Lactobacilli have significant scientific and economic value, but their extraordinary diversity means that they are not robustly classified. The 10 homogeneous genera/subgeneric entities that we identify here are characterized by uniform patterns of the presence/absence of specific sets of genes which offer potential as discovery tools for understanding differential biological features. Reclassification/subdivision of the genus Lactobacillus into more uniform taxonomic nuclei will also provide accurate molecular markers that will be enabling for regulatory approval applications. Reclassification will facilitate scientific communication related to lactobacilli and prevent misidentification issues, which are still the major cause of mislabeling of probiotic and food products reported worldwide.
KEYWORDS: Lactobacillus, taxonomy, phylogenomics, phylogeny, comparative genomics, reclassification
ABSTRACT
The genus Lactobacillus includes over 200 species that are widely used in fermented food preservation and biotechnology or that are explored for beneficial effects on health. Naming, classifying, and comparing lactobacilli have been challenging due to the high level of phenotypic and genotypic diversity that they display and because of the uncertain degree of relatedness between them and associated genera. The aim of this study was to investigate the feasibility of dividing the genus Lactobacillus into more homogeneous genera/clusters, exploiting genome-based data. The relatedness of 269 species belonging primarily to the families Lactobacillaceae and Leuconostocaceae was investigated through phylogenetic analysis (by the use of ribosomal proteins and housekeeping genes) and the assessment of the average amino acid identity (AAI) and the percentage of conserved proteins (POCP). For each subgeneric group that emerged, conserved signature genes were identified. Both distance-based and sequence-based metrics showed that the Lactobacillus genus was paraphyletic and revealed the presence of 10 methodologically consistent subclades, which were also characterized by a distinct distribution of conserved signature orthologues. We present two ways to reclassify lactobacilli: a conservative division into two subgeneric groups based on the presence/absence of a key carbohydrate utilization gene or a more radical subdivision into 10 groups that satisfy more stringent criteria for genomic relatedness.
IMPORTANCE Lactobacilli have significant scientific and economic value, but their extraordinary diversity means that they are not robustly classified. The 10 homogeneous genera/subgeneric entities that we identify here are characterized by uniform patterns of the presence/absence of specific sets of genes which offer potential as discovery tools for understanding differential biological features. Reclassification/subdivision of the genus Lactobacillus into more uniform taxonomic nuclei will also provide accurate molecular markers that will be enabling for regulatory approval applications. Reclassification will facilitate scientific communication related to lactobacilli and prevent misidentification issues, which are still the major cause of mislabeling of probiotic and food products reported worldwide.
INTRODUCTION
The genus Lactobacillus includes 232 species (as reported elsewhere [http://www.bacterio.net/lactobacillus.html]), a number which is rising continuously, as novel species are described every year. Lactobacilli are Gram-positive bacteria that are mostly nonmotile, catalase negative, non-spore forming, and rod shaped (although coccobacilli are observed). They populate nutrient-rich habitats associated with food, feed, soil, plants, animals (both vertebrates and invertebrates), and humans (1) and are mainly characterized by a fermentative metabolism, but they show some evidence of respiration (2), with lactic acid being the main product.
Lactobacilli are key players in industry, food, and human and animal health-related fields: they contribute to fermented food production, to food texture, and to food preservation; they deliver pure lactic acid from raw carbohydrates for onward conversion to bioplastics; and some strains are marketed as probiotics, meaning that they exhibit health benefits beyond the basic nutritional value. In addition, lactobacilli are also being explored as therapeutics and delivery systems for vaccines (1, 3, 4, 5).
From a food regulatory viewpoint, 84 Lactobacillus species are certified for safe, technological, and beneficial use by the European Food and Feed Cultures Association (6), 36 species have qualified presumption of safety (QPS) status according to the European Food Safety Authority (EFSA) (7), and 12 species are generally recognized as safe (GRAS) according to the U.S. Food and Drug Administration (FDA) (http://www.accessdata.fda.gov/scripts/fdcc/?set=GRASNotices) (8).
The economic value of lactobacilli is substantial: the probiotics and direct-fed microbials markets, in which lactobacilli play an essential role, are projected to reach a value of $64 billion and $1.4 billion, respectively, by 2022 (https://www.marketsandmarkets.com/Market-Reports/probiotic-market-advanced-technologies-and-global-market-69.html). Continued or, indeed, enhanced levels of economic exploitation of lactobacilli will benefit from a rigorous comparative genomics framework, such as the documentation of endogenous or transmissible antibiotic resistance elements across the genus (I. Campedelli, H. Mathur, E. Salvetti, S. Clarke, M. C. Rea, S. Torriani, R. P. Ross, C. Hill, and P. W. O'Toole, submitted for publication).
From a taxonomic perspective, the primary distinction between members of the genus Lactobacillus has historically been based on physiological characteristics, until the first proposal of introducing 16S rRNA gene sequence analysis in 1991 (9). Thus far, analysis of 16S rRNA gene similarity is combined with the analysis of the carbohydrate fermentation profile, according to which lactobacilli are divided into the homofermentative (use of hexose and production of lactic acid), facultatively heterofermentative (use of pentose/hexose and production of lactic acid and other products), and obligately heterofermentative (use of pentose/hexoses and production of lactic acid, side products, and CO2) groups (10). However, the expansion of the Lactobacillus genus since its first description and the presence of overlapping characteristics, together with the threshold ambiguity associated with 16S rRNA gene sequence comparison, have led to frequent taxonomic changes and misidentification issues for strains and species at a short phylogenetic range and for clade distinction for strains and species at a long phylogenetic range (11–14). Further, the comparative analysis of the genome sequences of almost all Lactobacillus type strains and historically related genera (3, 4) revealed an overall level of genomic diversity associated with that between members of a bacterial order, and the currently defined genus Lactobacillus sensu lato encompasses members of the genus Pediococcus (Lactobacillaceae family) and the genera Convivina, Fructobacillus, Leuconostoc, Weissella, and Oenococcus (family Leuconostocaceae).
The extreme diversity of the genus Lactobacillus and its polyphyletic structure strongly suggest that this taxonomic arrangement should be formally reevaluated. Hence, the aim of the present study was to understand the evolutionary relationships within the families Lactobacillaceae and Leuconostocaceae and to provide a robust genome-based framework for a novel taxonomic scheme for the genus Lactobacillus. Genomics provides bacterial taxonomists with powerful evolutionary information which has been successfully employed for the identification and classification of prokaryotic species as well as the elucidation of diagnostic components in different taxonomic groups (15, 16). Here we interrogated the genome sequences of 222 strains of Lactobacillus and associated genera through the application of distance-based metrics, viz., the average nucleotide identity (ANI), the average amino acid identity (AAI) (17), and the percentage of conserved proteins (POCP) (18), and sequence-based methods, namely, phylogenetic and network analyses based on 29 ribosomal proteins and 12 established phylogenetic markers. With respect to previous observations, which were based essentially on the maximum likelihood of 73 core genes (3), here we (i) integrated information derived from distance-based methods to obtain a consensus on delineated clades, (ii) reduced the number of genes for multilocus sequence analysis and deeply investigated the phylogenetic signal by means of split decomposition, and (iii) revealed the presence of clade-specific genes. The data obtained illustrate the feasibility and advisability of dividing the current genus Lactobacillus into a number of more homogeneous genera and provide the basis for the development of future taxonomic procedures, which should be robust and straightforward.
