Abstract
Objective
To measure maternal gut microbiome biodiversity in pregnancy.
Methods
In phase 1, maternal fecal samples were collected by rectal swab in twenty healthy pregnant women (14–28 weeks gestation) to measure bacterial abundance. In phase 2, fecal samples were collected from 31 women at enrollment (< 20 weeks gestation, baseline) and at 36–39 weeks gestation (follow-up). We assessed cluster analysis to assess bacterial community profiles at the phylum level longitudinally through pregnancy. DNA was extracted from swabs, followed by PCR of the bacterial 16s rRNA gene and multiplex high-throughput sequencing (Ion Torrent).
Results
In phase 1, 16 of 20 samples yielded usable data. White women (n=10) had greater abundance of Firmicutes (23 ± 0.15 vs 16% ± 0.75, p =0.007) and Bacteroidetes (24 ± 0.14 vs 19% ± 0.68, p = 0.015) compared to non-White women (n=6). In the 11 paired specimens, Bacteroidetes increased in abundance from baseline to follow-up. Compared with women who gained below the median gestational weight gain (GWG) (<15.4 kg), those who gained above the median GWG had increased abundance of Bacteroidetes (p=0.02) and other phyla (p=0.04).
Conclusion
Maternal microbiome biodiversity changes as pregnancy progresses and correlates with GWG.
Keywords: gut microbiome, pregnancy, gestational weight gain, intestinal microflora
Introduction
We are becoming increasingly aware that the gut microbiome plays a key role in healthy weight maintenance, nutritional absorption and development of obesity. The local composition of the gut microbiome impacts the efficiency of nutrient absorption from the colon.1,2 Obesity in mice is associated with changes in the relative abundance of the two dominant bacterial phyla, Firmicutes and Bacteroidetes.3,4 In mice, an obese ‘microbiome’ (greater presence of Firmicutes and less of Bacteroidetes) harvests more calories from the diet than a ‘lean microbiome’, and consequently, is more likely to cause weight gain and fat deposition.1 In studies of non-pregnant humans, altered gut microbiota has also been associated with excessive weight gain and obesity.5,6 In a study of 91 Finnish pregnant women, Koren et al. found that maternal gut microbiome diversity between mothers expanded dramatically between the first and third trimesters with overall increase in Proteobacteria and Actinobacteria in pregnancy and that individual women underwent a decrease in diversity.7 When microbiome was transferred to germ-free mice, the third trimester microbiota increased fat deposition, inflammation, and insulin insensitivity compared to first trimester microbiota. These findings suggest that microbiota changes over pregnancy may facilitate the metabolic alterations seen in late gestation. Most women in the above-mentioned Finnish study were White and had a pre-pregnancy body mass index (BMI) < 25 kg/m2 (58%), and therefore are likely not generalizable to U.S. women. The objective of our study was two-fold: 1) to define maternal gut microbiome in a diverse population of U.S. women by gestational age and race, and 2) to examine correlation between maternal gut microbiome changes over pregnancy with maternal (obesity and antibiotic use) and clinical [gestational weight gain (GWG) and infant birth weight) characteristics.
Materials and methods
Study Participants and Specimen Collection
This study was conducted in two phases. In phase 1, our objective was to define the maternal gut microbiota at the phylum level in our obstetric population. We conducted a cross-sectional study of twenty non-diabetic pregnant women enrolled between 14–28 weeks’ gestation. Race was self-reported. Maternal fecal samples were collected by inserting two sterile Dacron swabs 1–2 inches into the rectum and held for 15–20 seconds. The primary outcome for phase I was description of microbiota at the phylum levels at early (14–21 weeks) and late (22–28 weeks) second trimester gestational age, and by maternal race.
