapy employed in specific disease conditions to ‘restore’ wellness and ameliorating dysbiosis-associated diseases. Finally, we discuss future directions and research areas that require further elucidation in order to better understand the human microbiome and its relationship with the host.
The Gut Microbiome and Its Multifarious Functions
The symbiotic relationship between the gut microbiota and the host is regulated and stabilized by a complex network of interactions that encompass metabolic, immune, and neuroendocrine crosstalk between them. This crosstalk is potentially mediated by microbial-synthesized metabolites which exhibit pleiotropic effects, including acting as signaling molecules in regulating host neuro-immune-inflammatory axes that could physiologically link gut with other organ systems. The predominant functions of gut microbiota and the associated key metabolites in governing host wellness are depicted in the following subsections, with some other microbial metabolites being described in Table 1.
Table 1.
Metabolites contributed by gut microbiota and their respective functions.
| Metabolites | Functions | References |
|---|---|---|
| Short-chain fatty acids (SCFAs): |
|
Blottière et al., 2003; Backhed et al., 2004; Xiong et al., 2004; Samuel et al., 2008; Tolhurst et al., 2012; De Vadder et al., 2014 |
| E.g., Acetate, butyrate, propionate, hexanoate, valerate |
|
|
| Indole derivatives: |
|
Bendheim et al., 2002; Deguchi et al., 2002; Wikoff et al., 2009; Venkatesh et al., 2014 |
| E.g., Indole, indoxyl sulfate, indole-3-propionic acid (IPA) |
|
|
| Bile acid metabolites: E.g., Deoxycholic acid (DCA), lithocholic acid (LCA) |
|
Inagaki et al., 2006; Hylemon et al., 2009 |
| Choline metabolites: |
|
Dumas et al., 2006; Wang Z. et al., 2011 |
| E.g., Choline, trimethylamine N-oxide (TMAO) and betaine |
|
|
| Phenolic derivatives: |
|
Larrosa et al., 2006, 2010; Lee H.C. et al., 2006; Selma et al., 2009 |
| E.g., 4-OH phenylacetic acid, equol, urolithins, enterolactone, enterodiol, 8-prenylnaringenin, 2-(3,4-dihydroxyphenyl)acetic acid, 3-(4-hydroxyphenyl)propionic acid, 5-(3,4-dihydroxyphenyl)valeric acid |
|
|
| Vitamins: |
|
LeBlanc et al., 2011; Nicholson et al., 2012; Forster et al., 2017; Lerner et al., 2017b |
| E.g., Thiamine (B1), riboflavin (B2), niacin (B3), pyridoxine (B6), pantothenic acid (B5), biotin (B7), folate (B11–B9), cobalamin (B12), and menaquinone (K2) |
|
|
| Polyamines: |
|
Guo et al., 2003, 2005; Perez-Cano et al., 2010; Johnson et al., 2015; Rooks and Garrett, 2016 |
| E.g., putrescine, spermidine, and spermine |
|
Metabolism
Human fecal sample analysis using 16S ribosomal RNA and metagenomic sequencing techniques reveal significant enrichment in metabolism of polysaccharides, amino acids, xenobiotics and micronutrients conferred by gut microbiota, suggest that these indigenous microbes facilitate host energy harvesting and metabolic efficiency (Gill et al., 2006). These findings were further validated by germ-free (GF) mice experiments where it was found that germ-free mice had 40% lower epididymal fat and an additional 10–30% food consumption was needed to maintain the same body mass as mice with normal microbiota (Backhed et al., 2004). Gut microbiota is important in fermenting unabsorbed starch and soluble dietary fiber. The fermented end products exist in the form of a SCFAs. SCFA (such as butyrate, propionate, acetate and pentanoate) act as one of the energy substrates for the host (Salminen et al., 1998) thereby contributing an extra 10% daily dietary energy for utilization by the host for other metabolic processes (Payne et al., 2012). Microbial-synthesized SCFAs contribute 70% of ATP production in colon, with butyrate as the preferred fuel for colonocytes (Firmansyah et al., 1989; Donohoe et al., 2011). Butyrate-producing microbes rescued the deficit mitochondrial respiration, ATP synthesis and autophagy in colonocytes of germ-free mice (Donohoe et al., 2011), proving the importance of butyrate in colonic cellular respiration and energy production. Furthermore, SCFAs which are the ligands for G protein-coupled receptor 41 (GPR41) expressed by a subset of the gut epithelial enteroendocrine cells, had been shown to regulate energy homeostasis by stimulating GPR41-mediated leptin production in mouse adipocytes, in which this multifunctional circulating hormone, leptin exhibits pleiotropic effects on a vast range of host physiological functions such as energy metabolism, appetite, as well as sympathetic nerve activity and immune response, potentially giving rise to interactive host-microbe signaling and gut-brain axis immune-inflammatory crosstalk (Xiong et al., 2004; Samuel et al., 2008).
Besides SCFA, gut microbiota-synthesized micronutrients such as vitamins exhibit beneficial value for both microbial and host metabolisms. Vitamin-K-producing gut bacteria namely Bacteroides fragilis, Eubacterium lentum, Enterobacter agglomerans, Serratia marcescens, and Enterococcus faecium (Fernandez and Collins, 1987; Cooke et al., 2006) anaerobically synthesize vitamin K2 (menaquinone) which is essential in decreasing vascular calcification, elevating HDL and lowering cholesterol levels, contributing to lower risk of cardiovascular disorders such as atherosclerosis and coronary heart disease (Kawashima et al., 1997; Geleijnse et al., 2004). Gut microbiota also serves as an important source of vitamins B for the host (Salminen et al., 1998; Degnan et al., 2014). Among them, vitamins B5 and B12, which are exclusively synthesized by intestinal microbiota, act as coenzyme for extensive range of host biochemical processes including production of acetylcholine and cortisol which are required for normal functioning of the nervous system. Deficiency of vitamins B5 and B12 have been linked to several disorders such as gastrointestinal discomfort, insomnia, neuropsychological and hematological disorders (Andres et al., 2004; Gominak, 2016). However, the possible link between loss of vitamin-producing gut microbiota and disease onset has not yet been elucidated.
Gut microbiota also plays an important role in the co-metabolism of bile acids with the host. These cholesterol derivatives are synthesized in the liver, followed by conjugation with taurine or glycine prior to storage in the gall bladder and subsequent secretion into duodenum to aid digestion, cholesterol and lipid metabolisms. In humans, 95% of bile acids are reabsorbed at distal ileum (Staels and Fonseca, 2009). The 5% unabsorbed primary bile acids are then bioconverted or deconjugated to secondary bile acids (predominantly DCA and LCA) by bile salt hydrolases secreted by several colonic microbiota such as Clostridium perfringens and Clostridium scindens, followed by partly colonic reabsorption and transportation back to liver for conjugation (Gopal-Srivastava and Hylemon, 1988; Ajouz et al., 2014) and the unabsorbed secondary bile acids are excreted by the host (Sorg and Sonenshein, 2008). Both primary and secondary bile acids are able to activate host nuclear FXR signaling, which in turn regulates the bile acids production, glucose metabolism, and potentially hepatic autophagy (Lee et al., 2014; Nie et al., 2015). Secondary bile acids serve as a potent activator of TGR5 (a member of the Rhodopsin-like subfamily of GPCRs) which is widely expressed on different tissues including in gall bladder, liver, spleen, intestine, and the immune cells. Such GPCR activation will stimulate second messenger c-AMP production, and the subsequent c-AMP-dependent downstream signaling, inducing expressions of a multitude of genes, whereby its physiological importance remains to be fully elucidated, pointing toward impacts of secondary bile acid-TCR5 axis (Hylemon et al., 2009). Secondary bile acids also possess antimicrobial effects which alter microbial cell membrane integrity, causing spillage of intracellular contents, thus inhibiting growth of bile acid-intolerant microbes (Nie et al., 2015). Such antimicrobial properties may contribute in shaping the composition of the gut microbiota and protecting the host from an array of infectious pathogens.
Immunity and the Nervous System
Gut barrier which is composed of mucus layer and epithelial layer (containing several junctional protein structures that regulate barrier integrity and paracellular permeability), serves as the interface between the outside world and host internal environment. Disrupted gut barrier function will increase gut permeability to commensal microbes, microbial derived products (such as metabolites, virulence factors) as well as other luminal components, contributing to aberrant immune-inflammatory response such as inflammation, allergy, and autoimmune disorder mediated by molecular mimicry and dysregulated T-cell response (Barnaba and Sinigaglia, 1997). Such physical and immunological barrier’s function is cross-regulated by host-gut microbiota interactions. The regulation of gastrointestinal T-lymphocytes balance [regulatory T cell/T helper type 17 (Treg/TH17) ratio], which is crucial in maintaining intestinal homeostasis, discriminating between pathogens and commensal microbes via organizing “immune tolerance-productive immune response” status, is found to be involving the role of gut microbiota. Several commensal microbes such as Bacteroides fragilis, Bifidobacterium infantis, and Firmicutes are capable in inducing expansion of Treg cells such as FOXP3 expressing Treg and anti-inflammatory IL-10-producing Treg lymphocytes, which are crucial in suppressing pathological inflammation induced by aberrant effector T cells, hence fortifying gut barrier function (Paust et al., 2004; El Aidy et al., 2012; Lawley and Walker, 2013). Suppression of inflammatory response by Treg cells also play a critical role in inducing host tolerance in non-host cells, possibly enabling gut microbiota to construct their niches in host without being attacked by host immunity under normal circumstances. Furthermore, several microbiota-derived metabolites such as SCFAs had been shown to confer protection to the gastrointestinal barrier integrity against the disruptive effects of proinflammatory cytokines resulted by aberrant immune-inflammatory axis (Peng et al., 2007; Chen et al., 2017), whereby the mechanism remains unclear. On the other hand, commensal SFB was shown to induce intestinal TH17 cells and production of pro-inflammatory IL-17 and IL-22, enhancing antimicrobial defense and mucosal immunity in against of intestinal pathogen Citrobacter rodentium in gnotobiotic mice model (Ivanov et al., 2009). Besides, according to Atarashi et al. (2008), ATP which can be contributed by gut microbiota, is capable to activate differentiation of lamina propria cells (CD70 and CD11c) into TH17. In short, tight regulation of Treg/TH17 balance by healthy host-gut microbiota interactions are critical to prevent aberrant immune-inflammatory response.
Participation of gut microbiota in host immunity development and immune response regulation at both local and systemic levels had been demonstrated, whereby reversal of the differentiation-suppressed states of myeloid and lymphoid progenitor cells in GF mice was observed after colonization with normal microbiota, indicating gut microbiota facilitates maturation of haematopoiesis (innate immunity) and lymphocytopoiesis (adaptive immunity) (Khosravi et al., 2014). Intriguingly, a recent study by Erny et al. (2015) had revealed a novel function of host microbiota and their SCFAs in regulating maturation and functionality of microglia, the tissue macrophage of CNS. Colonization of GF mice with complex gut microbiota had shown partial reversal in the initial immature microglia phenotype in GF mice. Moreover, mice model with normal microbiota but deficit SCFAs receptor had mimicked the defective feature of microglia observed in GF mice, thus, indicating an important role of microbial-synthesized SCFAs in microglia homeostasis which is crucial in CNS health maintenance (Nishioku et al., 2010).
On the other hand, the emerging modulatory role of gut microbiota in the ENS had been documented in various studies. The ENS is crucial for life and is capable in autonomously regulating the physiology and function of the GIT, and bidirectionally communicate with the CNS via vagal pathways, forming the “gut-brain axis.” The major component of ENS is the EGC which resembles astrocyte in the CNS. EGCs form the enteric glial network that critically regulates a variety of GI functions including exocrine/endocrine secretions, gut motility, blood flow, and immune-inflammatory reactions, via a complex repertoire of calcium-dependent signaling (Ochoa-Cortes et al., 2015). Dysfunctions in ENS and EGC have been implicated in various gastrointestinal disorders (such as IBD, IBS, postoperative ileus), motility disorder (such as constipation), neurodegenerative disorder (such as PD) and infection-induced gut inflammation. Given the close proximity of gut microbiota and the ENS that located throughout the GIT, it is not surprised that gut microbiota can affect and modulate the development and function of ENS. In a study by Collins et al. (2014), region-specific significant reduction of myenteric nerve fiber density was observed in jejunum and ileum, but not duodenum of early postnatal GF mice compared to the SPF mice and ASF colonized mice. In addition, notably decrease in gut motility of GF mice compared with SPF and ASF counterparts was observed as well, reflecting the importance of gut microbiota in postnatal development of ENS in mid-to-distal small intestine, however, the reason underlies the region-specific myenteric ganglia reduction remains unclear. Another study by Kabouridis et al. (2015) had demonstrated marked reduction in mean number and density of mucosal EGCs in 8-week old GF mice compared to the conventionally-raised (CONV-R) counterparts, indicating the requirement of gut microbiota in normal development of mucosal enteric glial network. The same study also showed that gut microbiota-dependent EGC normal development does not restricted to critical early postnatal period as conventionalization of 4-week old GF mice demonstrated restoration of enteric glial network.
Toll like receptors (TLRs) from the microorganism-sensing PRR family serve a critical role in maintaining gut microbiota-host symbiotic relationship and intestinal homeostasis (Rakoff-Nahoum et al., 2004). Expression of TLR4 that recognizes the LPS of Gram-negative microbes, can be found on intestinal neurons and glial cells, possibly conferring ENS the ability to react straight to the stimulus derived from gut microbiota. The observation of decreased gut motility and lesser nNOS (neuronal NO synthase) neurons had been demonstrated in GF and antibiotic-treated mice, whereby such deficits are reproduced in TLR4-knockout mice as well as TLR signal transducer MyD88-knockout mice (Anitha et al., 2012), suggesting TLRs signaling might be the mediator between intestinal microbiota and ENS development. Furthermore, Soret et al. (2010) had reported a noteworthy elevation in ChAT but not nNOS-immunoreactive neuron, as well as increased cholinergic-mediated colonic circular muscle contractile reaction, induced by intracecal perfusion of butyrate, hence indicating potential regulatory effect of butyrate in ENS and gut motility.
Colonization Resistance
Another important role of human microbiota is colonization resistance where indigenous microbiota confers protection to host against colonization of pathogenic invader and prevention against overgrowth of pathogenic microbiota members (pathobionts). Although the molecular basis of colonization resistance remains to be elucidated, the postulated mechanisms of actions can be classified into (1) direct interaction between human microbiota and pathogens in competing for shared niches and nutrients, and (2) exploitation or enhancement of host defense machinery by human microbiota to suppress pathogen (refer to Table 2).
Table 2.
Colonization resistance mechanisms employed by gut microbiota.
| Colonization resistance categories | Potential mechanisms involved in resisting colonization of pathogens | Reference |
|---|---|---|
| Direct interaction of gut microbiota with pathogenic invader or pathobiont |
|
Cherrington et al., 1991; Britton and Young, 2012; Kamada et al., 2013; McNally and Brown, 2015 |
| Gut microbiota-mediated stimulation or enhancement of host defense mechanisms |
|
Ivanov et al., 2009; Lawley and Walker, 2013; Yiu et al., 2017 |
Dominant non-pathogenic gut microbiota members play a central role in occupying the niche and suppressing the growth and colonization of pathogens. However, during gut microbiome perturbation, decrease in dominant microbiota members reduces the colonization resistance capacity, allowing opportunistic pathogenic strains to invade or colonize the empty niches, leading to occurrence of infection. C. difficile is a classic example of pathobiont which we have discussed below.
Gut Microbiome and Diseases
External factors (such as antibiotic consumption, dietary component, psychological and physical stress) and host factors can induce dysbiosis in gut microbiome. Dysbiosis is likely to impair the normal functioning of gut microbiota in maintaining host wellness, and potentially induce selective-enumeration of certain microbiota member including pathobionts, leading to dysregulated production of microbial-derived products or metabolites which might be harmful to the host, causing diverse range of diseases on local, systemic or remote organ (refer to Table 3), with some of the notable diseases, along with their respective microbiome-based therapy being discussed as below.
Table 3.
