ABSTRACT
Selenocysteine (Sec), a rare genetically encoded amino acid with unusual chemical properties, is of great interest for protein engineering. Sec is synthesized on its cognate tRNA (tRNASec) by the concerted action of several enzymes. While all other aminoacyl-tRNAs are delivered to the ribosome by the elongation factor Tu (EF-Tu), Sec-tRNASec requires a dedicated factor, SelB. Incorporation of Sec into protein requires recoding of the stop codon UGA aided by a specific mRNA structure, the SECIS element. This unusual biogenesis restricts the use of Sec in recombinant proteins, limiting our ability to study the properties of selenoproteins. Several methods are currently available for the synthesis selenoproteins. Here we focus on strategies for in vivo Sec insertion at any position(s) within a recombinant protein in a SECIS-independent manner: (i) engineering of tRNASec for use by EF-Tu without the SECIS requirement, and (ii) design of a SECIS-independent SelB route.
KEYWORDS: Protein engineering; selenocysteine; selenoproteins; genetic code expansion, tRNA, SelA, SelB
Why make selenoproteins
Selenium (Se) is a trace element and a micronutrient that is both essential and toxic [1]. Insufficient levels of Se in human or animal diets can lead to a variety of disorders including liver necrosis, seizures, muscular dystrophy, cardiomyopathy, exudative diathesis and atherosclerosis [2]. The primary biological form of Se is selenocysteine (Sec), the 21st genetically encoded amino acid found in all three domains of life [3,4]. Disruption of Sec biosynthesis in mammals is lethal [5], but some groups of organisms lack Sec, such as fungi, higher plants, and many insects [6]. Some lineages, like nematodes, Drosophila flies, and plasmodium parasites, encompass species harboring the selenoprotein synthesis machinery and others that lack it [7–9], suggesting selective loss of Sec during evolution.
Selenoproteins play varied physiological roles. Many bacterial and archaeal selenoproteins are catabolic enzymes that only function in anaerobic environment [10,11], while most eukaryotic selenoproteins are engaged in redox signaling and oxidative stress response [12]. Out of 25 selenoproteins in human, five are glutathione peroxidases (Gpx), three are thioredoxin reductases (TrxR) and one is methionine-R-sulfoxide reductase (MsrB). In addition, Sec is involved in activation and deactivation of the thyroid hormones because all three human iodothyronine deiodinases are selenoproteins [12].
Sec shares many similarities with cysteine (Cys), such as high affinity to metals, nucleophilicity and the ability to form diselenide/disulfide bonds, but their use and distribution in the cell are very different [13]. Sec is the least abundant proteinogenic amino acid in eukaryotes and non-methanogenic bacteria [14]. Utilization of Sec is limited to a smaller number of proteins, but its presence often grants those proteins unique catalytic properties [2]. Sec-variants of mammalian methionine-R-sulfoxide reductases are 100- to 1000- fold more active than their Cys-variants although both versions co-exist and function in different cellular compartments [15]. Formate dehydrogenase, a natural selenoprotein from Escherichia coli (E. coli) has over 300-fold higher turnover (k cat), 20-fold higher K d, and 3-fold higher K m towards formate compared to its mutant variant with Cys as an active site residue [16]. In another example, Cys to Sec substitution in the active site of mammalian TrxR results in over 100-fold loss in k cat towards thioredoxin [17]. However, Cys-containing TrxR from Drosophila is an active enzyme that displays only 50% reduction in k cat compared to its mammalian homolog [18], indicating that the effect of thiol-to-selenol substitution greatly depends on the architecture of the enzyme's active site. Other selenoproteins have fully functional Cys-homologs [19]. In the case of selenophosphate synthetase, the specific activity of the Cys-containing enzyme from E. coli is even higher than of its Sec-containing homolog from Haemophilus influenza [20], suggesting that Sec can grant advantages not related to the kinetic properties of the enzyme.
Perhaps the most important property of Sec is the ability to recover from oxidative inactivation [21,22]. Comparison of variants of the same protein or its close homolog shows that selenoenzymes stay active in conditions when Cys-enzymes are not [23–25]. This is because seleninic or selenonic acid (products of Sec oxidation) can be reduced back to selenol by thiols [21,22], while oxidation of Cys to sulfinic or sulfonic acid is irreversible [26,27].
Together, these studies not only shed light on the role of selenoproteins in human health, but also demonstrate the potential for engineering new selenoproteins. Placing Sec in the active site of an enzyme can result in different catalytic activity [23]; for example, Cys-containing subtilisin is a protease, but Sec-subtilisin is a peroxidase [28]. Sec can be used as an effective redox-reactive tag for affinity purification due to its scarcity in the proteome [29]. A valuable biophysical probe in itself for X-ray, NMR and EPR, the unique reactivity of the selenol moiety also allows for site-specific post-translational modification of the targeted protein with various probes, such as fluorescence, or through cross-metathesis, a conjugation technique successfully used to install a molecular mimic of the epigenetic marker Nε-acetyl-l-lysine [30]. Strategically positioned diselenide bonds can assist protein folding [31, 32], trap folding intermediates [33] and make the product more resistant to proteases, like in case of selenoinsulin [34]. Ardent interest in properties of natural and artificial selenoproteins fuels demands production and study of recombinant proteins containing Sec.
How selenoproteins are made
Unlike other proteinogenic amino acids, Sec does not have a cognate aminoacyl-tRNA synthetase; instead, Sec biosynthesis occurs directly on its tRNA (Fig. 1A). First, tRNASec is acylated with serine (Ser) by seryl-tRNA synthetase (SerRS) [35]. In bacteria, Ser-tRNASec is recognized by a selenocysteine synthase (SelA) which directly replaces the hydroxyl group of Ser with a selenol group [36]. In eukaryotes and archaea, formation of Sec-tRNASec from Ser-tRNASec occurs in two steps. First, Ser-tRNASec is phosphorylated by O-phosphoseryl-tRNASec kinase (PSTK) [37]. Second, Sep (O-phosphoserine)-tRNA:Sec-tRNA synthase (SepSecS) forms Sec-tRNASec [38]. Despite the differences, certain components of Sec biosynthesis from different clades remain compatible; for example, bacterial Ser-tRNASec can be effectively phosphorylated by archaeal PSTK and further used as a substrate by human SepSecS [39].
Figure 1.
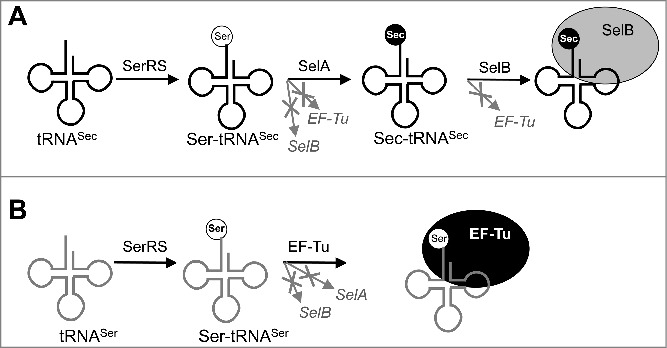
Steps in Sec biosynthesis. First, SerRS activates both tRNASec (A) and tRNASer (B). Ser-tRNASec is not a substrate for translation because it is rejected by EF-Tu and SelB. Ser-tRNASec undergoes Ser-to-Sec conversion catalyzed by SelA, followed by Sec-tRNASec recruitment to the ribosome by SelB. In contrast, Ser-tRNASer is directly recruited to the ribosome by EF-Tu and does not interact with SelA or SelB.