The purpose of phase 2 was to assess the change in maternal microbiome over pregnancy in our obstetrics population. We conducted a longitudinal study of 31 pregnant women, enrolled for the first study visit at less than 20 weeks gestation (baseline) with a second study visit at 36–39 weeks gestation (follow-up). We excluded women with any condition associated with immunosuppression (e.g., pre-existing diabetes, HIV/AIDS, inflammatory bowel disease, end stage renal disease, Crohn’s disease, etc.) or taking immunosuppressive medications (e.g., daily corticosteroid, immunomodulators). Maternal rectal swab samples were collected at both study visits. In phase 2, we excluded women from analysis for whom we did not obtain a follow-up sample because they delivered preterm prior to sample collection (n=7), had a second trimester miscarriage (n=1) or transferred to another facility (n=2). The primary outcome for phase II was phylum and genus level microbiota change over two collection time points, baseline and follow-up. We assessed phylum and genus level microbiota change by race/ethnicity, which was self-reported. Because the majority of our population were White women, we also assessed the change of phylum and genus level change over gestational age only among White women. We also assessed change in phylum and genus level microbiota abundance by those who gained above and below the median gestational weight gain (GWG) for this cohort, early pregnancy obesity (defined as maternal body mass index ≥ 30 kg/m2 at first visit), antibiotic exposure during pregnancy, and infant birth weight among singleton gestation. We describe the alpha diversity, the number of sequences in a sample; richness, the number of species present in a sample; and evenness, the relative abundance of different species that make up the richness in a sample. This study was reviewed and approved by UNC - Chapel Hill School of Medicine Institutional Review Board.
Microbiota Analysis
DNA Extraction
Rectal swabs were placed into a cryogenic vial containing PBS and were stored (immediately or within 60 minute timeframe) at −80 °C. Prior to DNA extraction, the cryogenic vial containing the swab was thawed to room temperature, then vortexed for 60 seconds to release all cells from the swab. DNA extraction was performed on the bacterial cells from the swabs using a modified protocol of the DNeasy Blood and Tissue kit (Qiagen). Cells suspended in PBS were pelleted by centrifugation, then the supernatant was removed followed by the addition of lysozyme (20 mg/ml), mutanolysin (25 ku/ml), and kit buffer ATL and incubation at 37 °C for 30 minutes. After incubation, proteinase K was added and the samples were incubated at 56 °C overnight. Bead-beating was performed on the samples using 0.5 mm stainless steel beads in a Bullet Blender (Next Advance, Averill Park, NY). The supernatant was removed from the beads and used to carry out the remainder of the extraction according to kit protocol.
Ion Torrent Library Preparation and Sequencing
For amplicon library preparation, we used fusion primers comprising Ion Torrent adapter 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′ for the forward primer and 5′-CCTCTCTATGGGCAGTCGGTGAT-3′ for the reverse primer, and universal bacterial primer 8F 5′-AGAGTTTGATCCTGGCTCAG-3′ and 338R 5′-GCTGCCTCCCGTAGGAGT-3′. The forward primer also included a 10bp IonXpress™ barcode, unique to each sample. Each bacterial DNA sample was run in duplicate in a 25 μL PCR reaction containing: 4 μL of 5× MyTaq Reaction Buffer (Bioline, London, UK); 0.6 μL each of 15 μM Forward Primer and 15 μM Reverse Primer (Integrated DNA Technologies, Coralville, Iowa); 0.5 μL MyTaq HS DNA Polymerase (Bioline); 100 ng template DNA; water to 25 μL. Samples were denatured at 94 °C for 5 minutes, followed by 35 cycles of 94 °C for 45 seconds, 55 °C for 45 seconds and 72 °C for 90, followed by an extension at 72 °C for 10 minutes and a 4 °C hold.
PCR visualization and purification were combined into a single step using the E-Gel Electrophoresis System and an E-Gel Size-Select Agarose Gel, 2% (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. Samples were quantified using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). The purified PCR product was then combined in equimolar amount concentration to create a library. The library was sequenced on the Ion Torrent PGM Instrument (Life Technologies, Carlsbad, CA) according to the manufacturer’s protocol.