RESULTS
MLSA and rMLSA define 10 discrete clades within the lactobacilli.
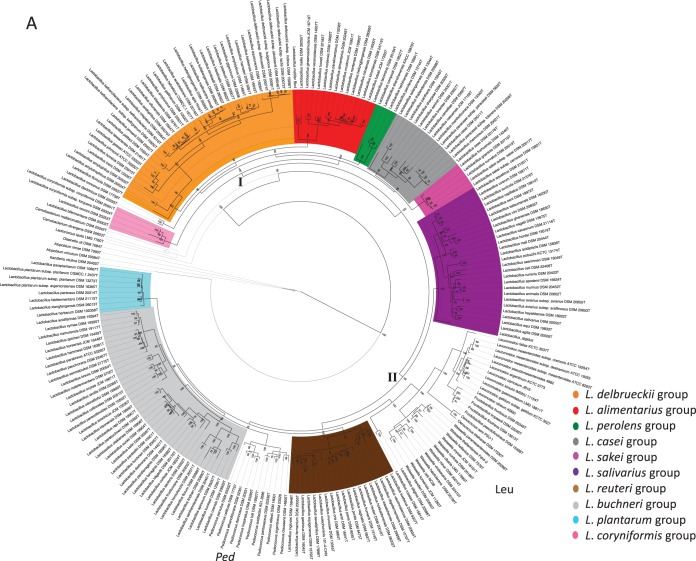
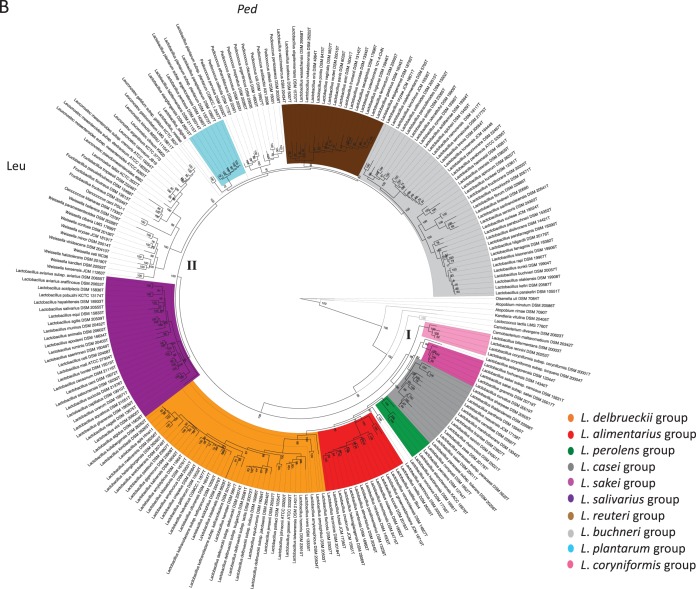
We constructed phylogenetic trees for selected strains belonging to the genus Lactobacillus and related genera based on multilocus sequence analysis (MLSA) of 29 ribosomal proteins (rMLSA) and 12 phylogenetic markers (MLSA), as shown in Fig. 1A and B, respectively. Both trees are characterized by high bootstrap values, which indicate that the proteins selected are reflective of robust evolutionary relatedness between taxa and clades. The trees show that lactobacilli branch in several clades (defined by the colors in both trees) and are intermixed with the genera Pediococcus, Fructobacillus, Leuconostoc, Oenococcus, and Weissella. This supports previous observations on the paraphyly of the genus Lactobacillus, which is taxonomically noncohesive.
FIG 1.
Phylogenetic trees based on the amino acid sequences of 29 ribosomal proteins (A) and 12 phylogenetic markers (B). Clusters I and II are indicated in the tree. Leu, Leuconostocaceae; Ped, Pediococcus. The phylogeny was inferred using the PROTCATWAG model in RAxML (v8.0.22) and rooted using Atopobium minutum DSM 20584T, Atopobium rimae DSM 7090T, Kandleria vitulina DSM 20405T, and Olsenella uli DSM 7084T. Bootstrapping was carried out using 100 replicates, and values are indicated on the nodes.
At a long phylogenetic range, the individual Lactobacillus species are split into cluster I (46% of all lactobacilli; bootstrap value, 100% in both trees) and cluster II (54% of lactobacilli; bootstrap value, 98% in rMLSA trees and 100% in MLSA trees) (Fig. 1A and B), which are consistent in branching order and composition across the two trees. Cluster I includes six highly supported phylogroups, whose nomenclature we assigned on the basis of their description in previous studies (3, 4, 11, 12), and are the following: (i) the Lactobacillus delbrueckii group (orange), (ii) the Lactobacillus alimentarius group (red), (iii) the Lactobacillus perolens group (green), (iv) the Lactobacillus casei group (gray), (v) the Lactobacillus sakei group (dark pink), and (vi) the Lactobacillus coryniformis group (light pink). Cluster II comprises four phylogroups, namely, (i) the Lactobacillus salivarius group (violet); (ii) the Lactobacillus reuteri and Lactobacillus vaccinostercus groups, which can be collapsed into a single phylogroup referred to as the Lactobacillus reuteri group (brown); (iii) the Lactobacillus fructivorans, Lactobacillus brevis, Lactobacillus buchneri, and Lactobacillus collinoides groups, which form a unique phylogroup that we designated the L. buchneri group (since L. buchneri was the first species described within this group) (light gray); and (iv) the Lactobacillus plantarum group (light blue). Remarkably, cluster II also includes the Leuconostocaceae family and the genus Pediococcus, which is a sister branch of the expanded L. buchneri group in both trees.
For those species not clustered in phylogroups, two couples emerged: Lactobacillus concavus-Lactobacillus dextrinicus, which are peripheral in cluster I, and Lactobacillus rossiae-Lactobacillus siliginis, which are associated with the Leuconostocaceae in cluster II, in both trees. Lactobacillus selangorensis represents a single line of descent, and it is the sole inconsistency between the two trees: it belongs to cluster I in both trees, but it is associated with the L. casei phylogroup in the ribosomal protein tree (Fig. 1A) or the L. sakei group in the other phylogenetic tree (Fig. 1B).
The paraphyletic nature of the Lactobacillus genus was also corroborated by the split decomposition analysis (see Fig. S1A and B in the supplemental material): the 10 phylogroups were recapitulated in both the phylogenetic structures, in which pediococci and leuconostocs were interspersed. Interconnecting networks were also revealed, indicating the occurrence of events more complicated than speciation in the evolution of the genus Lactobacillus and, more generally, of the families Lactobacillaceae and Leuconostocaceae.