Sec incorporation into proteins requires a number of specialized protein and RNA components. Sec-tRNASec is rejected by EF-Tu which delivers every other aminoacyl-tRNA (aa-tRNA) to the ribosome (Fig. 1) [40]. Instead, all natural selenoproteins rely on the Sec-specific elongation factor SelB or its eukaryotic homolog eEFSec that recognizes Sec-tRNASec and delivers it to the ribosome [3]. Co-evolution with SelA and SelB resulted in tRNASec acquiring a number of distinctive features in the variable arm, D- and acceptor stems, and antidiscriminator box (Fig. 2) [41]. SelB mediates Sec insertion at a UGA stop codon in response to the SECIS (SElenoCysteine Insertion Sequence) element in mRNA [3]. In vivo recoding of UGA is specific [42]. In bacteria, SECIS elements are located immediately downstream of Sec insertion sites. SECIS is directly recognized by the C-terminal domain 4 of SelB; this domain is absent in EF-Tu [43,44]. In eukaryotes and archaea, the SECIS element is typically located in the 3’ untranslated region of the mRNA, and its interaction with eEFSec requires additional protein cofactors [45,46]. Different requirements for Sec insertion further complicate expression of heterologous selenoproteins in E. coli [47].
Figure 2.
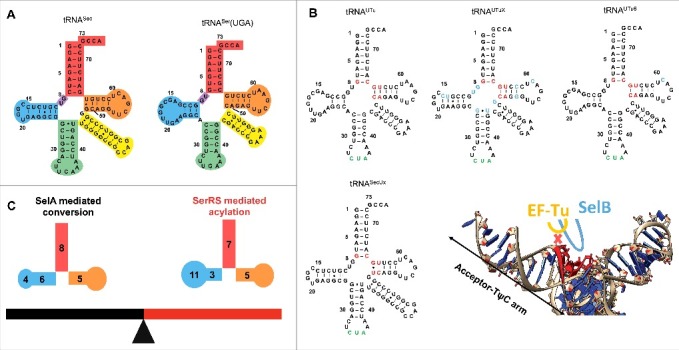
tRNAs for Sec incorporation in E. coli. (A) Cloverleaf models of tRNASec (left) and tRNASer(UGA) from E. coli (right). The acceptor arm is red, TψC-arm is orange, variable arm is yellow, anticodon arm is green, D-arm is blue, and AD linker is purple. The positions of nucleotides in the tRNAs are indicated. (B) Cloverleaf models of tRNASec variants including tRNAUTu, tRNAUTuX, tRNAUTuT6, and tRNASecUx. The antideterminant box, anticodon and nucleotides that are changed from the original tRNAUTu are colored red, green, and blue, respectively. Superposition of SelB and EF-Tu with tRNASec (PDB: 3W3S) reveals the extended loop in domain 3 of SelB is compatible with the antideterminant sequence in tRNASec, while the interaction between the short loop in EF-Tu and the antideterminant box is prevented by steric hindrance. The acceptor-TψC arm is indicated. (C) The “seesaw effect” in tRNA. The 8/5 fold, long D-stem and the tRNASec elbow are important for recognition by E. coli SelA but inhibit recognition by SerRS; while the 7/5 fold and D-arm of bacterial tRNASer allow for efficient acylation by SerRS but are not compatible with binding to SelA. Note tRNASec has a 6-bp D-stem and 4-nucleotide D-loop while tRNASer has a 3-bp D-stem and 11-nucleotide D-loop.
Partial chemical synthesis and native peptide ligation have been successfully used for Sec insertion into several proteins [48,49]. These methods are well suited for relatively small proteins (up to 200 amino acids) and can yield homogenous product in milligram quantities. Human selenoproteins M and W (SelM and SelW) were produced in such manner [48]. Larger proteins, such as Sec-containing RNase A, or human TrxR can be obtained via expressed-protein ligation, whereby the Sec-containing peptide is fused to the recombinantly expressed part(s) of the protein [48].
In vivo expression systems require less specialized equipment, but require subsequent separation of the desired product from other cellular components. Overall, bacterial expression platforms remain the most common way of recombinant selenoprotein production due to their versatility and low cost. Production of selenoproteins can be achieved by UAG-encoded incorporation of protected Sec followed by removal of the protection group. This strategy is attractive because incorporation of a non-canonical amino acid that can be converted into Sec is independent of Sec biosynthesis and insertion machinery. In addition, this strategy allows to avoid the consequences of high reactivity of Sec in the cells. Note that photocaged-Sec incorporation has been reported in yeast cells [50].
Incorporation of Sec into multiple positions within the same open reading frame, or into sites that cannot accommodate a SECIS structure, can be accomplished via one of the SECIS-independent approaches. One way to bypass the requirement of the SECIS is to evolve EF-Tu-compatible tRNASec variants [51–53]. This approach allows Sec insertion at any position of the protein with high specificity and efficiency [52]. Unwanted recoding of other translational stop codons to Sec can be avoided by using the engineered E. coli C321.ΔA strains having all 321 UAG (amber) stop codons in its genome changed to the UAA (ochre) stop codon, and the corresponding release factor 1 (RF1) deleted [54]. Together with changing the anticodon of tRNASec or its EF-Tu-compatible variants to pair with UAG, such strains allow unambiguous reassignment of the amber codon to Sec, further improving selenoprotein yields [52,55]. Alternatively, one can redesign E. coli SelB to work efficiently in a SECIS-independent manner. Such an approach has the benefits of preserving the natural efficiency and specificity of bacterial Sec incorporation, due to the unique ability of SelB to distinguish between Sec-tRNASec and its precursor, Ser-tRNASec [56].
Engineering tRNASec for EF-Tu-mediated selenoprotein synthesis
tRNASec evolved to interact with three protein factors: SerRS, SelA and SelB (Fig. 1B) [3]. Thus, any attempt to make tRNASec a better substrate for EF-Tu must be done with careful consideration of how these changes will affect tRNA utilization by SerRS and SelA. Below, we will summarize the key traits and structural characteristics of tRNASec, especially the bacterial type, which are important for its recognition by SerRS, SelA and translation elongation factors.
SerRS forms Ser-tRNASec
Charging of tRNASec or its variants by SerRS is a prerequisite for Sec biosynthesis. Although the structure of tRNASec in complex with E. coli SerRS still remains to be determined, understanding the molecular mechanism of Ser-tRNASec formation is much advanced via biochemical and structural studies. tRNASec is a close structural homolog of tRNASer. The common features of these two tRNAs in E. coli include the overall shape, the long variable arm, G73 as the discriminator base, the first three base pairs (bp) including the G2:C71 pair as a weak identity element, and nearly identical sequences in the D- and T-loops (Fig. 2A). Some of these conserved features shared by tRNASec and tRNASer are important for their recognition and activation by SerRS. For example, the orientation and the length, but not the specific sequence, of the long variable arm greatly contribute to the interactions between E. coli SerRS and tRNASec/tRNASer, as revealed by in vitro aminoacylation and electrophoretic mobility shift assays [57–60]. Consistent with these findings, the variable arm orientation of bacterial tRNASec resembles that of bacterial tRNASer as well as eukaryotic tRNASec [61–63]. Similar interactions between the variable arm and the N-terminal coiled-coil domain of SerRS are observed in crystal structures of bacterial tRNASec in complex with archaeal SerRS [61], the tRNASer and SerRS complex from bacteria [63], and the human tRNASec and SerRS complex [64]. In addition, the sequence-independent recognition of the variable arm by SerRS explains why E. coli SerRS can charge one tRNASec and five isoacceptor tRNASer species, which have different sequences in the variable arm. The 3’ end of the acceptor stems of tRNASec and tRNASer are also important for SerRS recognition, although less than the variable arm [58]. Mutations in the discriminator base at position 73 or the G2:C71 base pair of the E.coli tRNASer acceptor stem impair the ability of the corresponding tRNA variants to be efficiently charged by SerRS [59]. By systematically substituting the acceptor stem-TψC minihelices derived from tRNASer, base pairs G1:C72, G2:C71 and A/U3:U/A70 are found to be preferred by E. coli SerRS [65]. These highly conserved 3’ regions in tRNASer and tRNASec are required for discrimination against non-cognate tRNAs (e.g., tRNATyr and tRNALeu) by SerRS [60,66,67].