Gut microbiome-associated human diseases and their respective dysbiotic features.
| Disease categories | Specific diseases | Associated dysbiotic features | Reference |
|---|---|---|---|
| Immune-mediated/autoimmune diseases | inflammatory bowel disease (IBD) |
|
Sokol et al., 2008; Png et al., 2010; Willing et al., 2010; Machiels et al., 2014 |
| Irritable bowel syndrome (IBS) |
|
Duboc et al., 2012 | |
| Celiac disease |
|
Tjellström et al., 2005; Nadal et al., 2007; De Palma et al., 2010 | |
| Systemic lupus erythematosus (SLE) |
|
Shi et al., 2014; Luo et al., 2018 | |
| Type-1 diabetes |
|
Murri et al., 2013 | |
| Rheumatoid arthritis (RA) |
|
Liu et al., 2013; Scher et al., 2013 | |
| Atopic disease (E.g., childhood allergic asthma) |
|
Kalliomäki et al., 2001 | |
| Metabolic disorders/cardiovascular disorders | Obesity |
|
Turnbaugh et al., 2006, 2009a; Koliada et al., 2017 |
| Type-2 diabetes |
|
Qin et al., 2012; Sato et al., 2014 | |
| Hypertension |
|
Yang et al., 2015 | |
| Atherosclerosis |
|
Wiedermann et al., 1999; Koeth et al., 2013 | |
| Cancer | Colorectal cancer (CRC) |
|
Wang et al., 2012; Wu et al., 2013 |
| Neurop sy chi atri c | Autism spectrum disorder (ASD) |
|
Song et al., 2004; Adams et al., 2011 |
| Alzheimer’s disease |
|
Pistollato et al., 2016 | |
| Depression |
|
Kelly et al., 2016 | |
| Parkinson’s Disease |
|
Keshavarzian et al., 2015 | |
| Infectious disease | Clostridium difficile infection (CDI) |
|
Theriot et al., 2014 |
| Uremic disease | Chronic kidney disease |
|
Vaziri et al., 2013 |
Clostridium difficile Infection (CDI)
Clostridium difficile is a Gram-positive toxin- and spore-producing anaerobe. It is also one of the Firmicutes members in normal gut microbiota. Catalytic activity of C. difficile toxins A (TcdA) and B (TcdB) damages cytoskeleton and colonic epithelial barrier integrity, thereby inducing aberrant inflammatory response and cell death (Genth et al., 2006; Pruitt et al., 2012). C. difficile infection (CDI)-associated symptoms include diarrhea, pseudo-membranous colitis, sepsis and death in severe cases (Bartlett, 1982). In America, around 453,000 incidences of CDI with 453,000 mortality cases were observed in 2011 alone (Lessa et al., 2015). Intriguingly, antibiotic administration was found to be the major risk factor for CDI (Pear et al., 1994). Around 5–35% antibiotic-treated individuals developed diarrhea as a common side effect. Association of C. difficile with antibiotic-associated diarrhea is the most frequent, which accounts for 10–20% of total incidences, compared to other pathogens such as Staphylococcus aureus and Salmonella species (Song et al., 2008). It was observed that greater incidence of diarrhea was correlated with uptake of antibiotics with broad-spectrum antibacterial effect (Bartlett, 2002). Also, C. difficile acquisition of resistant genes toward broad spectrum clindamycin, erythromycin, chloramphenicol, and linezolid mediated by multiple horizontal gene transfer modes (potentially via mobilizable transposon, phage transduction, conjugative plasmids) within C. difficile strains and possibly among commensal microbes had been reported (Johanesen et al., 2015). A cohort study by Pépin et al. (2005) discovered that broad-spectrum fluoroquinolone appeared to be the most potent risk contributor to C. difficile-associated diarrhea compared to other antibiotics. Precise mechanism of this antibiotic-associated diarrhea remains unknown, however, its noteworthy correlation with CDI inspires research on the relationship between gut microbiome and its pathogenic member – C. difficile in healthy non-disease state.
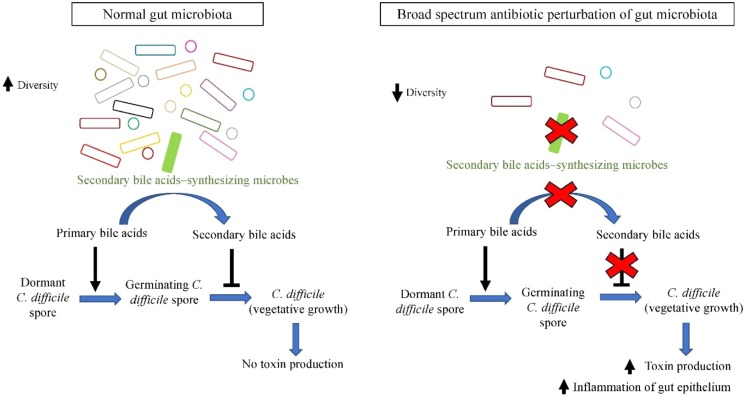
Currently it is postulated that dominant gut microbiota species confer protection to the host by employing colonization resistance mechanisms against overgrowth of C. difficile in normal microbiome. One of the proposed mechanisms is via the bio-conversion of primary bile acid to secondary bile acids. Primary bile acids (cholate derivatives) serves as germinant for C. difficile spores, whereas secondary bile acids (deoxycholate) inhibits vegetative growth of C. difficile (Sorg and Sonenshein, 2008). Antibiotic administration perturbs the gut microbial communities and reduces their diversity, especially secondary bile acids-synthesizing dominant microbes such as C. scindens (Antonopoulos et al., 2009). As a result, there is a significant reduction in microbial bioconversion of primary bile acids into antimicrobial secondary bile acids, leading to reduced inhibition of C. difficile vegetative growth, allowing C difficile outgrowth and colonization of the empty niches, leading to higher susceptibility of the host toward CDI (Samuel et al., 1973; Theriot et al., 2014), as illustrated in Figure 1. The increased toxin secretion by greater amount of vegetative C. difficile exerts greater damage on intestinal barrier, stimulating severe inflammatory response and causing impairment in intestinal ion absorption that leads to diarrhea.
FIGURE 1.
Secondary bile acids synthesized by members of gut microbiota (such as C. scindens) inhibit outgrowth of C. difficile, whilst broad spectrum antibiotic-induced dysbiosis favors C. difficile growth.
Improved understanding on antibiotic-induced microbiome dysbiosis in pathogenesis of CDI and its recurrence has given rise to development of new promising therapeutic approach that involves restoration of gut microbiota, such as FMT. This therapy has successfully restored gut homeostasis via re-introduction of gut microbiota from healthy donor feces. Patients received FMT showed long-lasting elevation of fecal microbial diversity and a high recovery rate (90%) compared to vancomycin (narrow-spectrum antibiotic) therapy (recovery rate = 60%) (van Nood et al., 2013). Besides, elevation in gut Bacteroidetes and Clostridium cluster IV and XIVa (Firmicutes), and reduction in Proteobacteria were observed post-FMT (van Nood et al., 2013), indicating the importance of dominant microbes such as Bacteroidetes and non-pathogenic Clostridia member in suppressing C. difficile outgrowth. Similar study by Konturek et al. (2016) also had demonstrated a high healing rate (94%) of CDI post FMT treatment, with no recurrent CDI observed across 16 months follow-up. Besides, marked reduction of proinflammatory cytokines (such as TNF-α, IL-1β, IL-6, IL-8, and IL-12), significant elevation in plasma level of human antimicrobial peptide LL-37, along with increase in beneficial bacteria (such as Lactobacillaceae, Ruminococcaceae, Desulfovibrionaceae, Sutterellaceae, and Porphyromonadaceae) were observed in successfully treated patients post FMT. Although the exact beneficial strains and the underlying mechanisms of FMT in conferring high cure rate of CDI have not yet been clarified, these studies undoubtedly demonstrate a strong association between gut microbiome and development of CDI, and shed a light on the potential widespread use of microbiota displacement therapy in targeting CDI.
Inflammatory Bowel Disease (IBD)
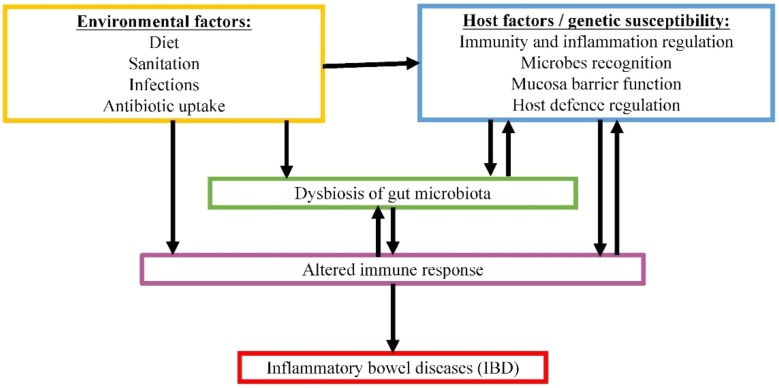
Another example of gut microbiome-associated disease is IBD. IBD is a group of multifactorial, idiopathic, persistent and recurring gastrointestinal inflammations. Two common forms of IBD are CD and UC (Lennard-Jones, 1989). In CD, inflammation can occur anywhere along the whole GIT, whereas UC is only restricted to the large intestine. Both forms are associated with relapsing diarrhea, fever and abdominal pain. The occurrence of IBD is rising at alarming rate worldwide, estimating 1.4 million and 2.2 million individuals in America and Europe, respectively (Loftus, 2004). IBD generally involves host factors combined with environmental factors (Franceschi et al., 1987; Gent et al., 1994; Hugot et al., 2001). Although our understanding on the mechanism of pathogenesis for this disease is still lacking, crosstalk between gut microbiota and host factors show great potential in contributing to the disease development, as demonstrated in Figure 2. The inappropriate host immune response against gastrointestinal microbiota in genetically predisposed individual is speculated to be the main culprit in causing severe inflammation.
FIGURE 2.
Proposed complex interactive relationships among gut microbiome, host factors and environmental factors in IBD pathogenesis.
One of the possible etiologies for IBD is due to the hyper-responsiveness of T-lymphocyte toward non-pathogenic antigens presented on gut microbiota. Several studies have observed the developments of antibodies against commensal microbial antigens and autoantigens such as anti-Saccharomyces cerevisiae, anti-OmpC, perinuclear anti-neutrophil cytoplasmic antibody and anti-Pseudomonas fluorescens-associated sequence 12 (Duchmann et al., 1999; Landers et al., 2002). Also, it has been reported that each of these antibodies response patterns highly correlates with distinctive clinical characteristics, disease onset and severity, suggesting loss of different microbiota species that would affect the gut barrier function and gut immunity, results in different degree of gut inflammation (Vasiliauskas et al., 2000). In short, selective loss of tolerance toward gut microbiota in IBD patient with aberrant immune response leads to dysbiosis and loss of microbiota that might be responsible in maintaining the gut mucus barrier integrity. Impaired barrier function subsequently increases the exposure between gut microbiota and epithelial cells, causing further stimulation of local immunity, contributing to severe gut inflammation. All the studies mentioned above clearly suggest that dysbiosis in gastrointestinal microbiome might be a secondary consequence of gastrointestinal inflammation.
Conversely, Nagao-Kitamoto et al. (2016) suggested that gut dysbiosis potentially contributes to IBD pathogenesis. In this study, elevated pro-inflammatory gene expression was observed in GF mice colonized by gut microbiota isolated from IBD patients. Colitis-prone genetically predisposed GF mice colonized by IBD-associated-microbiota developed severe colitis compared to those that were colonized by healthy human microbiota. Together these findings strongly indicate bidirectional relationship between such disease and gut dysbiosis, in which dysbiosis potentially contributes to the onset of IBD and also serves as a secondary consequence of gut inflammation. Example of the dysbiotic features that are commonly observed in IBD patients is the reduction in gut Firmicutes such as Faecalibacterium prausnitzii and Roseburia sp. (Sokol et al., 2008; Willing et al., 2010; Machiels et al., 2014). These bacteria play an important anti-inflammatory role in reducing pro-inflammatory cytokines (IL-12, IFN-γ) and increasing anti-inflammatory IL-10 (Sokol et al., 2008). Besides, Firmicutes is important producer of butyrate, the primary energy substrates for colonocytes. Therefore, reduction in Firmicutes could elicit or heighten local inflammation by decreasing anti-inflammatory cytokine, an important regulator of mucosal immunity, and/or by SCFA-deficiency-induced impairment in colonic barrier function (Peng et al., 2007; Machiels et al., 2014). As such, it may be interesting to explore the therapeutic use of probiotic F. prausnitzii in managing IBD. Another dysbiotic feature observed in IBD patients is the elevation of virulent gut microbes such as Enterobacteriaceae species and Bacteroides fragilis with both having high endotoxic LPS in their outer membranes (Ruseler-van Embden and Both-Patoir, 1983; Darfeuille-Michaud et al., 1998). High endotoxic LPS expressed by microbiota has shown to induce gut inflammation and colitis development in mice, possibly via suppression of regulatory T-lymphocyte and/or activation of effector helper-T (TH-1/TH-17) through host TLR4 signaling pathway (Gronbach et al., 2014). Inversely, colonization of GF mice with low endotoxic microbiota prevented experimentally induced colitis (Gronbach et al., 2014), indicating that an increase in gut microbes with high endotoxic LPS and decrease in microbes with low endotoxic LPS in gut microbiome may lead to pathogenesis of IBD.
Another suggestion regarding the IBD-gut microbiome link is the initial impairment in gut mucus barrier function (either due to dysbiosis or other factors), resulting in elevation of mucus-eating (mucolytic) gut microbial species, which in turn aggravates the barrier function and stimulates severe inflammatory response. Gastrointestinal mucus layer and antimicrobial peptides (such as human defensins) secreted from epithelium work together as a barrier in preventing direct contact of luminal gut microbiota with gastrointestinal epithelial cells and inhibit aberrant inflammation (Johansson et al., 2008; Salzman et al., 2010). Colon biopsy samples of patients with UC showed reduced mucus layer, defective epithelium and deep microbe- and leukocyte-infiltration in colon mucosa (Swidsinski et al., 2007). Moreover, reduction in antimicrobial peptides such as Paneth cells alpha-defensins in patients with ileal CD was reported (Wehkamp et al., 2005). These findings propose that the impaired host antimicrobial defense in mucus barrier reduces the initial suppression of certain commensal microbiota such as mucolytic bacteria. This is further supported by Png et al. (2010) where they observed a significant elevation of mucolytic Ruminococcus sp. in gut microbiome of IBDs patients. Decrease in microbial growth inhibition by human defensins could enable outgrowth of mucolytic species, resulting in increased mucolytic species that feast on the mucus glycans, leading to deeper penetration of the inner mucosa layer and microbial infiltration, consequently, inflammation ensues.
Since IBD involves heightened host inflammatory response, current therapeutic approaches generally target the aberrant pro-inflammatory immune response at intestinal mucosa. Anti-inflammation therapies such as antibodies targeting proinflammatory cytokines (anti-TNF-alpha, anti-IL12, anti-IL23) and employing α4β7-integrin antagonist to inhibit trafficking of T-lymphocytes to gut tissue (Feagan et al., 2013; Christensen et al., 2015). However, IBD tends to recur in long-term, and chronic treatment with immuno-suppressive agents will develop neurotoxic side effects (Hyde et al., 1998; Croft et al., 2013). Therefore, development of a safer and more effective novel treatment is in great need. Given the association of IBD patients with decreased anti-inflammatory cytokines IL-10, administration of genetically modified probiotic (beneficial microorganism) such as Lactococcus lactis to express IL-10 had demonstrated a significant remission of disease activity in CD patients during trial (Braat et al., 2006). In this study, 80% CD patients demonstrated clinical improvements, with 50% showing complete remission, and no severe side effect was observed, thus, indicating a promising and safer maintenance treatment strategy for chronic intestinal disease, as compared to sole administration of inflammatory cytokines. However, 40% patients developed CD relapse upon discontinuation of such therapy, indicating the need of long-term prescription of this medication, which in turn will impact its economic viability. Besides, administration of prebiotics (fermentable polysaccharides) to stimulate growth and metabolic activity of beneficial protective gut microbes such as butyrate-synthesizing bacteria might be an attractive concept to treat gastrointestinal inflammation in IBD (Joossens et al., 2012; De Preter et al., 2013). Positive outcome had been reported in a randomized controlled trial by De Preter et al. (2013), whereby around 40–50% of CD patients with mild to moderately active disease achieved remission post treatment with prebiotic OF-IN. OF-IN intake induced a significant reduction in mucolytic Ruminococcus gnavus and a marked elevation of beneficial Bifidobacterium longum, which the latter was strongly correlated with the clinical improvement in CD disease activity (Joossens et al., 2012). This may due to the anti-oxidant and anti-inflammatory effects of B. longum in neutralizing reactive oxygen/nitrogen species (ROS and RNS) at site of inflammation, reducing gastrointestinal discomfort and tissue injury. Despite these exciting novel approaches in treating or managing IBD, the elusive etiology of IBD in each patient renders individual-specific variation in therapeutic response, posing a great challenge in designing a curative treatment that is effective to all.