Selection of distance-based methods to assess genetic relatedness.
ANI, AAI, and POCP values were calculated across the 222 genome sequences to assess their genetic relatedness. The majority of ANI values obtained were below the 75 to 80% range (Fig. S2), meaning that the genomes are distantly related and indicating that ANI calculation was not appropriate for the current data set (16, 19). Thus, only AAI and POCP were considered in the present study, since they provide a much more robust resolution.
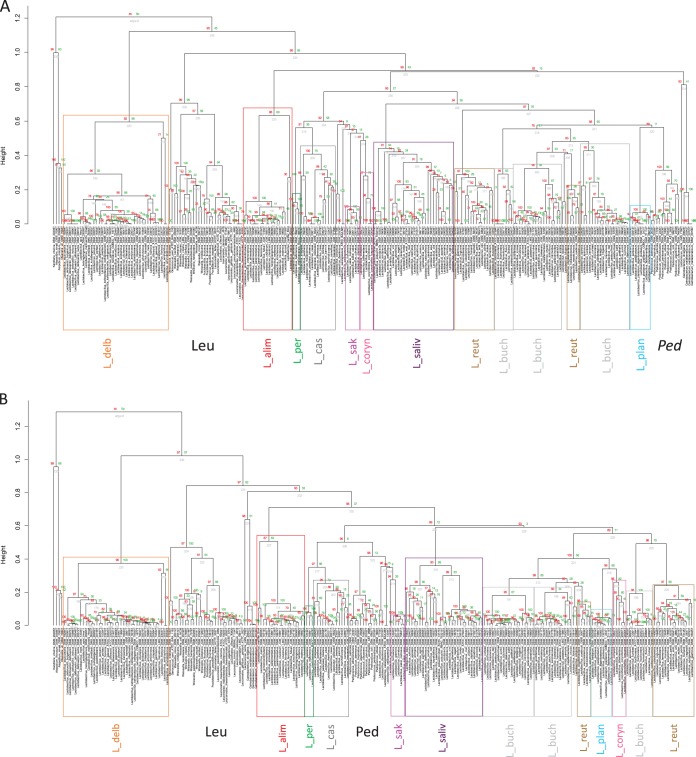
The AAI and POCP metrics support the phylogenetic analysis.
AAI and POCP clusterings are shown in Fig. 2. Their statistical robustness is supported by the high bootstrap values at the nodes. The dendrograms substantiate the conclusions from the phylogenetic analysis: the genus Pediococcus and the family Leuconostocaceae are clustered within the genus Lactobacillus; further, lactobacilli are branched in almost the same phylogroups observed in the phylogenetic trees. In detail, Lactobacillus species are split into two clusters in both the dendrograms: cluster I comprises just the L. delbrueckii phylogroup, while cluster II contains all the other species, including the Leuconostocaceae (which is peripheral in cluster II in both the graphics) and pediococci. In the dendrogram based on AAI values, the L. perolens, L. casei, L sakei, and L. coryniformis phylogroups form a single subclade in cluster II, while the L. salivarius phylogroup is associated with the L. reuteri-L. vaccinostercus, L. buchneri, and L. plantarum phylogroups and the Pediococcus genus (Fig. 2A). In the POCP dendrogram, the L. perolens, L. casei, and L. sakei phylogroups form a single clade together with the Pediococcus genus, while L. coryniformis is associated with the L. reuteri-L. vaccinostercus, L. buchneri, and L. plantarum phylogroups (Fig. 2B).
FIG 2.
Dendrograms depicting the genome relatedness based on the average amino acid identity (AAI) (A) and the percentage of conserved protein (POCP) (B) calculations. Colors refer to the same phylogroups indicated in Fig. 1. L_delb, L. delbrueckii group; L_alim, L. alimentarius group; L_per, L. perolens group; L_cas, L. casei group; L_sak, L. sakei group; L_coryn, L. coryniformis group; L_saliv, L. salivarius group; L_reut, L. reuteri group; L_buch, L. buchneri group; L_plan, L. plantarum group; Leu, Leuconostocaceae; Ped, Pediococcus. Statistics and visualization were carried out in R (v3.1.1) (https://www.r-project.org/), using the pvclust package (49). Red numbers are unbiased P values, green numbers are bootstrap probabilities, and gray numbers are node numbers.
In contrast to the phylogenetic analysis, the L. reuteri-L. vaccinostercus and L. buchneri groups are split into their original group composition and intermixed. L. concavus-L. dextrinicus and L. selangorensis are associated with the L. sakei phylogroup, while L. rossiae-L. siliginis are clustered with the L. vaccinostercus group in both dendrograms.
Identification of conserved signature genes within Lactobacillus phylogroups.
To investigate the functional differences in phylogroups established with distance-based (AAI, POCP) and sequence-based (MLSA) methods, a large-scale orthology analysis was performed. This led to the identification of 15 orthologs which were selected as putative clade-specific genes on the basis of their pattern of presence/absence among the phylogroups (Tables 1, 2, and S3). One of the key genes was the glycolytic phosphofructokinase (Pfk) gene (pfk, QTS_863), which is present in all the members of the L. delbrueckii, L. alimentarius, L. perolens, L. casei, L. sakei, L. salivarius, L. plantarum, and L. coryniformis phylogroups, in the L. concavus-L. dextrinicus group, and in the Pediococcus genus, while it is lacking in all the members of the L. reuteri-L. vaccinostercus group, the expanded L. buchneri group, the L. rossiae-L. siliginis group, and all the Leuconostocaceae. The presence/absence pattern of Pfk seems to have an impact on the carbohydrate metabolism of these species. In fact, members within the Pfk-lacking group (Table 2) were classified as obligately heterofermentative (3, 12), with the rest being facultatively heterofermentative or homofermentative. Taking the presence/absence pattern of Pfk as a reference, the distribution of nine other signature genes is distinct in species belonging to different phylogroups in the Pfk-positive group (Table 1). Four of them have been associated with a function, and they belong to different clusters of orthologous genes (COGs) (Table 1), while five of these genes are annotated as hypothetical proteins and lack conserved domains. Interestingly, QTS_569, a zinc-dependent peptidase, is present in all the Pfk-positive species, except members of the L. delbrueckii group, which, on the other hand, are the only species within the Pfk-positive group with QTS_2524, a hypothetical protein (Table 1, profile A). Furthermore, QTS_4707, another hypothetical protein, seems to be specific to the L. alimentarius group (profile B). The presence/absence profiles of these nine genes (reported in Table 1) are almost unique for each Pfk-positive phylogroup, the Pediococcus genus included; the only exception is the couple L. concavus-L. dextrinicus, which has the same profile as the L. sakei phylogroup (profile E), characterized by the presence of QTS_569, the zinc-dependent peptidase, and QTS_898, a protein annotated as a cell division inhibitor, and the absence of the rest of the genes.