The overall shape of bacterial tRNASec is maintained by contacts between the D- and T-arms, including the universally conserved G18:U55 bp and a unique set of interactions conserved across most bacterial tRNASec species: Y16:Y59 (Y is C or U), U20:G19:C56 and C15:G20a:G48 (Fig. 2A) [61,68,69]. The latter base triple is proposed to determine the orientation of the variable arm based on the structure of Aquifex aeolicus (A. aeolicus) tRNASec [61]. Despite the fact that the tertiary structures of tRNASec and tRNASer are held together by distinctive sets of contacts, they have similar overall shapes, important for their recognition by SerRS.
Although both tRNASec and tRNASer are SerRS substrates (Fig. 1), the enzyme favors tRNASer; the degree of this preference varies between different organisms. For example, E. coli SerRS displays a 100-fold higher catalytic efficiency for tRNASer compared with tRNASec in vitro [70]. However, human cytosolic SerRS prefers tRNASer around 6-fold over tRNASec, and mainly at the level of acylation rather than the initial interaction between the tRNA and the enzyme [71]. Delicate biochemical studies of several tRNASec mutants and tRNASer/tRNASec hybrids provide insights into the elements responsible for the differential recognition by SerRS. Compared to tRNASer and other canonical tRNAs, most tRNASec species, including the E. coli one, have an additional bp in the acceptor arm (Fig. 2A) [41, 72]. The 13-bp-long acceptor-TψC arm of tRNASec consists of the 8-bp acceptor stem and the 5-bp TψC arm (namely 8/5 fold) in bacteria or the 9-bp acceptor stem and the 4-bp TψC arm (namely 9/4 fold) in archaea and eukaryotes. Shortening of the acceptor arm by 1 bp makes both E. coli tRNASec and human tRNASec better SerRS substrates than the wild-type [70,71], indicating that the long tRNASec acceptor-TψC arm prevents efficient SerRS charging. Genes encoding a 7/5 fold tRNASec have been predicted in diverse bacterial lineages [41,73], opening the possibility that those novel tRNASec species can be better substrates for E. coli SerRS.
Other factors may contribute to poor charging of tRNASec compared with tRNASer. It has been proposed that some sequence-specific interactions might play a role in the discrimination against tRNASec after formation of the initial tRNA-SerRS complex, although the identities of the residues involved in such contacts remain to be determined [63,71]. The U20 of human tRNASec is solvent-exposed and is in proximity of the highly conserved Ser61 of human SerRS, while tRNASer has a buried nucleotide at position 20 [64]. Based on this observation, the conserved U20 of human tRNASec is thought to serve as the structural basis to distinguish itself from tRNASer for SerRS recognition [64]. Note that U20 is also found in E. coli tRNASec (Fig. 2A). Going forward, the crystal structure of E. coli SerRS in complex with tRNASec will be very useful in providing guidance for engineering tRNASec to be a better substrate for SerRS.
Interestingly, unlike the majority of aminoacyl-tRNA synthetases that recognize the anticodon nucleotides as one of the major tRNA identity elements for specific interactions, SerRS is not in contact with the anticodons of tRNASer and tRNASec [61,63,64]. This property of SerRS explains recent findings that all stop and most sense codons can be read by tRNASec or its variants [74,75]. Anticodon-independent tRNA recognition by SerRS has been employed by synthetic biologists for Sec incorporation by genetic code expansion. For instance, tRNASec variants utilized for EF-Tu mediated Sec incorporation have anticodons that match the amber codon [51–53,55].
SelA converts Ser-tRNASec to Sec-tRNASec
The efficiency of SelA-catalyzed Ser-tRNASec to Sec-tRNASec conversion determines the purity of selenoproteins in bacterial cells. Thus, it is essential to understand the features of tRNASec as a SelA substrate when designing tRNA to efficiently mediate selenoprotein production. The conversion occurs with high fidelity during natural Sec biosynthesis, as Ser-tRNASec must be strictly discriminated against Ser-tRNASer to prevent Sec incorporation at serine codons. The structure of A. aeolicus SelA complexed to Thermoanaerobacter tengcongensis tRNASec provides insights into this discrimination process [76]. It reveals that the SelA N-terminal domain specifically recognizes the tRNASec D-arm composed of a 6-bp stem and 4-nucleotide loop [76]. However, the tRNASer D-arm consisting of a 3-bp stem and 11-nucleotide loop would clash with the SelA N-terminal domain, as suggested by a docking model of SelA·tRNASer [76]. This idea guided evolution of tRNAUTu, a chimera of the E. coli tRNASer and tRNASec used for SECIS-independent Sec incorporation [51]. Introducing elements of the tRNASec D-arm into tRNAUTu resulted in tRNAUTuX, a variant that was a better substrate for SelA (Fig. 2B) [53]. Overall, the long D-stem and the tRNA elbow composed of the D-loop and the T-loop in tRNASec are important for its interactions with SelA [76].
The unique D-arm of tRNASec is not the only feature required for SelA-specific interaction, as some chimeric tRNAs containing the tRNASer D-arm (tRNAUTu and tRNAUTu6 in Fig. 2B) can be utilized by E. coli SelA [51,55]. The length of the acceptor-TψC arm may also contribute to the discrimination between tRNASec and tRNASer by SelA. Based on the structure of SelA in complex with tRNASec, the 13-bp acceptor-TψC arm of canonical tRNASec is complementary to the space formed between the N-terminal and C-terminal catalytic domains of A. aeolicus SelA [76], while a 12-bp acceptor-TψC arm of tRNASer is thought to be suboptimal. Consistent with this idea, the in vitro conversion rate of Ser-tRNASec to Sec-tRNASec is significantly impaired by a 1-bp deletion in the acceptor stem in E. coli tRNASec [70]. Interestingly, Sec incorporation has been confirmed in Aeromonas salmonicida which has a non-canonical tRNASec species with a 12-bp acceptor-TψC arm [74], suggesting the existence of a special type of bacterial SelA that can efficiently recognize a shorter acceptor arm of tRNASec. The mechanism of tRNA recognition by this novel type of SelA needs further characterization, and the intrinsic flexibility of the N-terminal domain of SelA may account for its ability to recognize this unusual tRNASec [76]. Taken together, the differences between the D-arms and acceptor-TψC arms of tRNASec and tRNASer explain why SelA can discriminate the two tRNAs in E. coli, suggesting that these elements should be retained or carefully modified during tRNASec engineering.
