Letter
Enterobacter cloacae is a clinically important gram-negative member of the Enterobacteriaceae, which has increasingly been recognised as a major pathogen in nosocomial infections. Despite this, knowledge about the population structure and the distribution of virulence factors and antibiotic resistance determinants of this species is scarce. In this study, we analysed a systematic collection of multidrug resistant E. cloacae isolated between 2001 and 2011 from bloodstream infections across hospitals in the UK and Ireland. We found that the population is characterized by the presence of multiple clones formed at widely different time periods in the past. The clones exhibit a high degree of geographical heterogeneity, which indicates extensive dissemination of these E. cloacae clones across the UK and Ireland. These findings suggest that a diverse, community-based, commensal population underlies multidrug resistant E. cloacae infections within hospitals.
Enterobacter cloacae is a non-motile bacterium that belongs to the Enterobacteriaceae family. E. cloacae have been recovered from natural environments 1–3. In addition, E. cloacae is an opportunistic pathogen that can cause a wide range of infections, in particular in immune-compromised individuals. E. cloacae has been increasingly recognized as a cause of hospital-acquired infections and outbreaks in the past few decades 4–6, as reported in hospitals 7,8, especially in the neonatal settings 9. The reservoirs for E. cloacae in hospitals are thought to include equipment, cleaning solutions and healthcare workers 9–11. Furthermore, endogenous infection from an established clone carried by individuals prior to admission to hospital has also been described. Multi-locus sequencing typing (MLST) has shown that the global E. cloacae population is highly diverse, and includes a limited number of clonal complexes that have spread internationally and independently acquired antibiotic resistance elements 12,13.
A number of antibiotic resistance genes, and to a lesser extent pathogenicity mechanisms, of E. cloacae have been elucidated to date 5,14–17. Multiple virulence factors and extended spectrum beta-lactamases (ESBLs) or the hyper-production of constitutive AmpC beta-lactamase have been shown to be involved in the development of infection and antimicrobial resistance, respectively 6. Furthermore previous genomic studies have shown that the E. cloacae genome harbours a large repertoire of accessory genetic elements that enable colonization of diverse environments and efficient metabolic adaptation to different niches and environmental stresses 6,18–20.
There is a growing recognition that E. cloacae is now the third major Enterobacteriaceae species involved in nosocomial infections after Escherichia coli and K. pneumoniae. To understand the spread of multidrug-resistant pathogens within the population it is necessary to determine their epidemiology and population structure. However, unlike other important pathogens, our knowledge about the population genomics of E. cloacae is limited. To fill this gap, we utilized whole genome sequencing of a large systematic collection of multidrug resistant (MDR) E. cloacae collected from various hospitals across the UK between 2001 and 2011.
Our collection contained 316 genomes composed of MDR E. cloacae collected from 37 hospitals in the UK and Ireland between 2001 and 2011. We first calculated the set of core genes (genes shared by every member of the population) and accessory genes (genes that were variably present between isolates). De-novo assembly and annotation of core and accessory genomes showed that the pan-genome of E. cloacae contained 44,235 genes, 1,567 and 490 of which were in the hard core (genes present in >99% of samples) and the soft core genome (genes present in >95% and <99% of samples), respectively. This large number of accessory genes indicates a very high degree of flexibility of the E. cloacae genome, which reflects the diverse environmental habitats that this bacterium occupies. The core genome was ~1.5 Mbp. We found a high degree of congruence between the pattern of the presence and absence of accessory genes and the genetic distance between the core genomes in our dataset (p < 0.001, Mantel test) suggesting that the MDR clones of which the data set comprises were each formed with a distinct subset of genes which have undergone little change since the clone emerged (Supplementary Figure 1 A).