TABLE 1.
Details of signature proteins for species with Pfk
| Gene | NCBI annotation | Locus taga | COGb | Presence of the gene in: |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. delbrueckii | L. alimentarius | L. perolens | L. casei | L. sakei | L. salivarius | L. plantarum | L. coryniformis | L. concavus-L. dextrinicus | L. selangorensis | Pediococcus | ||||
| QTS_863 | 6-Phosphofructokinase | lp_1898 | COG0205G | + | + | + | + | + | + | + | + | + | + | + |
| QTS_569 | Zn-dependent peptidase | lp_2306 | COG0612R | − | + | + | + | + | + | + | + | + | + | + |
| QTS_898 | Cell division inhibitor | lp_2316 | COG0850D | − | + | + | + | + | + | + | + | + | + | − |
| QTS_1754 | Transcription termination factor Rho | lp_0511 | COG1158K | − | − | − | − | − | + | + | + | − | − | + |
| QTS_2490 | Hypothetical protein | LBA0167 | ND | +c | −d | −e | − | − | − | − | − | − | + | − |
| QTS_2524 | Hypothetical protein | LBA0844 | ND | +c | − | − | − | − | − | − | − | − | − | − |
| QTS_2525 | S1 family RNA-binding protein | LBA0276 | COG1098R | + | + | +f | − | − | − | + | − | − | − | −g |
| QTS_3870 | Hypothetical protein | LSEI_1730 | ND | − | − | + | + | − | − | − | + | − | + | − |
| QTS_4397 | Hypothetical protein | LSEI_0696 | ND | − | − | − | + | − | − | − | + | − | + | − |
| QTS_4707 | Hypothetical protein | FC67_GL001143 | ND | − | + | − | − | − | − | − | − | − | − | − |
| Profile | A | B | C | D | E | F | G | H | E | I | L | |||
Locus tags lp_1898, lp_2306, lp_2316, and lp_0511 are for Lactobacillus plantarum WCFS1; locus tags LBA0167, LBA0844, and LBA0276 are for Lactobacillus acidophilus NCFM; locus tags LSEI_1730 and LSEI_0696 are for Lactobacillus paracasei ATCC 334; and locus tag FC67_GL001143 is for Lactobacillus alimentarius DSM 20249.
COGs are as follows: D, cell cycle control, cell division, and chromosome partitioning; G, carbohydrate transport and metabolism; K, transcription; R, general function prediction only. ND, not determined.
Absent in L. floricola.
Present in L. mellifer and L. mellis.
Present in L. composti.
Absent in L. composti.
Present in P. claussenii.
TABLE 2.
Details of signature proteins for species without Pfk
| Gene | NCBI annotation | Locus taga | COGb | Presence of the gene in: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| L. reuteri | L. vaccinostercus | L. fructivorans | L. brevis | L. buchneri | L. collinoides | L. rossiae-L. siliginis | Leuconostocaceae | ||||
| QTS_863 | 6-Phosphofructokinase | lp_1898 | COG0205G | − | − | − | − | − | − | − | − |
| QTS_494 | Thiamine biosynthesis protein Thil | LVIS_RS1765 | COG0301HJ | + | + | + | + | + | + | + | − |
| QTS_497 | tRNA methyltransferase | LVIS_RS18530 | COG0482J | + | + | + | + | + | + | + | − |
| QTS_502 | Transcriptional regulator NrdR | LVIS_RS16605 | COG1327K | + | + | + | + | + | + | + | − |
| QTS_509 | tRNA uridine 5-carboxymethylaminomethyl modification protein | LVIS_RS22810 | COG0445J | + | + | + | + | + | + | + | − |
| QTS_514 | DNA replication initiation control protein YabA | LVIS_RS14505 | COG4467L | + | + | + | + | + | + | + | − |
| QTS_898 | Cell division inhibitor | LVIS_RS17610 | COG0850D | − | − | + | + | + | + | − | − |
| QTS_2490 | Hypothetical protein | LVIS_RS11970 | ND | − | − | − | + | − | − | − | − |
| Profile | A | A | B | C | B | B | A | D | |||
Locus tag lp_1898 is for Lactobacillus plantarum WCFS1, and the other locus tags are for Lactobacillus brevis ATCC 367.
COGs are as follows: D, cell cycle control, cell division, and chromosome partitioning; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; R, general function prediction only. ND, not determined.
Regarding the Pfk-negative group, the differential distribution of seven genes uniquely describes the members of most of the groups (Table 2). Six out of the seven genes have been annotated and were found to belong to six COGs (Table 2), while only one gene has been annotated as encoding a hypothetical protein. Species belonging to the L. reuteri and L. vaccinostercus clades have the same pattern, one displayed also by L. rossiae-L. siliginis (Table 2, profile A), which is characterized by the absence of QTS_898, the cell division inhibitor, and QTS_2490, a hypothetical protein. Members of the L. fructivorans, L. buchneri, and L. collinoides groups display all the genes except QTS_2490 (profile B), which is, instead, present in L. brevis group members (profile C). Interestingly, the species belonging to the Leuconostocaceae family have a profile completely different from that of the other Pfk-negative groups, as they lack all the genes under consideration (profile D).
DISCUSSION
One of the overall aims of this study was to stop the never-ending expansion of Lactobacillus as a heterogeneous clade (1, 3, 4, 11, 12, 20). We used two methods with a phylogenetic component (MLSA of ribosomal proteins and a set of housekeeping genes) and two which were phylogeny independent (AAI and POCP analysis). MLSA affords a higher resolution of the phylogenetic relationships of species within a genus and genera within a family (16, 21) and successfully resolved the complex taxonomic structure of the genera Escherichia and Shigella and the family Enterobacteriaceae (22–24). The housekeeping protein-coding genes used for MLSA are believed to evolve at a low but constant rate and have a better resolution power than the 16S rRNA gene; ribosomal proteins are usually syntenic and colocated in the same genomic area, thus allowing binning errors which could perturb the geometry of the tree to be avoided (19, 21, 25). The phylogenetic trees that we generated confirmed the paraphyletic nature of the genus Lactobacillus (first observed with a 16S rRNA gene-based phylogeny and a smaller data set of genome sequences [11, 12, 13]), where Leuconostocaceae and pediococci branched from the lactobacilli as subgroups. The topologies of the trees obtained here confirmed the phylogenomic topology inferred from 73 core proteins (3) and from 172 core genes shared by 174 genomes of lactobacilli and pediococci (1, 4). Each phylogenomic reconstruction revealed the association of obligately heterofermentative lactobacilli with the Leuconostocaceae (displaying the same metabolism) and their separation from the homofermentative and facultatively heterofermentative Lactobacillus species (4). Ten historically recognized Lactobacillus subgroups could also be identified from our analysis (1, 3, 4, 11, 12, 26, 27), which updates the phylogroupings which we described with Sun and colleagues (3).
