Abstract
Background
Coagulase negative staphylococci (CoNS) are commensal bacteria on human skin. Staphylococcus lugdunensis is a unique CoNS which produces various virulence factors and may, like S. aureus, cause severe infections, particularly in hospital settings. Unlike other staphylococci, it remains highly susceptible to antimicrobials, and genome-based phylogenetic studies have evidenced a highly conserved genome that distinguishes it from all other staphylococci.
Results
We demonstrate that S. lugdunensis possesses a closed pan-genome with a very limited number of new genes, in contrast to other staphylococci that have an open pan-genome. Whole-genome nucleotide and amino acid identity levels are also higher than in other staphylococci. We identified numerous genetic barriers to horizontal gene transfer that might explain this result. The S. lugdunensis genome has multiple operons encoding for restriction-modification, CRISPR/Cas and toxin/antitoxin systems. We also identified a new PIN-like domain-associated protein that might belong to a larger operon, comprising a metalloprotease, that could function as a new toxin/antitoxin or detoxification system.
Conclusion
We show that S. lugdunensis has a unique genome profile within staphylococci, with a closed pan-genome and several systems to prevent horizontal gene transfer. Its virulence in clinical settings does not rely on its ability to acquire and exchange antibiotic resistance genes or other virulence factors as shown for other staphylococci.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-4978-1) contains supplementary material, which is available to authorized users.
Keywords: Staphylococcus lugdunensis, Comparative genomics, Pan genome, Core genome, Toxin/antitoxin, Restriction-modification, CRISPR
Background
Staphylococcus lugdunensis is a commensal bacterium found on human skin that has been reported as a cause of severe infections in hospital and community settings [1]. Its clinical virulence clearly distinguishes this coagulase-negative staphylococcus (CoNS) from others in the genus. It appears closest to S. aureus in terms of clinical significance and virulence; infection rates may reach 40% when, typically, hospital microbiology laboratories consider infection rates of 25% or less for other CoNS [2]. In vitro studies have revealed the existence of several putative virulence factors, such as haemolysins, adhesion proteins, and one protease that might constitute the cornerstone of S. lugdunensis virulence [3, 4]. Recent genomic studies have demonstrated some general characteristics of this particular CoNS. Its genome is closer to that of S. aureus than other CoNS, possessing several mobile genetic elements (MGEs) such as plasmids and prophages, which have been described at a genetic level in seven strains, although these do not seem to support the virulence profile of this bacterium [5, 6]. In contrast, MGEs in the form of plasmids, phages, phage-related chromosomal islands (PRCIs, including S. aureus pathogenicity islands SaPIs), transposons, staphylococcal cassette chromosomes (SCCs), integrative and conjugative elements, accounting for up to 25% of the genome of S. aureus, are also widespread in other CoNS, and play a crucial role in the modulation of their virulence [7]. Surprisingly, S. lugdunensis, in contrast to all other staphylococci, displays a highly conserved antibiotic sensitivity profile, and methicillin resistance is extremely rare even in hospital settings, notwithstanding the few SCC mec-bearing strains that have been described [3, 8, 9]. Prophages, plasmids, and SaPIs usually bear S. aureus pore-forming toxins and superantigen enterotoxin coding sequences [10]. The existence of such a large repertoire of MGEs in staphylococci is evidence of an open pan-genome with a constantly increasing collection of distinct genes [11–13]. This is exemplified in S. aureus and S. epidermidis, despite their core genome being limited and remarkably conserved, favoring their clonality. In contrast to sexual species such as Streptococcus pneumoniae, recombinations are very rare in S. aureus, and to a lesser extent in S. epidermidis. Similarly, Chassain et al. found that S. lugdunensis also presents a clonal population structure in multilocus sequence typing (MLST) studies with an allelic polymorphism even lower than in S. aureus and S. epidermidis [14, 15]. This observation, along with the highly conserved antibiotic susceptibility, probably indicates the existence of barriers to horizontal genetic transfer, and correlates with the difficulties experienced in transformation of S. lugdunensis [16–18].
Various genetic elements have been proposed that control genome stability in bacteria [19]. In S. aureus, whose genetic resistance to horizontal gene transfer (HGT) has long been noticed in laboratories, this relative “immunity” mainly relies on a strong restriction-modification (RM) system that also exists in CoNS such as S. epidermidis, and this noticeably impairs phage infectivity [20–23]. To date, four RM systems have been described in staphylococci—Types I, II, III, and IV—with Type II not observed in S. epidermidis [20, 24]. These systems comprise two enzyme factors, a restriction endonuclease and a methyltransferase, which may differ in their subunit composition, sequence recognition, cleavage position, cofactor requirements, and substrate specificity [24]. Heilbronner et al. showed that the S. lugdunensis strain N920143 possessed a functional Type I RM system (SluI), whose inactivation resulted in improved transformation with E. coli plasmid [16].
Clustered regularly interspaced short palindromic repeats (CRISPR) associated with Cas protein (CRISPR/Cas) systems have been described more recently in S. aureus and S. epidermidis, and constitute another strong barrier to foreign DNA uptake, particularly plasmid DNA [25–27]. In S. aureus and CoNS, Class 1 Type IIIA CRISPR/Cas systems have been predominantly identified, containing the universal cas1–2 genes in addition to cas6, and csm1 to csm6 [28, 29]. Class 2 Type IIC CRISPR/Cas systems have also been identified in staphylococci, containing the cas9 gene in addition to the cas1–2 genes. Rossi et al. screened 122 genomes from 15 species of CoNS and found that only 15% of them harbored complete CRISPR/Cas systems, mainly from Type IIIA (Cas6-associated system) and Type IIC (Cas9-associated system) [28]. It has been proposed that this low abundance of CRISPR/Cas systems in CoNS (compared to other bacteria among which 40 to 50% bear CRISPR/Cas systems) could be linked to their role as gene reservoirs for other staphylococci such as S. aureus, particularly for antibiotic resistance genes [29].
Toxin/antitoxin (T/AT) systems form a third group of systems that might prevent foreign DNA uptake in bacteria, including S. aureus and CoNS. If, as has been proposed, their role in controlling bacterial growth and metabolic processes is central, then these systems could protect their host from phages and other MGE acquisition [30, 31]. Currently, various models have been described in S. aureus, including some among Type I systems (SprA1/SprA1AS, SprF/SprG), Type II systems (MazEF, PemIK, YefM-YoeB, Omega/Epsilon/Zeta), and Type III systems (tenpIN). MazEF was originally described in E. coli and was the first chromosomal T/AT system reported in S. aureus. Since then, a MazEF system has also been characterized in S. equorum, and several orthologues have been described in Gram-positive bacteria, but not in CoNS (other than S. equorum), even though the presence of such systems might be expected considering their wide distribution [32]. To date, the MazEF system from S. aureus is the best characterized, particularly through the work of Schuster et al. [33–36].
The extremely conserved antibiotic sensitivity profile of S. lugdunensis, along with the existence of various MGEs in this pathogenic species, motivated our study to explore its core and pan-genome profiles through comparative genomics analysis, and to further research the presence of barriers to HGT.
Results
Comparative genomics
The core and pan-genome development plots of S. lugdunensis, S. aureus, and S. epidermidis are shown in Fig. 1 and Additional file 1. S. aureus and S. epidermidis possess an open pan-genome that constantly grows as new genomes are added, reaching 3864 genes for S. epidermidis after the inclusion of 13 genomes, and 3828 genes for S. aureus after the inclusion of 15 genomes. In contrast, S. lugdunensis seems to possess a closed pan-genome that rapidly plateaus at under 3000 genes even after the inclusion of 15 genomes. The core genome of the 3 species displays a similar evolution, and rapidly stagnates at close to 2000 genes. Core and pan-genome development extrapolations gave the same results, projecting a constantly increasing number of genes in the pan-genome of S. aureus and S. epidermidis while projecting S. lugdunensis to plateau at under 3000 genes (Fig. 1 and Additional file 1). Growth exponent value was 0.066 (95% confidence interval 0.065–0.067) for S. lugdunensis versus 0.217 (95% confidence interval 0.214–0.220) and 0.123 (95% confidence interval 0.121–0.124) for S. epidermidis and S. aureus, respectively (Additional file 1). Core genome trends are similar, rapidly becoming limited to about 2000 genes for the three species. The extrapolated core genome sizes were 1944 (95% confidence interval 1933–1956) for S. lugdunensis, 2099 (95% confidence interval 2091–2015) for S. aureus, and 1811 (95% confidence interval 1807–1817) genes for S. epidermidis, respectively (Additional file 1). As suggested by MLST studies, S. lugdunensis might even possess a conserved core genome with average nucleotide identity (ANI) ranging from 99.5 to 99.9%, whereas it ranges from 97.5 to 99.8% for S. aureus and 96.6 to 99.7% for S. epidermidis (detailed results in Additional file 2) [14, 15]. Average amino acid identity (AAI) ranges were similar, from 99.5 to 99.9% for S. lugdunensis, 98.5 to 99.9% for S. aureus and 98.3 to 99.7% for S. epidermidis.
Fig. 1.