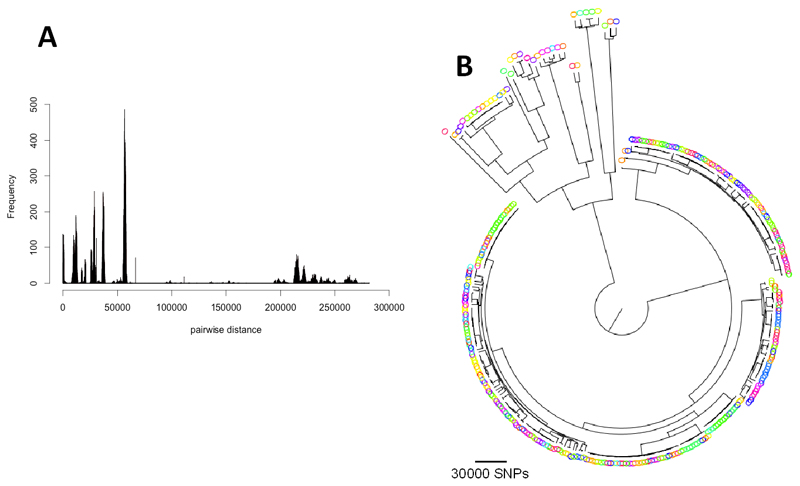
The distribution of pairwise SNP distances between isolates showed that the population of MDR isolates was highly structured and composed of closely-related clades together with strains that were far more distantly related to other isolates in the collection (Figure 1A and Figure 1B). In line with this, the phylogenetic tree constructed from the core genome variant sites showed that the population consisted of one large primary group with a number of closely related sub-populations (clones) nested within it, and a divergent clade that mainly contained individual isolates that, according to rpoB and hsp60 typing 21, represent separate sub-species in the E. cloacae complex including Enterobacter asburiae and Enterobacter kobei (Supplementary Figure 1 B and 1 C in Supplementary Material). We observed a low degree of geographic clustering in the tree, and the high level of diversity of the species was also reflected in the number of different MLST sequence types (STs) that made up the population, some of which are the most widespread STs in a global collection of E. cloacae 13. This suggests that isolates in our collection are part of a global circulation of these clones (Supplementary Figure 2 in Supplementary Material).
Figure 1.
A) Histogram of pair-wise SNP distance between the core genome sequences of isolates. B) Maximum Likelihood tree for the isolates. The colours of the isolates correspond to hospitals of isolation.
In order to investigate the recent evolution of E. cloacae in greater detail, we analysed the major clones in our collection. Utilizing the clustering method detailed in the methods section, we identified 11 distinct groups that contained more than 9 isolates (Figure 2A). We found that isolates in these groups could be adequately identified by MLST since most clusters contained a single sequence type (Figure 2B, Supplementary Notes 2 in Supplementary Material). As with the whole tree, the clones exhibited a high level of geographical diversity in that the majority of the clusters contain isolates originating from multiple hospitals (Figure 2C). Furthermore, and in line with this, the distribution of the pairwise geographical distances between isolates within each clone were as widespread as those within the whole collection for the majority of clones, and only for clones 14, 2 and 13 was the geographical clustering significant (p <0.01, one-sided t-test < 0.01) (Figure 2D). This implies that, prior to the time of the study, the dissemination of all the different E. cloacae MDR clones appeared to have already taken place across the country.
Figure 2.
A) Detected clusters (CLs) represented on the phylogenetic tree. The intensity of branch colour corresponds to the bootstrap support (SH support). B) MLST sequence type (ST) composition of each of the clusters. Each colour signifies one ST. C) Hospitals from which isolates in each cluster were identified. D) Distribution of pairwise geographical distance for each cluster compared with the distribution of the whole population. The notched area shows the 95% confidence interval around the median. E) Distribution of the time of isolation for isolates in each cluster. The boxes in D) and E) give the interquartile range, the whiskers indicate the boundary of 1.5 times the interquartile range, and the points beyond that are outliers.