16S rRNA Sequence Analysis and Statistical Analysis
The bacterial 16S rRNA sequences of V4 region were processed to remove low quality reads and analyzed in QIIME.8 Sequences were assigned to operational taxonomic units (OTUs) using the Greengenes database.9 The number of sequences assigned to each OTU was used to generate a data matrix that was used for downstream multivariate and microbiota diversity analyses. Raw counts were standardized, transformed (Log (X+1)), and used to build a resemblance matrix. We performed cluster analysis using PRIMER 7 (Lutton, Ivybridge, UK) to determine whether bacterial profiles differed by gestational age and longitudinally through pregnancy at two time points. Differentially abundant taxa between groups or time points were identified using Metastats which included correction for multiple testing.10 A corrected p-value <0.05 was considered statistically significant.
Results
In phase 1, sixteen (80%) samples had detectable DNA (Table 1). Eight (50%) were obtained at 14–21 weeks’ gestation and eight (50%) at 22–28 weeks’ gestation. Ten (63%) were from White women and six (37%) were from non-White women (six African-American). Among women in the early second trimester group (14–21 weeks), there was a greater abundance of Proteobacteria (20 ± 0.40 vs 16% ± 0.25, p=0.06) compared to the late second trimester group, however this difference was not statistically significant. There was no difference in the overall abundance of Bacteroidetes (21 ± 0.19 vs 22% ± 0.25, p = 0.9) or Firmicutes (20 ± 0.21 vs 21% ± 0.32, p = 0.45). When we compared early versus late second trimester group samples, there was no difference in microbiota evenness (J′=0.83 ± 0.012 vs 0.80 ± 0.021, p=0.20), richness (S=5 ± 0.33 vs 5.63 ± 0.36, p=0.17), or diversity (H′=1.32 ± 0.05 vs 1.35 ± 0.05, p=0.38) at the phylum level. When we compared microbiota characteristics by maternal race, we found that compared to Non-White women, White women have greater richness (N=7.1 ± 0.25 vs 2.34 ± 0.46, p=0.007) and diversity (H′=0.85 ± 0.03 vs 0.33 ± 0.06, p=0.01) but not evenness (J′= 0.57 ± 0.02 vs 0.28 ± 0.02, p=0.09) at the phylum level. White women had significantly higher abundance of Firmicutes bacteria (23 ± 0.15 vs 16% ± 0.75, p=0.007) and Bacteroidetes (24 ± 0.14 vs 19% ± 0.68, p=0.02) and lower abundance of Synergistetes (5.6 ± 0.42 vs 6.7% ± 1.10, p = 0.02) compared to non-White women (Table 2).
Table 1.
Total women enrolled and study samples available for analysis
| Phase 1 | |
| Total number of women enrolled | 20 |
| Undetectable DNA on sample | 4 |
| Total number of samples with DNA results | 16 |
| Phase 2 | |
| Total number of women enrolled at Baseline | 41 |
| Undetectable DNA at baseline | 24 |
| Total number of baseline samples with DNA results | 17 |
| Potential women available for follow-up | 41 |
| Women lost to follow-up | 16 |
| Total number of women with follow-up samples | 25 |
| Total number of women with singleton gestation with available follow-up samples | 24 |
| Total number of samples with DNA results matched at baseline and follow-up | 11 |
Table 2.
Phylum-Level Bacterial Abundance by maternal race from Phase 1 (n=16)
| White Abundance (%) |
Non-White Abundance (%) |
* p-value | |
|---|---|---|---|
| Actinobacteria | 16.50 ± 0.18 | 10.36 ± 0.84 | 0.054 |
| Bacteroidetes | 23.57 ± 0.14 | 18.83 ± 0.68 | 0.015 |
| Firmicutes | 23.20 ± 0.15 | 16.08 ± 0.75 | 0.007 |
| Fusobacteria | 2.77 ± 0.28 | 14.10 ± 1.71 | 0.055 |
| Proteobacteria | 17.85 ± 0.30 | 18.90 ± 1.11 | 0.548 |
| Synergistetes | 5.57 ± 0.42 | 6.70 ± 1.10 | 0.024 |
| Other | 10.54 ± 0.24 | 15.04 ± 1.53 | 0.211 |
Adjusted for multiple correction testing
In phase 2, eighteen women self-identified as White (58%), eight (26%) as African-American, and five as Hispanic (16%). The median pre-pregnancy BMI was 27.0 kg/m2 (interquartile range (IQR) 22, 36), median gestational weight gain (GWG) was 15.4 kg (IQR 17.5, 37.5), and median infant birth weight for singletons was 3291 g (IQR 2505, 4077). Three women had twin pregnancies. Ten women (32%) reported having had infections during pregnancy (urinary tract infection, pyelonephritis, gastroenteritis, influenza, chlamydia) and nine of these women received antibiotics during pregnancy. We enrolled women in early gestation and did not exclude women based on antibiotic use. While women did not collect a diet history, one woman reported a lacto-ovo vegetarian diet. No women used probiotics supplements during their pregnancy.