Pan-genome and core genome development plot projections for S. lugdunensis (Panel a and b), S. epidermidis (Panel c and d), and S. aureus (Panel e and f). All calculations were done using the EDGAR software platform. For pan-genome development plot extrapolation: the red curve shows the fitted exponential Heaps’ low function, and the blue and green curves indicate the upper and lower boundary of the 95% confidence interval. For Core genome development plot extrapolation: the red curve shows the fitted exponential decay function, and the blue and green curves indicate the upper and lower boundary of the 95% confidence interval
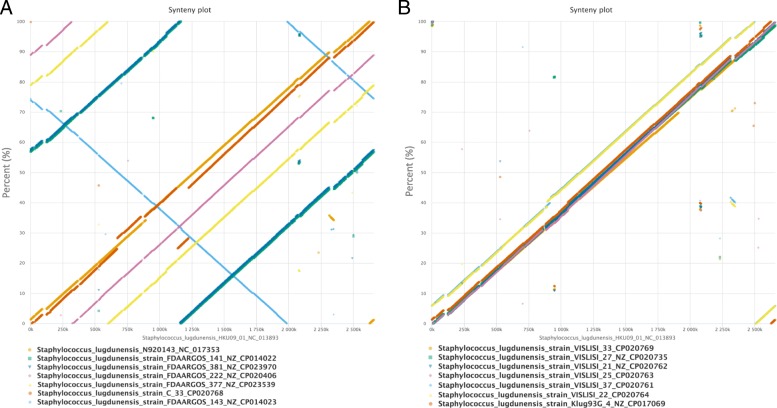
A phylogenetic tree created for all three species together shows a clear separation of S. aureus, S. epidermis, and S. lugdunensis (see Fig. 2). They all form monophyletic branches that are clearly separated from each other. Phylogenetic distances were very small in general, with the S. lugdunensis branch showing the lowest distances between within-species branches. Structural genomic analysis of S. lugdunensis genomes gave further evidence of the highly conserved genomes of the S. lugdunensis species. With the exception of small translocations in strains C33, VISLISI 37 and VISLISI 22, all 15 compared genomes show a highly conserved gene order, with no signs of larger genomic rearrangements. This again demonstrates the genomic stability of S. lugdunensis (Fig. 3).
Fig. 2.
Phylogenetic tree illustrating genetic relationships between strains. The phylogenetic tree was built based on the complete core genome of the analyzed strains. Alignments of the each individual core gene set were generated using MUSCLE and subsequently concatenated to one large supermatrix. Fasttree was used to infer a maximum likelihood tree from this core gene alignment. Shimodaira-Hasegawa support values. Calculations were made for the three species and show that they form three clearly separated clusters within the phylogenetic tree
Fig. 3.
Structural genomic comparison of S. lugdunensis reference strain HKU0901 with the 14 other complete S. lugdunensis genomes. Synteny plots were produced by using EDGAR web server, showing the stop positions of orthologous gene pairs in different genomes. S. lugdunensis HKU09101 genome was compared with the 14 other genomes by splitting the analysis in two for better visual representation (panel a and b). Although some genomes have differing start positions, they all show a high degree of co-linearity, indicating a high level of relatedness and a low level of genomic rearrangement activity within the nalayzed set
Functional analysis
To compare core genome functional categories, we used functional assignments from the COG database. Results are shown in Fig. 4. The core gene category repartition was highly similar among the 3 species, exceptions being that S. epidermidis lacks any genes involved in chromatin structure and dynamics, and both S. lugdunensis and S. aureus lack any cytoskeleton category genes.
Fig. 4.
COG functional categories from the core genome of S. lugdunensis, S. aureus, and S. epidermidis strains. Gene lists were predicted using the EDGAR web server, and COG categories obtained by loading them into the WebMGA web server. COG categories are as follows: for cellular processes and signaling, d is cell cycle control, cell division, and chromosome partitioning; m is cell wall/membrane/envelope biogenesis; n is cell motility; o is post-translational modification, protein turnover, and chaperones; t is signal transduction mechanisms; u is intracellular trafficking, secretion, and vesicular transport; v is defense mechanisms; and z is cytoskeleton. For information storage and processing, b is chromatin structure and dynamics; j is translation, ribosomal structure, and biogenesis; k is transcription; and l is replication, recombination, and repair. For metabolism, c is energy production and conversion; e is amino acid transport and metabolism; f is nucleotide transport and metabolism; g is carbohydrate transport and metabolism; h is coenzyme transport and metabolism; i is lipid transport and metabolism; p is inorganic ion transport and metabolism; and q is secondary metabolite biosynthesis, transport, and catabolism. r is for general function prediction only, and s for unknown function
Identification of barriers to HGT in MGEs
S. lugdunensis genome length ranged from 2.5 to 2.7 Mb, with GC content constituting between 33.7 and 33.9% (Table 1). All genomes contained 2397–2584 coding sequences, with 46–60 tRNA, 4–19 rRNA, and all strains displayed one tmRNA. In addition to the phages previously identified in the VISLISI strains, we identified 3 additional prophages. One additional plasmid was retrieved from the strain FDAARGOS_381. We did not identify pathogenicity islands in any of the 15 published genomes for S. lugdunensis. Seven complete prophages were identified (Table 2). Length ranged from 37.7–57 kb, and GC content varied from 33.8 to 35.2%. All are close to known prophages previously identified in S. aureus, S. epidermidis, and S. hominis. Four of the 7 prophages exhibited a Zn2+ carboxy peptidase gene sequence, but no sequences for antibiotic resistance genes or T/AT systems. We identified a CRISPR-associated gene cas2 in the phage from VISLISI_22, but without either CRISPR-associated genes or CRISPR sequence. None of the plasmid sequences retrieved from the GenBank database carried any loci coding for protease, PIN-like domain, T/AT, or CRISPR/Cas. A Type II RM system was identified in C33 pVISLISI_5, and sequence analysis is detailed below.
Table 1.
S. lugdunensis whole genome sequence content in comparison with S. aureus and S. epidermidis
| Strain | Size (Mb) | GC (%) Content | Gene | Protein | rRNA | tRNA | tmRNA | Plasmids | Phages | PRCIs1 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S. lugdunensis | HKU0901 | 2.7 | 33.9 | 2567 | 2425 | 19 | 61 | 1 | 0 | 1 | 0 |
| N920143 | 2.6 | 33.8 | 2498 | 2383 | 16 | 55 | 1 | 0 | 1 | 0 | |
| FDAARGOS_141 | 2.6 | 33.8 | 2465 | 2350 | 19 | 60 | 1 | 0 | 0 | 0 | |
| FDAARGOS_143 | 2.6 | 33.9 | 2515 | 2347 | 19 | 60 | 1 | 0 | 1 | 0 | |
| FDAARGOS_222 | 2.5 | 33.8 | 2414 | 2261 | 16 | 59 | 1 | 0 | 0 | 0 | |
| Klug93G-4 | 2.6 | 33.8 | 2501 | 2358 | 20 | 69 | 1 | 0 | 0 | 0 | |
| FDAARGOS_377 | 2.6 | 33.8 | 2479 | 2351 | 19 | 60 | 1 | 0 | 0 | 0 | |
| FDAARGOS_381 | 2.6 | 33.8 | 2482 | 2344 | 19 | 60 | 1 | 1 | 0 | 0 | |
| VISLISI_21 | 2.5 | 33.7 | 2437 | 2344 | 6 | 46 | 1 | 0 | 0 | 0 | |
| VISLISI_22 | 2.6 | 33.8 | 2459 | 2353 | 6 | 59 | 1 | 1 | 1 | 0 | |
| VISLISI_25 | 2.5 | 33.8 | 2397 | 2293 | 4 | 48 | 1 | 0 | 0 | 0 | |
| VISLISI_27 | 2.6 | 33.7 | 2508 | 2391 | 7 | 59 | 1 | 1 | 0 | 0 | |
| VISLISI_33 | 2.7 | 33.7 | 2584 | 2465 | 6 | 55 | 1 | 1 | 2 | 0 | |
| VISLISI_37 | 2.6 | 33.7 | 2482 | 2391 | 6 | 52 | 1 | 0 | 1 | 0 | |
| C33 | 2.5 | 33.9 | 2405 | 2292 | 5 | 52 | 1 | 2 | 0 | 0 | |
| S. aureus | MW2 | 2.8 | 32.8 | 2934 | 2778 | 19 | 60 | 1 | 0 | 2 | 1 |
| S. epidermidis | ATCC12228 | 2.6 | 32.0 | 2545 | 2378 | 19 | 60 | 1 | 2 | 0 | 1 |
1 Phage Related Chromosomal Islands (including S. aureus pathogenicity islands)
Table 2.