Sec-tRNASec interacts with SelB but not EF-Tu
The partition of the substrates between EF-Tu and SelB is achieved through recognition of specific determinants within the tRNAs [40], as well as the aminoacyl group. tRNASec has a unique sequence (the antideterminant box) that simultaneously prevents interactions with EF-Tu and promotes interactions with SelB through contacts with the domain 3 extended loop (Fig. 2B) [77,78]. The corresponding loop in the EF-Tu domain 3 is shorter and contacts canonical tRNA in a sequence-specific manner [79,80]. However, the interaction between this loop and the antideterminant box of tRNASec is prevented by steric hindrance, as revealed by structural superposition (Fig. 2B) [77]. Another tRNASec structural element that contacts SelB but not EF-Tu is the variable arm. In a model of the ribosome complexed with Sec-tRNASec, SelB, and GTP, the variable arm of tRNASec is in close proximity to the linker region connecting the SelB domains 3 and 4 [77]. EF-Tu cannot establish such contacts as it lacks domain 4 and the corresponding linker.
Finally, Sec itself contributes to discrimination of EF-Tu against Sec-tRNASec. The amino acid binding pocket of EF-Tu is lined with negatively charged residues [81]. The selenol group of Sec is also negatively charged under physiological conditions [16], and therefore its binding to EF-Tu is disfavored. In contrast, the aminoacyl-binding pocket of SelB contains several highly conserved arginine and tyrosine residues that interact and stabilize the selenol moiety [77, 82]. Inspired by these findings, transplantation of conserved residues of the amino acid binding pocket from SelB to EF-Tu generates variants with improved Sec incorporation efficiency [83].
Current progress and challenges in tRNASec engineering
Knowing the pivotal role of the antideterminant box in tRNASec, mutation or replacement of this region with the tRNASer sequence resulted in variants that are good EF-Tu substrates (Fig. 2B) [51-53,55]. These tRNAs contain the anticodon sequence to match the amber codon, and can be divided into two structural types: tRNASer-like and tRNASec-like molecules. tRNAUTu and its derivatives tRNAUTuX and tRNAUTuT6 are mainly derived from tRNASer with the acceptor stem from tRNASec (Fig. 2A–B) [51,53,55]. tRNAUTu mediates robust suppression of amber codons in an EF-Tu-dependent manner, but the resulting protein products are a mixture of Sec- and Ser-containing populations, presumably due to incomplete conversion of Ser-tRNAUTu to Sec-tRNAUTu. tRNAUTu was further evolved into tRNAUTuX and tRNAUTuT6 which are better substrates for SelA and can mediate synthesis of homogenous Sec-containing proteins (Fig. 2B) [52]. The class of tRNASec-like EF-Tu substrates is represented by tRNASecUx (Fig. 2B) [52]. tRNASecUx retains most of the sequence of tRNASec and was selected from a library generated by full randomization of the antideterminant box [52]. As tRNASecUx closely resembles the natural substrate of SelA, the conversion of Ser-tRNASecUx to Sec-tRNASecUx is efficient, and the products resulting from amber suppression are homogenous selenoproteins. The examples of tRNAUTuX, tRNAUTuT6 and tRNASecUx demonstrate a certain degree of flexibility in the Sec biosynthesis pathway, as similar outcomes were obtained by different engineering strategies. Nevertheless, the yields of selenoproteins synthesized with the help of tRNASecUx and tRNAUTuT6 are low, likely due to the competition with the translation termination factor RF1 for amber codons. In agreement with this notion, Sec insertion into proteins mediated by tRNASecUx or tRNAUTuT6 was improved in E. coli strain C321.ΔA that lacks RF1 [52,55].
Further engineering of tRNA will be important in producing pure selenoproteins at high yields. However, there is a ‘fine opposing line’ between the efficiencies of tRNASec-dependent acylation by SerRS and Ser-tRNASec-dependent Ser to Sec conversion by SelA (Fig. 2C). For instance, the 8/5 fold and the unique D-arm of bacterial tRNASec, features essential for E. coli SelA recognition, inhibit its acylation by SerRS [70,76]. Conversely, transplantation of tRNASer features into tRNASec results in tRNA variants that are better substrates for SerRS, but interferes with their recognition by SelA [51,53,71]. Moreover, because tRNASec is moved by 3.4 Å and rotated by ∼33° when compared to tRNASer [71,77], pairing efficiency between the codon and the anticodon may be reduced for these engineered tRNAs with a 13-bp acceptor-TψC arm.
Advantages of SelB-mediated Sec incorporation
Partial overlapping of the recognition sites on tRNASec for SerRS, SelA and SelB has an important implication for tRNASec engineering: modifying its structure for efficient recognition by one protein factor inevitably affects the interactions with the others. One may attempt to further evolve SelA and SerRS to improve their recognition of EF-Tu-friendly tRNASec variants, while preserving the orthogonality of the system towards other components of translation. Alternatively, the problems associated with tRNASec engineering can be avoided altogether by modifying SelB to work without the SECIS and leaving tRNASec intact. Converting SelB into a SECIS-independent elongation factor presents several advantages. First, SelB binds Sec-tRNASec with exceptionally high affinity (Kd = 0.2 pM) and discriminates against all other aa-tRNAs [56,84]. Most importantly, SelB has a much lower affinity for Ser-tRNASec (Kd = 0.2 µM), an on-pathway precursor of Sec-tRNASec, providing a critical checkpoint in selenoprotein quality control [56]. Second, the nucleotide-binding properties and GTP hydrolysis rates of SelB are superior to those of EF-Tu. SelB binds Sec-tRNASec not only in GTP-bound form, but also in GDP-bound and apo-form [56,84]. SelB hydrolyzes GTP one order of magnitude faster than EF-Tu, and dissociates from tRNA rapidly thereafter, ensuring fast accommodation of Sec-tRNASec in the A site of the ribosome (Fig. 3) [56]. For all other elongator aa-tRNAs, the rate-limiting step in the formation of the peptide bond is the release of the elongation factor after hydrolysis of GTP [85]. Because SelB binds GTP with high affinity and releases GDP fast, it does not require a nucleotide-exchange factor [86]. Finally, tight binding to SelB likely protects Sec-tRNASec from hydrolysis. Hydrolysis of Sec-tRNASec releases free Sec into cytosol, where it can take the place of Cys. Recently, recoding of amber stop to Sec mediated by SelB in a SECIS-independent manner was observed in C321.ΔA strain (although with low efficiency), showing the potential of SelB-based systems [87].
Figure 3.
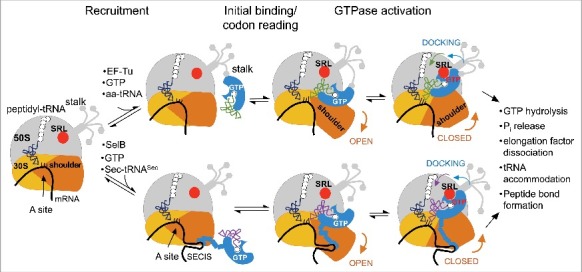
Schematic of aa-tRNA delivery to the A-site by EF-Tu (top) and SelB (bottom). Adopted with modifications from Rodnina et al [95]. The large ribosomal subunit with its stalk is grey; the small ribosomal subunit is yellow and its mobile shoulder domain is orange. During recruitment, the ternary complex does not form sequence-specific contacts with the translating ribosome; EF-Tu is recruited via the L7/L12 stalk and SelB is recruited via the SECIS stem-loop on mRNA. Contacts with the SECIS element are maintained in all subsequent steps; whether EF-Tu maintains contacts with the stalk after recruitment is not clear. During the initial binding, the 30S shoulder opens and allows tRNA entry; tRNA attempts to read the codon. SRL (red) does not make contact with GTP-binding domains of EF-Tu/SelB; instead, it is bound to the elbow region of aa-tRNA. The structure of the ternary complex on the ribosome during codon recognition is similar and is not shown separately. Codon recognition causes tRNA movement towards the codon and away from SRL; the 30S shoulder is open. No structures of initial binding or codon reading are available for EF-Tu, but the structure of SelB-assisted codon reading by Sec-tRNASec is available [77]. Finally, codon recognition causes local closure of the decoding center, allowing for large-scale rotation of the 30S shoulder. EF-Tu/SelB move together with the shoulder domain towards SRL. Docking on SRL activates GTPase domains of EF-Tu/SelB.