Eleven women (35%) had paired samples (baseline and follow-up) available for analysis (Table 1). A total of twenty women (65%) had one sample available for analysis (six baseline and fourteen follow-up).
Matched samples
Among the 11 paired samples, 5 distinct phyla were observed (Bacteroidetes 30%, Firmicutes 37%, Proteobacteria 18%, Actinobacteria 9%, Fusobacteria 3%, Other 3%). The “Other” category is made up of phyla that contributed <1% of the overall bacterial abundance namely: Acidobacteria, Cyanobacteria, Spirochaetes, Synergistetes, Tenericutes, and Verrucomicrobia. At the phylum level, there was no difference in overall richness at baseline compared to follow-up (d= 2.07 ± 0.09 vs 1.94 ± 0.07; p=0.27) while evenness (J=0.66 ± 0.04 vs 0.79 ± 0.02, p=0.01) and diversity (H=1.12 ± 0.05 vs 1.34 ± 0.03; p=0.002) were increased from baseline to follow-up. Between baseline and follow-up samples, there was an increase in relative abundance of Actinobacteria, Proteobacteria and Fusobacteria and a decrease in relative abundance of Bacteroidetes and Firmicutes, although these trends were not statistically significant (Table 3).
Table 3.
Phylum changes in maternal gut microbiome among matched samples from baseline (<20 weeks) and follow-up (36–39 weeks) (n=11)
| Phylum | Baseline Abundance (%) |
Follow-up Abundance (%) |
* p-value |
|---|---|---|---|
| Actinobacteria | 4.9 ± 0.21 | 12.2 ± 0.32 | 0.08 |
| Bacteroidetes | 31.7 ± 0.25 | 29.0 ± 0.23 | 0.30 |
| Firmicutes | 41.7 ± 0.29 | 33.8 ± 0.21 | 0.39 |
| Fusobacteria | 2.6 ± 0.23 | 3.1 ± 0.29 | 0.53 |
| Proteobacteria | 17.5 ±0.46 | 18.6 ± 0.37 | 0.82 |
| Other | 1.6 ± 0.07 | 3.3 ± 0.18 | 0.28 |
Adjusted for multiple correction testing
There were also significant differences in the relative abundance of several taxa between baseline and follow-up, including Actinomyces (phylum Actinobacteria), Finegoldia (phylum Firmicutes), Anaerococcus (phylum Firmicutes), and Eggerthella (phylum Firmicutes), (Table 4). Some taxa (Acidaminococcus (phylum Firmicutes), Pseudomonas (phylum Proteobacteria), Ralstonia (phylum Proteobacteria)) were absent at baseline but markedly present at follow-up.
Table 4.