Celiac Disease
Furthermore, evidence has pointed to dysbiosis as the possible risk factor of autoimmune disorders, such as celiac disease. Other examples of gut microbiome-associated autoimmune disorders such as Type 1 diabetes, SLE and RA are depicted in Table 3. Celiac disease is a multifactorial chronic immune-mediated disorder in small intestine characterized by permanent intolerance to dietary gluten (such as gliadin peptides) and prolamines in genetically predisposed individuals expressing human leukocyte antigen (HLA)-DQ2 and/or HLA-DQ8 (Sanz et al., 2011). Celiac disease serves as one of the most prevalent lifelong disease in Europe, affecting 1% of the general population of all ages (Mustalahti et al., 2010), contributing to absolute mortality rate of 10.4 per 1000 person-years in celiac disease (Ludvigsson et al., 2009). In celiac patients, gluten triggers the activation of adaptive immune response in mucosa, inducing TH1 and TH17 mediated production of pro-inflammatory cytokines [for example IL-21, interferon (IFN)-gamma, TNF-alpha]; as well as innate immune response through stimulation of IL-15 synthesis, consequently leading to NKG2D-mediated enterocyte killing (Stepniak and Koning, 2006). Gliadin-mediated zonulin (a type of tight junction protein) signaling activation also had been reported, whereby it causes cytoskeleton rearrangement, reduced occludin – ZO-1 protein–protein interaction and increased gut permeability, allowing trafficking of luminal antigens to the submucosa (Drago et al., 2006). The etiology of celiac disease remains to be fully elucidated, with several factors including genetic predisposition, immunological factors (mucosal IgA), and environmental factors including gluten consumption, breast-feeding duration as well as gut infections (for example rotavirus infection), have been postulated to serve a concerted effort in contributing the risk and timing of celiac disease onset (Akobeng et al., 2006; Mulder and Mulder-Bos, 2006; Stene et al., 2006). These factors also play a critical role in shaping and altering gut microbiota composition, therefore potentially modulating gut microbiota roles in gut barrier function and immunity development.
Gut dysbiosis has been largely reported in celiac disease patients, in which the active phase of celiac disease is characterized by a remarkable decrease in Gram-positive bacteria, whereby this bacterial population is partly restored in patients that receive gluten-free diet, suggesting the initial reduction of the recovered species might be the secondary consequences of the disease. Besides, inconsistent observation encompasses of increase, decrease and indifferent abundance changes in commensal Atopobium, Eubacterium rectale–Clostridium coccoides, Clostridium histolyticum, Clostridium lituseburense, Faecalibacterium prausnitzii, Escherichia coli, Lactobacillus-Enterococcus, and Staphylococcus in celiac patients across different studies had been demonstrated (Nadal et al., 2007; De Palma et al., 2010; Marasco et al., 2016), suspecting existence of confounding factors (such as disease severity, disease duration, diets, age and sex-related factors of the experiment subject, and the experimental settings and sample types) which remain elusive. Despite that, a significant decrease in total Gram-positive to Gram-negative bacteria ratio was observed in all phases of celiac disease patients as compared to that of healthy controls, with marked reduction in “health-promoting” Bifidobacteria, and elevation in virulent Gram-negative Bacteroides–Prevotella groups being universally observed in multiple studies, reflecting possible pivotal role of Bifidobacteria and Bacteroides–prevotella in celiac disease (Nadal et al., 2007; De Palma et al., 2010; Marasco et al., 2016). Together, it is suggested that dysbiosis potentially plays both secondary and primary roles in celiac disease pathogenesis. Although such dysbiotic feature and its mechanistic link with celiac disease manifestation remain elusive, it is suspected that overgrowth of these pro-inflammatory endotoxin-secreting Gram-negative pathobionts may contribute to the gluten intolerance as well as the onset and/or progression of celiac disease, through the enhancement of the pro-inflammatory responses such as increased IFN-gamma and TNF-alpha production, which will impair gut barrier integrity and normal functioning. The resulting increased gut permeability could favor the infiltration of more luminal antigens (gluten and microbial products) to the submucosa, leading to amplification or perpetuation of inflammatory reactions. Intriguingly, despite the notable elevation in Bacteroides–Prevotella proportions, mucosal first line of defense mediated by secretory IgA was shown to be weaker in celiac disease patients, in which the IgA-coated Bacteroides–Prevotella was significantly lower than that of healthy control (De Palma et al., 2010). This finding postulates the possible defect in mucosal barrier of celiac disease patient which fails to stabilize gut microbiota, and loses its protective effect against invasion of noxious antigens and pathogens.
Another possible role of gut microbiota in contributing to celiac disease is their capability in synthesizing mTgs, which are the homologs of human-synthesized tissue transglutaminases (tTgs). The mTg gene, which is potentially associated with microbial survivability, is encoded in the genome of many commensal bacteria, with vast majority belongs to Firmicutes phylum. Transglutaminase, which is also the autoantigen in celiac disease, is a protein cross-linker in which it transamidates or deamidates gliadin peptides to form stable neo-compounds in a process called PTMP (Lerner et al., 2017a). The Tg-linked proteins are found to be autoimmunogenic in celiac disease patients, whereby induction of anti-tTG autoantibodies serves as a remarkable serological marker of celiac disease. Similar observation toward its microbial counterpart also had been documented, in which specific anti-mTg and mTg-gliadin neo complex antibodies were found in the serum of celiac disease patients but absent in healthy controls (Lerner and Matthias, 2015). Furthermore, mTgs which are extensively used as industrial food additives is capable in enhancing leakage of the epithelial tight junction, which is the major component of gut epithelial barrier in regulating the equilibrium between immune tolerance and immune response to non-self antigens. Tight junction dysfunction and “leaky gut” are commonly observed in autoimmune diseases including celiac disease, as the resulting increased gut permeability promotes entry of luminal immunogenic antigens, eliciting both local and systemic immune reactions, and the pathological autoimmune signaling cascade (Lerner and Matthias, 2015). Together, these findings speculate the endogenous mTgs synthesized by dysbiotic gut microbiota (especially dysregulation in Firmicutes/Bacteroidetes ratio) potentially abrogate the tight junction barrier integrity and drive autoimmunity through the intestinal luminal PTMP, in which the additional dietary source of mTgs may potentiate or exacerbate such deleterious effects.
Besides the aforementioned mTgs-mediated PTMP of dietary gluten by Firmicutes, PTMP-catalyzing ability had been reported in other commensal microbes, including those potentially virulent strains associated in the dysbiotic feature of celiac disease. Bacteroides fragilis of Bacteroidetes phylum was found to encode protein-modifier ubiquitin, suggesting potential aberrant ubiquitin especially in a dysbiotic configuration (such as over-representation of Bacteroides in celiac disease) might cross-react and interfere human ubiquitin activity, and its downstream signaling, as well as induce inappropriate activation of host immune (Patrick et al., 2011). Gram-negative bacteria, especially those pathogenic strains exhibit secretion systems to synthesize effector proteins with several enzymatic properties to manipulate host cellular signaling, for example via modification of host phosphoproteome to facilitate microbial survival and niche establishment in host (Grishin et al., 2015). Linking the dysbiotic features, microbial-mediated PTMP activities with the aberrant immune-inflammatory response in celiac disease, it is hypothesized that dysbiosis-mediated dysregulated PTMP activities causing both functional and conformational transformations of dietary and host proteins, generating novel aberrant epitopes which are potentially immunogenic and capable to interfere critical host cellular signaling, hence, triggering manifestation of autoimmune disease such as celiac disease (Lerner et al., 2016).
Alteration in SCFAs compositions and the SCFA total abundance in celiac children were also been documented by Tjellström et al. (2005). Significant increase in acetic, i-butyrate, i-valeric acid and the total SCFAs level were observed in celiac disease patients compared to that of healthy controls. Intriguingly, no significant difference of SCFAs composition and the total SCFAs was observed in the untreated celiac patient and gluten-free diet treated celiac patient, indicating the altered gut microbiota, particularly those SCFA-producing commensal microbes that responsible for such SCFAs composition profiles in celiac patients, might be long established in the genetic predisposed host before the onset of the disease, rather than a mere secondary consequence of celiac disease that can be corrected by eliminating gluten the “trigger” of celiac disease symptoms. However, the unknown bacterial origin in contributing such SCFA patterns in celiac patient renders a missing link between SCFA and the aforementioned key dysbiotic microbial communities.
Gluten-free diet has been largely employed to ameliorate the deleterious disease symptoms in celiac patient. However, gluten-free diet exhibits therapeutic limitation which it allows only a partial recovery of gut microbiota in celiac disease patients, both microbiologically (for example reduction in certain potentially pathogenic strains, incomplete restoration of beneficial Bifidobacteria) and metabolically (for example indifferent SCFA pattern in treated and non-treated patient), possibly due to the genetic factor of the patient, or the selective-enumeration of gut microbiota mediated by the absence of dietary gluten, which the underlying reason remains unclear. Strict adherence to total gluten-free diet appears challenging to the patient as well due to the vast application of gluten in processed food, hence alternative measures using microbiome-based therapy such as probiotic might be a promising option to manage celiac disease. Several in vivo, animal studies as well as human studies had demonstrated various beneficial effects of probiotics Bifidobacterium and Lactobacillus strains in ameliorating epithelial-gliadin induced deleterious effects. For example, Bifidobacterium lactis exerts inhibitory effect on the increased gut permeability induced by gliadin, and it protects the healthy curvy appearance of epithelial tight junctions from the unfavorable gliadin-induced “straightening” effect on tight junction in human colon Caco-2 cells (Lindfors et al., 2008). B. longum was shown to be capable in enhancing anti-inflammatory response by stimulating markedly high Treg cell mediated production of IL-10, suppressing pro-inflammatory TH1 cytokine IFN-gamma induced by fecal microbiota of celiac patient in a cell culture-based study (Medina et al., 2008). B. longum and Lactobacillus casei also have been reported to exert protective effect on animal model from gliadin-induced enteropathy, whereby B. longum can downregulate the production of pro-inflammatory TNF-alpha and reduce CD4+ T-cell mediated immune reactions (Laparra et al., 2012); whilst L. casei can induce complete recovery of gliadin-induced villus blunting, recover the basal TNF-alpha level, rescue homeostasis of GALT (gut-associated lymphoid tissue) (D’arienzo et al., 2011). Administration of probiotics Bifidobacterium in children with newly diagnosed celiac disease together with gluten-free diet had significantly reduced the potentially harmful Bacteroides fragilis abundance, decreased peripheral CD3+ T cells, and improved the disease symptoms (Olivares et al., 2014). Together, despite complete disease remission is not observed thus far, these studies demonstrate great therapeutic value of probiotics Bifidobacterium and Lactobacillus in ameliorating gliadin-induced toxic effects and improving celiac disease symptoms, indicating promising approaches in managing celiac disease.
Obesity
In recent years, the newly identified factor – gut microbiome which is largely involved in host metabolism regulation, has been integrated into crosstalk studies between genetic factors, behavior and environmental factors as a possible contributor to obesity. Obesity is a global health hazard affecting more than 600 million people worldwide in 2014 (World Health Organization [WHO], 2016). It is associated with elevated energy intake and decreased energy expenditure, causing excessive fat accumulation with raised body mass index (BMI ≥ 30kg/m2), and is linked to metabolic syndrome, posing obese individuals to have a higher risk of developing obesity-associated disorders (for example cardiovascular disease, type-II diabetes, and liver abnormalities), low-grade inflammation, and premature mortality (Donnelly et al., 2005; Cani et al., 2007).
Metagenomics studies had discovered a significant increase in butyrate-producing Firmicutes and generally a decrease in Bacteroidetes were observed in distal colonic microbiome of obese patients and genetically obese mice, compared to their normal lean counterparts. These obesity-associated dysbiosis features were also accompanied by elevation in starch-degrading glycoside hydrolase and SCFAs (butyrate and acetate), and increased energy harvest capability as evidenced by markedly decreased fecal energy in obese mice (Ley et al., 2006; Turnbaugh et al., 2006), speculating significant elevation in host metabolism-related microbial communities would abnormally increase energy harvest, thus increasing the risk of developing obesity. This speculation is supported by gnotobiotic mouse model, whereby noteworthy elevations in body fat, fasting glucose and insulin levels, and development of insulin resistance that mimic physiological alterations in obesity patients, have been observed in GF mice post-colonization with a single saccharolytic gut bacterial species (Bacteroides thetaiotaomicron) and with gut microbiota from obese mice respectively (Backhed et al., 2004; Turnbaugh et al., 2006). Elevations in monosaccharides delivered to liver and liver triglycerides level due to enhanced starch metabolism facilitated by gut microbiota indicate a strong association between gut microbiota and host metabolism in glucose homeostasis and lipogenesis. These findings are consistent with previous studies where lipogenic substrates (such as monosaccharides and SCFAs) were able to trigger the expression of hepatic fatty acids-synthesizing enzymes like ACC-1 and FAS, promoting hepatic DNL (Higuchi et al., 2008). The subsequent storing of triacyl glycerides in adipocytes also has been shown to be possibly mediated by gut microbiota via suppressing of lipoprotein lipase-inhibitors (Backhed et al., 2004; Solinas et al., 2006). Together, this data suggests that over-representation of saccharolytic gut microbiota facilitates augmentation in food digestion, leading to higher energy harvest and increased fat deposition, thus contributing to obesity development.
Moreover, several studies suggest that increase in endotoxic LPS of Gram-negative gut bacteria contributes to obesity-associated metabolic syndrome. For instance, it was reported that elevated LPS led to obesity-associated insulin resistance and low-grade inflammation as demonstrated by Cani et al. (2007). Similarly, plasma LPS (metabolic endotoxemia) showed a marked increase in high-fat-fed mice, along with reduction of Bifidobacteria, a potential down-regulator of intestinal endotoxin (Griffiths et al., 2004). In investigation of the association between metabolic endotoxemia with obesity-associated metabolic disorders, continuous-LPS-infused mice were shown to develop fasting glycaemia, insulinemia, hepatic insulin resistance, increased hepatic and whole-body fat gains, similar to high-fat-fed mice phenotypes (Cani et al., 2007). In addition, absence of LPS-receptor resisted these adverse features regardless of LPS-infusion or high-fat diet treatments as shown in CD14 (LPS-receptor) knockout mice, indicating that the LPS/CD14 system mediates insulin insensitivity, thus inducing the onset of obesity and obesity-related metabolic disorders (Cani et al., 2007).
Since obesity-related metabolic disorder has been associated with gut microbiota dysbiosis, selective modulation of microbial composition via dietary intervention such as administration of prebiotics or probiotics might be a promising therapeutic approach. Administration of bacterial-synthesized CLA or CLA-producing bacteria such as Lactobacillus rhamnosus has shown a significant decrease in plasma cholesterol, triacyl glycerides and white adipose tissue in animal models (Koba et al., 2002; Lee H.Y. et al., 2006). Prebiotics such as inulin-type fructans that selectively nourish Roseburia and Clostridium cluster XIVa, and arabinoxylan that increases the abundance of Bifidobacterium, Roseburia, and Bacteroides, have demonstrated anti-adipogenic effect in high-fat-induced obese mice (Dewulf et al., 2011; Neyrinck et al., 2011). These findings demonstrate promising application of prebiotics and probiotics in obesity management, however, more supportive data from human models and clinical trials are required to justify these treatment strategies and their respective success rates.