Only five Lactobacillus species remained outside the phylogroups: two couples, namely, L. rossiae-L. siliginis and L. concavus-L. dextrinicus, and L. selangorensis. These species were not clustered within any other Lactobacillus phylogroups using other data sets ranging from 16S rRNA genes to core genes (1, 3, 4, 12). Interestingly, L. dextrinicus was first described as Pediococcus dextrinicus (28), while L. selangorensis constituted the sole species of the genus Paralactobacillus (29). Both species were later reclassified as Lactobacillus species based on MLSA of the 16S rRNA gene and other housekeeping genes (30, 31).
Furthermore, 10 consistent subgroups were defined, namely, (i) L. delbrueckii (named after the type species of Lactobacillus), which also comprises the peripheral species Lactobacillus amylophilus, Lactobacillus amylotrophicus, and Lactobacillus floricola; (ii) L. alimentarius; (iii) L. perolens: (iv) L. casei; (v) L. sakei (without L. selangorensis); (vi) L. coryniformis; (vii) L. salivarius; (viii) L. plantarum; (ix) L. reuteri, which also includes L. vaccinostercus-related species; and (x) L. buchneri, which encompasses members of the L. brevis, L. fructivorans, and L. collinoides groups (the group was given the name L. buchneri, since it was the first species described within the phylogroup).
The inferred subgroups were largely corroborated by AAI and POCP analyses, which were rigorously applied to lactobacilli in the present project. AAI analysis has shown excellent potential to improve the classification of higher taxa (e.g., the Enterobacteriaceae family [32]); POCP analysis was proposed by Qin and colleagues (18) as a complementary approach to AAI analysis, and POCP is calculated using all the proteins of the genomes to be compared. ANI analysis was also applied to the data set since it has been officially recommended as a substitute for DNA-DNA hybridization and has been used in more than 30 classifications (19), but most of the ANI values fell below the 75 to 80% range (as also observed by Zheng and colleagues [4]), showing the extremely wide genetic diversity of the strains under study and making this method unreliable for the present data set. This method gives robust resolution to genomes that have ANI values of 80 to 100% and/or that share at least 30% of their gene content, a scenario which typically occurs within species belonging to the same genus (but it is clearly not applicable to lactobacilli); if two strains have a distant genetic relationship, only a small proportion of the whole-genome DNA sequence is considered for ANI calculation and the majority of DNA information is discarded due to the lack of homology (18, 33). In fact, such strains could then be ascribed to different genera, as the low values render comparison essentially impossible.
Despite the relatively high intragroup AAI and POCP values, some inconsistencies in the phylogenetic trees among the obligately heterofermentative groups emerged. Specifically, the L. vaccinostercus-related species were separated from the L. reuteri group and the L. buchneri group was split into its original subclades (L. fructivorans, L. brevis, L. collinoides, and L. buchneri groups). In the light of this incongruence, genome sequences were further explored to identify signature genes which could assist in the definition of the supported Lactobacillus subgroups. A set of 15 genes whose presence/absence pattern was specific for the 10 phylogroups was thus identified. The most discriminative gene was the phosphofructokinase gene (pfk), which was present in all the homofermentative and facultatively heterofermentative lactobacilli and absent in the obligately heterofermentative lactobacilli (and Leuconostocaceae). Production of CO2 differentiates obligately from facultatively heterofermentative metabolism (13). The pfk gene distribution represents the first element in Lactobacillus taxonomy in which phylogenetic clustering, genome-based analysis, and phenotypic (metabolic) analysis come to an agreement. The other retrieved genes could not be attributed to specific functions or to unambiguous phenotypic traits. Nevertheless, they represent a biological signature, which, together with robust phylogenetic groupings, can be used for the definition of cohesive taxonomic entities within the genus Lactobacillus and thus used as diagnostic tools. Furthermore, given their crucial position at the branch points that occurred during the evolution of lactobacilli, they provide a resource to be functionally explored and from which new important information on these bacteria may be uncovered (32, 34).
A summary of the data from the sequence-based and distance-based methods (Table 3) combining the analysis of orthologous gene presence/absence crystallizes two scenarios for the formal reclassification of the Lactobacillus genus. The first scenario consists of splitting the genus into two groups on the basis of the presence/absence of pfk. These groups are relatively consistent with the phylogenetic trees based on ribosomal proteins, housekeeping genes, and core genes and congruent with carbohydrate fermentation profiles. However, these two subgeneric groups are still characterized by POCP and AAI values that would not meet the criteria for genus delineation (species should share at least 55 to 60% AAI and 50% POCP to be considered within the same genus [18, 33]). A second scenario envisages the proposal of the 10 subgroups that emerged from the phylogenetic analysis as nuclei of novel genera within lactobacilli: the subgroups are consistent in the different trees; they were mainly recapitulated by 16S rRNA-based sequence analysis (including also species for which a genome sequence is not available [see Fig. S3 in the supplemental material]); most of them share values of POCP and AAI higher than 50% and 55 to 60%, respectively; and they are also characterized by distinct gene distributions (Table 3). In this scenario, some questions remain unanswered: the first challenge regards the L. delbrueckii, L. alimentarius, and L. perolens groups, whose intragroup diversity changes when peripheral species are considered. For instance, the exclusion of L. floricola, L. amylophilus, and L. amylotrophicus from the L. delbrueckii group increases intragroup AAI and POCP values from 52.1 and 46.4% to 59.3 and 52.9%, respectively, thus allowing this group to meet the criteria suggested for genus delineation based on distance-based metrics (the same situation applies for the L. perolens and L. alimentarius groups). For the clade composed of members of the expanded L. buchneri group (L. fructivorans, L. brevis, L. buchneri, and L. collinoides members), a consistent phylogenetic inference faces unmet criteria in distance-based methods (particularly POCP, which is 45.9%) and a differential distribution of clade-specific genes (i.e., members of L. brevis have a gene presence/absence pattern different from that of the other species).
TABLE 3.