How E. coli SelB interacts with SECIS
To rid SelB of its dependence on the SECIS, we need to take a closer look into the molecular details of their interaction. In bacteria, the SECIS element interacts directly with the C-terminal domain 4 of SelB, which is connected by a flexible linker to the tRNA-binding portion of the protein [43,88]. SelB domains 1, 2 and 3 are homologues of the corresponding EF-Tu domains in their structure and function [82,89,90]. Truncated SelB lacking domain 4 cannot support selenoprotein synthesis, even though it still binds Sec-tRNASec. Conversely, the C-terminal domain of SelB is sufficient for binding to the SECIS element, and when expressed separately, competitively inhibits Sec insertion into proteins in vivo [43]. These data suggest that recruitment of the SelB·GTP·Sec-tRNASec ternary complex to the SECIS element occurs before a translating ribosome approaches the Sec insertion site on an mRNA template (Fig. 3) [44]. A minimal RNA segment that can bind SelB is comprised of a 17-nt motif with a conserved stem-loop structure separated by 11–12 nt from the Sec insertion codon, with critical determinants located in the bulged and looped out regions [42,91,92].
The RNA-binding domain of SelB is not well conserved. In bacteria it is comprised of four winged-helix motifs, but only the ultimate C-terminal motif makes direct contacts with the SECIS in the crystal structure [93]. Whether binding to the SECIS element affects accommodation of Sec-tRNASec on the ribosome or the nucleotide-exchange properties of SelB remains controversial. The linker connecting RNA-binding domain 4 and the rest of SelB appears to be flexible, making communication between them and concerted conformational changes unlikely. The first reported structure of domain 4 from Moorella thermoacetica bound to the minimal SECIS RNA substrate did not reveal any significant changes in the conformation of the protein compared to the RNA-free form [93], while the analysis of the complex formed by E. coli partial domain 4 of SelB led authors to conclude that binding to the SECIS does cause small changes in interdomain interactions [94]. It remains unclear if and how these changes in the mRNA-binding part of SelB affect its tRNA-binding properties. The ribosome model based on single-particle cryo-EM images of the various intermediates along the pathway of Sec insertion into the peptide supports the notion that the SECIS exerts its effect primarily by bringing SelB into close proximity of the ribosome. In the model, the flexible linker between domain 4 and domains 1–3 of SelB uncouples their movements: the mRNA-binding domain remains bound to the SECIS while the section of SelB bound to tRNA follows the movements of the small ribosomal subunit (Fig. 3) [77]. Thus, the SECIS element facilitates Sec insertion by positioning the SelB·GTP·Sec-tRNASec ternary complex for initial binding, rather than by triggering long-distance conformational changes in SelB. Importantly, the overall conformation of domains 1–3 of SelB bound to the ribosome appeared remarkably similar to the one of EF-Tu.
How EF-Tu and SelB interact with the ribosome
Despite many similarities between the two, SelB is not simply a version of EF-Tu with an extra RNA-binding motif. Removal of domain 4 results in loss of the ability of SelB to mediate Sec insertion into proteins (unpublished data, D. Söll lab). This indicates that SelB lacks critical determinants that allow EF-Tu to interact with the ribosome directly. Comparison of cryo-EM snapshots of tRNA accommodation and GTP hydrolysis by SelB and EF-Tu revealed that their overall structures and the ways the two translational factors interact with the ribosome are similar [95]. Both factors follow the ribosome dynamics during initial codon binding and reading, culminating in GTP hydrolysis (Fig. 3). Two ribosome components are required for successful decoding: the shoulder of the small ribosomal subunit, and the sarcin-ricin loop (SRL) of the 23S ribosomal RNA [96,97]. Docking of EF-Tu/SelB on the SRL triggers GTPase activation, an irreversible step that must only occur in cognate complexes [96]. Conformational dynamics of the small subunit are proposed to be pivotal to cognate decoding [97]. It is the movement of the shoulder that guides the ternary complex from an inactive conformation, with SRL bound to the tRNA elbow prior to decoding, towards EF-Tu/SelB docking on the SRL in response to correct codon-anticodon interaction (Fig. 3). Comparison of EF-Tu and SelB active sites shows that GTP hydrolysis occurs by essentially the same mechanism [95]. Small changes in the way SRL interacts with a catalytic histidine moiety might contribute to the differences in GTP hydrolysis rates between EF-Tu and SelB, but do not explain why the latter requires the SECIS element.
The differences in the interactions between the small subunit shoulder and domains 2 of EF-Tu and SelB were not analyzed in great detail. A high-resolution crystal structure of the Thermus thermophilus ribosome bound to the ternary complex revealed a single salt bridge between E249 of EF-Tu and the K119 residue of the small ribosomal protein S12 (E. coli numbering here and throughout) [98]. However, neither the significance of the salt bridge between EF-Tu and S12, nor whether alternative contacts might form between SelB and S12 have been demonstrated. In addition, domains 2 of EF-Tu and SelB show sequence variations in the two loops that contact the 30S shoulder in the structure of the ribosome complexed with EF-Tu ternary complex [98]. This region of EF-Tu undergoes the most significant movement upon binding to the ribosome [98]. Together, these findings can inform further studies into whether altered interactions with the 30S shoulder contribute to the dependency of SelB on the SECIS.
Alternatively, the inability of SelB to promote Sec insertion in the absence of the SECIS might be due to unproductive binding in the earlier steps of interaction with the translating ribosome [95]. Structures corresponding to initial tRNA binding and early codon reading recently became available for SelB, but not for EF-Tu, thus making a direct comparison between the two impossible. Despite the fact that early non-specific interactions between EF-Tu and the ribosome are short-lived, they appear to be crucial for proper transition to the later steps of aa-tRNA insertion. Moreover, the initial binding site on the ribosome for EF-Tu and many other elongation factors displays considerable conformational flexibility and thus is poorly resolved in the available structures of the ribosome. The binding site is located on the stalk of the large ribosomal subunit and consists of two L7/L12 dimers (Fig. 3) [99]. The primary sequences of L7 and L12 are identical except that L7 is acylated on its N-terminus. The N-terminal domain of L7/L12 is responsible for dimerization and binding to the ribosome through L10, while the mobile C-terminal domains protrude outwards in a tentacle-like fashion (Fig. 3). Interestingly, removal of L7/L12 has little effect on physical association of EF-Tu to the ribosome, but reduces its GTPase activity 2500 fold [100]. Interactions with L7/L12 are proposed to stimulate EF-Tu activity indirectly, by inducing a catalytically active conformation of its GTPase domain, but the exact nature of those conformational changes is unknown. Mutational analysis revealed several E and L residues in the D-helix of EF-Tu that are important for rapid A-site tRNA binding [101]. Different modes of recruitment to the ribosome likely allow EF-Tu and SelB to effectively partition their roles in protein synthesis, although EF-Tu appears to be able to share its binding platform with other translational factors, such as IF2, EF-G and RF3 [99, 100,102]. We are intrigued by the possibility that restoring interactions between the ribosomal stalk and SelB, together with utilization of an amber-less strain could lead to development of a highly efficient SECIS-independent system for selenoprotein synthesis.