Genus changes in maternal gut microbiome among matched samples from baseline (<20 weeks) and follow-up (36–39 weeks) (n=11)
| Genus | Baseline % | Followup % | * p-value |
|---|---|---|---|
| Actinomyces | 0.44 ± 0.002 | 6.89 ± 0.147 | 0.011 |
| Anaerococcus | 7.66 ± 0.045 | 10.68 ± 0.094 | 0.008 |
| Butyricimonas | 0.10 ± 0.001 | 0 | 0.001 |
| Eggerthella | 0.02 ± 0.0002 | 0.06 ± 0.001 | 0.007 |
| Facklamia | 0.17 ± 0.0009 | 5.50 ± 0.201 | 0.020 |
| Finegoldia | 2.54 ± 0.0107 | 18.12 ± 0.204 | 0.002 |
| Propionibacterium | <0.00 | 7.72 ± 0.299 | 0.001 |
| Pseudomonas | <0.01 | 3.89 ± 0.153 | 0.001 |
| Ralstonia | <0.01 | 1.23 ± 0.048 | 0.001 |
| Shewanella | <0.01 | 6.55 ± 0.257 | 0.001 |
| Tepidimonas | <0.01 | 2.01 ± 0.079 | 0.001 |
Adjusted for multiple correction testing
Maternal and clinical characteristics
Race
Compared to White women, non-White women had a decrease in relative abundance of Proteobacteria (22 ± 0.34 vs 7.5% ± 0.29, p = 0.01). While not statistically significant, the relative increase in Fusobacteria among non-White women (9%) compared to White women (0.3%) seems to account for this relative change in abundance.
Because of the majority of women in our paired samples were White (n=8, 72%), we also analyzed change in gestational age among only White women. There were no significant differences between baseline and follow-up period at the phylum level. However, at the genus level, there were significant differences including Anaerococcus (phylum Firmicutes), Butryricimonas (phylum Bacteroides), Eggerthella (phylum Actinobacteria), Finegoldia (phylum Firmicutes), Propionibacterium (phylum Firmicutes), Pseudomonas (phylum Proteobacteria), Roseburia (phylum Firmicutes) Shewanella (phylum Proteobacteria) and Tepidimonas (phylum Proteobacteria) (all p <0.05) (Figure 2). Most notably, Roseburia had a 10 fold reduction from 53% relative abundance at baseline to 5.9% (p = 0.027).
Figure 2.
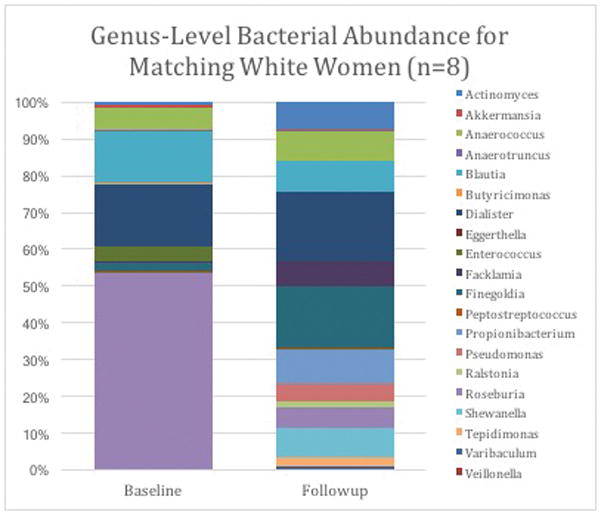
Genus changes in maternal gut microbiome among matched samples from baseline (<20 weeks) and follow-up (36–39 weeks) among White women (n=8)
Gestational Weight Gain
We assessed microbiome change among women who gained above or below the median GWG for our sample (15.4 kg). Compared with those who gained below the median GWG, women who gained above the median GWG had increased relative abundance of Bacteroidetes (p=0.02) and other phyla (p=0.04) from baseline to follow-up. While the difference in Fusobacteria abundance was not statistically significant, this phylum was only seen in 0.3% of women with less than the median GWG compared to 5% of women above the median GWG (p=0.14). For women with above median GWG, there was increased bacterial diversity (p= 0.04) compared to those who gained less than the median GWG.
Maternal Obesity
When we compared obese and non-obese women at baseline, there was no difference between relative abundance of Bacteroidetes (31 ± 0.42 vs 33% ± 0.3, p=0.78) or Firmicutes (41 ± 0.4 vs 42% ± 0.48, p=0.75). At follow-up, there was also no difference in any phylum. While not statistically significant, obese women had a markedly decreased abundance of Fusobacteria (baseline 4.6 ± 0.42 vs 0.03% ± 0.002, p=0.32; follow-up 7.1 ± 0.8 vs 0.72% ± 0.06, p=0.38).