Combination of distance-based and sequence-based data with the analysis of signature proteins for each phylogroup
| Phylogroup | No. of species | AAI (%)g | POCPg | Presence of the following gene: |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pfk | QTS_569 | QTS_898 | QTS_1754 | QTS_2490 | QTS_2425 | QTS_2525 | QTS_3870 | QTS_4397 | QTS_4707 | QTS_494 | QTS_497 | QTS_502 | QTS_509 | QTS_514 | QTS_898 | QTS_2490 | ||||
| L. delbrueckii | 35 | 52.1 (59.3a) | 46.4 (52.9a) | + | − | − | − | + | + | + | − | − | − | |||||||
| L. alimentarius | 21 | 52.8 (68.4b) | 44.6 (62.4b) | + | + | + | − | − | − | + | − | − | + | |||||||
| L. perolens | 4 | 55.9 (72.9c) | 48 (67.8c) | + | + | + | − | − | − | + | − | − | + | |||||||
| L. casei | 16 | 59.3 | 55.2 | + | + | + | − | − | − | − | + | + | − | |||||||
| L. sakei | 4 | 76.7 | 75.2 | + | + | + | − | − | − | − | − | − | − | |||||||
| L. plantarum | 9 | 76.5 | 76 | + | + | + | + | − | − | + | − | − | − | |||||||
| L. coryniformis | 5 | 62.5 | 61.1 | + | + | + | + | − | − | − | + | + | − | |||||||
| L. salivarius | 27 | 56.1 (61.1d) | 53.5 (59.3d) | + | + | + | + | − | − | − | − | − | − | |||||||
| L. concavus- L. dextrinicus | 2 | 72.7 | 70.9 | + | + | + | − | − | − | − | − | − | − | |||||||
| L. selangorensis | 1 | + | + | + | − | + | − | − | + | + | − | |||||||||
| L. reuteri | 23 | 63.2 (57.6e) | 62 (51e) | − | + | + | + | + | + | − | − | |||||||||
| L. vaccinostercus | 68.9 | 69 | − | + | + | + | + | + | − | − | ||||||||||
| L. fructivorans | 48 | 58.3 (56.1f) | 58.3 (45.9f) | − | + | + | + | + | + | + | − | |||||||||
| L. brevis | 74.6 | 70.8 | − | + | + | + | + | + | + | + | ||||||||||
| L. buchneri | 63.3 | 55.6 | − | + | + | + | + | + | + | − | ||||||||||
| L. collinoides | 62.07 | 62.2 | − | + | + | + | + | + | + | − | ||||||||||
| L. rossiae- L. siliginis | 2 | 73.7 | 67.3 | − | + | + | + | + | + | − | − | |||||||||
AAI and POCP values for the L. delbrueckii group without considering peripheral species (L. amylophilus, L. amylotrophicus, L. floricola).
AAI and POCP values for the L. alimentarius group without considering peripheral species (Lactobacillus mellifer, Lactobacillus mellis).
AAI and POCP values for the L. perolens group without considering peripheral species (Lactobacillus composti).
AAI and POCP values for the L. salivarius group without considering peripheral species (Lactobacillus algidus).
AAI and POCP values considering members of the L. reuteri and L. vaccinostercus groups.
AAI and POCP values considering members of the L. fructivorans, L. brevis, L. buchneri, and L. collinoides groups.
Numbers in bold are values of >55 to 60% ANI and >50% POCP, which are the thresholds empirically taken as genus delineation. Lower percentages are found within a single phylogroup.
Those challenges suggest that, besides the improvements that genome analyses deliver, genomics-derived thresholds should not be used in isolation or be applied agnostically. Indeed, formal reclassifications should be proposed on the basis of the results of a polyphasic study (10) to ensure that the diversity of taxa is coherently described by names at the different taxonomic ranks. De facto, thresholds (i.e., AAI and POCP) are useful to uniformly delineate taxonomic ranks among phylogenetic lineages, but they should be applied flexibly, and other factors, such as other genomic markers (e.g., clade-specific proteins or conserved amino acids within essential protein sequences [51]), the phenotype (e.g., the carbohydrate fermentation pattern or chemotaxonomic markers [35]), the ecology, and the niche adaptation, should be included in the analysis of all taxonomic ranks, including species (1, 36). A valuable case toward this perspective is given by Zhang and colleagues, who showed a clear link between the Lactobacillus phylogenetic clusterings, their vancomycin-sensitive/resistant phenotype, and the sequence composition of Ddl dipeptide ligase enzyme (51).
Notwithstanding these caveats, the data reported here represent a significant further step toward the splitting of the genus Lactobacillus into more homogeneous genera: they demonstrate a very robust evolutionary backbone at the basis of a possible renovated classification scheme, and this is of utmost importance to guarantee the stability of names of future taxa, once they are delineated, as this is one of the essential points in nomenclature (37). Indeed, until a complete revaluation of the phenotypic coherence of the groups proposed here is performed, no reclassification is advisable; principle 1 of the Bacteriological Code (37) suggests avoiding the useless creation of names, a condition that could occur if genomic thresholds are strictly applied (for instance, if all the peripheral species of the groups in Table 3 were unhelpfully proposed as novel genera) and if the broad effect that this reclassification could have for the scientific community and Lactobacillus users, such as legislative bodies, regulatory agencies, microbial safety assessors (Campedelli et al., unpublished), and probiotic and fermented food manufacturers, is not considered.
The pragmatic genome-based approach applied here to the genus Lactobacillus sheds light on the feasibility of creating a renovated taxonomic scheme in which at least 10 homogeneous genera/clusters could accommodate the existing species and those still to be discovered. An open discussion among other experts, such as the lactic acid bacteria scientific and industrial community and members of the Subcommittee of Taxonomy of the Genus Lactobacillus (35), is now advocated in order to proceed toward the formal proposal of the reclassification of the genus Lactobacillus.
MATERIALS AND METHODS
Data set.
The 222 strains belonging to the genus Lactobacillus and related genera whose genome sequences were used in the present study are described in Table S1 in the supplemental material. A further 47 strains for which the genome sequences were not available were included on the basis of their 16S rRNA gene sequences (Table S1).
Multilocus sequence analysis based on 29 ribosomal proteins and 12 phylogenetic markers and phylogenetic tree construction.
A maximum likelihood phylogeny was built from 29 ribosomal proteins and 12 housekeeping markers, which were chosen on the basis of their use in published multilocus sequence typing schemes and their presence in the genomes of the 222 strains (Table S2) (38).
Amino acid sequences were aligned and concatenated, and the phylogeny was inferred using the PROTCATWAG model in RAxML (v8.0.22) and rooted using Atopobium minutum DSM 20584T, Atopobium rimae DSM 7090T, Kandleria vitulina DSM 20405T, and Olsenella uli DSM 7084T. Bootstrapping was carried out using 100 replicates.
SplitsTree4 (39) was applied to detect conflicting signals (possible horizontal gene transfer events), which were then displayed as networks instead of bifurcating trees.
16S rRNA gene-based phylogeny.
16S rRNA gene phylogenetic analysis for each subgroup was carried out with the MEGA (v7.0.26) (40) software package using the Jukes-Cantor model as the distance model. The neighbor-joining (41) and minimum-evolution (42) methods were used for tree reconstruction. The statistical reliability of the phylogenetic tree topology was evaluated using bootstrapping with 1,000 replicates (43).
Distance-based methods: ANI, AAI, and POCP analyses.