S. lugdunensis complete prophages and identification of putative barriers to HGT
| Strain | Complete Prophage | Length (kb) | GC% | Total Proteins | Common Phage | Peptidases | T/AT1 | RM2 | CRISPR/Cas Systems |
|---|---|---|---|---|---|---|---|---|---|
| HKU0901 | 1 | 37.7 | 34.57 | 58 | PHAGE_Staphy_PH153 | 0 | 0 | 0 | 0 |
| N920143 | 1 | 49.4 | 34.42 | 63 | PHAGE_Staphy_TEM1234 | Zn2+ carboxy peptidase | 0 | 0 | 0 |
| FDAARGOS_143 | 1 | 57.0 | 35.24 | 55 | PHAGE_Staphy_CNPx_NC_0312415 | 0 | 0 | 0 | |
| VISLISI_22 | 1 | 49.4 | 34.3 | 53 | PHAGE_Staphy_StB126 | Zn2+ carboxy peptidase | 0 | 0 | cas2 |
| VISLISI_33 | 1 | 44.4 | 33.8 | 52 | PHAGE_Staphy_StB12 | Zn2+ carboxy peptidase | 0 | 0 | 0 |
| 1 | 28.1 | 34.4 | 52 | PHAGE_Staphy_PH15 | 0 | 0 | 0 | 0 | |
| VISLISI_37 | 1 | 47.0 | 34.35 | 58 | PHAGE_Staphy_StB12 | Zn2+ carboxy peptidase | 0 | 0 | 0 |
1 T/AT systems
2 RM systems
3 PHAGE_Staphy_PH15: S. epidermidis phage
4 PHAGE_Staphy_TEM123: S. aureus phage
5 PHAGE_Staphy_CNPx_NC_031241: S. epidermidis phage
6 PHAGE_Staphy_StB12: S. hominis phage
Identification of CRISPR/Cas systems
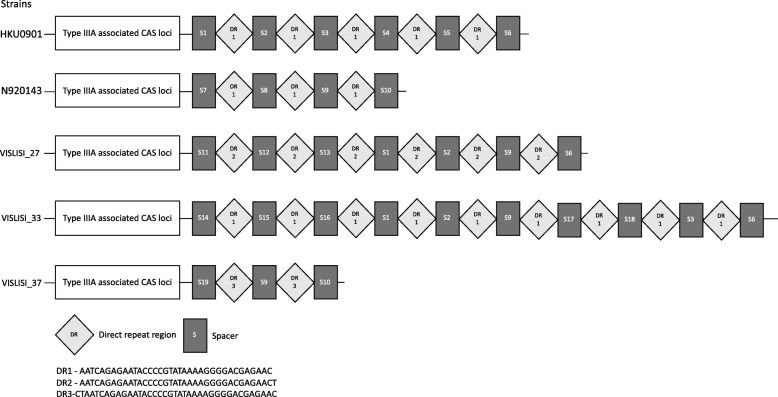
CRISPRFinder identified several CRISPR structures in the 15 S. lugdunensis genomes, some being confirmed CRISPR sequences, others being questionable as CRISPR either because of their small size (with only 2 or 3 direct repeat (DR) sequences), or because the repeat motifs in the CRISPR were not 100% identical. The complete list of all CRISPRs recovered is available in Additional file 3. Overall, 6 confirmed CRISPR/Cas systems were identified in 6 different genomes from the strains: HKU0901, N920143, VISLISI_27, VISLISI_33, VISLISI_37, and C33. Several questionable CRISPRs were identified in all 15 strains, with a total number ranging from 4 to 11 per genome. The genetic environment of all CRISPR sequences was analyzed using ARTEMIS (v.16.0.0), and we identified Cas genes in association with the 6 confirmed CRISPR sequences. The CRISPR/Cas systems from HKU0901, N920143, VISLISI_27, VISLISI_33, and VISLISI_37 corresponded to a Class 1 Type IIIA system according to the classification of Koonin et al., with the conserved modular organization of this family [29]. The adaptation module comprised cas1 and cas2, followed by the small subunit loci csm1 to csm6, the cas6 gene, which is the endonuclease that belongs to the effector module, and finally the CRISPR sequences. Unlike in S. aureus, the CRISPR sequences are located downstream of the cas6 gene, as seen in most other Type IIIA CRISPR/Cas systems in CoNS [26, 28]. These 5 CRISPR/Cas systems were aligned using Easyfig (v.2.2.2), and showed 100% sequence identity regarding the Cas coding sequences (Additional file 4). The CRISPR regions showed variable sequence identity levels, ranging from 71 to 100%. We identified 19 distinct spacers and 3 DRs (Fig. 5). Spacer sequence details are available in Additional file 3. No known origin was found in BLAST for any of the 12 spacers, whereas putative matches were found for 7 of them with sequences that might originate from known MGEs. Results are detailed in Table 3. We also observed that DRs are highly conserved and nearly identical in all 5 strains. In particular, the core region included a CCCC region separated by 8 nucleotides from a GGGG pattern; these could interact to form the typical hairpin structure involved in the initial processing of the CRISPR transcript. This conserved motif is also present in other CoNS and S. aureus [26, 28].
Fig. 5.
DR sequences and spacers in type IIIA CRISPR/Cas systems from S. lugdunensis
Table 3.
Origin of the spacers of the 5 Type IIIA CRISPR/Cas systems from S. lugdunensis strains HKU0901, N920143, VISLISI_27, VISLISI_33, and VISLISI_37
| Spacers | BLAST Match with Known MGEs | Cov1 | ID2 | Score3 |
|---|---|---|---|---|
| S1 | Campylobacter phage CP220 | 81% | 90% | 41 |
| S2 | Clostridium botulinum plasmid pND7 | 74% | 92% | 39 |
| S4 | Lactobacillus plasmid | 78% | 92% | 37 |
| S5 | Bacillus thuringiensis plasmid pAM65–52-2-350 K | 87% | 85% | 39 |
| S6 | Lactobacillus salivarius ZL5006 plasmid | 77% | 89% | 37 |
| S7 | Streptococcus phage IPP55 | 77% | 93% | 39 |
| S15 | Staphylococcus phage vB_SepS_SEP9 | 100% | 89% | 48 |
| S3, S8–14, S16–19 | None |
1 Coverage level
2 Identity level
3 BLAST score
Unlike in the other 5 strains, the C33 complete CRISPR sequence was not associated with Type IIIA Cas coding loci but with Class 2 Type IIC cas genes (as classified according to Koonin et al. [29]), including the cas1 and cas2 genes from the adaptation module, and the cas9 gene, which is the effector of this CRISPR/Cas system type. CRISPR sequences were located upstream of the cas9 gene that displayed several stop codons, making it a pseudogene and the whole operon probably ineffective. Nevertheless, when analyzing the possible BLAST matches of the 11 spacers, only 1 match for the second spacer was observed, for a Bacillus phage sequence (coverage 66%, identity 100%, score 39).
Finally, we performed BLAST searches for the 91 questionable CRISPR sequences that were not associated with any cas loci, since several CRISPRs described in the literature are not associated with cas genes, and conversely, several cas genes might be isolated [37]. We identified one recurrent DR sequence that does not match any of the 3 DRs identified in the Type IIIA complete CRISPR/Cas systems of S. lugdunensis, and that notably lacks the highly conserved CCCC-GGGG motif, which might confer loss of function. Nevertheless, we also identified 1 recurrent spacer that gives an interesting BLAST hit with the S. aureus pathogenicity island SaPI2 from the strain RN3994 (coverage 100%, identity 100%, score 79) [38]. Thus, most of these sequences are probably real orphan CRISPRs that have lost their function and are not misidentified repeated sequences (false CRISPRs).
Identification of T/AT systems
T/AT identification was performed on the available annotations of the 15 annotated S. lugdunensis genomes, and on the de novo annotations that were generated using PROKKA (v1.12). As described for S. equorum and S. aureus, we identified a complete MazEF-rsbUVW-sigB T/AT system in the 15 strains with a conserved operon organization at about 100% BLAST identity (Additional file 5). The mazEF genes are located upstream the of the sigB locus that comprises rsbU, rsbV, rsbW and sigB. We also identified an alanine racemase rac that belongs to this operon (whose role has not been clearly determined to date). MazEF has been reported in the genomes of several Gram-positive bacteria, but among these, the only CoNS representative is S. equorum KM1031 (NCBI accession number NZ_CP013980.1). We therefore extended the search for this system in S. aureus MW2, and 3 other CoNS: S. xylosus strain S170 (NCB accession number NZ_CP013922.1), S. capitis FDAARGOS_378 (NCBI accession number NZ_CP023966.1), and S. epidermidis strain ATCC12228 (NCBI accession number) (Additional file 5). All genes from the MazEF operon of the 15 S. lugdunensis genomes have conserved open reading frames except the strain FDARRGOS_143 in which the MazF coding sequence consisted of nonsense mutations.