Infant Birth weight
We assessed results for all samples (n=28) from singleton pregnancies to evaluate the relationship between mother’s microbiome and infant birthweight. Median infant birth weight among singletons was 3291 g (IQR 2505,4077). There was also moderate correlation between infant birth weight and Proteobacteria abundance (r = 0.4, p=0.04) and a strong negative correlation with the bacteria in the “Other” category (r=−0.6, p=0.001) (Figure 1).
Figure 1.
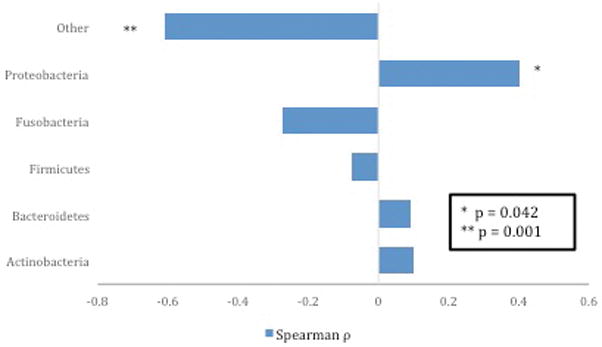
Correlation between singleton infant weight and relative abundance of bacteria at phylum level (n=24)
Antibiotic use
When comparing matched samples for women who received antibiotics for infections during pregnancy, there were three samples for women who received antibiotics and eight samples of women who did not receive antibiotics. Women who received antibiotics had a relative decrease in abundance in Bacteroidetes compared to women who did not receive antibiotics (25 ± 0.21 vs 31% ± 0.21, p < 0.001) (Table 5).
Table 5.
Phylum difference in maternal gut microbiome from follow up samples stratified by antibiotic use (n=10)
| Antibiotics % Abundance |
No Antibiotics % Abundance |
* p-value | |
|---|---|---|---|
| Actinobacteria | 21.45 ± 0.54 | 12.10 ± 0.25 | 0.1159 |
| Bacteroidetes | 24.74 ± 0.21 | 30.99 ± 0.21 | 0.0002 |
| Firmicutes | 35.34 ± 0.32 | 34.22 ± 0.21 | 0.5323 |
| Fusobacteria | 4.39 ± 0.30 | 2.41 ± 0.18 | 0.8816 |
| Proteobacteria | 12.07 ± 0.45 | 17.56 ± 0.32 | 0.3596 |
| Other | 2.01 ± 0.11 | 2.72 ± 0.11 | 0.5621 |
Adjusted for multiple correction testing
Discussion
Our study found that maternal gut microbiome changes over gestation in a population of diverse US women. Over the course of a pregnancy, richness decreased and diversity and evenness increased from early pregnancy (≤ 20 weeks) to late third trimester, similar to the findings of Koren et al.7 Women who gained above the median GWG had an increase in bacterial diversity compared to those who gained less than the median. In the entire cohort, some taxa were absent at baseline but markedly present at follow-up, suggesting the bacterial shifts are associated with changes in gestational age. Our results indicate gut microbiome is dynamic between and among pregnant women and correlates with clinically important characteristics including race, maternal obesity, GWG and infant birth weight. Our study corroborates the findings of others that the maternal gut microbiome shifts over the course of pregnancy, however, whether these changes reflect normal physiological change or dysbiosis leading to adverse outcomes remains unknown.
To date, we have contradictory information regarding changes in the microbiome during pregnancy. Koren et al. observed increased abundance of Proteobacteria and Actinobacteria from trimester 1 to trimester 3.7 In our Phase 1 results, we found that Proteobacteria was correlated with earlier gestation (≤ 21 weeks) and Black race. In our phase 2 results, we found that Proteobacteria was not different between baseline and follow-up. In phase 2, we did find that Actinobacteria increased between the time points but results did not reach statistical significance. In Koren’s study, Proteobacteria was associated with increased inflammatory markers. While we did not assess markers of inflammation in our study, in studies of non-pregnant individuals, high abundance of Proteobacteria has been associated with individuals with gastrointestinal disease compared to healthy individuals.11,12 In pregnancy, both Black race and early gestation are associated with increased pro-inflammatory states,13 a potential explanation for the increased abundance of Proteobacteria in our study. Our study did not find a correlation between maternal obesity and relative abundance of bacteria. In contrast, Collado et al. found that overweight pregnant women had significantly higher levels of Bacteroides and Staphylococcus, while Santacruz et al. detected an increase in Staphylococcus in overweight pregnant women and a decrease in Bifidobacterium (phylum Actinobacteria) and Bacteroides (phylum Bacteroidetes).14,15 Currently, the available literature regarding the maternal gut microbiome is contradictory. Conclusions about the normal or abnormal constitution or changes over pregnancy are premature.