Colorectal Cancer (CRC)
Colorectal cancer (CRC) is the fourth leading cause of cancer-related mortality worldwide (Arnold et al., 2017). Similar to other cancer forms, CRC is a multifaceted diseases associated with genetic and environmental factors. Recent findings have suggested that gut microbiota plays a role in the intersection of these factors, probably through shaping a tumor-promoting environment. Colorectal carcinogenesis of the gut has been demonstrated by Li et al. (2012) using mouse model of APC-related CRC. In this study, GF mice displayed a significantly lower colonic tumor incidence and reduced tumor load as compared to that of conventionally raised mice, which the latter also exhibited other distinctive phenotypes of CRC such as rectal bleeding and anemia with a massive infiltration of inflammatory cells arising from a dysfunctional intestinal epithelial barrier at older ages, thereby suggesting gut microbiome and host factor (such as age and genetic predisposition) exert a concerted effort in contributing to CRC growth and progression. Conversely, tumor outgrowth into gut lumen would damage the intestinal barrier, resulting in increased infiltration of gut microbiota and potentially harmful luminal substrates into deeper tissue, leading to further stimulation of immune-inflammatory response and myeloid cell recruitment, which could in turn perturb the gut microbiome (Li et al., 2012). Together, gut microbiome and CRC display a bidirectional self-feeding relationship.
16S rRNA sequencing study of fecal microbiota of CRC patients has revealed a significant enrichment of Bacteroides fragilis (Wang et al., 2012; Wu et al., 2013). Despite the low detection limit of some metagenomics studies in providing strain-level information, it is suspected that possible bloom of enterotoxigenic strains of B. fragilis (ETBF) contributes to CRC through enhanced production of oncogenic B. fragilis enterotoxin (BFT). This speculation is consistent with the findings of Ulger Toprak et al. (2006) in which the bft gene expression was significantly higher in CRC patients than healthy controls. Wu et al. (2009) had demonstrated that colonization of tumor-prone mice with ETBF strains can trigger TH17-dependent colon tumorigenesis, possibly facilitated by its major virulence factor – BFT. BFT is cytopathic to intestinal epithelial cells and can cause colitis and colonic tumor, owing to its capability of catalyzing proteolytic degradation of tight-junction proteins ZO-1 and E-cadherin, leading to disruption of intestinal paracellular barriers (Moncrief et al., 1998; Wu et al., 1998). Moreover, BFT-mediated loss of membrane-associated E-cadherin could in turn activate T-cell factor–dependent Wnt (Wingless-Int)/β-catenin nuclear signaling in intestinal epithelial cells, stimulating oncogene c-Myc expression that results in persistent cellular proliferation (Wu et al., 2003). Furthermore, combined action of BFT and IL-17 on colonic epithelial cell can induce myeloid differentiation into pro-tumoral monocytic-myeloid-derived suppressor cells (MO-MDSCs), leading to selective upregulation of Arg1 and NO synthase 2 (Nos2) thereby stimulating NO synthesis that could promote vascular endothelial growth factor (VEGF)-mediated tumor angiogenesis (Cianchi et al., 2003), and suppression of anti-tumor immunity (e.g., inhibit CD8+ T-cell proliferation) (Orberg et al., 2017).
On the other hand, non-colitogenic Fusobacterium nucleatum which is markedly enriched in CRC patients, is capable in inducing intestinal tumorigenesis in murine models, suggesting overrepresentation of the gut microbiota may be the driver for CRC that is non-associated with colitis or IBD (Castellarin et al., 2012; Wang et al., 2012; Wu et al., 2013). Although the causality and underlying mechanisms remain to be elucidated, it has been demonstrated that FadA adhesin of F. nucleatum could promote bacterial attachment and invasion into E-cadherin expressing CRC cells, subsequently activating β-catenin-regulated transcriptions, as shown by upregulation of lymphoid enhancer factor/T cell factor, host NF-κB, oncogene c-Myc and cyclin D1, consequently promoting CRC cell proliferation and inflammation (Rubinstein et al., 2013). Besides, F. nucleatum outer membrane protein, Fap2 also has been shown to mediate specific Fusobacterial attachment to Gal-GalNAc overexpressed in CRC, facilitating F. nucleatum invasion and colonization of CRC cells (Abed et al., 2016). In addition, involvement of TLR4/phosphorylated-p21-activated kinase 1/phosphorylated-β-catenin S675 cascade was found to be employed by invasive F. nucleatum as well (Wu et al., 2018). Briefly, both oncogenic ETBF and F. nucleatum along with their respective unique molecular signatures serve as potential biomarkers in identifying the risk of developing ETBF- and F. nucleatum-related CRC, and predicting malignancy progression. Antibiotics specifically targeting these pathobionts as well as the application of pharmacological inhibitors for pathogenic signaling pathways may be a promising approach to be ventured in. Further longitudinal studies are therefore recommended to assess the feasibility of such applications.
Microbial metabolites such as butyrate have been reported to inhibit proliferation and survival of tumor cells as evidenced by butyrate-induced upregulation of pro-apoptotic Bax, Fas, cell cycle checkpoint regulator p21 and p27, downregulation of anti-apoptotic Bcl-2, Bcl-xL, cyclin D1, and possible inhibition of LPS-induced activation of pro-inflammatory and pro-tumorigenic NF-κB signaling, via intracellular action of butyrate as HDAC inhibitor that could epigenetically regulate gene expression, and/or its extracellular action as a ligand that targets G-protein coupled receptor GPR109A-expressing cancer cell (Hassig et al., 1997; Thangaraju et al., 2009; Wei et al., 2016). Butyrate also exhibits chemoprotective effects in which colonocytes pre-treated with butyrate demonstrate higher resistance against hydrogen peroxide-induced oxidative stress and DNA damage, possibly via butyrate-mediated enhanced expression of GST M2 in colonocytes, which is a detoxifying isoenzyme specific for substrates that arise from oxidative stress, thereby reducing the impact of certain genotoxic CRC risk factors (Abrahamse et al., 1999; Ebert et al., 2003). Together with the observation of significantly reduced butyrate-producers Faecalibacterium and Roseburia in gut microbiota of CRC patients (Wang et al., 2012; Wu et al., 2013), it is speculated that the subsequent reduction in colonic butyrate could partly impair the anticancer immunosurveillance and potentiate tumorigenesis. Hence, the concept of increasing colonic butyrate either by supplementation of butyrogenic dietary fermentable carbohydrates (high fiber diet) or butyrate-producing probiotics exhibits great intervention potential on the prevention and therapy of CRC, whereby further studies are required to evaluate the therapeutic success rate, as well as to determine the effective concentration of butyrate in vivo, required to attain desirable clinical benefit. In a study by Femia et al. (2002), anti-tumorigenic activity of prebiotic OF-IN on azoxymethane-induced colon cancer rat model was demonstrated, as evidenced by significant reduction of tumor loads (adenoma and cancer) compared to control rat. However, serum metabolome such as butyrate level and the gut microbiota compositional change post-prebiotic treatment were not determined in this study. This makes it difficult to decipher which key microbiota and microbial products actually confer such anti-tumorigenic effect.
Accumulating data has also demonstrated protective effect of probiotics using lactic acid-producing bacteria (Lactobacillus and Bifidobacterium) against experimentally induced colon cancer, as shown by reduced tumor incidence and tumor loads. The underlying mechanisms are suggested to involve direct anti-proliferative effect on tumor cells, prevention of carcinogen-induced DNA damage, partly via inhibition of carcinogen and mutagen formation, reduction of pro-carcinogenic enzyme (ornithine decarboxylase) activity, and elevation of detoxifying enzyme activity, as well as enhancement of anti-tumor immunity as evidenced by increased natural killer cells, MHC class II antigen presenting cells, and CD4–CD8+ T cells post-probiotic treatment (Goldin et al., 1996; Pool-Zobel et al., 1996; Singh et al., 1997; Wollowski et al., 1999; Lee et al., 2004). Moreover, the gut microbiome is pivotal in influencing therapeutic efficacy of anti-cancer immunotherapy employing immune-checkpoint inhibitors. The anti-tumor effect of antibodies targeting CTLA-4, a major negative regulator of T-cell activation, is compromised in both GF mice and broad-spectrum antibiotics-treated mice, confirmed in both melanoma and colon cancer models, along with the observations of significantly reduced CD4+ T-cell and tumor-infiltrating lymphocytes. Such defects can be overcome by the administration of non-enterotoxin-producing strains of B. fragilis, immunization with immunostimulatory B. fragilis polysaccharide, or by adoptive transfer of B. fragilis-specific T-cells, in which the mechanisms underlie the restoration of therapeutic response to CTLA-4 blockade is suggested to be via induction of the IL-12-dependent TH1 immune response in tumor-draining lymph nodes, and stimulation of intratumoral dendritic cells (DCs) maturation (Vétizou et al., 2015). Similarly, Sivan et al. (2015) reported that Bifidobacterium could facilitate PD-L1 blockade efficacy via augmenting DCs function, enhancing CD8+ T-cell priming and accumulation at the tumor microenvironment level. The concept of combination treatment that encompasses anticancer immunotherapy and commensal microbes holds great promise for future CRC treatment.
Autism Spectrum Disorder (ASD)
In recent years, increasing number of studies have explored a possible link between the gut microbiome and brain- or neuro-developmental disorders, such as ASD, with symptoms like mood disorder (depression), neurodegenerative Alzheimer’s and PDs (Table 3). ASD is a multifarious group of neurobiological disorders affecting 1 in 68, 8-year-old children in America (Christensen et al., 2016) and 1 in 160 children globally (World Health Organization [WHO], 2017). It is characterized by social and communication deficits, and repetitive, stereotyped behaviors (Constantino et al., 2000; Kim and Lord, 2012). Despite the poorly understood causative factor of ASD, several studies had observed a significant association between ASD, gastrointestinal dysfunction and dysbiosis in gut microbiota. Comorbidity analysis of ASD patients revealed high occurrence of gastrointestinal symptoms, with the severity positively associated with that of ASD (Wang L.W. et al., 2011). This co-occurrence is speculated to be implicated by impaired tyrosine kinase MET signaling which is important in brain development, gastrointestinal health and immune response regulation (Ieraci et al., 2002; Okunishi et al., 2005). This hypothesis is supported by the observation of impaired MET-signaling in ASD patients especially those that comorbid with familial gastrointestinal disorder (Campbell et al., 2009; Jackson et al., 2009). Also, clinical data has shown that physical health problems like chronic abdominal pain caused by gastrointestinal disorders aggravate self-injury and tantrum in ASD patients (Carr and Owen-Deschryver, 2007), thereby suggesting that treating the gastrointestinal dysfunction will possibly alleviate such ASD-associated symptoms.
During the investigation of microbiota composition in ASD patients, dysbiotic features such as increased Clostridium sp., Bacteroidetes, Lactobacillus, Desulfovibrio, and reduced Bifidobacteria were reported (Song et al., 2004; Adams et al., 2011), suggesting a possible link between dysbiosis and ASD. In a study by Shultz et al. (2015), mice treated with PPA via intra-cerebroventricular injection had shown to develop brain abnormalities and ASD-like symptoms. PPA is a type of SCFA, synthesized by enteric bacteria include the aforementioned Clostridium, Bacteroidetes, and Desulfovibrio species, but PPA level usually exist at low levels in normal healthy individuals. This suggests that possible bloom in PPA-producing gut microbes leads to elevated neurotoxic PPA concentration in blood, contributing to neuropsychiatric disorders. However, to date, dysbiosis-associated elevation in microbial-synthesized PPA in plasma of ASD patients has not been reported. Future studies in examining plasma PPA level in ASD patients are required. Surprisingly, treatment targeting gut microbiome has demonstrated beneficial effects in alleviating ASD-like symptoms in murine model. Desbonnet et al. (2010) reported that probiotic Bifidobacterium infantis improved the behavioral abnormalities and stress-associated gastrointestinal dysfunction which were initially induced in mice suffering MS. Reduction in Bifidobacterium is commonly observed in ASD patients, positive outcome post-probiotic treatment indicates a potential protective role of Bifidobacterium in neuro-immune aspects, suggesting probiotic B. infantis as a potential ASD therapy. Despite the unclear underlying mechanisms, antioxidant properties of Bifidobacterium are suggested to have a neutralizing neurotoxic effect, induced by excess NO in ASD (Yui et al., 2016). Further studies that mimic the human physiological environment are very much required to assess the therapeutic success and underlying mechanism of probiotic B. infantis in managing ASD.
Perspectives and Future Direction
Emerging research on human microbiome and its role in human wellness and disease are currently making it to the limelight, by virtue of which, researchers of diverse clinical specialty areas are attempting to fit this potential “missing piece of puzzle” into existing disease models, particularly those with unknown etiology. Tremendous data has demonstrated strong association of the gut microbiome with host metabolism, immune and neuroendocrine homeostasis, and the possible dysregulation or alteration of gut microbiome. Perturbation of such delicate equilibrium will in turn contribute to disease manifestation. However, the causal or correlation link remains debatable due to lack of direct evidence and mechanistic details. Our understanding on gut microbiome is still at a very preliminary stage, whereby there are several limitations and research gaps that are worth further exploration to better justify the link. For instance, large-scale longitudinal studies that account for subject-specific variations in characterizing the human microbiome are strongly required. On the other hand, another important component of human microbiome – the human virome that is dominated by bacteriophage (viruses that infect bacteria), is relatively less established compared to that of commensal bacteriome research. Only recently the presence of bacteriophage (or phage) in the human microbiome has been highlighted. Their potential role in modulating immune homeostasis has been suggested via (1) indirect mechanisms such as through regulating bacterial microbiome composition (phage-mediated lytic killing of host bacteria) and function (phage-mediated introduction of novel functional genes to lysogenic host such as production of endotoxin and antibiotic resistance), and (2) direct interference with human immune-inflammatory responses such as downregulation of the pro-inflammatory NF-κB pathway, inhibition of excessive ROS synthesis, induction of anti-inflammatory IL-10 production, and T4 phage adhesin gp12 - mediated counteraction to LPS-induced inflammation, as evidenced by downregulation of inflammation markers IL-1α and IL-6, and diminished leukocyte infiltration (Górski et al., 2016; Miernikiewicz et al., 2016). In addition, simultaneous increase in pro-inflammatory markers such as IL-1β, IL-1α, along with upregulation in anti-inflammatory IL-1 receptor antagonist and SOCS3 have been observed in P. aeruginosa and S. aureus phages treated cells, and the authors postulate that at least some phages have evolved anti-inflammatory properties to aid their survival and propagation, while retaining some pro-inflammatory properties (Van Belleghem et al., 2017). On the contrary to the largely reported anti-inflammatory properties of phage, significantly increased abundance of bacteriophages and changes in phage communities composition in CD patients compared to healthy controls have indicated a possible contributory role of phage in aberrant immune response (Lepage et al., 2008; Wagner et al., 2013). Undeniably, integrating human virome studies into the existing human bacterial microbiome will provide a bigger and clearer picture on how such transkingdom interactions could impact human health, thus, facilitating disease etiology studies. However, overcoming the obstacles such as limitations in virome sequencing techniques, uninformative database for characterization of novel viral genomes and the difficulties in identifying origin of virus from patient sample followed by deciphering the underlying mechanisms are urgently required.