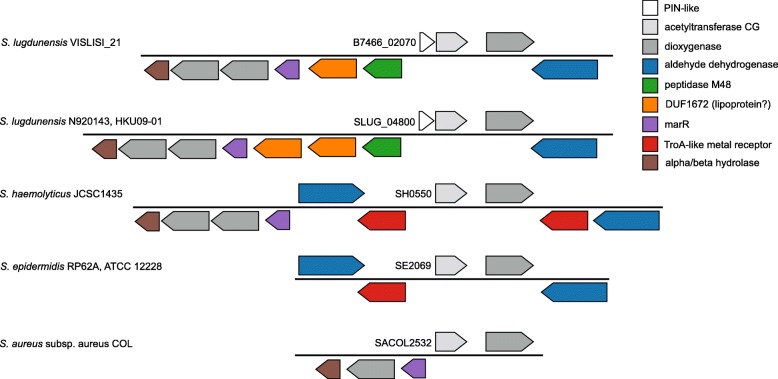
We did not identify any of the other T/AT systems that have been previously described in S. aureus as detailed in the material and methods section. Nevertheless, we found one locus with a predicted PIN-like domain in 13 of the 15 S. lugdunensis genomes. This locus was systematically associated with 1 metalloprotease coding sequence that might belong to the M48 family (according to the MEROPS database), and 1 N-acetyl transferase coding sequence, leading us to hypothesize the presence of a possible T/AT system (Fig. 6) [39]. These 3 loci, and the 10 kb upstream and downstream nucleotide sequences, display 100% sequence identity in the 13 genomes. The PIN-like domain locus could be regarded as a pseudogene rather than a functional PIN gene, since the length of its coding sequence was 41 amino acids, while the minimum length for the PIN fold is ~ 100 amino acids. Also, its protein product lacks a cluster of positively charged residues that would be necessary for nucleic acid binding, and consequently might fail to work as a functional nuclease, as expected for PIN-domain containing proteins. The downstream gene encodes a member of the minimal acetyltransferase CG family (GCN5-related N-acetyl-transferase, Pfam family PF14542). In an article reporting its crystal structure in an S. aureus member of this family, it was suggested that a second protein providing a substrate-binding region must combine with it to yield fully functional N-acetyltransferase [40]. It could be that in S. lugdunensis the binding partner for the GNAT-like protein is the deteriorated PIN protein, and the acetylation target could be the MarR-like (of the HTH fold) protein, encoded on the other strand.
Fig. 6.
Genomic context of the PIN-like domain coding sequence of S. lugdunensis and other staphylococci
Identification of RM systems in the main chromosome of S. lugdunensis.
All 15 genomes were examined for the presence of RM systems using the REBASE database, resulting in the identification of multiple Type I and Type II RM systems (Table 4 and Additional file 6 for genomic coordinates) [23],. Six nearly identical Type I RM systems were identified in HKU0901, N920143, FDAARGOS_143, VISLISI_27, VISLISI_33, and VISLISI_37. The methylase coding sequence displayed 90 to 98% aminoacid identity level with the methylase Sau18 from S. aureus strain C18, a draft S. aureus genome with 123 contigs (NCBI accession number GCA_001921685.1). The methylase target specificity remains unknown. All 6 operons comprised 3 consecutive genes as described for this RM Type: hsdR (restriction locus), hsdM (modification locus), and hsdS (specificity locus). A BLAST alignment performed with Easyfig (v.2.2.2) found nearly 100% sequence identity among all 6. We extended the comparison with S. aureus strain MW2, and with E. coli strain K-12 (GenBank accession number SC000913.3), which bears the canonical Type I RM, EcoKI [41]. We found a very low level of identity between EcoKI and S. lugdunensis VISLISI_33 hsd subunit amino acid sequences, with identity scores ranging from 16 to 25% (according to EMBOSS Needle pairwise sequence alignment). Conversely, we observed higher levels of identity between S. lugdunensis VILSISI_33 and S. aureus MW2 loci, with scores of 71, 58, and 35% for hsdM, hsdR, and hsdS, respectively. Alignment files are available in Additional file 7. Interestingly, despite low AAI levels between hsdR from S. aureus and that from S. lugdunensis, we observed that the essential motifs for DNA cleavage and translocation were highly conserved according to the amino acid sites identified by Roberts et al. This could support the hypothesis that both systems belong to the same subfamily [41]. The strains FDAARGOS_143 and VISLISI_27 have 2 other distinct Type I RM systems, with their methylase coding sequences displaying 97% AAI level with methylase Sca9557 from S. caprae strain C18, and 94% AAI level with SauMSSIII from S. aureus, respectively. Type II RM systems were identified in 4 strains, FDAARGOS_222, VISLISI_22, VISLISI_25, and the plasmid sequence pVISLIS_5 from C33. pVISLISI_5 is a mobilizable plasmid with a repA replication gene, whose closest homologous plasmid is VRSAp from S. aureus (NCBI accession number NC_002774.1) [5]. The closest homologue found for the methylase of pVISLISI_5 (60% identity) was EfaPC41 from Enterococcus faecium strain PC4.1 (and there are no RM systems in VRSAp). The methylase from FDAARGOS_222 and VISLISI_22 showed 82% identity with ShaJ from S. haemolyticus, and the nucleotide sequence specificity was known (GATC). Finally, the Type II RM system from VISLISI_25 was unique among S. lugdunensis strains. The methylase gene was duplicated as seen in its closest homologue, Sep60 from S. epidermidis, and the sequence specificity was known (GGTGA).
Table 4.
RM systems identified in S. lugdunensis using the REBASE database, and homology analysis of methylase
| S. lugdunensis | RM System | Closest Methylase | ||||
|---|---|---|---|---|---|---|
| ID1 | ID score | REBASE score | Strain origin | Nucleotide specificity | ||
| HKU0901 | Type I | M.SauC18 | 98% | 1127 | S. aureus C18 | Unknown |
| N920143 | Type I | M.SauC18 | 91% | 1128 | S. aureus C18 | Unknown |
| FDAARGOS_141 | None | |||||
| FDAARGOS_143 | Type I | M.SauC18 | 90% | 1121 | S. aureus C18 | Unknown |
| Type I | Sca9557 | 97% | 1170 | S. caprae 9557 | Unknown | |
| FDAARGOS_222 | Type II | M.ShaJ | 82% | 844 | S. haemolyticus JCSJ1435 | GATC |
| VISLISI_22 | Type II | M.ShaJ | 82% | 844 | S. haemolyticus JCSJ1435 | GATC |
| VISLISI_25 | Type II | Methylase 1 | ||||
| M2.Sep60 | 66% | 329 | S. epidermidis BCM-HMP0060 | GGTGA | ||
| Methylase 2 | ||||||
| M1.Sep60 | 66% | 558 | S. epidermidis BCM-HMP0060 | GGTGA | ||
| VISLISI_27 | Type I | M.SauC18 | 91% | 1128 | S. aureus C18 | Unknown |
| Type I | M.SauMSSIII | 94% | 1346 | S. aureus MSSA476 | TAAYNNNNNNNTCNNC | |
| VISLISI_33 | Type I | M.SauC18 | 91% | 1128 | S. aureus C18 | Unknown |
| VISLISI_37 | Type I | M.SauC18 | 91% | 1128 | S. aureus C18 | Unknown |
| C33 pVISLISI_5 | Type II | M.EfaPC41 | 60% | 312 | E. faecium PC4.1 | Unknown |
1 Identity
Discussion
This study presents the first comparative genomic analysis of S. lugdunensis, a species emerging as a significant nosocomial pathogen [3]. Pan-genome and core genome analyses revealed that S. lugdunensis displays a closed pan-genome in contrast to all other staphylococci studied to date and to most commensal and pathogenic human bacteria [11, 42–46]. This wholly unexpected observation could be explained by the concomitant identification of several barriers to HGT, namely, CRISPR/Cas, RM, and T/AT loci that constitute specialized systems preventing HGT, particularly through MGEs. Although RM systems are widespread in staphylococci, according to the REBASE database, the identification of T/AT systems in 100% of the 15 S. lugdunensis genomes, and of complete CRISPR/Cas systems in 33% of them, is more surprising. Indeed, in 2017 Rossi et al. found that only 15% of 122 genomes in 15 different CoNS species harbored complete CRISPR/Cas systems [28]. In addition, T/AT systems have been described in only 1 CoNS species, S. equorum. The characterization of multiple HGT prevention systems in a single species, S. lugdunensis, is consistent with the presence of a closed pan-genome. Besides specialized elements such as phages and plasmids, homologous recombination constitutes another frequent modality for HGT, but it is seldom seen in staphylococci, even though such a mechanism may have had an impact on the evolutionary history of lineage separation. Indeed, Meric et al. found evidence that homologous recombination might have changed 40 and 24% of the core genome of S. epidermidis and S. aureus, respectively [13]. However, over a short time scale, such events are extremely rare, and the core genome remains mostly conserved. The high values we found for AAI and ANI from S. lugdunensis genomes, higher even than for S. epidermidis and S. aureus, also suggest that genomic diversity of this species is lower than for other staphylococci, and this observation clearly correlates with the previously reported highly clonal population structure of this species [14, 15].