Concluding remarks
Our understanding of the Sec incorporation pathway has been significantly enhanced by available genetic, biochemical, and structural studies. Driven by experiments on selenoproteins related to human health and the development of enzymes with new and improved catalytic properties, new ways for selenoprotein production have emerged. Perhaps the most valuable lesson we learned is that there might not be a single best method that fits all needs when it comes to production of selenoproteins. Depending on the size of the desired protein, the number and position(s) of Sec residues, the required yield and purity, and the individual characteristics of the target, different approaches may provide the best outcome. We envision that development of systems for efficient and site-specific incorporation of Sec by state-of-the-art laboratory evolution techniques combined with deep sequence search and comprehensive analyses of sequence databases will lead to novel types of tRNAs and enzymes useful for engineering selenoprotein synthesis.
Funding Statement
This work was supported by the US National Institutes of Health under R01GM022854 and R35GM122560 (to D.S.). The Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the Department of Energy supported the genetic experiments (DE-FG02-98ER20311, to D.S.).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgment
The authors are grateful to Takahito Mukai, Noah M. Reynolds, Corwin Miller, Kyle Hoffman, Anna Merkuryev, and Oscar Vargas-Rodriguez for discussions and a critical reading of the manuscript.
References
- [1].Jukes TH. Selenium, an “essential poison”. J Appl Biochem. 1983;5:233–4. [PubMed] [Google Scholar]
- [2].Reich HJ, Hondal RJ. Why Nature Chose Selenium. ACS Chem Biol. 2016;11:821–41. doi: 10.1021/acschembio.6b00031 [DOI] [PubMed] [Google Scholar]
- [3].Böck A, Thanbichler M, Rother M, et al.. Selenocysteine. In: Ibba M, Francklyn CS, Cusack S, eds. Aminoacyl-tRNA Synthetases. Georgetown: TX: Landes Bioscience; 2005. Landes Bioscience. [Google Scholar]
- [4].Cone JE, Del Rio RM, Davis JN, et al.. Chemical characterization of the selenoprotein component of clostridial glycine reductase: identification of selenocysteine as the organoselenium moiety. Proc Natl Acad Sci U S A. 1976;73:2659–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bösl MR, Takaku K, Oshima M, et al.. Early embryonic lethality caused by targeted disruption of the mouse selenocysteine tRNA gene (Trsp). Proc Natl Acad Sci U S A. 1997;94:5531–4. doi: 10.1073/pnas.94.11.5531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lobanov AV, Hatfield DL, Gladyshev VN. Selenoproteinless animals: selenophosphate synthetase SPS1 functions in a pathway unrelated to selenocysteine biosynthesis. Protein Sci. 2008;17:176–82. doi: 10.1110/ps.073261508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lobanov AV, Delgado C, Rahlfs S, et al.. The Plasmodium selenoproteome. Nucleic Acids Res. 2006;34:496–505. doi: 10.1093/nar/gkj450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Otero L, Romanelli-Cedrez L, Turanov AA, et al.. Adjustments, extinction, and remains of selenocysteine incorporation machinery in the nematode lineage. RNA. 2014;20:1023–34. doi: 10.1261/rna.043877.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chapple CE, Guigó R. Relaxation of selective constraints causes independent selenoprotein extinction in insect genomes. PLoS One. 2008;3:e2968. doi: 10.1371/journal.pone.0002968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Peng T, Lin J, Xu YZ, et al.. Comparative genomics reveals new evolutionary and ecological patterns of selenium utilization in bacteria. ISME J. 2016;10:2048–59. doi: 10.1038/ismej.2015.246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang Y, Gladyshev VN. High content of proteins containing 21st and 22nd amino acids, selenocysteine and pyrrolysine, in a symbiotic deltaproteobacterium of gutless worm Olavius algarvensis. Nucleic Acids Res. 2007;35:4952–63. doi: 10.1093/nar/gkm514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Lobanov AV, Hatfield DL, Gladyshev VN. Eukaryotic selenoproteins and selenoproteomes. Biochim Biophys Acta. 2009;1790:1424–8. doi: 10.1016/j.bbagen.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jacob C, Giles GI, Giles NM, et al.. Sulfur and selenium: the role of oxidation state in protein structure and function. Angew Chem Int Ed Engl. 2003;42:4742–58. doi: 10.1002/anie.200300573 [DOI] [PubMed] [Google Scholar]
- [14].Kryukov GV, Gladyshev VN. The prokaryotic selenoproteome. EMBO Rep. 2004;5:538–43. doi: 10.1038/sj.embor.7400126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Kim HY, Gladyshev VN. Methionine sulfoxide reduction in mammals: characterization of methionine-R-sulfoxide reductases. Mol Biol Cell. 2004;15:1055–64. doi: 10.1091/mbc.E03-08-0629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Axley MJ, Böck A, Stadtman TC. Catalytic properties of an Escherichia coli formate dehydrogenase mutant in which sulfur replaces selenium. Proc Natl Acad Sci U S A. 1991;88:8450–4. doi: 10.1073/pnas.88198450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhong L, Holmgren A. Essential role of selenium in the catalytic activities of mammalian thioredoxin reductase revealed by characterization of recombinant enzymes with selenocysteine mutations. J Biol Chem. 2000;275:18121–8. doi: 10.1074/jbc.M000690200 [DOI] [PubMed] [Google Scholar]
- [18].Kanzok SM, Fechner A, Bauer H, et al.. Substitution of the thioredoxin system for glutathione reductase in Drosophila melanogaster. Science. 2001;291:643–6. doi: 10.1126/science.291.5504.643 [DOI] [PubMed] [Google Scholar]
- [19].Lobanov AV, Hatfield DL, Gladyshev VN. Reduced reliance on the trace element selenium during evolution of mammals. Genome Biol. 2008;9:R62. doi: 10.1186/gb-2008-9-3-r62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lacourciere GM, Stadtman TC. Catalytic properties of selenophosphate synthetases: comparison of the selenocysteine-containing enzyme from Haemophilus influenzae with the corresponding cysteine-containing enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1999;96:44–8. doi: 10.1073/pnas.96144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hondal RJ, Marino SM, Gladyshev VN. Selenocysteine in thiol/disulfide-like exchange reactions. Antioxid Redox Signal. 2013;18:1675–89. doi: 10.1089/ars.2012.5013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Snider GW, Ruggles E, Khan N, et al.. Selenocysteine confers resistance to inactivation by oxidation in thioredoxin reductase: comparison of selenium and sulfur enzymes. Biochemistry. 2013;52:5472–81. doi: 10.1021/bi400462j [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Boschi-Muller S, Muller S, Van Dorsselaer A Bock A, et al.. Substituting selenocysteine for active site cysteine 149 of phosphorylating glyceraldehyde 3-phosphate dehydrogenase reveals a peroxidase activity. FEBS Lett. 1998;439:241–5. doi: 10.1016/S0014-5793(98)01377-5 [DOI] [PubMed] [Google Scholar]