Utilizing the variation within the time of isolation within each cluster (Figure 2E), we estimated the substitution rate for the genome for those clusters where there was a detectable signal to be between 0.5 and 3 SNPs per genome per year (an average of 1.5 x 10-7 SNPs per site per year across the genome) (Supplementary Figures 3 A and S 4 in Supplementary Material). These calculations indicate that these clusters were established between ~10 and ~150 years ago (Supplementary Figure 3 B). Using the average substitution rate, and applying it to the other major clusters for which we could not detect a temporal signal, we calculated the root age for these clusters to be between 50 and 250 years ago (Supplementary Figure 3 B and 4 in Supplementary Material). We conclude that drug resistant E. cloacae clones have been continuously arising and expanding over the past several decades. Subsequently, we used these substitution rates to estimate the dates and numbers of putative recent relationships of isolates in different hospitals. To this end we extracted all the pairs of isolates that were less than 40 SNPs apart (which corresponds to ~20 years). We found that these isolates originated from different hospitals across the country, and when we compared the geographical distances between these linked hospitals with the distribution of pairwise geographical distance for the whole collection we found no significant difference (p > 0.05, one-sided t-test) (Supplementary Figure 3 C in Supplementary Material). The results demonstrate that inter-hospital connectivity between genetically similar isolates exists, but that this is not necessarily between geographically close hospitals (Supplementary Figure 3 C in Supplementary Material). To further examine this finding, we reconstructed the networks for all SNP cutoffs since the most recent clone (clone 7) emerged in the population. Our results show that although the proportion of inter-hospital relationships decreased with stricter cut-off values, the number of isolates in the network also declined rapidly (Supplementary Figure 5 in Supplementary Material). The findings show that even with 316 samples the population is still so diverse that only a very few recent close relationships can be detected.
Within the population, we were able to identify the differential presence of multiple plasmids and putative virulence factors, some of which were clade specific, implying that the population of E. cloacae has developed both conserved and variable virulence mechanisms (Supplementary Notes 4 and 5 in Supplementary Material). Furthermore our antimicrobial resistance analysis demonstrated various patterns of emergence of antimicrobial resistance determinants in the collection, where resistance determinants were acquired independently throughout the population (Supplementary Notes S6 in Supplementary Material). This implies a potential for rapid adaptability in E. cloacae. The observations of the antimicrobial resistance analysis are particularly important since the emergence of antibiotic resistance for the currently effective antibiotics such as imipenem, tigecycline and ciprofloxacin raises concerns that the resistance profile may extend in the future, in particular through mobile genetic elements. Moreover, the commensal nature of the bacterial population documented here will allow rapid spread of resistant E. cloacae throughout the human population, outside of hospital-based infection control, from which it can readily be imported into hospitals.
The dissemination of E. cloacae raises questions about the underlying reservoirs and routes of the spread of pathogens between hospitals. A diverse range of sources, including healthcare staff and equipment, waste waster and food have been shown to serve as reservoirs of E. cloacae 2,3,22. Despite being the largest genomic collection of E. cloacae studied to date, our collection only contained clinical isolates so we are not able to draw a definitive conclusion about the sources and potential reservoirs of E. cloacae. However the relationship between our isolates and previously published E. cloacae genomes indicates that clinical and non-clinical strains originating from the USA, India and China are scattered throughout our phylogeny, both within and outside the clones identified here (Supplementary Figure 17). Clinical isolates recovered from a systematic screen of infections in a single hospital in the USA 23 were also spread across the tree, although mainly in the rarer sub-species. An environmental isolate recovered from oil-contaminated soil in India 24, a food borne isolate from pepper 19 in Hong Kong and an isolate of plant infection origin 20 were also mainly in the rarer sub-species. Together, this suggests that the broad diversity of E. cloacae causing infections in the UK may be part of a global population transmitting via different routes involving both clinical and non-clinical sources. To fully elucidate this, a more comprehensive collection, containing isolates of different non-clinical and clinical origins is required, which would serve as the basis for future genomic studies on E. cloacae.