MGEs are able directly and rapidly (within hours, even) to modify the Staphylococcus accessory genome in vivo via any genetic exchange occurring between S. aureus and S. epidermidis [7]. MGEs are not to be underestimated in their ability to reshape the whole bacterial genome, even in what are usually considered as “immune” species, such as staphylococci (which display a small genome size that reflects evolutionary constraints that probably fitted them to a limited number of hosts). Interestingly, a pan-genome study of the sexual species S. pneumoniae, which is highly susceptible to HGT through homologous recombination, showed a relatively limited pan-genome size that plateaued at under 5000 genes, placing this species on the boundary between an open and closed pan-genome [43]. The openness of the pan-genomes of S. aureus and S. epidermidis obviously relies on MGEs and, if their core genome is highly conserved, their dispensable genomes offer an extremely large repertoire of genes that confer specific advantages in a defined host under particular environmental conditions, and from a clinical point of view, support their virulence [11]. Additionally, in staphylococci, MGEs allow the occurrence of inter-species genetic exchange, providing real potential for CoNS to act as gene reservoirs facilitating the transfer of methicillin resistance to S. aureus, especially since S. aureus has recently been exposed as a putative gene reservoir for CoNS [47, 48]. In this context, the existence of a closed pan-genome in S. lugdunensis (a species emerging as a significant pathogen) and a putative relative immunity to HGT cannot be clearly understood in terms of evolutionary advantage. MGEs facilitate the acquisition of genes conferring antibiotic resistance and thus confer evolutionary advantage in staphylococci such as S. epidermidis and S. aureus, which easily and frequently acquire various resistance genes (one example being methicillin resistance through SCC mec). However, S. lugdunensis remains highly susceptible to most antibiotics, and identification of SCC mec-bearing strains is rare; another illustration of its apparent immunity to HGT.
Another hypothesis could be that S. lugdunensis speciation has occurred only recently, and we are only now experiencing the start of the emergence of new S. lugdunensis clones whose genomes have been augmented by various MGEs originating from other CoNS or S. aureus; and yet, S. lugdunensis has been studied for several years now, even in clinical settings where such genetic exchange should have occurred. In addition, our study included genomes from strains originating not just from one location but from multiple countries and various settings (nosocomial, community, infective, and contaminant strains) (see Additional file 8). Since its first description in 1988 by Freney et al., several phylogenetic studies have suggested that S. lugdunensis always appears to occupy a unique cluster group, whatever the method used for phenotyping (16S rRNA, housekeeping genes, whole genome sequences) [49–51].
The role of the MazEF T/AT system in S. lugdunensis has to be interpreted in the light of its particular location on the bacterial chromosome. If the roles of plasmid T/AT systems have been only partially elucidated, those of chromosomal T/AT are even less well understood (Fernández-García et al. 2016; Lee & Lee 2016; Schuster & Bertram 2016) [33, 49, 50]. It has been suggested that such elements lead to genetic stabilization of various MGEs as prophages or pathogenicity islands, or impact the stress response functions of modular elements of bacterial growth and death [49, 51]. Interestingly, Saavedra De Bast et al. also showed that chromosomal T/AT systems could efficiently act as anti-addiction modules by protecting bacteria against post-segregation killing, a mechanism by which plasmid-encoded T/AT systems favour plasmid maintenance by eliminating daughter bacteria that do not receive a plasmid copy [30]. The widespread occurrence of the MazEF system and its highly conserved nucleic acid sequence do not support the hypothesis that it is a simple remnant of past evolutionary events, and its role needs now to be phenotypically elucidated. By searching T/AT systems, we identified a PIN-associated locus with an undetermined role, although the genetic environment might help us to formulate a hypothesis. Perhaps the PIN protein used to work as a toxin in a toxin-antitoxin system, as a partner for another helix-turn-helix (HTH) folded transcription factor. HTH transcription factors are typical antitoxins for PIN-like toxins. The MarR protein could work as a transcription factor, regulating transcription of other genes involved in that pathway. However, the system could also be independent of the PIN protein, since the homologous operon is absent in other Staphylococcus species (Fig. 6), and acetylation could be performed on a metabolite or an antibiotic [52]. Other genes in the genomic neighborhood would support this hypothesis; they encode dioxygenases, aldehyde dehydrogenases, alpha/beta hydrolases and some metal-binding receptors, which could work in a concerted way to detoxify a specific molecule.
Regarding CRISPR/Cas loci, we identified a complete Type IIIA CRISPR/Cas system in 5 strains among the 15 studied, whereas such systems have been identified in only 15% of CoNS. CRISPR/Cas systems can efficiently prevent plasmid conjugation and transformation, as well as phage infection. This specific observation has been reported in S. epidermidis and S. aureus Type IIIA CRISPR/Cas systems [26]. Our genetic findings need functional confirmation, but a similar observation with S. lugdunensis would be expected. Additionally, we identified several orphan CRISPR sequences with a repeated spacer that might correspond to an extract from the sequence of S. aureus pathogenicity island SaPI2. The significance of such sequences is unknown, but they probably constitute remnants of past genomic events involving MGEs, and their role in S. lugdunensis might be limited or even non-existent due to the absence of functional DR sequences. Isolated CRISPRs can be orphans, though it has been shown that they may be functional in combination with distant cas loci, even where the median distance between CRISPR and corresponding type cas genes is 268 bp for type IIIA, and 103 bp for Type II [37]. Additionally, questionable CRISPRs can also be false CRISPR sequences, corresponding to other kinds of repeated element such as tandem repeats, S. aureus repeat (STAR) elements, or even simple repeat elements [53, 54]. Regarding isolated cas loci, they are widespread in bacterial genomes as remnants of lost CRISPR/Cas complete systems, now without any immune function; however, they could play a role in the maintenance of other functions, such as DNA repair [37].
Finally, we identified several Type I and Type II RM systems in 10 strains among the 15 studied, and 1 Type II RM system in a plasmid sequence. RM systems have many features in common with T/AT systems, one being cell killing in the case of foreign DNA invasion which, in this case, is based on epigenetic identities (methylation level) [55]. RM systems, particularly Type I, are one of the major mechanisms by which S. aureus prevents HGT. Phage- and plasmid-mediated HGT between S. aureus strains from different lineages is strictly controlled by RM systems, particularly Type I [41]. Identification of such systems in CoNS is exceptionally rare; we found only one complete report involving the presence of an RM system in S. epidermidis [20]. The presence of an RM system in S. lugdunensis constitutes a novel barrier for HGT, a situation identified by Heilbronner et al. as the main obstacle for transformation with E. coli using a hsdR mutant [16].
We reported in a previous study the whole genome sequence of seven S. lugdunensis strains and the presence of MGE: plasmids and prophages which genetic content suggested the existence of HGT with CoNS and S. aureus [5]. The results of the present study suggest that such HGT might remain scarce and, if they can mobilize genetic elements between those species, and enrich the whole genome, they are probably too rare to enrich significantly S. lugdunensis pan-genome.
Our study is limited by the number of S. lugdunensis genomes included even if pan and core genome size extrapolation tool that uses a Heaps’ law function, gave concordant results. Among 15 S. lugdunensis genomes, seven originated from a unique location over a 3 year period (VISLISI clinical trial), which might have limited, de facto, the genetic diversity of the genomes. In addition, we did not bring any evidence of a causative link between the presence of several barriers to HGT and a closed pan-genome, we only observed their co-occurrence which is only very suggestive.
Conclusions
S. lugdunensis displays a closed pan-genome, a striking observation for a human pathogenic bacterium, and particularly for a Staphylococcus. This trait is co-occurring with the presence of multiple and dispersed mechanisms that could prevent HGT by MGEs then suggesting their implication in such an unusual pan-genome profile. Functional analysis using knockout mutants is now needed to prove that all the described operons are operational. Also, the presence of such systems in S. aureus, and also more rarely in other CoNS that display an open pan-genome, lead us to hypothesize the existence of other mechanisms. Identification of PIN-like domain-encoding loci, and of several putative nucleases, constitute new pathways that need to be explored.
Methods
The 15 S. lugdunensis genome sequences used in this study, and their associated plasmid sequences, were taken from the GenBank database. S. lugdunensis strain HKU0901 (NCBI accession number NC_013893), whose complete genome sequence was first published in 2010 by Tse et al., was used as a reference in the comparative analyses [56]. Clinical and geographical origins of all 15 strains are listed in Additional file 8. Regarding S. aureus, we randomly selected 1 genomes from the 299 complete sequences in the GenBank database, where 8450 genome assemblies are available, most being draft sequences. The genome of S. aureus strain DSM 20231 (NCBI accession number NZ_CP011526), served as a reference in the comparative analyses. The complete genome of this type strain was determined in 2015 by PacBio single-molecule real-time technology by Shiroma et al., and proposed as a reference strain to perform comparative genomic studies due to its genotypic and phenotypic characteristics [57]. Thirteen complete genomes of S. epidermidis among the 532 genome assemblies available in the GenBank database were included in this study (a fourteenth genome from strain GTH 12 is also available but with no annotation). S. epidermidis strain ATCC_12228 (NCBI accession number NZ_CP022247), a non-biofilm-forming, non-infection-associated strain, was selected as a reference for the comparative analyses. NCBI accession numbers of all strains are listed in Additional file 8. We excluded draft sequences from S. lugdunesis (eight additional genomes) and S. epidermidis (518 additional genomes) for which only scaffolds are available and no finished bacterial chromosome. Those genomes might present several gaps, no or uncomplete annotations, and no error correction steps during assembly process. The use of draft genomes in comparative genomics is questionable, even not recommended, particularly for synteny studies, and we favored stringency in this study. Every gap in draft sequences may split, truncate or completely mask a gene which may add bias to the EDGAR software platform analyses [58, 59].
Whole genome sequence analysis and comparative genomics.