- [24].Maller C, Schröder E, Eaton P. Glyceraldehyde 3-phosphate dehydrogenase is unlikely to mediate hydrogen peroxide signaling: studies with a novel anti-dimedone sulfenic acid antibody. Antioxid Redox Signal. 2011;14:49–60. doi: 10.1089/ars.2010.3149 [DOI] [PubMed] [Google Scholar]
- [25].Choudhury SB, Pressler MA, Mirza SA, et al.. Structure and redox chemistry of analogous nickel thiolato and selenolato complexes: implications for the nickel sites in hydrogenases. Inorg Chem. 1994;33:4831–9. doi: 10.1021/ic00100a005 [DOI] [Google Scholar]
- [26].Finlayson AJ, MacKenzie SL, Finlay JW. Reaction of alanine-3-sulfinic acid with 2-mercaptoethanol. Can J Chem. 1979;57:2023–77. doi: 10.1139/v79-332 [DOI] [Google Scholar]
- [27].Biteau B, Labarre J, Toledano MB. ATP-dependent reduction of cysteine-sulphinic acid by S. cerevisiae sulphiredoxin. Nature. 2003;425:980–4. doi: 10.1038/nature02075 [DOI] [PubMed] [Google Scholar]
- [28].Bell IM, Fisher ML, Wu ZP, et al.. Kinetic studies on the peroxidase activity of selenosubtilisin. Biochemistry. 1993;32:3754–62. doi: 10.1021/bi00065a030 [DOI] [PubMed] [Google Scholar]
- [29].Johansson L, Chen C, Thorell JO, et al.. Exploiting the 21st amino acid-purifying and labeling proteins by selenolate targeting. Nat Methods. 2004;1:61–6. doi: 10.1038/nmeth707 [DOI] [PubMed] [Google Scholar]
- [30].Lin YA, Boutureira O, Lercher L, et al.. Rapid cross-metathesis for reversible protein modifications via chemical access to Se-allyl-selenocysteine in proteins. J Am Chem Soc. 2013;135:12156–9. doi: 10.1021/ja403191g [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Metanis N, Hilvert D. Strategic use of non-native diselenide bridges to steer oxidative protein folding. Angew Chem Int Ed Engl. 2012;51:5585–8. doi: 10.1002/anie.201109129 [DOI] [PubMed] [Google Scholar]
- [32].Steiner AM, Woycechowsky KJ, Olivera BM, et al.. Reagentless oxidative folding of disulfide-rich peptides catalyzed by an intramolecular diselenide. Angew Chem Int Ed Engl. 2012;51:5580–4. doi: 10.1002/anie.201200062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pegoraro S, Fiori S, Cramer J, et al.. The disulfide-coupled folding pathway of apamin as derived from diselenide-quenched analogs and intermediates. Protein Sci. 1999;8:1605–13. doi: 10.1110/ps.8.8.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Arai K, Takei T, Okumura M, et al.. Preparation of selenoinsulin as a long-lasting insulin analogue. Angew Chem Int Ed Engl. 2017;56:5522–6. doi: 10.1002/anie.201701654 [DOI] [PubMed] [Google Scholar]
- [35].Leinfelder W, Zehelein E, Mandrand-Berthelot MA, et al.. Gene for a novel tRNA species that accepts L-serine and cotranslationally inserts selenocysteine. Nature. 1988;331:723–5. doi: 10.1038/331723a0 [DOI] [PubMed] [Google Scholar]
- [36].Forchhammer K, Böck A. Selenocysteine synthase from Escherichia coli. Analysis of the reaction sequence. J Biol Chem. 1991;266:6324–8. [PubMed] [Google Scholar]
- [37].Carlson BA, Xu XM, Kryukov GV, et al.. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci U S A. 2004;101:12848–53. doi: 10.1073/pnas.0402636101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Araiso Y, Palioura S, Ishitani R, et al.. Structural insights into RNA-dependent eukaryal and archaeal selenocysteine formation. Nucleic Acids Res. 2008;36:1187–99. doi: 10.1093/nar/gkm1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Agamy O, Ben Zeev B, Lev D, et al.. Mutations disrupting selenocysteine formation cause progressive cerebello-cerebral atrophy. Am J Hum Genet. 2010;87:538–44. doi: 10.1016/j.ajhg.2010.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Förster C, Ott G, Forchhammer K, et al.. Interaction of a selenocysteine-incorporating tRNA with elongation factor Tu from E.coli. Nucleic Acids Res. 1990;18:487–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Santesmasses D, Mariotti M, Guigó R. Computational identification of the selenocysteine tRNA (tRNASec) in genomes. PLoS Comput Biol. 2017;13:e1005383. doi: 10.1371/journal.pcbi.1005383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Heider J, Baron C, Böck A. Coding from a distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO J. 1992;11:3759–66. doi: 10.1002/j.1460-2075.1992.tb05461.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kromayer M, Wilting R, Tormay P, et al.. Domain structure of the prokaryotic selenocysteine-specific elongation factor SelB. J Mol Biol. 1996;262:413–20. doi: 10.1006/jmbi.1996.0525 [DOI] [PubMed] [Google Scholar]
- [44].Ringquist S, Schneider D, Gibson T, et al.. Recognition of the mRNA selenocysteine insertion sequence by the specialized translational elongation factor SelB. Genes Dev. 1994;8:376–85. doi: 10.1101/gad.8.3.376 [DOI] [PubMed] [Google Scholar]
- [45].Berry MJ, Banu L, Harney JW, et al.. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–22. doi: 10.1002/j.1460-2075.1993.tb06001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tujebajeva RM, Copeland PR, Xu XM, et al.. Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 2000;1:158–63. doi: 10.1038/sj.embor.embor604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tormay P, Böck A. Barriers to heterologous expression of a selenoprotein gene in bacteria. J Bacteriol. 1997;179:576–82. doi: 10.1128/jb.179.3.576-582.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Dery L, Reddy PS, Dery S, et al.. Accessing human selenoproteins through chemical protein synthesis. Chem Sci. 2017;8:1922–6. doi: 10.1039/c6sc04123j [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Hondal RJ. Using chemical approaches to study selenoproteins-focus on thioredoxin reductases. Biochim Biophys Acta. 2009;1790:1501–12. doi: 10.1016/j.bbagen.2009.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rakauskaitė R, Urbanavičiūtė G, Rukšėnaitė A, et al.. Biosynthetic selenoproteins with genetically-encoded photocaged selenocysteines. Chem Commun (Camb). 2015;51:8245–8. doi: 10.1039/c4cc07910h [DOI] [PubMed] [Google Scholar]
- [51].Aldag C, Bröcker MJ, Hohn MJ, et al.. Rewiring translation for elongation factor Tu-dependent selenocysteine incorporation. Angew Chem Int Ed Engl. 2013;52:1441–5. doi: 10.1002/anie.201207567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Thyer R, Robotham SA, Brodbelt JS, et al.. Evolving tRNASec for efficient canonical incorporation of selenocysteine. J Am Chem Soc. 2015;137:46–9. doi: 10.1021/ja510695g [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Miller C, Bröcker MJ, Prat L, et al.. A synthetic tRNA for EF-Tu mediated selenocysteine incorporation in vivo and in vitro. FEBS Lett. 2015;589:2194–9. doi: 10.1016/j.febslet.2015.06.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lajoie MJ, Rovner AJ, Goodman DB, et al.. Genomically recoded organisms expand biological functions. Science. 2013;342:357–60. doi: 10.1126/science.1241459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Fan Z, Song J, Guan T, et al.. Efficient expression of glutathione peroxidase with chimeric tRNA in amber-less Escherichia coli. ACS Synth Biol. 2017;7(1):249–57. doi: 10.1021/acssynbio.7b00290 [DOI] [PubMed] [Google Scholar]