Enterobacter infections can be acquired within a hospital or originate from a pathogen within an already colonized patient. Although there are reports tracing infections to the contamination of hospital devices, most of the infections reported previously do not appear to have a common nosocomial source and may be accounted for by previously established clones within patients 25,26. Our findings are consistent with diverse sources of E. cloacae in any individual hospital. The close genetic relationship between isolates from different geographic regions that was frequently observed suggest that individuals that have already been colonised before entering hospital introduce multiple strains into hospitals, some of which may be involved in subsequent intra-hospital transmissions. Isolates from hospital infections therefore represent samplings from the diversity of commensal E. cloacae in the community. In this sense the apparent spread of E. cloacae between the hospitals found here may only mirror the dissemination of this species throughout the UK and Ireland. The presence of some isolates from the known major globally circulating STs, as well as the close genetic distance between isolates of different source and country of origins, indicate that the geographical distribution of E. cloacae lineages goes beyond UK and Ireland and belongs to a global population 7. From this, it is apparent that controlling E. cloacae infections may be more difficult than other nosocomial pathogens, where within-hospital transmission is often the primary source of new infections.
Methods
Isolates and antimicrobial susceptibility testing
The study was approved by the National Research Ethics Service (ref: 12/EE/0439) and the CUH Research and Development (R&D) Department. We studied a national collection of 316 E. cloacae isolates that were collected as part of the British Society for Antimicrobial Chemotherapy (BSAC) Resistance Surveillance project (www.bsacsurv.org). The collection was part of a bacteraemia surveillance programme (all isolates were from blood of patients with blood infection) that ran between 2001-2011 in 37 hospitals across the United Kingdom and Ireland (UK&I). The strains were identified to species level using API20E strips and the species were further confirmed using Kraken 27. Selection for the collection was based on the presence of phenotypic resistance to at least 4 antimicrobial groups. We provide a list of isolates in Supplementary Table 1.
Antimicrobials that were tested in this study were beta-lactams (penicillin: amoxicillin, cephalosporins: cefuroxime, cefoxitin, cefotaxime, ceftazidime, and piperacillin-tazobactam and amoxicilin-clavulanic acid), tetracyclines (minocycline, tetracycline and tigecycline), aminoglycosides (gentamicin) and fluoroquinolones (ciprofloxacin). We defined Multidrug resistance (MDR) as acquired non-susceptibility to at least one agent in three or more antimicrobial categories, as it is proposed in 28. We then selected Isolates based on the presence of phenotypic resistance to at least one antimicrobial in three of the antimicrobial groups: penicillins, carbapenems, cephalospsorins, tetracyclines, aminoglycoside and fluoroquinolones.
The minimum inhibitory concentration (MIC) for each antimicrobial agent was determined using the BSAC agar dilution method. To obtain an insight into the distribution of MIC values, we compared the distribution of MIC values of our samples with those from the European Committee on Antimicrobial Susceptibility Testing (EUCAST). We obtained these distributions and also the clinical susceptibility breakpoints to determine resistance from the EUCAST website (www.eucast.org) on 02/02/2016.
Sequencing, pan-genome and phylogenetic analysis
DNA was extracted using a QIAxtractor (QIAgen), according to the manufacturer’s instructions. Sequencing libraries were constructed as described by the manufacturer (http://www.illumina.com/techniques/sequencing/ngs-library-prep). The sequencing was conducted on an Illumina HiSeq2000 with a read length of 2X100 bp. 96 samples were multiplexed per lane to give an average depth of coverage of 71-fold. The assembled data and raw sequences were deposited in the ENA under the accession numbers provided in Supplementary Table 1. We assembled paired end sequence reads using an in-house assembly and improvement pipeline 29, based on Velvet 30. Subsequently we annotated the assemblies with Prokka, which takes fragmented de novo assemblies 31. We then used the annotated assemblies produced by Prokka as input to Roary, a tool that rapidly builds large-scale pan-genomes, to construct the core and accessory genome 32 (One isolate was excluded from pan-genome analysis, as it fell outside Roary’s similarity cut-offs). To estimate a Maximum Likelihood tree, we employed FastTree version 2.1.3 with generalized time-reversible model to reconstruct the phylogenetic tree from the core genome alignment 33. We assessed the confidence values through SH-like support. To visualize trees and associated metadata, we used FigTree (http://tree.bio.ed.ac.uk/software/figtree/), Dendroscope 3 34 and in-house tools.