Identification of the core genome and pan-genome was performed using the EDGAR software platform [59]. Several tools have been made available recently as stand-alone, open source, or web-based tools [60]. EDAGR 2.2 is a powerful tool that uses several publicly available genomes but also accommodates customized projects and genomes. The orthology analyses for pan-genome and core genome calculations in EDGAR are performed using BLAST score ratio values (SRV) with an orthology threshold calculated from the analyzed data rather than a fixed cut-off [61]. All calculations are made starting with 1 reference genome. The software allows calculation of pan genome and core genome subsets, as well as statistical extrapolation of core- and pan-genome sizes for a larger number of genomes.
For the statistical extrapolation, it uses non-linear least-squares curve fitting of the observed core and pan genome sizes as function of the number of analyzed genomes. For the core genome extrapolation an exponential decay function is used as described by Tettelin et al., where c is the amplitude of the function, n is the genome number, Ω is the extrapolated size of the core genome for n → ∞, and τ is the decay constant indicating the speed at which f converges to Ω [62].
For the pan genome, a Heaps’ power law function is used, where n is the number of compared genomes, c is a proportionality constant and γ the growth exponent that indicates at which speed the pan genome is growing [63].
Results were compared among S. lugdunensis, S. aureus, and S. epidermidis.
For EDGAR phylogeny analyses, the pipeline uses the complete core genome. Every set of orthologous genes found in all genomes are separately aligned using the multiple alignment tool MUSCLE phylogenetic tree using the maximum likelihood approach as implemented in Fasttree 2 [64]. The trees calculated by Fasttree provide local support values calculated using the Shimodaira-Hasegawa test [65]. The phylogenetic trees were produced from newick file by using Phylogenetic Tree Viewer from ETE Toolkit [66]. Synteny and rearrangements in S. lugdunensis genomes were explored by using EDGAR. S. ludgunensis strain HKU0901 were chosen as a reference to create synteny plots. ANI and AAI matrices were calculated for the 3 species using the EDGAR interface. For AAI matrix calculation, all needed sequence similarity information is available from the BLAST step underlying the EDGAR orthology estimation method. Average nucleotide identity values are computed as described by and as implemented in the popular JSpecies package [67, 68].
Functional analyses
Functional categories of the putative proteins encoded by S. lugdunensis, S. aureus, and S. epidermidis were compared using the clusters of orthologous groups of proteins (COG) database. COG categories were retrieved using the WebMGA software platform, with an e-value cut-off of 0.001 for prediction [69, 70]. We compared the putative functions of the proteins encoded by the core genome of these three species as issued by EDGAR.
Identification of MGEs
MGEs from S. lugdunensis were searched to identify elements potentially controlling genome stability (such as CRISPR/Cas and T/AT systems). From previous studies, we had characterized several MGEs (prophages and plasmids) in S. lugdunensis strains coming from the VISLISI clinical trial [3, 5], and these were retrieved first. The analysis was extended to all complete S. lugdunensis genomes available in the GenBank database with the same methodology [5]. Briefly, prophage searches and annotations were performed using PHASTER (Phage Search Tool Enhanced Release) [71]. Plasmids were retrieved from GenBank database. Pathogenicity islands were identified through IslandViewer4 [72].
Identification of CRISPR/Cas systems in S. lugdunensis
Several tools exist for CRISPR/Cas identification in whole genome sequences [73]. CRISPRFinder is a web server that offers a regularly updated database of CRISPR sequences that may be searched within an entire genome [74]. It does not focus on the genetic environment of the CRISPR sequences, and thus it does not identify the cas genes. To do this, we loaded the annotations from the GenBank database and the PROKKA de novo annotated genomes into Artemis software (v.16.0.0) to retrieve the CRISPR sequences identified with CRISPRFinder, and to analyze their genetic context [75]. All open reading frames surrounding the CRISPR sequences were also manually verified using the Uniprot and BLAST databases. We set the limits to 15 kb upstream and downstream of the CRISPR sequences, since cas genes were expected to be found in very close proximity, and because CRISPR/Cas system operons are not expected to be larger than 15 kb, particularly type IIIA and II [26, 28, 29]. All CRISPR/Cas sequences identified were aligned using Easyfig (v.2.2.2) to generate a BLAST alignment figure, with a minimum length of 100 bp, maximum e-value of 0.001, and minimum identity value of 90 [76]. CRISPR spacer origins were analyzed using the BLAST and Uniprot databases to search for known homologies.
Identification of T/AT systems in S. lugdunensis
To identify T/AT systems in S. lugdunensis, we loaded the GenBank annotated genomes and the PROKKA de novo annotated genomes into the Artemis software (v.16.0.0) and searched all gene names, qualifier values, and keys that comprised the term “toxin.” To ensure that new candidate T/AT systems were not missed, we searched for VapBC and MazEF systems with BLASTP (E-value < 0.01, against previously identified PIN-like sequences belonging to potential toxin families) and HMMER (E-value < 0.01, against PF04014 and PF02452 models), respectively [77].
Identification of RM systems in S. lugdunensis
Several tools have been developed to identify such coding sequences, the most complete being the REBASE database that provides an extensive in silico analysis of several bacterial genomes available in GenBank and allows identification and localization of RM systems [23]. Methylase specificity, nucleotide specificity, and closest neighbors were analyzed for each S. lugdunensis RM system.
Additional files
Pan-genome and core genome development projections for S. lugdunensis (A), S. epidermidis (B), and S. aureus (C). (DOCX 19 kb)
ANI and average AAI of S. lugdunensis (A-B), S. aureus (C-D), and S. epidermidis (E-F) using the EDGAR interface. ANIs were calculated as the mean identity of all BLASTN matches that showed more than 30% overall sequence identity over at least 70% of an alignable region. (PPTX 667 kb)
CRISPRs identified in 15 S. lugdunensis genomes using CRISPRFinder (available at: http://crispr.i2bc.paris-saclay.fr/). (DOCX 22 kb)
Type IIIA CRISPR/Cas system alignments from S. lugdunensis strains HKU0901, N920143, VISLISI_27, VISLISI_33, and VISLISI_37. Nucleotide BLAST alignments were performed using Easyfig (v.2.2.2). (PDF 225 kb)
MazEF operon comparison in 15 S. lugdunensis genomes, and in five other Staphylococcus species. All nucleotide BLAST comparisons were performed using Easyfig (v.2.2.2). (PDF 398 kb)
Genomic coordinates of RM systems identified in 15 S. lugdunensis strains according to the REBASE database. (DOCX 16 kb)
Pairwise alignment results according to EMBOSS Needle for the 3 loci of the Type I RM systems identified in S. lugdunensis VISLISI_33 and S. aureus MW2. (DOCX 574 kb)
Genome accession numbers from GenBank database of staphylococci. Clinical and geographical origins of S. lugdunensis strains. (DOCX 18 kb)
Acknowledgments
We thank Enago for the English language review.
Funding
Funding to D.M. and K.G was provided by the Polish National Science Centre [2014/15/B/NZ1/03357].
Availability of data and materials
The data set supporting the conclusions of this article are available in the GenBank database, https://www.ncbi.nlm.nih.gov/genbank/, and all genome accession numbers are listed in Additional file 8.
Abbreviations
- AAI
Average amino acid identity
- ANI
Average nucleotide identity
- COG
Clusters of orthologous groups
- CoNS
Coagulase-negative Staphylococci
- CRISPR
Clustered regularly interspaced short palindromic repeats
- DR
Direct repeat
- HGT
Horizontal gene transfer
- MGE
Mobile genetic element
- MLST
Multilocus sequence typing
- PRCI
Phage-related chromosomal island
- RM
Restriction-modification
- SaPI
Staphylococcus aureus pathogenicity island
- SCC
Staphylococcal cassette chromosome
- SRV
Score ratio values
- T/AT
Toxin/antitoxin
Authors’ contributions
XA, YH, GP, and PR designed the study; XA performed whole genome comparisons, MGE and RM systems analysis; DM and KG searched T/AT systems; JL, SD and MPC analyzed phylogenic implications of the results; JB managed EDGAR software platform and its bioinformatic parameters for the tools used and also provided phylogenetic trees and synteny plots; XA, MPC, and DM wrote the manuscript. All authors read, revised extensively, and gave final approval of the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xavier Argemi, Phone: (+0033) 609220088, Email: xavier_argemi@hjotmail.com, Email: xavier_argemi@hotmail.com.
Dorota Matelska, Email: d.matelska@cent.uw.edu.pl.
Krzysztof Ginalski, Email: k.ginalski@cent.uw.edu.pl.
Philippe Riegel, Email: philippe.riegel@chru-strasbourg.fr.
Yves Hansmann, Email: yves.hansmann@chru-strasbourg.fr.
Jochen Bloom, Email: edgar@computational.bio.uni-giessen.de.
Martine Pestel-Caron, Email: martine.pestel@chu-rouen.fr.
Sandrine Dahyot, Email: sandrine.dahyot@chu-rouen.fr.
Jérémie Lebeurre, Email: jeremie.lebeurre@hotmail.fr.
Gilles Prévost, Email: prevost@unistra.fr.