Our study had challenges. In our study, 80% of rectal samples had detectable DNA. We attribute the lack of results in 20% of our sample to our technique of using rectal swabs as opposed to fecal samples, which has been used by others.7,14,15 This may have impacted our ability to obtain detectable DNA in some samples. We chose to use rectal swabs as we could obtain them directly in clinic and not rely on women providing a stool sample. We believe that the smaller amount of fecal material obtained using a rectal swab as opposed to a fecal sample likely explains the relatively high proportion of women with samples that had undetectable DNA on rectal swab. Another possibility explanation for the frequency of undetectable DNA is the presence of PCR inhibitors preventing the sample from amplifying.16,17 In designing studies of future gut microbiome in pregnancy, researchers should be aware that rectal swab technique may lead to a relatively high percentage of women who did not have samples which yielded useable data. Only one woman in our cohort had gestational diabetes (GDM). In Koren’s study, women with GDM had a diminished microbial richness compared to those without GDM, although the microbiota did not differ significantly in composition.
Our most significant limitation was the relatively small number of women in our matched sample. One of our primary objectives was to define maternal gut microbiome in a diverse population of U.S. women race. Of our 20 participants, 70% self-identified as White. While we did detect a difference in relative abundance of Proteobacteria between White and non-White women, we were underpowered to detect other differences in gut microbiome composition, thus these results should be interpreted with caution. We are also limited in our ability to assess if race is a confounder in our analysis of obesity. As described in our results, we found no significant difference among women with BMI ≥ 30 kg/m2. We lacked sufficient sample size to examine class 3 obesity (BMI ≥ 40 kg/m2). Future directions for studies of maternal microbiome with larger sample sizes could address if race and obesity are confounders of maternal microbiome changes. Based on the results of this pilot study, we suggest that larger studies of racially and ethnically diverse pregnant women are needed to characterize the inter and intra- racial and ethnic gut microbiome variability. Because of our relatively small sample size, we may be underpowered to detect differences at the phylum level, however we were able to detect difference at the genus level. We also did not collect detailed dietary logs. While no women reported highly restrictive diets, we did not have detailed dietary data. Diet is known to rapidly affect gut microbiome.18,19 Because of our small sample size, we are also not able to assess difference that could be attributable to seasonal variation.20 While we did assess the effect of antibiotic use in pregnancy, our sample size was small and these results are intended to provide exploratory data. Approximately 30% of pregnant women receive a course of antibiotics during pregnancy,21 future studies should also explore on the impact of antibiotics on maternal microbiome.
In our cohort, the median GWG was 15.4 kg, which corresponds to the high GWG group (16 kg) in Cedergren’s study – one of the key studies correlating adverse maternal and fetal outcomes. Importantly, high GWG does not only affect obese women. In fact, the relative risk of LGA infants and cesarean delivery is higher among women with non-obese pre-pregnancy BMI.21 Reducing fetal overgrowth among obese and non-obese pregnant women could both lessen immediate infant delivery complications and mitigate the intergenerational transmission of obesity.