Divergence analysis and cluster analysis
In addition to phylogenetic analysis we reconstructed the network of relatedness between the isolates using the SeqTrack algorithm function in R package adegenet 35,36. The algorithm reconstructs ancestries between the sequences, by considering the genetic distances and isolation dates, to obtain a tree with maximum parsimony. In Supplementary Figure 3, we included the network in which the edges that correspond to >40 SNPs, i.e. the number of variant sites in the core genome alignment, were excluded. Given the estimated substitution rate of E. cloacae, 40 SNPs is equivalent to SNPs accumulated over approximately ~20 years (see main text). To identify the clusters in the phylogenetic tree, we used the clustering algorithm detailed in the adegenet package. The distance-based clustering algorithm takes the pairwise SNP distance matrix and a SNP cut-off to identify the clones. We tested various SNP cut-off values from 0, producing 315 clusters, to very large values, which collapses the entire sample set into one cluster. To find the best cut-off for clustering we first excluded 14 outliers, i.e. the isolates that were very distantly related to other isolates in the collection, and then varied the SNP cutoffs while measuring the number of clusters identified. The longest plateau where the clustering did not change extended for 3356 SNPs, and this yielded 88 clusters We then used the 11 clusters with more than 9 members, including 194 isolates in total, for downstream analysis.
To assess the output of the clustering method, we compared the result with the output of HierBAPS, which performs Bayesian analysis of population structure to cluster similar genomic sequences, using 2 clustering iterations and 10, 30 and 50 expected number of clusters (k) as input parameters. Our clusters are strongly in agreement with the clusters of the second iteration of BAPS clustering (Supplementary Figure 18).
Phylogenetic analysis and substitution rate calculation
After identifying major clusters on the tree we attempted to estimate the recent nucleotide substitution rate. For the nucleotide substitution rate estimation, we first employed Gubbins 37 to remove high SNP density regions, which signified putative recombination events, from the multiple alignment for each cluster. We then reconstructed the tree for each cluster and plotted root-to-tip distance versus year of isolation for each sample. Subsequently we conducted 10,000 bootstraps with re-sampled years to obtain a distribution for R-squared values. We then compared the real R-squared values with the distribution to check the significance of the temporal signal in each cluster. We found a strong temporal signal for cluster 8 and weaker signals for clusters 7, 18 and 31. Subsequently we used the BEAST v1.7 package to obtain the substitution rate and the age of the root node for the clusters with a signal 38.
We tested various models including a strict molecular clock (uniform prior distribution for the clock rate model) and a log-normal model with constant population model. For each parameter set we performed 100 million generations with sampling every 10 generations for three times. Besides checking that different runs had converged on similar values, we also assessed the convergence for each model by ensuring that ESS values were greater than 200 for key parameters. A burn-in of 10 million states was discarded from each of the three runs. Since the log-normal clock model always converged, we included the results of this model in the manuscript. We took the age of the root as the height of the root of the Maximum Clade Credibility (MCC) tree reconstructed by combining trees using tree annotator program from the BEAST package.