References
- 1.Argemi X, Hansmann Y, Riegel P, Prévost G. Is Staphylococcus lugdunensis significant in clinical samples? J Clin Microbiol. 2017;55(11):3167–3174. doi: 10.1128/JCM.00846-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argemi X, Riegel P, Lavigne T, Lefebvre N, Grandpré N, Hansmann Y, et al. Implementation of MALDI-TOF MS in routine clinical laboratories improves identification of coagulase negative staphylococci and reveals the pathogenic role of Staphylococcus lugdunensis. J Clin Microbiol. 2015;53(7):2030–2036. doi: 10.1128/JCM.00177-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argemi X, Prévost G, Riegel P, Keller D, Meyer N, Baldeyrou M, et al. VISLISI trial, a prospective clinical study allowing identification of a new metalloprotease and putative virulence factor from Staphylococcus lugdunensis. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2017;23:334. doi: 10.1016/j.cmi.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Foster TJ, Geoghegan JA, Ganesh VK, Höök M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62. doi: 10.1038/nrmicro3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argemi X, Martin V, Loux V, Dahyot S, Lebeurre J, Guffroy A, et al. Whole-genome sequencing of seven strains of Staphylococcus lugdunensis allows identification of mobile genetic elements. Genome Biol Evol. 2017;9 [DOI] [PMC free article] [PubMed]
- 6.Heilbronner S, Holden MTG, van Tonder A, Geoghegan JA, Foster TJ, Parkhill J, et al. Genome sequence of Staphylococcus lugdunensis N920143 allows identification of putative colonization and virulence factors: Staphylococcus lugdunensis genome sequence. FEMS Microbiol Lett. 2011;322:60–67. doi: 10.1111/j.1574-6968.2011.02339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol. 2014;6:2697–2708. doi: 10.1093/gbe/evu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang S-C, Lee M-H, Yeh C-F, Liu T-P, Lin J-F, Ho C-M, et al. Characterization of two novel variants of staphylococcal cassette chromosome mec elements in oxacillin-resistant Staphylococcus lugdunensis. J Antimicrob Chemother. 2017;72:3258–3262. doi: 10.1093/jac/dkx291. [DOI] [PubMed] [Google Scholar]
- 9.Yen T-Y, Sung Y-J, Lin H-C, Peng C-T, Tien N, Hwang K-P, et al. Emergence of oxacillin-resistant Staphylococcus lugdunensis carrying staphylococcal cassette chromosome mec type V in Central Taiwan. J Microbiol Immunol Infect Wei Mian Yu Gan Ran Za Zhi. 2016;49:885–891. doi: 10.1016/j.jmii.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 10.Sato’o Y, Omoe K, Ono HK, Nakane A, Hu D-L. A novel comprehensive analysis method for Staphylococcus aureus pathogenicity islands. Microbiol Immunol. 2013;57:91–99. doi: 10.1111/1348-0421.12007. [DOI] [PubMed] [Google Scholar]
- 11.Bosi E, Monk JM, Aziz RK, Fondi M, Nizet V, Palsson BØ. Comparative genome-scale modelling of Staphylococcus aureus strains identifies strain-specific metabolic capabilities linked to pathogenicity. Proc Natl Acad Sci. 2016;113:E3801–E3809. doi: 10.1073/pnas.1523199113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Post V, Harris LG, Morgenstern M, Mageiros L, Hitchings MD, Méric G, et al. A comparative genomics study of Staphylococcus epidermidis from orthopedic device-related infections correlated with patient outcome. J Clin Microbiol. 2017;55(10):3089–3103. doi: 10.1128/JCM.00881-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Méric G, Miragaia M, de Been M, Yahara K, Pascoe B, Mageiros L, et al. Ecological overlap and horizontal gene transfer in Staphylococcus aureus and Staphylococcus epidermidis. Genome Biol Evol. 2015;7:1313–1328. doi: 10.1093/gbe/evv066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chassain B, Lemee L, Didi J, Thiberge J-M, Brisse S, Pons J-L, et al. Multilocus sequence typing analysis of Staphylococcus lugdunensis implies a clonal population structure. J Clin Microbiol. 2012;50:3003–3009. doi: 10.1128/JCM.00988-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Didi J, Lemée L, Gibert L, Pons J-L, Pestel-Caron M. Multi-virulence locus sequence typing of Staphylococcus lugdunensis is consistent with clonal structure and reliable for epidemiological typing. J Clin Microbiol. 2014;52(10):3624–3632. doi: 10.1128/JCM.01370-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heilbronner S, Hanses F, Monk IR, Speziale P, Foster TJ. Sortase a promotes virulence in experimental Staphylococcus lugdunensis endocarditis. Microbiol Read Engl. 2013;159:2141–2152. doi: 10.1099/mic.0.070292-0. [DOI] [PubMed] [Google Scholar]
- 17.Heilbronner S, Monk IR, Foster TJ. The phage integrase vector pIPI03 allows RecA-independent, site-specific labelling of Staphylococcus lugdunensis strains. Plasmid. 2013;70:377–384. doi: 10.1016/j.plasmid.2013.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Marlinghaus L, Becker K, Korte M, Neumann S, Gatermann SG, Szabados F. Construction and characterization of three knockout mutants of the fbl gene in Staphylococcus lugdunensis: CHARACTERIZATION OF ISOGENIC MUTANTS OF FBL. APMIS. 2012;120:108–116. doi: 10.1111/j.1600-0463.2011.02819.x. [DOI] [PubMed] [Google Scholar]
- 19.Darmon E, Leach DRF. Bacterial genome instability. Microbiol Mol Biol Rev. 2014;78:1–39. doi: 10.1128/MMBR.00035-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monk IR, Shah IM, Xu M, Tan M-W, Foster TJ. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio. 2012;3 [DOI] [PMC free article] [PubMed]
- 21.Monk IR, Foster TJ. Genetic manipulation of Staphylococci—breaking through the barrier. Front Cell Infect Microbiol. 2012;2:49. doi: 10.3389/fcimb.2012.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomas CM, Nielsen KM. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 23.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—a database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasu K, Nagaraja V. Diverse functions of restriction-modification Systems in Addition to cellular defense. Microbiol Mol Biol Rev MMBR. 2013;77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Xie X, Yin K, Tang Y, Zhou X, Chen Y, et al. Characterization of CRISPR-Cas system in clinical Staphylococcus epidermidis strains revealed its potential association with bacterial infection sites. Microbiol Res. 2016;193:103–110. doi: 10.1016/j.micres.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Cao L, Gao C-H, Zhu J, Zhao L, Wu Q, Li M, et al. Identification and functional study of type III-A CRISPR-Cas systems in clinical isolates of Staphylococcus aureus. Int J Med Microbiol IJMM. 2016;306:686–696. doi: 10.1016/j.ijmm.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 27.Louwen R, Staals RHJ, Endtz HP, van Baarlen P, van der Oost J. The role of CRISPR-Cas Systems in Virulence of pathogenic Bacteria. Microbiol Mol Biol Rev MMBR. 2014;78:74–88. doi: 10.1128/MMBR.00039-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rossi CC, Souza-Silva T, Araújo-Alves AV, Giambiagi-deMarval M. CRISPR-Cas Systems Features and the Gene-Reservoir Role of Coagulase-Negative Staphylococci. Front Microbiol. 2017;8:1545. doi: 10.3389/fmicb.2017.01545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koonin EV, Makarova KS, Zhang F. Diversity, classification and evolution of CRISPR-Cas systems. Curr Opin Microbiol. 2017;37:67–78. doi: 10.1016/j.mib.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bast MSD, Mine N, Melderen LV. Chromosomal toxin-antitoxin systems may act as Antiaddiction modules. J Bacteriol. 2008;190:4603–4609. doi: 10.1128/JB.00357-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sberro H, Leavitt A, Kiro R, Koh E, Peleg Y, Qimron U, et al. Discovery of functional toxin/antitoxin systems in bacteria by shotgun cloning. Mol Cell. 2013;50:136–148. doi: 10.1016/j.molcel.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mittenhuber G. Occurrence of mazEF-like antitoxin/toxin systems in bacteria. J Mol Microbiol Biotechnol. 1999;1:295–302. [PubMed] [Google Scholar]
- 33.Schuster CF, Bertram R. Toxin-Antitoxin Systems of Staphylococcus aureus. Toxins. 2016;8 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4885055/ [DOI] [PMC free article] [PubMed]
- 34.Schuster CF, Mechler L, Nolle N, Krismer B, Zelder M-E, Götz F, et al. The MazEF toxin-antitoxin system alters the β-lactam susceptibility of Staphylococcus aureus. PLoS One. 2015;10:e0126118. doi: 10.1371/journal.pone.0126118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster CF, Bertram R. Toxin-antitoxin systems are ubiquitous and versatile modulators of prokaryotic cell fate. FEMS Microbiol Lett. 2013;340:73–85. doi: 10.1111/1574-6968.12074. [DOI] [PubMed] [Google Scholar]
- 36.Schuster CF, Park J-H, Prax M, Herbig A, Nieselt K, Rosenstein R, et al. Characterization of a mazEF toxin-antitoxin homologue from Staphylococcus equorum. J Bacteriol. 2013;195:115–125. doi: 10.1128/JB.00400-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Ye Y. Not all predicted CRISPR–Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinformatics. 2017;18:92. doi: 10.1186/s12859-017-1512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subedi A, Ubeda C, Adhikari RP, Penadés JR, Novick RP. Sequence analysis reveals genetic exchanges and intraspecific spread of SaPI2, a pathogenicity island involved in menstrual toxic shock. Microbiol Read Engl. 2007;153:3235–3245. doi: 10.1099/mic.0.2007/006932-0. [DOI] [PubMed] [Google Scholar]