Another noteworthy aspect is that the observed association of human microbiome with the host in healthy and disease states in fact reflects a far more complicated interactive relationship, rather than a mere unidirectional “cause and effect.” Hence, experimental design should be revised and tailored accordingly to comprehensively investigate at which point of the disease (for example: disease onset, early disease stage, disease progression, active or latent disease) does the human microbiota play a role in, as this will reveal either the primary role, secondary role, or both roles undertaken by human microbiota in disease manifestation. Besides, varying observations such as dysbiotic features in specific diseases, and host response toward introduced stimuli across published literatures on this topic, suspecting the existence of confounding factors, for example, subject-specific differences or distinctive experimental designs across studies becomes important. Subject-specific factors such as genetic, age, gender, lifestyle, diet, infection, disease and postnatal exposure to maternal and environmental microflora serve a crucial role in shaping the unique composition of human microbiota in each individual. Also the commensal microbial composition will in turn affect the development, maturation and normal functioning of the host factors (such as the neuro-immune network development and maturation), will contribute toward issues on varying disease susceptibility. Interconnection of these factors make it much more challenging to design constructive and conclusive experiments to test the role of the microbiome in targeted disease and establishing the true human microbiome-host relationship in multifactorial disease conditions. Whilst for the experimental design, differences across studies, standardization of research protocols are required to enable effective comparison of findings across studies, reduce data variance and biases, and producing sound and robust findings for better explanation of human microbiome-disease relationships.
A better understanding of human microbiome and how the commensal microbes interact with the host is undeniably useful to delineate the etiology and pathophysiological aspects of several human diseases, as well as developing a more effective therapeutic option to counteract the limitation of currently existing treatments. Application of FMT, probiotic and prebiotics derived from advances in gut microbiome studies to ‘rectify’ or ‘restore’ the altered gut microbiota in the dysbiosis-associated disease state back to the ‘healthy’ equilibrium, holds great promise as the alternative therapeutic option in several symptomatic disease management. However, the beneficial therapeutic effect of microbiome-based therapy is largely dependent on the role of dysbiosis in contributing to the nature of the disease. Accurate identification of key microbiota members, intricate selection of microbial strains used in probiotics, or types of prebiotics administered to selectively enumerate the desirable commensal, have definitely added extra challenges to the extensive application of microbiome-based therapy to future clinical practices. In addition, majority of the probiotics are generally regarded as safe but it does not warrant permanent safety. Several potential risks include possible transfer of antibiotic resistance gene or virulent genes among microbes. Although severe systemic side effects post-microbiome therapy are not observed to date, minor gastrointestinal discomforts such as abdominal pain, vomiting, nausea are commonly reported. This subsequently leads to withdrawal or non-compliance by patients during field trials. Therefore, improvement on treatment regime, route of administration, and effective communication with the patients are strongly encouraged. In short, Human Microbiome field of research is still relatively new but rapidly growing, showing several preliminary but promising studies on the modulatory role of human microbiome in human wellness and disease. Future applications of microbiome-based disease diagnosis, prognosis monitoring, prophylaxis and treatments which exhibit great potential in revolutionizing the current measures in disease management and treatment, are definitely worth anticipating.
Author Contributions
ZK conducted the literature search and compiled the information. SL contributed academic assistance and edited the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
- ACC-1
acetyl-CoA carboxylase
- APC
adenomatous polyposis coli
- Arg1
arginase 1
- ASD
autism spectrum disorder
- ASF
altered Schaedler flora
- ATP
adenosine triphosphate
- BFT
Bacteroides fragilis enterotoxin
- BMI
body mass index
- c-AMP
cyclic adenosine monophosphate
- CD
Crohn’s disease
- CD14
cluster of differentiation 14
- CDI
Clostridium difficile infection
- chAT
choline acetyltransferase
- CLA
conjugated linoleic acid
- CNS
central nervous system
- CoA
coenzyme A
- CRC
colorectal cancer
- CTLA-4
cytotoxic T lymphocyte protein 4
- DCA
deoxycholic acid
- DNL
de novo lipogenesis
- EGC
enteric glial cell
- ENS
enteric nervous system
- ETBF
enterotoxigenic Bacteroides fragilis
- FAS
fatty acid synthase
- Fiaf
fasting-induced adipose factor
- FMT
fecal microbiota transplant
- FOXP3
forkhead box P3
- FXR
farnesoid X receptor
- GABA
γ-aminobutyric acid
- GF
germ free
- GIT
gastrointestinal tract
- GLP
glucagon-like peptide
- GPCR
G protein-coupled receptor
- GST
glutathione S-transferase
- HDAC
histone deacetylase
- HDL
high-density lipoprotein
- IBD
inflammatory bowel disease
- IBS
irritable bowel syndrome
- IFN-γ
interferon gamma
- IgA
immunoglobulin A
- IL
interleukin
- IPA
indole-3-propionic acid
- LCA
lithocholic acid
- LPS
lipopolysaccharide
- MET
mesenchymal epithelial transition
- MHC
major histocompatibility complex
- MS
maternal separation
- mTg
microbial transglutaminase
- NF-κB
nuclear factor-kappa B
- NKG2D
natural-killer group 2 member D
- NLR
nod-like receptor
- NO
nitric oxide
- NOS
nitric oxide synthase
- OF-IN
oligofructose-enriched inulin
- OmpC
Escherichia coli outer membrane protein
- PD
Parkinson’s disease
- PD-L1
programmed cell death protein 1 ligand 1
- PPA
propionic acid
- PRR
pattern recognition receptor
- PTMP
post-translational modification of proteins
- PXR
pregnane X receptor
- RA
rheumatoid arthritis
- RNA
ribonucleic acid
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SCFA
short-chain fatty acid
- SFB
segmented filamentous bacteria
- SLE
systemic lupus erythematosus
- SOCS3
suppressor of cytokine signaling 3
- sp
species
- SPF
specific-pathogen free
- TcdA
Clostridium difficile toxin A
- TcdB
Clostridium difficile toxin B
- Tg
transglutaminase
- TH
helper T-cell
- TLR
toll-like receptor
- TMAO
trimethylamine N-oxide
- TNF
tumor necrosis factor
- Treg
regulatory T cell
- tTg
tissue transglutaminase
- UC
ulcerative colitis
- WGS
whole genome sequencing
- WHO
World Health Organization
- Wnt
Wingless Int
- ZO-1
zonula occludens-1
Footnotes
Funding. This publication was supported by funding from the Tropical Medicine and Biology Platform.
References
- Abed J., Emgård J. E., Zamir G., Faroja M., Almogy G., Grenov A., et al. (2016). Fap2 mediates Fusobacterium nucleatum colorectal adenocarcinoma enrichment by binding to tumor-expressed Gal-GalNAc. Cell Host Microbe 20 215–225. 10.1016/j.chom.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamse S. L., Pool-Zobel B. L., Rechkemmer G. (1999). Potential of short chain fatty acids to modulate the induction of DNA damage and changes in the intracellular calcium concentration by oxidative stress in isolated rat distal colon cells. Carcinogenesis 20 629–634. 10.1093/carcin/20.4.629 [DOI] [PubMed] [Google Scholar]
- Adams J. B., Johansen L. J., Powell L. D., Quig D., Rubin R. A. (2011). Gastrointestinal flora and gastrointestinal status in children with autism–comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 11:22. 10.1186/1471-230x-11-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajouz H., Mukherji D., Shamseddine A. (2014). Secondary bile acids: an underrecognized cause of colon cancer. World J. Surg. Oncol. 12:164. 10.1186/1477-7819-12-164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akobeng A. K., Ramanan A. V., Buchan I., Heller R. F. (2006). Effect of breast feeding on risk of coeliac disease: a systematic review and meta-analysis of observational studies. Arch. Dis. Child. 91 39–43. 10.1136/adc.2005.082016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres E., Loukili N. H., Noel E., Kaltenbach G., Abdelgheni M. B., Perrin A. E., et al. (2004). Vitamin B12 (cobalamin) deficiency in elderly patients. Can. Med. Assoc. J. 171 251–259. 10.1503/cmaj.1031155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anitha M., Vijay-Kumar M., Sitaraman S. V., Gewirtz A. T., Srinivasan S. (2012). Gut microbial products regulate murine gastrointestinal motility via Toll-like receptor 4 signaling. Gastroenterology 143 1006–1016. 10.1053/j.gastro.2012.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonopoulos D. A., Huse S. M., Morrison H. G., Schmidt T. M., Sogin M. L., Young V. B. (2009). Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77 2367–2375. 10.1128/iai.01520-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M., Sierra M. S., Laversanne M., Soerjomataram I., Jemal A., Bray F. (2017). Global patterns and trends in colorectal cancer incidence and mortality. Gut 66 683–691. 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- Atarashi K., Nishimura J., Shima T., Umesaki Y., Yamamoto M., Onoue M., et al. (2008). ATP drives lamina propria TH17 cell differentiation. Nature 455 808–812. 10.1038/nature07240 [DOI] [PubMed] [Google Scholar]
- Backhed F., Ding H., Wang T., Hooper L. V., Koh G. Y., Nagy A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101 15718–15723. 10.1073/pnas.0407076101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnaba V., Sinigaglia F. (1997). Molecular mimicry and T cell–mediated autoimmune disease. J. Exp. Med. 185 1529–1532. 10.1084/jem.185.9.1529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J. G. (1982). Antimicrobial agents implicated in Clostridium difficile toxin-associated diarrhea or colitis. Obstet. Gynecol. Surv. 37:46 10.1097/00006254-198201000-00019 [DOI] [PubMed] [Google Scholar]
- Bartlett J. G. (2002). Clinical practice. Antibiotic-associated diarrhea. N. Engl. J. Med. 346 334–339. 10.1056/NEJMcp011603 [DOI] [PubMed] [Google Scholar]
- Bendheim P. E., Poeggeler B., Neria E., Ziv V., Pappolla M. A., Chain D. G. (2002). Development of indole-3-propionic acid (OXIGON™) for Alzheimer’s disease. J. Mol. Neurosci. 19 213–217. 10.1007/s12031-002-0036-0 [DOI] [PubMed] [Google Scholar]
- Blottière H. M., Buecher B., Galmiche J. P., Cherbut C. (2003). Molecular analysis of the effect of short-chain fatty acids on intestinal cell proliferation. Proc. Nutr. Soc. 62 101–106. 10.1079/PNS2002215 [DOI] [PubMed] [Google Scholar]
- Braat H., Rottiers P., Hommes D. W., Huyghebaert N., Remaut E., Remon J. P., et al. (2006). A phase I trial with transgenic bacteria expressing interleukin-10 in Crohn’s disease. Clin. Gastroenterol. Hepatol. 4 754–759. 10.1016/j.cgh.2006.03.028 [DOI] [PubMed] [Google Scholar]
- Britton R. A., Young V. B. (2012). Interaction between the intestinal microbiota and host in Clostridium difficile colonization resistance. Trends Microbiol. 20 313–319. 10.1016/j.tim.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. B., Buie T. M., Winter H., Bauman M., Sutcliffe J. S., Perrin J. M., et al. (2009). Distinct genetic risk based on association of MET in families with co-occurring autism and gastrointestinal conditions. Pediatrics 123 1018–1024. 10.1542/peds.2008-0819 [DOI] [PubMed] [Google Scholar]
- Cani P. D., Amar J., Iglesias M. A., Poggi M., Knauf C., Bastelica D., et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes Metab. Res. Rev 56 1761–1772. 10.2337/db06-1491 [DOI] [PubMed] [Google Scholar]
- Carr E. G., Owen-Deschryver J. S. (2007). Physical illness, pain, and problem behavior in minimally verbal people with developmental disabilities. J. Autism. Dev. Disord. 37 413–424. 10.1007/s10803-006-0176-0 [DOI] [PubMed] [Google Scholar]
- Castellarin M., Warren R. L., Freeman J. D., Dreolini L., Krzywinski M., Strauss J., et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 22 299–306. 10.1101/gr.126516.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T., Kim C. Y., Kaur A., Lamothe L., Shaikh M., Keshavarzian A., et al. (2017). Dietary fibre-based SCFA mixtures promote both protection and repair of intestinal epithelial barrier function in a Caco-2 cell model. Food Funct. 8 1166–1173. 10.1039/c6fo01532h [DOI] [PubMed] [Google Scholar]
- Cherrington C. A., Hinton M., Pearson G. R., Chopra I. (1991). Short-chain organic acids at pH 5.0 kill Escherichia coli and Salmonella spp. without causing membrane perturbation. J. Appl. Microbiol. 70 161–165. [DOI] [PubMed] [Google Scholar]
- Christensen D. L., Baio J., Braun K. V., Bilder D., Charles J., Constantino J. N., et al. (2016). Prevalence and characteristics of autism spectrum disorder among children aged 8 years - autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill. Summ. 65 1–23. 10.15585/mmwr.ss6503a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen K. R., Steenholdt C., Buhl S. S., Ainsworth M. A., Thomsen O. Ø, Brynskov J. (2015). Systematic information to health-care professionals about vaccination guidelines improves adherence in patients with inflammatory bowel disease in anti-tnfα therapy. Am. J. Gastroenterol. 110 1526–1532. 10.1038/ajg.2015.162 [DOI] [PubMed] [Google Scholar]
- Cianchi F., Cortesini C., Fantappiè O., Messerini L., Schiavone N., Vannacci A., et al. (2003). Inducible nitric oxide synthase expression in human colorectal cancer: correlation with tumor angiogenesis. Am. J. Pathol. 162 793–801. 10.1016/S0002-9440(10)63876-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J., Borojevic R., Verdu E. F., Huizinga J. D., Ratcliffe E. M. (2014). Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol. Motil. 26 98–107. 10.1111/nmo.12236 [DOI] [PubMed] [Google Scholar]
- Constantino J. N., Przybeck T., Friesen D., Todd R. D. (2000). Reciprocal social behavior in children with and without pervasive developmental disorders. J. Dev. Behav. Pediatr. 21 2–11. 10.1097/00004703-200002000-00002 [DOI] [PubMed] [Google Scholar]
- Cooke G., Behan J., Costello M. (2006). Newly identified vitamin K-producing bacteria isolated from the neonatal faecal flora. Microb. Ecol. Health Dis. 18 133–138. 10.3402/mehd.v18i3-4.7681 [DOI] [Google Scholar]
- Croft A., Walsh A., Doecke J., Cooley R., Howlett M., Radford-Smith G. (2013). Outcomes of salvage therapy for steroid-refractory acute severe ulcerative colitis: cyclosporine vs. Infliximab. Aliment. Pharmacol. Ther. 38 294–302. 10.1111/apt.12375 [DOI] [PubMed] [Google Scholar]
- Darfeuille-Michaud A., Neut C., Barnich N., Lederman E., Martino P. D., Desreumaux P., et al. (1998). Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohns disease. Gastroenterol. 115 1405–1413. 10.1016/s0016-5085(98)70019-8 [DOI] [PubMed] [Google Scholar]
- D’arienzo R., Stefanile R., Maurano F., Mazzarella G., Ricca E., Troncone R., et al. (2011). Immunomodulatory effects of Lactobacillus casei administration in a mouse model of gliadin-sensitive enteropathy. Scand. J. Immunol. 74 335–341. 10.1111/j.1365-3083.2011.02582.x [DOI] [PubMed] [Google Scholar]