Molecular Typing of E. cloacae complex, in silico MLST analysis and identification of antimicrobial resistance determinants, virulence factors and plasmids
Using the srst2 package 39, we mapped the paired-end reads to resistance, and plasmid replicon genes in the databases detailed in the srst2 package (coverage cut-off 90%). We also used srst2 to find virulence genes in the Virulence Factors Database (VFDB) (http://www.mgc.ac.cn/VFs/main.htm) in the isolates. We then visualized the genes across the phylogenetic tree using an in-house tool. We determined the STs of the samples using an in silico MLST in-house script that uses the assemblies as input. Furthermore, we conducted the rpoB and hsp60 (groL) typing of E. cloacae complex to detect sub-species of E. cloacae within our dataset 21. To this end, gene sequences for rpoB and hsp60 (groL) were extracted from the assembled genomes. These were then trimmed and compared to the reference set 21. After phylogenetic estimation with RAxML using a general time reversible (GTR) evolutionary model and a gamma correction for among site rate variation 40, isolates were allocated to the different clusters based on the reference dataset. This grouping was then compared back to the whole-genome phylogeny and clustering.
Regression analysis to identify antimicrobial resistance determinants
We developed three linear regression models to identify genetic determinants that are strongly associated with resistance. To this end we used MIC values as a continuous dependent variable in the linear regression models, instead of the categorical resistant/susceptible status. This is because MIC values enabled us to identify the effect of different genetic elements even when all samples are classed as resistant. Moreover, the MIC values have a higher variance than the categorical resistance status and therefore provide more information and allow greater statistical confidence in the results.
In the first linear regression model, we took the MIC values for different antimicrobials as a continuous dependent variable and the presence/absence of individual accessory genes as a categorical predictor variable. By doing so we aimed to identify genes in the accessory genome that are strongly associated (p-value<10-5) with resistance level.
We performed two types of SNP based regression analysis. We first identified genes in which the number of non-synonymous SNPs strongly correlates with resistance levels and identified individual SNPs, the presence of which are strongly associated with MIC levels. To this end, we mapped the short reads against the reference genome E. cloacae ATCC 13047 (accession number: CP001918) using SMALT v0.7.4 (https://www.sanger.ac.uk/resources/software/smalt/). The annotation of SNPs was done with an in-house script that combines SAMtools mplieup 41 and BCFtools as detailed in 42 (7 isolates were excluded from the SNP analysis). The SNP association regression model identified known/putative resistance SNPs that were strongly correlated with MIC values (p-value<10-5) primarily for ciprofloxacin. Samples in which SNPs were absent or unknown were excluded from the analysis for each site. We also excluded the sites in which more than 10% of nucleotides across the population were unknown. In the linear regression model we considered MIC values as continuous dependent variable and the presence/absence of individual SNPs as a categorical predictor variable. In our second SNP-based analysis, we aimed to find genes with a strong correlation between their non-synonymous variation pattern and MIC. To this end we developed a linear regression model in which the MIC values for different antimicrobials were considered as the continuous dependent variable and the number of non-synonymous SNPs in individual genes was used as the continuous predictor variable. In all of the above regression models, we accounted for the population structure by including the MLST information as the categorical predictor variable into the regression models. We set a stringent p-value of 10-5 to define strong associations and then screened the hit list of strongly associated genes/SNPs for putative resistance genes/SNPs.
We have provided the output files of pan-genome analysis, the input and R code for the three regression models in a public repository (https://data.mendeley.com/datasets/vg6cnk9bc5/1). We have also included the hit lists for the three regression models as Supplementary Tables 2, 3 and 4. The sequence data for the isolates from this study have been submitted to the European Nucleotide Archive (ENA; http://www.ebi.ac.uk/ena) under accession numbers PRJEB5065 and ERP004424.
Supplementary Material
Acknowledgements
We thank Hayley Brodrick and Kim Judge for their laboratory assistance, and the library construction, sequencing and core informatics teams at the Wellcome Trust Sanger Institute. We thank the BSAC for allowing the use of isolates from the BSAC Resistance Surveillance Project. This publication presents independent research supported by the Health Innovation Challenge Fund (HICF-T5-342 and WT098600), a parallel funding partnership between the UK Department of Health and Wellcome Trust. The views expressed in this publication are those of the authors and not necessarily those of the Department of Health, Public Health England or the Wellcome Trust. This project was also funded by a grant awarded to the Wellcome Trust Sanger Institute (098051).