The ANI, AAI, and POCP values across the genomes were calculated according to methods proposed by Konstantinidis and colleagues (17, 44) and Qin et al. (18). In detail, the ANI between two genomes was calculated as the mean identity of all matches obtained by analysis with the BLASTN (v2.2.26+) program based on 1-kb fragments which showed more than 30% overall sequence identity over an alignable region of at least 70% of the total length (45). We used a command line version of the AAI software (http://enve-omics.ce.gatech.edu/aai/) that takes two FASTA files of predicted genes as input, identifies reciprocal best hits by analysis with the BLAST program, and calculates the AAI score on the basis of these orthologs (17). For POCP, an in-house script was written following the formula of Qin et al. (18), which uses two-way BLAST analysis to calculate a POCP score: [(C1 + C2)/(T1 + T2)] · 100, where C is the number of conserved proteins (identity, ≥40%; aligned length of query, ≥50%) and T is the total number of proteins; 1 and 2 refer to input files 1 and 2, respectively (18). The in-house script has been deposited on figshare with the following digital object identifier: https://doi.org/10.6084/m9.figshare.4577953.v1. The amino acid sequences used in the AAI and POCP analyses were predicted using a combination of three software, Glimmer3 (v3.02) (46), GeneMark.HMM (v1.1) (47), and MetaGene (48), where a gene sequence predicted by at least one software was included in the data set. Statistics and visualization were carried out in R (v3.1.1; https://www.r-project.org/) using the pvclust package (49).
Ortholog prediction and identification of clade-specific genes.
Orthologs were predicted using QuartetS, where two sequences from separate genomes were considered to be orthologs if they were bidirectional best hits (BBH) of each other and had ≥30% identity and ≥25% alignment length. QuartetS also differentiates paralogs from orthologs by building quartet gene trees that include two sequences from a third genome. The output from QuartetS was a table with 222 genomes as columns and 34,257 clusters of orthologs as rows, where the presence of a sequence for a particular ortholog was represented as 1 and its absence was represented as 0. This table therefore provided a sequence presence/absence distribution for each ortholog that was used to predict clade-specific genes. The Random Forest algorithm (50) was used to predict clade-specific genes from the R package randomForest. The software was run in an iterative manner using default parameters, where all orthologs having a Gini index of 0 at each iteration were removed. The remaining 90 genes gave an out-of-bag error rate of 0, which is Random Forest's internal method of cross-validation. This suggested that the subset of orthologs contained potential clade-specific genes. These clade-specific genes were identified in R, and further manual assessment was carried out to exclude potential false positives, including the alignment of sequences back to genomes using the TBLASTN program.
Supplementary Material
ACKNOWLEDGMENTS
This project has received funding from the European Union's Horizon 2020 research and innovation program under Marie Skłodowska-Curie grant agreement no. 659801. Work in P.W.O.'s laboratory is funded in part by the Science Foundation Ireland (APC/SFI/12/RC/2273) in the form of a research center, the APC Microbiome Institute.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00993-18.
[This article was published on 17 August 2018 with an incorrect email address for the corresponding author. This was updated in the version posted on 21 August 2018.]
REFERENCES
- 1.Duar RM, Lin XB, Zheng JZ, Martino ME, Grenier T, Pérez-Muñoz ME, Leulier F, Gänzle M, Walter J. 2017. Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41:S27–S48. doi: 10.1093/femsre/fux030. [DOI] [PubMed] [Google Scholar]
- 2.Zotta T, Parente E, Ricciardi A. 2017. Aerobic metabolism in the genus Lactobacillus: impact on stress response and potential applications in the food industry. J Appl Microbiol 122:857–869. doi: 10.1111/jam.13399. [DOI] [PubMed] [Google Scholar]
- 3.Sun Z, Harris HM, McCann A, Guo C, Argimón S, Zhang W, Yang X, Jeffery IB, Cooney JC, Kagawa TF, Liu W, Song Y, Salvetti E, Wrobel A, Rasinkangas P, Parkhill J, Rea MC, O'Sullivan O, Ritari J, Douillard FP, Ross RP, Yang R, Briner AE, Felis GE, de Vos WM, Barrangou R, Klaenhammer TR, Caufield PW, Cui Y, Zhang H, O'Toole PW. 2015. Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6:8322. doi: 10.1038/ncomms9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng J, Ruan L, Sun M, Gänzle M. 2015. A genomic view of lactobacilli and pediococci demonstrates that phylogeny matches ecology and physiology. Appl Environ Microbiol 81:7233–7243. doi: 10.1128/AEM.02116-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stefanovic E, Fitzgerald G, McAuliffe O. 2017. Advances in the genomics and metabolomics of dairy lactobacilli: a review. Food Microbiol 61:33–49. doi: 10.1016/j.fm.2016.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Bourdichon F, Casaregola S, Farrokh C, Frisvad JC, Gerds ML, Hammes WP, Harnett J, Huys G, Laulund S, Ouwehand A, Powell IB, Prajapati JB, Seto Y, Ter Schure E, Van Boven A, Vankerckhoven V, Zgoda A, Tuijtelaars S, Hansen EB. 2012. Food fermentations: microorganisms with technological beneficial use. Int J Food Microbiol 154:87–97. doi: 10.1016/j.ijfoodmicro.2011.12.030. [DOI] [PubMed] [Google Scholar]
- 7.Ricci A, Allende A, Bolton D, Chemaly M, Davies R, Girones R, Herman L, Koutsoumanis K, Lindqvist R, Nørrung B, Robertson L, Ru G, Sanaa M, Simmons M, Skandamis P, Snary E, Speybroeck N, Ter Kuile B, Threlfall J, Wahlström H, Cocconcelli PS, Klein G, Prieto Maradona M, Querol A, Peixe L, Suarez JE, Sundh I, Vlak JM, Aguilera-Gómez M, Barizzone F, Brozzi R, Correia S, Heng L, Istace F, Lythgo C, Fernandéz Escaméz PS. 2017. Scientific opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA. EFSA J 15:4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salvetti E, O'Toole PW. 2017. When regulation challenges innovation: the case of genus Lactobacillus. Trends Food Sci Technol 66:187–194. doi: 10.1016/j.tifs.2017.05.009. [DOI] [Google Scholar]
- 9.Collins MD, Rodrigues U, Ash C, Aguirre M, Farrow JAE, Martinez-Murcia A, Phillips BA, Williams AM, Wallbanks S. 1991. Phylogenetic analysis of the genus Lactobacillus and related lactic acid bacteria as determined by reverse transcriptase sequencing of 16S rRNA. FEMS Microbiol Lett 77:5–12. doi: 10.1111/j.1574-6968.1991.tb04313.x. [DOI] [Google Scholar]
- 10.Vandamme P, Pot B, Gillis M, de Vos P, Kersters K, Swings J. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev 60:407–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felis GE, Dellaglio F. 2007. Taxonomy of lactobacilli and bifidobacteria. Curr Issues Intestinal Microbiol 8:44–61. [PubMed] [Google Scholar]
- 12.Salvetti E, Torriani S, Felis GE. 2012. The genus Lactobacillus: a taxonomic update. Probiotics Antimicrob Proteins 4:217–226. doi: 10.1007/s12602-012-9117-8. [DOI] [PubMed] [Google Scholar]