- De Palma G., Nadal I., Medina M., Donat E., Ribes-Koninckx C., Calabuig M., et al. (2010). Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 10:63. 10.1186/1471-2180-10-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Preter V., Joossens M., Ballet V., Shkedy Z., Rutgeerts P., Vermeire S., et al. (2013). Metabolic profiling of the impact of oligofructose-enriched inulin in Crohn’s disease patients: a double-blinded randomized controlled trial. Clin. Transl. Gastroenterol. 4:e30. 10.1038/ctg.2012.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vadder F., Kovatcheva-Datchary P., Goncalves D., Vinera J., Zitoun C., Duchampt A., et al. (2014). Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156 84–96. 10.1016/j.cell.2013.12.016 [DOI] [PubMed] [Google Scholar]
- Degnan P. H., Taga M. E., Goodman A. L. (2014). Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 20 769–778. 10.1016/j.cmet.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi T., Ohtsuki S., Otagiri M., Takanaga H., Asaba H., Mori S., et al. (2002). Major role of organic anion transporter 3 in the transport of indoxyl sulfate in the kidney. Kidney Int. 61 1760–1768. 10.1046/j.1523-1755.2002.00318.x [DOI] [PubMed] [Google Scholar]
- Desbonnet L., Garrett L., Clarke G., Kiely B., Cryan J., Dinan T. (2010). Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience 170 1179–1188. 10.1016/j.neuroscience.2010.08.005 [DOI] [PubMed] [Google Scholar]
- Dewulf E. M., Cani P. D., Neyrinck A. M., Possemiers S., Holle A. V., Muccioli G. G., et al. (2011). Inulin-type fructans with prebiotic properties counteract GPR43 overexpression and PPARγ-related adipogenesis in the white adipose tissue of high-fat diet-fed mice. J. Nutr. Biochem. 22 712–722. 10.1016/j.jnutbio.2010.05.009 [DOI] [PubMed] [Google Scholar]
- Donnelly K. L., Smith C. I., Schwarzenberg S. J., Jessurun J., Boldt M. D., Parks E. J. (2005). Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. J. Clin. Invest. 115 1343–1351. 10.1172/jci200523621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D. R., Garge N., Zhang X., Sun W., Oconnell T. M., Bunger M. K., et al. (2011). The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 13 517–526. 10.1016/j.cmet.2011.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drago S., El Asmar R., Di Pierro M., Grazia Clemente M., Sapone A. T. A., Thakar M., et al. (2006). Gliadin, zonulin and gut permeability: effects on celiac and non-celiac intestinal mucosa and intestinal cell lines. Scand. J. Gastroenterol. 41 408–419. 10.1080/00365520500235334 [DOI] [PubMed] [Google Scholar]
- Duboc H., Rainteau D., Rajca S., Humbert L., Farabos D., Maubert M., et al. (2012). Increase in fecal primary bile acids and dysbiosis in patients with diarrhea-predominant irritable bowel syndrome. Neurogastroenterol. Motil. 24 513–520 10.1111/j.1365-2982.2012.01893.x [DOI] [PubMed] [Google Scholar]
- Duchmann R., May E., Heike M., Knolle P., Neurath M., zum Bschenfelde K. M. (1999). T cell specificity and cross reactivity towards Enterobacteria, Bacteroides, Bifidobacterium, and antigens from resident intestinal flora in humans. Gut 44 812–818. 10.1136/gut.44.6.812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumas M. E., Barton R. H., Toye A., Cloarec O., Blancher C., Rothwell A., et al. (2006). Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice. Proc. Natl. Acad. Sci. U.S.A. 103 12511–12516. 10.1073/pnas.0601056103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert M. N., Klinder A., Peters W. H., Schäferhenrich A., Sendt W., Scheele J., et al. (2003). Expression of glutathione S-transferases (GSTs) in human colon cells and inducibility of GSTM2 by butyrate. Carcinogenesis 24 1637–1644. 10.1093/carcin/bgg122 [DOI] [PubMed] [Google Scholar]
- El Aidy S., van Baarlen P., Derrien M., Lindenbergh-Kortleve D. J., Hooiveld G., Levenez F., et al. (2012). Temporal and spatial interplay of microbiota and intestinal mucosa drive establishment of immune homeostasis in conventionalized mice. Mucosal Immunol. 5 567–579. 10.1038/mi.2012.32 [DOI] [PubMed] [Google Scholar]
- Erny D., de Angelis A. L. H., Jaitin D., Wieghofer P., Staszewski O., David E., et al. (2015). Host microbiota constantly control maturation and function of microglia in the CNS. Nat. Neurosci. 18 965–977. 10.1038/nn.4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith J. J., Rey F. E., O’donnell D., Karlsson M., McNulty N. P., Kallstrom G., et al. (2010). Creating and characterizing communities of human gut microbes in gnotobiotic mice. ISME J. 4 1094–1098. 10.1038/ismej.2010.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagan B. G., Rutgeerts P., Sands B. E., Hanauer S., Colombel J. F., Sandborn W. J., et al. (2013). Vedolizumab as induction and maintenance therapy for ulcerative colitis. N. Engl. J. Med. 369 699–710. 10.1056/NEJMoa1215734 [DOI] [PubMed] [Google Scholar]
- Femia A. P., Luceri C., Dolara P., Giannini A., Biggeri A., Salvadori M., et al. (2002). Antitumorigenic activity of the prebiotic inulin enriched with oligofructose in combination with the probiotics Lactobacillus rhamnosus and Bifidobacterium lactis on azoxymethane-induced colon carcinogenesis in rats. Carcinogenesis 23 1953–1960. 10.1093/carcin/23.11.1953 [DOI] [PubMed] [Google Scholar]
- Fernandez F., Collins M. D. (1987). Vitamin K composition of anaerobic gut bacteria. FEMS Microbiol. Lett. 41 175–180. 10.1016/0378-1097(87)90233-3 [DOI] [Google Scholar]
- Firmansyah A., Penn D., Lebenthal E. (1989). Isolated colonocyte metabolism of glucose, glutamine, n-butyrate, and β-hydroxybutyrate in malnutrition. Gastroenterology 97 622–629. 10.1016/0016-5085(89)90633-1 [DOI] [PubMed] [Google Scholar]
- Forster V. J., McDonnell A., Theobald R., McKay J. A. (2017). Effect of methotrexate/vitamin B12 on DNA methylation as a potential factor in leukemia treatment-related neurotoxicity. Epigenomics 9 1205–1218. 10.2217/epi-2016-0165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi S., Panza E., Vecchia C. L., Parazzini F., Decarli A., Porro G. B. (1987). Nonspecific inflammatory bowel disease and smoking. Am. J. Epidemiol. 125 445–452. 10.1093/oxfordjournals.aje.a114550 [DOI] [PubMed] [Google Scholar]
- Geleijnse J. M., Vermeer C., Grobbee D. E., Schurgers L. J., Knapen M. H., van der Meer I. M., et al. (2004). Dietary intake of menaquinone is associated with a reduced risk of coronary heart disease: the rotterdam study. J. Nutr. 134 3100–3105. 10.1093/jn/134.11.3100 [DOI] [PubMed] [Google Scholar]
- Gent A., Hellier M., Grace R., Swarbrick E., Coggon D. (1994). Inflammatory bowel disease and domestic hygiene in infancy. Lancet 343 766–767. 10.1016/s0140-6736(94)91841-4 [DOI] [PubMed] [Google Scholar]
- Genth H., Huelsenbeck J., Hartmann B., Hofmann F., Just I., Gerhard R. (2006). Cellular stability of Rho-GTPases glucosylated by Clostridium difficile toxin B. FEBS Lett. 580 3565–3569. 10.1016/j.febslet.2006.04.100 [DOI] [PubMed] [Google Scholar]
- Gill S. R., Pop M., Deboy R. T., Eckburg P. B., Turnbaugh P. J., Samuel B. S., et al. (2006). Metagenomic analysis of the human distal gut microbiome. Science 312 1355–1359. 10.1126/science.1124234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin B. R., Gualtieri L. J., Moore R. P. (1996). The effect of Lactobacillus GG on the initiation and promotion of DMH-induced intestinal tumors in the rat. Nutr. Cancer 25 197–204. 10.1080/01635589609514442 [DOI] [PubMed] [Google Scholar]
- Gominak S. (2016). Vitamin D deficiency changes the intestinal microbiome reducing B vitamin production in the gut. The resulting lack of pantothenic acid adversely affects the immune system, producing a “pro-inflammatory” state associated with atherosclerosis and autoimmunity. Med. Hypotheses 94 103–107. 10.1016/j.mehy.2016.07.007 [DOI] [PubMed] [Google Scholar]
- Gopal-Srivastava R., Hylemon P. B. (1988). Purification and characterization of bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 29 1079–1085. [PubMed] [Google Scholar]
- Górski A., Miȩdzybrodzki R., Weber-Dąbrowska B., Fortuna W., Letkiewicz S., Rogóż P., et al. (2016). Phage therapy: combating infections with potential for evolving from merely a treatment for complications to targeting diseases. Front. Microbiol. 7:1515. 10.3389/fmicb.2016.01515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E. A., Duffy L. C., Schanbacher F. L., Qiao H., Dryja D., Leavens A., et al. (2004). In vivo effects of Bifidobacteria and lactoferrin on gut endotoxin concentration and mucosal immunity in balb/c mice. Dig. Dis. Sci. 49 579–589. 10.1023/b:ddas.0000026302.92898.ae [DOI] [PubMed] [Google Scholar]
- Grishin A. M., Beyrakhova K. A., Cygler M. (2015). Structural insight into effector proteins of Gram-negative bacterial pathogens that modulate the phosphoproteome of their host. Protein Sci. 24 604–620. 10.1002/pro.2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronbach K., Flade I., Holst O., Lindner B., Ruscheweyh H. J., Wittmann A., et al. (2014). Endotoxicity of lipopolysaccharide as a determinant of T-cell-mediated colitis induction in mice. Gastroenterology 146 765–775. 10.1053/j.gastro.2013.11.033 [DOI] [PubMed] [Google Scholar]
- Guo X., Rao J. N., Liu L., Zou T., Keledjian K. M., Boneva D., et al. (2005). Polyamines are necessary for synthesis and stability of occludin protein in intestinal epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 288 G1159–G1169. 10.1152/ajpgi.00407.2004 [DOI] [PubMed] [Google Scholar]
- Guo X., Rao J. N., Liu L., Zou T. T., Turner D. J., Bass B. L., et al. (2003). Regulation of adherens junctions and epithelial paracellular permeability: a novel function for polyamines. Am. J. Physiol. Cell Physiol. 285 C1174–C1187. 10.1152/ajpcell.00015.2003 [DOI] [PubMed] [Google Scholar]
- Hassig C. A., Tong J. K., Schreiber S. L. (1997). Fiber-derived butyrate and the prevention of colon cancer. Chem. Biol. 4 783–789. 10.1016/S1074-5521(97)90111-3 [DOI] [PubMed] [Google Scholar]
- Higuchi N., Kato M., Shundo Y., Tajiri H., Tanaka M., Yamashita N., et al. (2008). Liver X receptor in cooperation with SREBP-1c is a major lipid synthesis regulator in nonalcoholic fatty liver disease. Hepatol. Res. 38 1122–1129. 10.1111/j.1872-034x.2008.00382.x [DOI] [PubMed] [Google Scholar]
- Hooper L. V., Gordon J. I. (2001). Commensal host-bacterial relationships in the gut. Science 292 1115–1118. 10.1126/science.1058709 [DOI] [PubMed] [Google Scholar]
- Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cezard J. P., Belaiche J., et al. (2001). Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411 599–603. 10.1038/35079107 [DOI] [PubMed] [Google Scholar]
- Hyde G. M., Thillainayagam A. V., Jewell D. P. (1998). Intravenous cyclosporin as rescue therapy in severe ulcerative colitis. Eur. J. Gastroenterol. Hepatol. 10 411–414. 10.1097/00042737-199805000-00010 [DOI] [PubMed] [Google Scholar]
- Hylemon P. B., Zhou H., Pandak W. M., Ren S., Gil G., Dent P. (2009). Bile acids as regulatory molecules. J. Lipid Res. 50 1509–1520. 10.1194/jlr.R900007-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ieraci A., Forni P. E., Ponzetto C. (2002). Viable hypomorphic signaling mutant of the met receptor reveals a role for hepatocyte growth factor in postnatal cerebellar development. Proc. Natl. Acad. Sci. U.S.A. 99 15200–15205. 10.1073/pnas.222362099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki T., Moschetta A., Lee Y. K., Peng L., Zhao G., Downes M., et al. (2006). Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl. Acad. Sci. U.S.A. 103 3920–3925. 10.1073/pnas.0509592103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Atarashi K., Manel N., Brodie E. L., Shima T., Karaoz U., et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139 485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson P. B., Boccuto L., Skinner C., Collins J. S., Neri G., Gurrieri F., et al. (2009). Further evidence that the rs1858830 C variant in the promoter region of theMETgene is associated with autistic disorder. Autism Res. 2 232–236. 10.1002/aur.87 [DOI] [PubMed] [Google Scholar]
- Johanesen P. A., Mackin K. E., Hutton M. L., Awad M. M., Larcombe S., Amy J. M., et al. (2015). Disruption of the gut microbiome: Clostridium difficile infection and the threat of antibiotic resistance. Genes 6 1347–1360. 10.3390/genes6041347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson M. E. V., Phillipson M., Petersson J., Velcich A., Holm L., Hansson G. C. (2008). The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc. Natl. Acad. Sci. U.S.A. 105 15064–15069. 10.1073/pnas.0803124105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. H., Dejea C. M., Edler D., Hoang L. T., Santidrian A. F., Felding B. H., et al. (2015). Metabolism links bacterial biofilms and colon carcinogenesis. Cell Metab. 21 891–897. 10.1016/j.cmet.2015.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joossens M., De Preter V., Ballet V., Verbeke K., Rutgeerts P., Vermeire S. (2012). Effect of oligofructose-enriched inulin (OF-IN) on bacterial composition and disease activity of patients with Crohn’s disease: results from a double-blinded randomised controlled trial. Gut 61:958. 10.1136/gutjnl-2011-300413 [DOI] [PubMed] [Google Scholar]
- Kabouridis P. S., Lasrado R., McCallum S., Chng S. H., Snippert H. J., Clevers H., et al. (2015). Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron 85 289–295. 10.1016/j.neuron.2014.12.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalliomäki M., Kirjavainen P., Eerola E., Kero P., Salminen S., Isolauri E. (2001). Distinct patterns of neonatal gut microflora in infants in whom atopy was and was not developing. J. Allergy Clin. Immunol. 107 129–134. 10.1067/mai.2001.111237 [DOI] [PubMed] [Google Scholar]
- Kamada N., Chen G. Y., Inohara N., Núñez G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14 685–690. 10.1038/ni.2608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima H., Nakajima Y., Matubara Y., Nakanowatari J., Fukuta T., Mizuno S., et al. (1997). Effects of vitamin K2 (menatetrenone) on atherosclerosis and blood coagulation in hypercholesterolemic rabbits. Jpn. J. Pharmacol. 75 135–143. 10.1254/jjp.75.135 [DOI] [PubMed] [Google Scholar]
- Kelly J. R., Borre Y., O’Brien C., Patterson E., El Aidy S., Deane J., et al. (2016). Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 82 109–118. 10.1016/j.jpsychires.2016.07.019 [DOI] [PubMed] [Google Scholar]
- Keshavarzian A., Green S. J., Engen P. A., Voigt R. M., Naqib A., Forsyth C. B., et al. (2015). Colonic bacterial composition in Parkinson’s disease. Mov. Disord. 30 1351–1360. 10.1002/mds.26307 [DOI] [PubMed] [Google Scholar]
- Khosravi A., Yáñez A., Price J. G., Chow A., Merad M., Goodridge H. S., et al. (2014). Gut microbiota promote hematopoiesis to control bacterial infection. Cell Host Microbe 15 374–381. 10.1016/j.chom.2014.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. H., Lord C. (2012). New autism diagnostic interview-revised algorithms for toddlers and young preschoolers from 12 to 47 months of age. J. Autism Dev. Disord. 42 82–93. 10.1007/s10803-011-1213-1 [DOI] [PubMed] [Google Scholar]
- Koba K., Akahoshi A., Yamasaki M., Tanaka K., Yamada K., Iwata T., et al. (2002). Dietary conjugated linolenic acid in relation to CLA differently modifies body fat mass and serum and liver lipid levels in rats. Lipids 37 343–350. 10.1007/s11745-002-0901-7 [DOI] [PubMed] [Google Scholar]
- Koeth R. A., Wang Z., Levison B. S., Buffa J. A., Org E., Sheehy B. T., et al. (2013). Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19 576–585. 10.1038/nm.3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliada A., Syzenko G., Moseiko V., Budovska L., Puchkov K., Perederiy V., et al. (2017). Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 17:120. 10.1186/s12866-017-1027-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konturek P. C., Koziel J., Dieterich W., Haziri S., Wirtz I., Glowczyk K., et al. (2016). Successful therapy of Clostridium difficile infection with fecal microbiota transplantation. J. Physiol. Pharmacol. 67 859–866. [PubMed] [Google Scholar]