- 39.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–D509. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cort JR, Ramelot TA, Murray D, Acton TB, Ma L-C, Xiao R, et al. Structure of an acetyl-CoA binding protein from Staphylococcus aureus representing a novel subfamily of GCN5-related N-acetyltransferase-like proteins. J Struct Funct Genom. 2008;9:7–20. doi: 10.1007/s10969-008-9041-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts GA, Houston PJ, White JH, Chen K, Stephanou AS, Cooper LP, et al. Impact of target site distribution for type I restriction enzymes on the evolution of methicillin-resistant Staphylococcus aureus (MRSA) populations. Nucleic Acids Res. 2013;41:7472–7484. doi: 10.1093/nar/gkt535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conlan S, Mijares LA, NISC comparative sequencing program. Becker J, Blakesley RW, Bouffard GG, et al. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol. 2012;13:R64. doi: 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Donati C, Hiller NL, Tettelin H, Muzzi A, Croucher NJ, Angiuoli SV, et al. Structure and dynamics of the pan-genome of Streptococcus pneumoniae and closely related species. Genome Biol. 2010;11:R107. doi: 10.1186/gb-2010-11-10-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gordienko EN, Kazanov MD, Gelfand MS. Evolution of pan-genomes of Escherichia coli, Shigella spp., and Salmonella enterica. J Bacteriol. 2013;195:2786–2792. doi: 10.1128/JB.02285-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.den Bakker HC, Cummings CA, Ferreira V, Vatta P, Orsi RH, Degoricija L, et al. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2010;11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rouli L, Merhej V, Fournier P-E, Raoult D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect. 2015;7:72–85. doi: 10.1016/j.nmni.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nanoukon C, Argemi X, Sogbo F, Orekan J, Keller D, Affolabi D, et al. Pathogenic features of clinically significant coagulase-negative staphylococci in hospital and community infections in Benin. Int J Med Microbiol. 2017;307:75–82. doi: 10.1016/j.ijmm.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Otto M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection. BioEssays News Rev Mol Cell Dev Biol. 2013;35:4–11. doi: 10.1002/bies.201200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freney J, Brun Y, Bes M, Meugnier H, Grimont F, Grimont PAD, et al. Staphylococcus lugdunensis sp. nov. and Staphylococcus schleiferi sp. nov., two species from human clinical specimens. Int J Syst Bacteriol. 1988;38:168–172. doi: 10.1099/00207713-38-2-168. [DOI] [Google Scholar]
- 50.Lamers RP, Muthukrishnan G, Castoe TA, Tafur S, Cole AM, Parkinson CL. Phylogenetic relationships among Staphylococcus species and refinement of cluster groups based on multilocus data. BMC Evol Biol. 2012;12:171. doi: 10.1186/1471-2148-12-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takahashi T, Satoh I, Kikuchi N. Phylogenetic relationships of 38 taxa of the genus Staphylococcus based on 16S rRNA gene sequence analysis. Int J Syst Bacteriol. 1999;49(Pt 2):725–728. doi: 10.1099/00207713-49-2-725. [DOI] [PubMed] [Google Scholar]
- 52.Fernández-García L, Blasco L, Lopez M, Bou G, García-Contreras R, Wood T, et al. Toxin-antitoxin Systems in Clinical Pathogens. Toxins. 2016;8:227. doi: 10.3390/toxins8070227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee K-Y, Lee B-J. Structure, biology, and therapeutic application of toxin–antitoxin Systems in Pathogenic Bacteria. Toxins. 2016;8:305. doi: 10.3390/toxins8100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Engelberg-Kulka H, Hazan R, Amitai S. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J Cell Sci. 2005;118:4327–4332. doi: 10.1242/jcs.02619. [DOI] [PubMed] [Google Scholar]
- 55.Warrier T, Kapilashrami K, Argyrou A, Ioerger TR, Little D, Murphy KC, et al. N-methylation of a bactericidal compound as a resistance mechanism in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2016;113:E4523–E4530. doi: 10.1073/pnas.1606590113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Purves J, Blades M, Arafat Y, Malik SA, Bayliss CD, Morrissey JA. Variation in the genomic locations and sequence conservation of STAR elements among staphylococcal species provides insight into DNA repeat evolution. BMC Genomics. 2012;13:515. doi: 10.1186/1471-2164-13-515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Belkum A. Short sequence repeats in microbial pathogenesis and evolution. Cell Mol Life Sci CMLS. 1999;56:729–734. doi: 10.1007/s000180050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mruk I, Kobayashi I. To be or not to be: regulation of restriction-modification systems and other toxin-antitoxin systems. Nucleic Acids Res. 2014;42:70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tse H, Tsoi HW, Leung SP, Lau SKP, Woo PCY, Yuen KY. Complete genome sequence of Staphylococcus lugdunensis strain HKU09-01. J Bacteriol. 2010;192:1471–1472. doi: 10.1128/JB.01627-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shiroma A, Terabayashi Y, Nakano K, Shimoji M, Tamotsu H, Ashimine N, et al. First complete genome sequences of Staphylococcus aureus subsp. aureus Rosenbach 1884 (DSM 20231T ), determined by PacBio single-molecule real-time technology. Genome Announc. 2015;3:e00800–e00815. doi: 10.1128/genomeA.00800-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fraser CM, Eisen JA, Nelson KE, Paulsen IT, Salzberg SL. The value of complete microbial genome sequencing (you get what you pay for) J Bacteriol. 2002;184:6403–6405. doi: 10.1128/JB.184.23.6403-6405.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Blom J, Kreis J, Spänig S, Juhre T, Bertelli C, Ernst C, et al. EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic Acids Res. 2016;44:W22–W28. doi: 10.1093/nar/gkw255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Guimarães LC, Florczak-Wyspianska J, de Jesus LB, Viana MVC, Silva A, Ramos RTJ, et al. Inside the Pan-genome - methods and software overview. Curr Genomics. 2015;16:245–252. doi: 10.2174/1389202916666150423002311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 66.Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of Phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:444. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertelli C, Laird MR, Williams KP, Simon Fraser University Research Computing Group. Lau BY, Hoad G, et al. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 2017;45(W1):W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prykhozhij SV, Rajan V, Gaston D, Berman JN. CRISPR multitargeter: a web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS One. 2015;10:e0119372. doi: 10.1371/journal.pone.0119372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. doi: 10.1186/1471-2105-8-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grissa I, Vergnaud G, Pourcel C. CRISPRcompar: a website to compare clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2008;36:W145–W148. doi: 10.1093/nar/gkn228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Carver T, Berriman M, Tivey A, Patel C, Böhme U, Barrell BG, et al. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sullivan MJ, Petty NK, Beatson SA. Easyfig: a genome comparison visualizer. Bioinforma Oxf Engl. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Matelska D, Steczkiewicz K, Ginalski K. Comprehensive classification of the PIN domain-like superfamily. Nucleic Acids Res. 2017;45:6995–7020. doi: 10.1093/nar/gkx494. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Pan-genome and core genome development projections for S. lugdunensis (A), S. epidermidis (B), and S. aureus (C). (DOCX 19 kb)
ANI and average AAI of S. lugdunensis (A-B), S. aureus (C-D), and S. epidermidis (E-F) using the EDGAR interface. ANIs were calculated as the mean identity of all BLASTN matches that showed more than 30% overall sequence identity over at least 70% of an alignable region. (PPTX 667 kb)
CRISPRs identified in 15 S. lugdunensis genomes using CRISPRFinder (available at: http://crispr.i2bc.paris-saclay.fr/). (DOCX 22 kb)
Type IIIA CRISPR/Cas system alignments from S. lugdunensis strains HKU0901, N920143, VISLISI_27, VISLISI_33, and VISLISI_37. Nucleotide BLAST alignments were performed using Easyfig (v.2.2.2). (PDF 225 kb)
MazEF operon comparison in 15 S. lugdunensis genomes, and in five other Staphylococcus species. All nucleotide BLAST comparisons were performed using Easyfig (v.2.2.2). (PDF 398 kb)
Genomic coordinates of RM systems identified in 15 S. lugdunensis strains according to the REBASE database. (DOCX 16 kb)
Pairwise alignment results according to EMBOSS Needle for the 3 loci of the Type I RM systems identified in S. lugdunensis VISLISI_33 and S. aureus MW2. (DOCX 574 kb)
Genome accession numbers from GenBank database of staphylococci. Clinical and geographical origins of S. lugdunensis strains. (DOCX 18 kb)
Data Availability Statement
The data set supporting the conclusions of this article are available in the GenBank database, https://www.ncbi.nlm.nih.gov/genbank/, and all genome accession numbers are listed in Additional file 8.