- 13.Salvetti E, Fondi M, Fani R, Torriani S, Felis GE. 2013. Evolution of lactic acid bacteria in the order Lactobacillales as depicted by analysis of glycolysis and pentose phosphate pathways. Syst Appl Microbiol 36:291–305. doi: 10.1016/j.syapm.2013.03.009. [DOI] [PubMed] [Google Scholar]
- 14.Pot B, Felis GE, De Bruyne K, Tsakalidou E, Papadimitriou K, Leisner J, Vandamme P. 2014. The genus Lactobacillus, p 249–353. In Holzapfel WH, Wood EJB (ed), Lactic acid bacteria: biodiversity and taxonomy. John Wiley & Sons, Hoboken, NJ. [Google Scholar]
- 15.Thompson CC, Chimetto L, Edwards RA, Swings J, Stackebrandt E, Thompson FL. 2013. Microbial genomic taxonomy. BMC Genomics 14:913. doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 17.Konstantinidis KT, Tiedje JM. 2005. Towards a genome-based taxonomy for prokaryotes. J Bacteriol 187:6258–6264. doi: 10.1128/JB.187.18.6258-6264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ. 2014. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosselló-Móra R, Amann R. 2015. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol 38:209–216. doi: 10.1016/j.syapm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Salvetti E, O'Toole PW. 2017. The genomic basis of lactobacilli as health-promoting organisms. Microbiol Spectr 3(3):BAD-0011-2016. doi: 10.1128/microbiolspec.BAD-0011-2016. [DOI] [PubMed] [Google Scholar]
- 21.Glaeser SP, Kämpfer P. 2015. Multilocus sequence analysis (MLSA) in prokaryotic taxonomy. Syst Appl Microbiol 38:237–245. doi: 10.1016/j.syapm.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 22.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguénec C, Lescat M, Mangenot S, Martinez-Jéhanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Médigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 1:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaureguy F, Landraud L, Passet V, Diancourt L, Frapy E, Guigon G, Carbonnelle E, Lortholary O, Clermont O, Denamur E, Picard B, Nassif X, Brisse S. 2008. Phylogenetic and genomic diversity of human bacteremic Escherichia coli strains. BMC Genomics 9:560. doi: 10.1186/1471-2164-9-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brady C, Cleenwerck I, Venter S, Vancanneyt M, Swings J, Coutinho T. 2008. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst Appl Microbiol 31:447–460. doi: 10.1016/j.syapm.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 26.Hammes WP, Hertel C. 2003. The genera Lactobacillus and Carnobacterium. In Dworkin M. (ed), The prokaryotes. Springer, Heidelberg, Germany. [Google Scholar]
- 27.Dellaglio F, Felis GE. 2005. Taxonomy of lactobacilli and bifidobacteria, p 25–50. In Tannock GW. (ed), Probiotics and prebiotics: scientific aspects. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 28.Coster E, White HR. 1964. Further studies of the genus Pediococcus. J Gen Microbiol 37:15–31. doi: 10.1099/00221287-37-1-15. [DOI] [PubMed] [Google Scholar]
- 29.Leisner JJ, Vancanneyt M, Goris J, Christensen H, Rusul G. 2000. Description of Paralactobacillus selangorensis gen. nov., sp. nov., a new lactic acid bacterium isolated from chili bo, a Malaysian food ingredient. Int J Syst Evol Microbiol 50:19–24. doi: 10.1099/00207713-50-1-19. [DOI] [PubMed] [Google Scholar]
- 30.Haakensen M, Dobson CM, Hill JE, Ziola B. 2009. Reclassification of Pediococcus dextrinicus (Coster and White 1964) Back 1978 (Approved Lists 1980) as Lactobacillus dextrinicus comb. nov., and emended description of the genus Lactobacillus. Int J Syst Evol Microbiol 59:615–621. doi: 10.1099/ijs.0.65779-0. [DOI] [PubMed] [Google Scholar]
- 31.Haakensen M, Pittet V, Ziola B. 2011. Reclassification of Paralactobacillus selangorensis Leisner et al. 2000 as Lactobacillus selangorensis comb. nov. Int J Syst Evol Microbiol 61:2979–2983. doi: 10.1099/ijs.0.027755-0. [DOI] [PubMed] [Google Scholar]
- 32.Alnajar S, Gupta RS. 2017. Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infect Genet Evol 54:108–127. doi: 10.1016/j.meegid.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-R LM, Konstantinidis KT. 2014. Bypassing cultivation to identify bacterial species. Microbe 9:111–118. [Google Scholar]
- 34.Gribaldo S, Brochier-Armanet C. 2012. Time for order in microbial systematics. Trends Microbiol 20:209–210. doi: 10.1016/j.tim.2012.02.006. [DOI] [PubMed] [Google Scholar]
- 35.Mattarelli P, Holzapfel W, Franz CM, Endo A, Felis GE, Hammes W, Pot B, Dicks L, Dellaglio F. 2014. Recommended minimal standards for description of new taxa of the genera Bifidobacterium, Lactobacillus and related genera. Int J Syst Evol Microbiol 64:1434–1451. doi: 10.1099/ijs.0.060046-0. [DOI] [PubMed] [Google Scholar]
- 36.Whitman WB. 2015. Genome sequences as the type material for taxonomic descriptions of prokaryotes. Syst Appl Microbiol 38:217–222. doi: 10.1016/j.syapm.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Parker CT, Tindall BJ, Garrity GM. 2015. International Code of Nomenclature of Prokaryotes. Int J Syst Evol Microbiol. doi: 10.1099/ijsem.0.000778. [DOI] [Google Scholar]
- 38.Bottari B, Felis GE, Salvetti E, Castioni A, Campedelli I, Torriani S, Bernini V, Gatti M. 2017. Effective identification of Lactobacillus casei group species: genome-based selection of the gene mutL as the target of a novel multiplex PCR assay. Microbiology 163:950–960. doi: 10.1099/mic.0.000497. [DOI] [PubMed] [Google Scholar]
- 39.Huson DH, Bryant D. 2006. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- 40.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. 1987. The neighbour-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- 42.Nei M, Kumar S. 2000. Molecular evolution and phylogenetics. Oxford University Press, New York, NY. [Google Scholar]
- 43.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 38:791–793. [DOI] [PubMed] [Google Scholar]
- 44.Konstantinidis KT, Tiedje JM. 2007. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol 10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 45.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 46.Delcher AL, Bratke KA, Powers EC, Salzberg SL. 2007. Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinformatics 23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Besemer J, Lomsadze A, Borodovsky M. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res 29:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Noguchi H, Park J, Takagi T. 2006. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 19:5623–5630. doi: 10.1093/nar/gkl723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suzuki R, Shimodaira H. 2006. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 50.Breiman L. 2001. Random forests. Mach Learn 45:5–32. [Google Scholar]
- 51.Zhang S, Oh J-H, Alexander LM, Özçam M, van Pijkeren J-P. 2018. d-Alanyl-d-alanine ligase as a broad-host-range counterselection marker in vancomycin-resistant lactic acid bacteria. J Bacteriol 13:607–617. doi: 10.1128/JB.00607-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.