There are knowledge gaps whether optimization of maternal gut microbiome can improve pregnancy outcomes. In pregnancy, there is significant inter-and intra-individual gut microbiota variability, particularly at the genus level. Our study did not find a change in the Bacteroidetes and Firmicutes phyla but there was significantly higher contribution of Proteobacteria in early second trimester and higher contribution of Fusobacteria in late second trimester. Among non-White pregnant women, there was a higher proportion of Firmicutes compared to White women. While we were able to correlate changes in maternal microbiome abundance with GWG, infant weight and BMI, the full impact of microbiome diversity on maternal and infant outcomes is unknown. Manipulation of maternal gut microbiota could represent an innovative strategy to optimize gestational weight gain and maternal and fetal outcomes, however more data is needed to assess if pregnancy itself accounts for the changes seems in microbiome and whether these changes are physiological or pathological. We suggest that larger studies of US women are needed to fully characterize the maternal gut microbiome by maternal characteristics including race, pre-pregnancy BMI, and antibiotic use during pregnancy and to assess if changes over time correlate with pregnancy outcomes. Once maternal gut microbiome change is better understood, we propose that potential interventional trials for probiotic and/or prebiotic supplementation and/or diet alterations can be properly designed and conducted.
Acknowledgments
We would like to thank the UNC Microbiome Core Facility for Ion Torrent sequencing and analysis. We would also like to acknowledge Kristen McGreevy for initial analysis of pilot data. This work was supported by the UNC Center for Gastrointestinal Biology and Disease (NIH P30 DK 034987).
Footnotes
The authors report no conflict of interest.
References
- 1.Turnbaugh PJ, Ley RE, Mahowald MA, et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 2.Jumpertz R, Le DS, Turnbaugh PJ, et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94(1):58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley RE, Backhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 5.Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arora T, Sharma R. Fermentation potential of the gut microbiome: implications for energy homeostasis and weight management. Nutrition reviews. 2011;69(2):99–106. doi: 10.1111/j.1753-4887.2010.00365.x. [DOI] [PubMed] [Google Scholar]
- 7.Koren O, Goodrich JK, Cullender TC, et al. Host remodeling of the gut microbiome and metabolic changes during pregnancy. Cell. 2012;150(3):470–480. doi: 10.1016/j.cell.2012.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol. 2009;5(4):e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology. 2014;146(6):1449–1458. doi: 10.1053/j.gastro.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pflughoeft KJ, Versalovic J. Human microbiome in health and disease. Annual Review of Pathology: Mechanisms of Disease. 2012;7:99–122. doi: 10.1146/annurev-pathol-011811-132421. [DOI] [PubMed] [Google Scholar]
- 13.Picklesimer AH, Jared HL, Moss K, et al. Racial differences in C-reactive protein levels during normal pregnancy. Am J Obstet Gynecol. 2008;199(5):523.e521–526. doi: 10.1016/j.ajog.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collado MC, Isolauri E, Laitinen K, Salminen S. Distinct composition of gut microbiota during pregnancy in overweight and normal-weight women. Am J Clin Nutr. 2008;88(4):894–899. doi: 10.1093/ajcn/88.4.894. [DOI] [PubMed] [Google Scholar]
- 15.Santacruz A, Collado MC, Garcia-Valdes L, et al. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr. 2010;104(1):83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 16.Budding AE, Grasman ME, Eck A, et al. Rectal swabs for analysis of the intestinal microbiota. PLoS One. 2014;9(7):e101344. doi: 10.1371/journal.pone.0101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Araújo-Pérez F, McCoy AN, Okechukwu C, et al. Differences in microbial signatures between rectal mucosal biopsies and rectal swabs. Gut microbes. 2012;3(6):530–535. doi: 10.4161/gmic.22157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Filippo C, Cavalieri D, Di Paola M, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences. 2010;107(33):14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davenport ER, Mizrahi-Man O, Michelini K, et al. Seasonal variation in human gut microbiome composition. PLoS One. 2014;9(3):e90731. doi: 10.1371/journal.pone.0090731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.DeVader SR, Neeley HL, Myles TD, Leet TL. Evaluation of Gestational Weight Gain Guidelines for Women With Normal Prepregnancy Body Mass Index. Obstet Gynecol. 2007;110(4):745–751. doi: 10.1097/01.AOG.0000284451.37882.85. doi:710.1097/1001.AOG.0000284451.0000237882.0000284485. [DOI] [PubMed] [Google Scholar]