Footnotes
Contributions
S.J.P., D.M. and J.P. designed the study. D.M. analysed the data. S.R. performed the in silico rpoB and hsp60 typing. V.M., S.J.P. and J.P. contributed materials and data. S.J.P and J.P. completed ethical approvals. J.P. and S.J.P. were responsible for management of the study.
References
- 1.Hua X, et al. Degradation of hexadecane by Enterobacter cloacae strain TU that secretes an exopolysaccharide as a bioemulsifier. Chemosphere. 2010;80:951–6. doi: 10.1016/j.chemosphere.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 2.White L, et al. Carbapenemase-producing Enterobacteriaceae in hospital wastewater: a reservoir that may be unrelated to clinical isolates. J Hosp Infect. 2016;93:145–51. doi: 10.1016/j.jhin.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Al-Kharousi ZS, Guizani N, Al-Sadi AM, Al-Bulushi IM, Shaharoona B. Hiding in Fresh Fruits and Vegetables: Opportunistic Pathogens May Cross Geographical Barriers. Int J Microbiol. 2016;2016 doi: 10.1155/2016/4292417. 4292417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Davin-Regli A, Pages JM. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Frontiers in Microbiology. 2015;6 doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sanders WE, Sanders CC. Enterobacter spp.: Pathogens poised to flourish at the turn of the century. Clinical Microbiology Reviews. 1997;10:220. doi: 10.1128/cmr.10.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mezzatesta ML, Gona F, Stefani S. Enterobacter cloacae complex: clinical impact and emerging antibiotic resistance. Future Microbiol. 2012;7:887–902. doi: 10.2217/fmb.12.61. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Baca V, et al. Molecular epidemiological typing of Enterobacter cloacae isolates from a neonatal intensive care unit: three-year prospective study. J Hosp Infect. 2001;49:173–82. doi: 10.1053/jhin.2001.1053. [DOI] [PubMed] [Google Scholar]
- 8.Pestourie N, et al. Outbreak of AmpC beta-lactamase-hyper-producing Enterobacter cloacae in a neonatal intensive care unit in a French teaching hospital. Am J Infect Control. 2014;42:456–8. doi: 10.1016/j.ajic.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Dalben M, et al. Investigation of an outbreak of Enterobacter cloacae in a neonatal unit and review of the literature. J Hosp Infect. 2008;70:7–14. doi: 10.1016/j.jhin.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 10.Dugleux G, Le Coutour X, Hecquard C, Oblin I. Septicemia caused by contaminated parenteral nutrition pouches: the refrigerator as an unusual cause. JPEN J Parenter Enteral Nutr. 1991;15:474–5. doi: 10.1177/0148607191015004474. [DOI] [PubMed] [Google Scholar]
- 11.Wang SA, et al. Enterobacter cloacae bloodstream infections traced to contaminated human albumin. Clinical Infectious Diseases. 2000;30:35–40. doi: 10.1086/313585. [DOI] [PubMed] [Google Scholar]
- 12.Miyoshi-Akiyama T, Hayakawa K, Ohmagari N, Shimojima M, Kirikae T. Multilocus Sequence Typing (MLST) for Characterization of Enterobacter cloacae. Plos One. 2013;8 doi: 10.1371/journal.pone.0066358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Izdebski R, et al. MLST reveals potentially high-risk international clones of Enterobacter cloacae. J Antimicrob Chemother. 2015;70:48–56. doi: 10.1093/jac/dku359. [DOI] [PubMed] [Google Scholar]
- 14.Potron A, Poirel L, Rondinaud E, Nordmann P. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill. 2013;18 doi: 10.2807/1560-7917.es2013.18.31.20549. [DOI] [PubMed] [Google Scholar]
- 15.Empel J, et al. Molecular survey of beta-lactamases conferring resistance to newer beta-lactams in Enterobacteriaceae isolates from Polish hospitals. Antimicrob Agents Chemother. 2008;52:2449–54. doi: 10.1128/AAC.00043-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barnes AI, Ortiz C, Paraje MG, Balanzino LE, Albesa I. Purification and characterization of a cytotoxin from Enterobacter cloacae. Can J Microbiol. 1997;43:729–33. doi: 10.1139/m97-105. [DOI] [PubMed] [Google Scholar]