- Landers C. J., Cohavy O., Misra R., Yang H., Lin Y. C., Braun J., et al. (2002). Selected loss of tolerance evidenced by Crohns disease–associated immune responses to auto- and microbial antigens. Gastroenterology 123 689–699. 10.1053/gast.2002.35379 [DOI] [PubMed] [Google Scholar]
- Laparra J. M., Olivares M., Gallina O., Sanz Y. (2012). Bifidobacterium longum CECT 7347 modulates immune responses in a gliadin-induced enteropathy animal model. PLoS One 7:e30744. 10.1371/journal.pone.0030744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larrosa M., González-Sarrías A., García-Conesa M. T., Tomás-Barberán F. A., Espín J. C. (2006). Urolithins, ellagic acid-derived metabolites produced by human colonic microflora, exhibit estrogenic and antiestrogenic activities. J. Agric. Food Chem. 54 1611–1620. 10.1021/jf0527403 [DOI] [PubMed] [Google Scholar]
- Larrosa M., González-Sarrías A., Yáñez-Gascón M. J., Selma M. V., Azorín-Ortuño M., Toti S., et al. (2010). Anti-inflammatory properties of a pomegranate extract and its metabolite urolithin-A in a colitis rat model and the effect of colon inflammation on phenolic metabolism. J. Nutr. Biochem. 21 717–725. 10.1016/j.jnutbio.2009.04.012 [DOI] [PubMed] [Google Scholar]
- Lawley T. D., Walker A. W. (2013). Intestinal colonization resistance. Immunology 138 1–11. 10.1111/j.1365-2567.2012.03616.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J. G., Laiño J. E., del Valle M. J., Vannini V., Van Sinderen D., Taranto M. P., et al. (2011). B-Group vitamin production by lactic acid bacteria–current knowledge and potential applications. J. Appl. Microbiol. 111 1297–1309. 10.1111/j.1365-2672.2011.05157.x [DOI] [PubMed] [Google Scholar]
- Lee H. C., Jenner A. M., Low C. S., Lee Y. K. (2006). Effect of tea phenolics and their aromatic fecal bacterial metabolites on intestinal microbiota. Res. Microbiol. 157 876–884. 10.1016/j.resmic.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Park J.-H., Seok S.-H., Baek M.-W., Kim D.-J., Lee K.-E., et al. (2006). Human originated bacteria, Lactobacillus rhamnosus PL60, produce conjugated linoleic acid and show anti-obesity effects in diet-induced obese mice. Biochim. Biophys. Acta 1761 736–744. 10.1016/j.bbalip.2006.05.007 [DOI] [PubMed] [Google Scholar]
- Lee J. M., Wagner M., Xiao R., Kim K. H., Feng D., Lazar M. A., et al. (2014). Nutrient-sensing nuclear receptors coordinate autophagy. Nature 516 112–115. 10.1038/nature13961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Kim E. H., Yim I. B., Joo H. G. (2004). Immunomodulatory and antitumor effects in vivo by the cytoplasmic fraction of Lactobacillus casei and Bifidobacterium longum. J. Vet. Sci. 5 41–48. [PubMed] [Google Scholar]
- Lennard-Jones J. E. (1989). Classification of inflammatory bowel disease. Scand. J. Gastroenterol. 24 2–6. 10.3109/00365528909091339 [DOI] [PubMed] [Google Scholar]
- Lepage P., Colombet J., Marteau P., Sime-Ngando T., Doré J., Leclerc M. (2008). Dysbiosis in inflammatory bowel disease: a role for bacteriophages? Gut 57 424–425. 10.1136/gut.2007.134668 [DOI] [PubMed] [Google Scholar]
- Lerner A., Aminov R., Matthias T. (2016). Dysbiosis may trigger autoimmune diseases via inappropriate post-translational modification of host proteins. Front. Microbiol. 7:84. 10.3389/fmicb.2016.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner A., Matthias T. (2015). Changes in intestinal tight junction permeability associated with industrial food additives explain the rising incidence of autoimmune disease. Autoimmun. Rev. 14 479–489. 10.1016/j.autrev.2015.01.009 [DOI] [PubMed] [Google Scholar]
- Lerner A., Aminov R., Matthias T. (2017a). Transglutaminases in dysbiosis as potential environmental drivers of autoimmunity. Front. Microbiol. 8:66. 10.3389/fmicb.2017.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner A., Neidhöfer S., Matthias T. (2017b). The gut microbiome feelings of the brain: a perspective for non-microbiologists. Microorganisms 5:E66. 10.3390/microorganisms5040066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessa F. C., Mu Y., Bamberg W. M., Beldavs Z. G., Dumyati G. K., Dunn J. R., et al. (2015). Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372 825–834. 10.1056/NEJMoa1408913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Bäckhed F., Turnbaugh P., Lozupone C. A., Knight R. D., Gordon J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102 11070–11075. 10.1073/pnas.0504978102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R. E., Turnbaugh P. J., Klein S., Gordon J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444 1022–1023. 10.1038/4441022a [DOI] [PubMed] [Google Scholar]
- Li Y., Kundu P., Seow S. W., de Matos C. T., Aronsson L., Chin K. C., et al. (2012). Gut microbiota accelerate tumor growth via c-jun and STAT3 phosphorylation in APC Min/+ mice. Carcinogenesis 33 1231–1238. 10.1093/carcin/bgs137 [DOI] [PubMed] [Google Scholar]
- Lindfors K., Blomqvist T., Juuti-Uusitalo K., Stenman S., Venäläinen J., Mäki M., et al. (2008). Live probiotic Bifidobacterium lactis bacteria inhibit the toxic effects induced by wheat gliadin in epithelial cell culture. Clin. Exp. Immunol. 152 552–558. 10.1111/j.1365-2249.2008.03635.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zou Q., Zeng B., Fang Y., Wei H. (2013). Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr. Microbiol. 67 170–176. 10.1007/s00284-013-0338-1 [DOI] [PubMed] [Google Scholar]
- Lloyd-Price J., Abu-Ali G., Huttenhower C. (2016). The healthy human microbiome. Genome Med. 8:51. 10.1186/s13073-016-0307-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus E. V. (2004). Clinical epidemiology of inflammatory bowel disease: incidence, prevalence, and environmental influences. Gastroenterology 126 1504–1517. 10.1053/j.gastro.2004.01.063 [DOI] [PubMed] [Google Scholar]
- Ludvigsson J. F., Montgomery S. M., Ekbom A., Brandt L., Granath F. (2009). Small-intestinal histopathology and mortality risk in celiac disease. JAMA 302 1171–1178. 10.1001/jama.2009.1320 [DOI] [PubMed] [Google Scholar]
- Luo X. M., Edwards M. R., Mu Q., Yu Y., Vieson M. D., Reilly C. M., et al. (2018). Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl. Environ. Microbiol. 84:e2288-17. 10.1128/AEM.02288-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiels K., Joossens M., Sabino J., De Preter V., Arijs I., Eeckhaut V., et al. (2014). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63 1275–1283. 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- Mahe S., Corthier G., Dubos F. (1987). Effect of various diets on toxin production by two strains of Clostridium difficile in gnotobiotic mice. Infect. Immun. 55 1801–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marasco G., Di Biase A. R., Schiumerini R., Eusebi L. H., Iughetti L., Ravaioli F., et al. (2016). Gut microbiota and celiac disease. Dig. Dis. Sci. 61 1461–1472. 10.1007/s10620-015-4020-2 [DOI] [PubMed] [Google Scholar]
- McNally L., Brown S. P. (2015). Building the microbiome in health and disease: niche construction and social conflict in bacteria. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370:20140298. 10.1098/rstb.2014.0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M., De Palma G., Ribes-Koninckx C., Calabuig M., Sanz Y. (2008). Bifidobacterium strains suppress in vitro the pro-inflammatory milieu triggered by the large intestinal microbiota of coeliac patients. J. Inflamm. 5:19. 10.1186/1476-9255-5-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernikiewicz P., Kłopot A., Soluch R., Szkuta P., Kȩska W., Hodyra-Stefaniak K., et al. (2016). T4 phage tail adhesin gp12 counteracts LPS-induced inflammation in vivo. Front. Microbiol. 7:1112. 10.3389/fmicb.2016.01112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncrief J. S., Duncan A. J., Wright R. L., Barroso L. A., Wilkins T. D. (1998). Molecular characterization of the fragilysin pathogenicity islet of enterotoxigenic Bacteroides fragilis. Infect. Immun. 66 1735–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder S. J., Mulder-Bos G. C. (2006). Most probable origin of coeliac disease is low immune globulin A in the intestine caused by malfunction of Peyer’s patches. Med. Hypotheses 66 757–762. 10.1016/j.mehy.2005.09.050 [DOI] [PubMed] [Google Scholar]
- Murri M., Leiva I., Gomez-Zumaquero J. M., Tinahones F. J., Cardona F., Soriguer F., et al. (2013). Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. 11:46. 10.1186/1741-7015-11-46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustalahti K., Catassi C., Reunanen A., Fabiani E., Heier M., McMillan S., et al. (2010). The prevalence of celiac disease in Europe: results of a centralized, international mass screening project. Ann. Med. 42 587–595. 10.3109/07853890.2010.505931 [DOI] [PubMed] [Google Scholar]
- Nadal I., Donant E., Ribes-Koninckx C., Calabuig M., Sanz Y. (2007). Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J. Med. Microbiol. 56 1669–1674. 10.1099/jmm.0.47410-0 [DOI] [PubMed] [Google Scholar]
- Nagao-Kitamoto H., Shreiner A. B., Gilliland M. G. I. I. I., Kitamoto S., Ishii C., Hirayama A., et al. (2016). Functional characterization of inflammatory bowel disease–associated gut dysbiosis in gnotobiotic mice. Cell. Mol. Gastroenterol. Hepatol. 2 468–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neyrinck A. M., Possemiers S., Druart C., Wiele T. V. D., Backer F. D., Cani P. D., et al. (2011). Prebiotic effects of wheat arabinoxylan related to the increase in Bifidobacteria, Roseburia and Bacteroides/Prevotella in diet-induced obese mice. PLoS One 6:e20944. 10.1371/journal.pone.0020944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson J. K., Holmes E., Kinross J., Burcelin R., Gibson G., Jia W., et al. (2012). Host-gut microbiota metabolic interactions. Science 336 1262–1267. 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- Nie Y. F., Hu J., Yan X.-H. (2015). Cross-talk between bile acids and intestinal microbiota in host metabolism and health. J. Zhejiang Univ. Sci. B 16 436–446. 10.1631/jzus.b1400327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioku T., Matsumoto J., Dohgu S., Sumi N., Miyao K., Takata F., et al. (2010). Tumor necrosis factor-α mediates the blood–brain barrier dysfunction induced by activated microglia in mouse brain microvascular endothelial cells. J. Pharmacol. Sci. 112 251–254. 10.1254/jphs.09292sc [DOI] [PubMed] [Google Scholar]
- Ochoa-Cortes F., Turco F., Linan-Rico A., Soghomonyan S., Whitaker E., Wehner S., et al. (2015). Enteric glial cells: a new frontier in neurogastroenterology and clinical target for inflammatory bowel diseases. Inflamm. Bowel Dis. 22 433–449. 10.1097/MIB.0000000000000667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okunishi K., Dohi M., Nakagome K., Tanaka R., Mizuno S., Matsumoto K., et al. (2005). A novel role of hepatocyte growth factor as an immune regulator through suppressing dendritic cell function. J. Immunol. 175 4745–4753. 10.4049/jimmunol.175.7.4745 [DOI] [PubMed] [Google Scholar]
- Olivares M., Castillejo G., Varea V., Sanz Y. (2014). Double-blind, randomised, placebo-controlled intervention trial to evaluate the effects of Bifidobacterium longum CECT 7347 in children with newly diagnosed coeliac disease. Br. J. Nutr. 112 30–40. 10.1017/S0007114514000609 [DOI] [PubMed] [Google Scholar]
- Orberg E. T., Fan H., Tam A. J., Dejea C. M., Shields C. D., Wu S., et al. (2017). The myeloid immune signature of enterotoxigenic Bacteroides fragilis-induced murine colon tumorigenesis. Mucosal Immunol. 10 421–433. 10.1038/mi.2016.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick S., Jobling K. L., O’Connor D., Thacker Z., Dryden D. T., Blakely G. W. (2011). A unique homologue of the eukaryotic protein-modifier ubiquitin present in the bacterium Bacteroides fragilis, a predominant resident of the human gastrointestinal tract. Microbiology 157 3071–3078. 10.1099/mic.0.049940-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paust S., Lu L., Mccarty N., Cantor H. (2004). Engagement of B7 on effector T cells by regulatory T cells prevents autoimmune disease. Proc. Natl. Acad. Sci. U.S.A. 101 10398–10403. 10.1073/pnas.0403342101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne A. N., Chassard C., Banz Y., Lacroix C. (2012). The composition and metabolic activity of child gut microbiota demonstrate differential adaptation to varied nutrient loads in an in vitro model of colonic fermentation. FEMS Microbiol. Ecol. 80 608–623. 10.1111/j.1574-6941.2012.01330.x [DOI] [PubMed] [Google Scholar]
- Pear S. M., Williamson T. H., Bettin K. M., Gerding D. N., Galgiani J. N. (1994). Decrease in nosocomial Clostridium difficile–associated diarrhea by restricting clindamycin use. Ann. Intern. Med. 120 272–277. 10.7326/0003-4819-120-4-199402150-00003 [DOI] [PubMed] [Google Scholar]
- Peng L., He Z., Chen W., Holzman I. R., Lin J. (2007). Effects of butyrate on intestinal barrier function in a caco-2 cell monolayer model of intestinal barrier. Pediatr. Res. 61 37–41. 10.1203/01.pdr.0000250014.92242.f3 [DOI] [PubMed] [Google Scholar]
- Pépin J., Saheb N., Coulombe M.-A., Alary M.-E., Corriveau M.-P., Authier S., et al. (2005). Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin. Infect. Dis. 41 1254–1260. 10.1086/496986 [DOI] [PubMed] [Google Scholar]
- Perez-Cano F. J., Gonzalez-Castro A., Castellote C., Franch A., Castell M. (2010). Influence of breast milk polyamines on suckling rat immune system maturation. Dev. Comp. Immunol. 34 210–218. 10.1016/j.dci.2009.10.001 [DOI] [PubMed] [Google Scholar]
- Pistollato F., Sumalla Cano S., Elio I., Masias Vergara M., Giampieri F., Battino M. (2016). Role of gut microbiota and nutrients in amyloid formation and pathogenesis of Alzheimer disease. Nutr. Rev. 74 624–634. 10.1093/nutrit/nuw023 [DOI] [PubMed] [Google Scholar]
- Png C. W., Lindén S. K., Gilshenan K. S., Zoetendal E. G., Mcsweeney C. S., Sly L. I., et al. (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 105 2420–2428. 10.1038/ajg.2010.281 [DOI] [PubMed] [Google Scholar]
- Pool-Zobel B. L., Neudecker C., Domizlaff I., Ji S., Schillinger U., Rumney C., et al. (1996). Lactobacillus-and Bifidobacterium-mediated antigenotoxicity in the colon of rats. Nutr. Cancer 26 365–380. 10.1080/01635589609514492 [DOI] [PubMed] [Google Scholar]
- Pruitt R. N., Chumbler N. M., Rutherford S. A., Farrow M. A., Friedman D. B., Spiller B., et al. (2012). Structural determinants of Clostridium difficile toxin a glucosyltransferase activity. J. Biol. Chem. 287 8013–8020. 10.1074/jbc.M111.298414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J., Li Y., Cai Z., Li S., Zhu J., Zhang F., et al. (2012). A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490 55–60. 10.1038/nature11450 [DOI] [PubMed] [Google Scholar]
- Rakoff-Nahoum S., Paglino J., Eslami-Varzaneh F., Edberg S., Medzhitov R. (2004). Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell 118 229–241. 10.1016/j.cell.2004.07.002 [DOI] [PubMed] [Google Scholar]
- Rooks M. G., Garrett W. S. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16 341–352. 10.1038/nri.2016.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M. R., Wang X., Liu W., Hao Y., Cai G., Han Y. W. (2013). Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 14 195–206. 10.1016/j.chom.2013.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruseler-van Embden J. G. H., Both-Patoir H. C. (1983). Anaerobic gram-negative faecal flora in patients with Crohn’s disease and healthy subjects. Antonie Van Leeuwenhoek 49 125–132. 10.1007/BF00393670 [DOI] [PubMed] [Google Scholar]