- 17.Stuber K, Frey J, Burnens AP, Kuhnert P. Detection of type III secretion genes as a general indicator of bacterial virulence. Molecular and Cellular Probes. 2003;17:25–32. doi: 10.1016/s0890-8508(02)00108-1. [DOI] [PubMed] [Google Scholar]
- 18.Ren Y, et al. Complete genome sequence of Enterobacter cloacae subsp. cloacae type strain ATCC 13047. J Bacteriol. 2010;192:2463–4. doi: 10.1128/JB.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu WY, et al. Complete genome sequence of the endophytic Enterobacter cloacae subsp. cloacae strain ENHKU01. J Bacteriol. 2012;194:5965. doi: 10.1128/JB.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coulson TJ, Patten CL. Complete Genome Sequence of Enterobacter cloacae UW5, a Rhizobacterium Capable of High Levels of Indole-3-Acetic Acid Production. Genome Announc. 2015;3 doi: 10.1128/genomeA.00843-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann H, Roggenkamp A. Population genetics of the nomenspecies Enterobacter cloacae. Appl Environ Microbiol. 2003;69:5306–18. doi: 10.1128/AEM.69.9.5306-5318.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raymond F, et al. The initial state of the human gut microbiome determines its reshaping by antibiotics. ISME J. 2016;10:707–20. doi: 10.1038/ismej.2015.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roach DJ, et al. A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota. PLoS Genet. 2015;11:e1005413. doi: 10.1371/journal.pgen.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A, Chettri B, Langpoklakpam JS, Singh AK, Chattopadhyay D. Draft Genome Sequence of Hydrocarbon-Degrading Enterobacter cloacae Strain S1:CND1, Isolated from Crude Oil-Contaminated Soil from the Noonmati Oil Refinery, Guwahati, Assam, India. Genome Announc. 2016;4 doi: 10.1128/genomeA.00367-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McConkey SJ, Coleman DC, Falkiner FR, McCann SR, Daly PA. Enterobacter cloacae in a haematology/oncology ward--first impressions. J Hosp Infect. 1989;14:277–84. doi: 10.1016/0195-6701(89)90067-4. [DOI] [PubMed] [Google Scholar]
- 26.Kuhn I, Aylingsmith B, Tullus K, Burman LG. The Use of Colonization Rate and Epidemic Index as Tools to Illustrate the Epidemiology of Fecal Enterobacteriaceae Strains in Swedish Neonatal Wards. Journal of Hospital Infection. 1993;23:287–297. doi: 10.1016/0195-6701(93)90146-q. [DOI] [PubMed] [Google Scholar]
- 27.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 29.Page A, De Silva N, Hunt M, Quail M, Parkhill J, Harris S, Otto T, Keane J. Robust high throughput prokaryote de novo assembly and improvement pipeline for Illumina data. Microbial Genomics. 2016 doi: 10.1099/mgen.0.000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 32.Page AJ, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3. doi: 10.1093/bioinformatics/btv421. 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huson DH, et al. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 2007;8:460. doi: 10.1186/1471-2105-8-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jombart T, Eggo RM, Dodd PJ, Balloux F. Reconstructing disease outbreaks from genetic data: a graph approach. Heredity. 2011;106:383–390. doi: 10.1038/hdy.2010.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jombart T, Ahmed I. adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics. 2011;27:3070–1. doi: 10.1093/bioinformatics/btr521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43:e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inouye M, et al. SRST2: Rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris SR, et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–74. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.