- Salminen S., Bouley C., Boutron M.-C., Cummings J. H., Franck A., Gibson G. R., et al. (1998). Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 80 147–171. 10.1079/bjn19980108 [DOI] [PubMed] [Google Scholar]
- Salzman N. H., Hung K., Haribhai D., Chu H., Karlsson-Sjöberg J., Amir E., et al. (2010). Enteric defensins are essential regulators of intestinal microbial ecology. Nat. Immunol. 11 76–83. 10.1038/ni.1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel B. S., Shaito A., Motoike T., Rey F. E., Backhed F., Manchester J. K., et al. (2008). Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. U.S.A. 105 16767–16772. 10.1073/pnas.0808567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel P., Holtzman C. M., Meilman E., Sekowski I. (1973). Effect of neomycin and other antibiotics on serum cholesterol levels and on 7α-dehydroxylation of bile acids by the fecal bacterial flora in man. Circ. Res. 33 393–402. 10.1161/01.res.33.4.393 [DOI] [PubMed] [Google Scholar]
- Sanz Y., Palma G. D., Laparra M. (2011). Unraveling the ties between celiac disease and intestinal microbiota. Int. Rev. Immunol. 30 207–218. 10.3109/08830185.2011.599084 [DOI] [PubMed] [Google Scholar]
- Sato J., Kanazawa A., Ikeda F., Yoshihara T., Goto H., Abe H., et al. (2014). Gut dysbiosis and detection of “live gut bacteria” in blood of Japanese patients with type 2 diabetes. Diabetes Care 37 2343–2350. 10.2337/dc13-2817 [DOI] [PubMed] [Google Scholar]
- Scher J. U., Sczesnak A., Longman R. S., Segata N., Ubeda C., Bielski C., et al. (2013). Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2:e01202. 10.7554/eLife.01202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selma M. V., Espin J. C., Tomas-Barberan F. A. (2009). Interaction between phenolics and gut microbiota: role in human health. J. Agric. Food Chem. 57 6485–6501. 10.1021/jf902107d [DOI] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. (2016). Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14:e1002533. 10.1371/journal.pbio.1002533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., Zhang Z., Angela M. Y., Wang W., Wei Z., Akhter E., et al. (2014). The SLE transcriptome exhibits evidence of chronic endotoxin exposure and has widespread dysregulation of non-coding and coding RNAs. PLoS One 9:e93846. 10.1371/journal.pone.0093846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shultz S. R., Aziz N. A., Yang L., Sun M., Macfabe D. F., O’Brien T. J. (2015). Intracerebroventricular injection of propionic acid, an enteric metabolite implicated in autism, induces social abnormalities that do not differ between seizure-prone (FAST) and seizure-resistant (SLOW) rats. Behav. Brain Res. 278 542–548. 10.1016/j.bbr.2014.10.050 [DOI] [PubMed] [Google Scholar]
- Silva A. M., Bambirra E. A., Oliveira A. L., Souza P. P., Gomes D. A., Vieira E. C., et al. (1999). Protective effect of bifidus milk on the experimental infection with Salmonella enteritidis subsp. typhimurium in conventional and gnotobiotic mice. J. Appl. Microbiol. 86 331–336. 10.1046/j.1365-2672.1999.00674.x [DOI] [PubMed] [Google Scholar]
- Singh J., Rivenson A., Tomita M., Shimamura S., Ishibashi N., Reddy B. S. (1997). Bifidobacterium longum, a lactic acid-producing intestinal bacterium inhibits colon cancer and modulates the intermediate biomarkers of colon carcinogenesis. Carcinogenesis 18 833–841. 10.1093/carcin/18.4.833 [DOI] [PubMed] [Google Scholar]
- Sivan A., Corrales L., Hubert N., Williams J. B., Aquino-Michaels K., Earley Z. M., et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 350 1084–1089. 10.1126/science.aac4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermúdez-Humarán L. G., Gratadoux J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U.S.A. 105 16731–16736. 10.1073/pnas.0804812105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas G., Summermatter S., Mainieri D., Gubler M., Montani J. P., Seydoux J., et al. (2006). Corticotropin-releasing hormone directly stimulates thermogenesis in skeletal muscle possibly through substrate cycling between de novo lipogenesis and lipid oxidation. Endocrinology 147 31–38. 10.1210/en.2005-1033 [DOI] [PubMed] [Google Scholar]
- Song H. J., Shim K.-N., Jung S.-A., Choi H. J., Lee M. A., Ryu K. H., et al. (2008). Antibiotic-associated diarrhea: candidate organisms other than Clostridium difficile. Korean J. Intern. Med. 23 9–15. 10.3904/kjim.2008.23.1.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Liu C., Finegold S. M. (2004). Real-time PCR quantitation of Clostridia in feces of autistic children. Appl. Environ. Microbiol. 70 6459–6465. 10.1128/aem.70.11.6459-6465.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soret R., Chevalier J., De Coppet P., Poupeau G., Derkinderen P., Segain J. P., et al. (2010). Short-chain fatty acids regulate the enteric neurons and control gastrointestinal motility in rats. Gastroenterology 138 1772–1782. 10.1053/j.gastro.2010.01.053 [DOI] [PubMed] [Google Scholar]
- Sorg J. A., Sonenshein A. L. (2008). Bile salts and glycine as cogerminants for Clostridium difficile spores. J. Bacteriol. 190 2505–2512. 10.1128/jb.01765-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staels B., Fonseca V. A. (2009). Bile acids and metabolic regulation: mechanisms and clinical responses to bile acid sequestration. Diabetes Care 32(Suppl. 2), S237–S245. 10.2337/dc09-s355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stedtfeld R. D., Chai B., Crawford R. B., Stedtfeld T. M., Williams M. R., Xiangwen S., et al. (2017). Modulatory influence of segmented filamentous bacteria on transcriptomic response of gnotobiotic mice exposed to TCDD. Front. Microbiol. 8:1708. 10.3389/fmicb.2017.01708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stene L. C., Honeyman M. C., Hoffenberg E. J., Haas J. E., Sokol R. J., Emery L., et al. (2006). Rotavirus infection frequency and risk of celiac disease autoimmunity in early childhood: a longitudinal study. Am. J. Gastroenterol. 101 2333–2340. 10.1111/j.1572-0241.2006.00741.x [DOI] [PubMed] [Google Scholar]
- Stepniak D., Koning F. (2006). Celiac disease—sandwiched between innate and adaptive immunity. Hum. Immunol. 67 460–468. 10.1016/j.humimm.2006.03.011 [DOI] [PubMed] [Google Scholar]
- Swidsinski A., Loening-Baucke V., Theissig F., Engelhardt H., Bengmark S., Koch S., et al. (2007). Comparative study of the intestinal mucus barrier in normal and inflamed colon. Gut 56 343–350. 10.1136/gut.2006.098160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tap J., Mondot S., Levenez F., Pelletier E., Caron C., Furet J. P., et al. (2009). Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11 2574–2584. 10.1111/j.1462-2920.2009.01982.x [DOI] [PubMed] [Google Scholar]
- Thangaraju M., Cresci G. A., Liu K., Ananth S., Gnanaprakasam J. P., Browning D. D., et al. (2009). GPR109A is a G-protein–coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res. 69 2826–2832. 10.1158/0008-5472.CAN-08-4466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Human Microbiome Project Consortium. (2012). A framework for human microbiome research. Nature 486 215–221. 10.1038/nature11209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theriot C. M., Koenigsknecht M. J., Carlson P. E., Hatton G. E., Nelson A. M., Li B., et al. (2014). Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 5:3114. 10.1038/ncomms4114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellström B., Stenhammar L., Högberg L., Fälth-Magnusson K., Magnusson K. E., Midtvedt T., et al. (2005). Gut microflora associated characteristics in children with celiac disease. Am. J. Gastroenterol. 100 2784–2788. 10.1111/j.1572-0241.2005.00313.x [DOI] [PubMed] [Google Scholar]
- Tolhurst G., Heffron H., Lam Y. S., Parker H. E., Habib A. M., Diakogiannaki E., et al. (2012). Short-chain fatty acids stimulate glucagon-like peptide-1 secretion via the G-protein–coupled receptor FFAR2. Diabetes Metab. Res. Rev. 61 364–371. 10.2337/db11-1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Hamady M., Yatsunenko T., Cantarel B. L., Duncan A., Ley R. E., et al. (2009a). A core gut microbiome in obese and lean twins. Nature 457 480–484. 10.1038/nature07540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ridaura V. K., Faith J. J., Rey F. E., Knight R., Gordon J. I. (2009b). The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 1:6ra14. 10.1126/scitranslmed.3000322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P. J., Ley R. E., Mahowald M. A., Magrini V., Mardis E. R., Gordon J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444 1027–1031. 10.1038/nature05414 [DOI] [PubMed] [Google Scholar]
- Ulger Toprak N., Yagci A., Gulluoglu B. M., Akin M. L., Demirkalem P., Celenk T., et al. (2006). A possible role of Bacteroides fragilis enterotoxin in the aetiology of colorectal cancer. Clin. Microbiol. Infect. 12 782–786. 10.1111/j.1469-0691.2006.01494.x [DOI] [PubMed] [Google Scholar]
- Van Belleghem J. D., Clement F., Merabishvili M., Lavigne R., Vaneechoutte M. (2017). Pro-and anti-inflammatory responses of peripheral blood mononuclear cells induced by Staphylococcus aureus and Pseudomonas aeruginosa phages. Sci. Rep. 7:8004. 10.1038/s41598-017-08336-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nood E., Vrieze A., Nieuwdorp M., Fuentes S., Zoetendal E. G., De Vos W. M. D., et al. (2013). Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med. 368 407–415. 10.1056/nejmoa1205037 [DOI] [PubMed] [Google Scholar]
- Vasiliauskas E. A., Kam L. Y., Karp L. C., Gaiennie J., Yang H., Targan S. R. (2000). Marker antibody expression stratifies Crohns disease into immunologically homogeneous subgroups with distinct clinical characteristics. Gut 47 487–496. 10.1136/gut.47.4.487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri N. D., Wong J., Pahl M., Piceno Y. M., Yuan J., DeSantis T. Z., et al. (2013). Chronic kidney disease alters intestinal microbial flora. Kidney Int. 83 308–315. 10.1038/ki.2012.345 [DOI] [PubMed] [Google Scholar]
- Venkatesh M., Mukherjee S., Wang H., Li H., Sun K., Benechet A. P., et al. (2014). Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 41 296–310. 10.1016/j.immuni.2014.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vétizou M., Pitt J. M., Daillère R., Lepage P., Waldschmitt N., Flament C., et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350 1079–1084. 10.1126/science.aad1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J., Maksimovic J., Farries G., Sim W. H., Bishop R. F., Cameron D. J., et al. (2013). Bacteriophages in gut samples from pediatric Crohn’s disease patients: metagenomic analysis using 454 pyrosequencing. Inflamm. Bowel Dis. 19 1598–1608. 10.1097/MIB.0b013e318292477c [DOI] [PubMed] [Google Scholar]
- Wang L. W., Tancredi D. J., Thomas D. W. (2011). The prevalence of gastrointestinal problems in children across the United States with autism spectrum disorders from families with multiple affected members. J. Dev. Behav. Pediatr. 32 351–360. 10.1097/dbp.0b013e31821bd06a [DOI] [PubMed] [Google Scholar]
- Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., et al. (2012). Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6 320–329. 10.1038/ismej.2011.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Klipfell E., Bennett B. J., Koeth R., Levison B. S., DuGar B., et al. (2011). Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 472 57–63. 10.1038/nature09922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehkamp J., Salzman N. H., Porter E., Nuding S., Weichenthal M., Petras R. E., et al. (2005). Reduced paneth cell α-defensins in ileal Crohns disease. Proc. Natl. Acad. Sci. U.S.A. 102 18129–18134. 10.1073/pnas.0505256102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Sun W., Yu S., Yang Y., Ai L. (2016). Butyrate production from high-fiber diet protects against lymphoma tumor. Leuk. Lymphoma 57 2401–2408. 10.3109/10428194.2016.1144879 [DOI] [PubMed] [Google Scholar]
- Wiedermann C. J., Kiechl S., Dunzendorfer S., Schratzberger P., Egger G., Oberhollenzer F., et al. (1999). Association of endotoxemia with carotid atherosclerosis and cardiovascular disease: prospective results from the Bruneck Study. J. Am. Coll. Cardiol. 34 1975–1981. 10.1016/S0735-1097(99)00448-9 [DOI] [PubMed] [Google Scholar]
- Wikoff W. R., Anfora A. T., Liu J., Schultz P. G., Lesley S. A., Peters E. C., et al. (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U.S.A. 106 3698–3703. 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willing B. P., Dicksved J., Halfvarson J., Andersson A. F., Lucio M., Zheng Z., et al. (2010). A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139 1844.e1–1854.e1. 10.1053/j.gastro.2010.08.049 [DOI] [PubMed] [Google Scholar]
- Wollowski I., Ji S. T., Bakalinsky A. T., Neudecker C., Pool-Zobel B. L. (1999). Bacteria used for the production of yogurt inactivate carcinogens and prevent DNA damage in the colon of rats. J. Nutr. 129 77–82. 10.1093/jn/129.1.77 [DOI] [PubMed] [Google Scholar]
- World Health Organization [WHO] (2016). Obesity and Overweight. Available at: http://www.who.int/mediacentre/factsheets/fs311/en/ [accessed 1 Jul. 2017]. [Google Scholar]
- World Health Organization [WHO] (2017). Autism Spectrum Disorders. Available at: http://www.who.int/mediacentre/factsheets/autism-spectrum-disorders/en/ [accessed 1 Jul. 2017]. [Google Scholar]
- Wu N., Yang X., Zhang R., Li J., Xiao X., Hu Y., et al. (2013). Dysbiosis signature of fecal microbiota in colorectal cancer patients. Microb. Ecol. 66 462–470. 10.1007/s00248-013-0245-9 [DOI] [PubMed] [Google Scholar]
- Wu S., Lim K. C., Huang J., Saidi R. F., Sears C. L. (1998). Bacteroides fragilis enterotoxin cleaves the zonula adherens protein, E-cadherin. Proc. Natl. Acad. Sci. U.S.A. 95 14979–14984. 10.1073/pnas.95.25.14979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Morin P. J., Maouyo D., Sears C. L. (2003). Bacteroides fragilis enterotoxin induces c-Myc expression and cellular proliferation. Gastroenterology 124 392–400. 10.1053/gast.2003.50047 [DOI] [PubMed] [Google Scholar]
- Wu S., Rhee K. J., Albesiano E., Rabizadeh S., Wu X., Yen H. R., et al. (2009). A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 15 1016–1022. 10.1038/nm.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wu J., Chen T., Li Q., Peng W., Li H., et al. (2018). Fusobacterium nucleatum potentiates intestinal tumorigenesis in mice via a toll-like receptor 4/p21-activated kinase 1 cascade. Dig. Dis. Sci. 63 1210–1218. 10.1007/s10620-018-4999-2 [DOI] [PubMed] [Google Scholar]
- Xiong Y., Miyamoto N., Shibata K., Valasek M. A., Motoike T., Kedzierski R. M., et al. (2004). Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc. Natl. Acad. Sci. U.S.A. 101 1045–1050. 10.1073/pnas.2637002100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T., Santisteban M. M., Rodriguez V., Li E., Ahmari N., Carvajal J. M., et al. (2015). Gut dysbiosis is linked to hypertension. Hypertension 65 1331–1340. 10.1161/HYPERTENSIONAHA.115.05315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu J. H., Dorweiler B., Woo C. W. (2017). Interaction between gut microbiota and toll-like receptor: from immunity to metabolism. J. Mol. Med. 95 13–20. 10.1007/s00109-016-1474-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yui K., Kawasaki Y., Yamada H., Ogawa S. (2016). Oxidative stress and nitric oxide in autism spectrum disorder and other neuropsychiatric disorders. CNS Neurol. Disord. Drug Targets 15 587–596. 10.2174/1871527315666160413121751 [DOI] [PubMed] [Google Scholar]