- [56].Paleskava A, Konevega AL, Rodnina MV. Thermodynamic and kinetic framework of selenocysteyl-tRNASec recognition by elongation factor SelB. J Biol Chem. 2010;285:3014–20. doi: 10.1074/jbc.M109.081380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wu XQ, Gross HJ. The long extra arms of human tRNA(Ser)Sec and tRNASer function as major identify elements for serylation in an orientation-dependent, but not sequence-specific manner. Nucleic Acids Res. 1993;21:5589–94. doi: 10.1093/nar/21.24.5589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Sampson JR, Saks ME. Contributions of discrete tRNASer domains to aminoacylation by E.coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res. 1993;21:4467–75. doi: 10.1093/nar/21.19.4467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Asahara H, Himeno H, Tamura K, et al.. Escherichia coli seryl-tRNA synthetase recognizes tRNASer by its characteristic tertiary structure. J Mol Biol. 1994;236:738–48. doi: 10.1006/jmbi.1994.1186 [DOI] [PubMed] [Google Scholar]
- [60].Himeno H, Hasegawa T, Ueda T, et al.. Conversion of aminoacylation specificity from tRNATyr to tRNASer in vitro. Nucleic Acids Res. 1990;18:6815–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Itoh Y, Sekine S, Suetsugu S, et al.. Tertiary structure of bacterial selenocysteine tRNA. Nucleic Acids Res. 2013;41:6729–38. doi: 10.1093/nar/gkt321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Itoh Y, Chiba S, Sekine S, et al.. Crystal structure of human selenocysteine tRNA. Nucleic Acids Res. 2009;37:6259–68. doi: 10.1093/nar/gkp648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Biou V, Yaremchuk A, Tukalo M, et al.. The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science. 1994;263:1404–10. doi: 10.1126/science.8128220 [DOI] [PubMed] [Google Scholar]
- [64].Wang C, Guo Y, Tian Q, et al.. SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res. 2015;43:10534–45. doi: 10.1093/nar/gkv996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Saks ME, Sampson JR. Variant minihelix RNAs reveal sequence-specific recognition of the helical tRNASer acceptor stem by E.coli seryl-tRNA synthetase. EMBO J. 1996;15:2843–9. doi: 10.1002/j.1460-2075.1996.tb00645.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Breitschopf K, Gross HJ. The exchange of the discriminator base A73 for G is alone sufficient to convert human tRNALeu into a serine-acceptor in vitro. EMBO J. 1994;13:3166–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Lesjak S, Weygand-Durasevic I. Recognition between tRNASer and archaeal seryl-tRNA synthetases monitored by suppression of bacterial amber mutations. FEMS Microbiol Lett. 2009;294:111–8. doi: 10.1111/j.1574-6968.2009.01560.x [DOI] [PubMed] [Google Scholar]
- [68].Abe T, Ikemura T, Sugahara J, et al.. tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res. 2011;39:D210–3. doi: 10.1093/nar/gkq1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Mukai T, Vargas-Rodriguez O, Englert M, et al.. Transfer RNAs with novel cloverleaf structures. Nucleic Acids Res. 2017;45:2776–85. doi: 10.1093/nar/gkw898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Baron C, Böck A. The length of the aminoacyl-acceptor stem of the selenocysteine-specific tRNASec of Escherichia coli is the determinant for binding to elongation factors SELB or Tu. J Biol Chem. 1991;266:20375–9. [PubMed] [Google Scholar]
- [71].Holman KM, Puppala AK, Lee JW, et al.. Insights into substrate promiscuity of human seryl-tRNA synthetase. RNA. 2017;23:1685–99. doi: 10.1261/rna.061069.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Commans S, Böck A. Selenocysteine inserting tRNAs: an overview. FEMS microbiology reviews. 1999;23:335–51. doi: 10.1111/j.1574-6976.1999.tb00403.x [DOI] [PubMed] [Google Scholar]
- [73].Cravedi P, Mori G, Fischer F, et al.. Evolution of the selenoproteome in Helicobacter pylori and Epsilonproteobacteria. Genome Biol Evol. 2015;7:2692–704. doi: 10.1093/gbe/evv177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Mukai T, Englert M, Tripp HJ, et al.. Facile recoding of selenocysteine in Nature. Angew Chem Int Ed Engl. 2016;55:5337–41. doi: 10.1002/anie.201511657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Bröcker MJ, Ho JM, Church GM, et al.. Recoding the genetic code with selenocysteine. Angew Chem Int Ed Engl. 2014;53:319–23. doi: 10.1002/anie.20130858410.1002/anie.201308584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Itoh Y, Bröcker MJ, Sekine S, et al.. Decameric SelA•tRNASec ring structure reveals mechanism of bacterial selenocysteine formation. Science. 2013;340:75–8. doi: 10.1126/science.1229521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Fischer N, Neumann P, Bock LV, et al.. The pathway to GTPase activation of elongation factor SelB on the ribosome. Nature. 2016;540:80–5. doi: 10.1038/nature20560 [DOI] [PubMed] [Google Scholar]
- [78].Rudinger J, Hillenbrandt R, Sprinzl M, et al.. Antideterminants present in minihelixSec hinder its recognition by prokaryotic elongation factor Tu. EMBO J. 1996;15:650–7. doi: 10.1002/j.1460-2075.1996.tb00397.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Nissen P, Thirup S, Kjeldgaard M, et al.. The crystal structure of Cys-tRNACys-EF-Tu-GDPNP reveals general and specific features in the ternary complex and in tRNA. Structure (London, England : 1993). 1999;7:143–56. doi: 10.1016/S0969-2126(99)80021-5 [DOI] [PubMed] [Google Scholar]
- [80].Schrader JM, Chapman SJ, Uhlenbeck OC. Understanding the sequence specificity of tRNA binding to elongation factor Tu using tRNA mutagenesis. J Mol Biol. 2009;386:1255–64. doi: 10.1016/j.jmb.2009.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Knudsen CR, Mansilla F, Pedersen GN, et al.. Point mutants of elongation factor Tu from E. Coli impaired in binding aminoacyl-tRNA. In: Barciszewski J, Clark BFC, eds. RNA Biochemistry and Biotechnology. Dordrecht: Springer; 1999. p. 169–73. [Google Scholar]
- [82].Leibundgut M, Frick C, Thanbichler M, et al.. Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 2005;24:11–22. doi: 10.1038/sj.emboj.7600505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Haruna K, Alkazemi MH, Liu Y, et al.. Engineering the elongation factor Tu for efficient selenoprotein synthesis. Nucleic Acids Res. 2014;42:9976–83. doi: 10.1093/nar/gku691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Forchhammer K, Leinfelder W, Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–6. doi: 10.1038/342453a0 [DOI] [PubMed] [Google Scholar]
- [85].Pape T, Wintermeyer W, Rodnina MV. Complete kinetic mechanism of elongation factor Tu-dependent binding of aminoacyl-tRNA to the A site of the E. coli ribosome EMBO J. 1998;17:7490–7. doi: 10.1093/emboj/17.24.7490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Thanbichler M, Böck A, Goody RS. Kinetics of the interaction of translation factor SelB from Escherichia coli with guanosine nucleotides and selenocysteine insertion sequence RNA. J Biol Chem. 2000;275:20458–66. doi: 10.1074/jbc.M002496200 [DOI] [PubMed] [Google Scholar]
- [87].Cheng Q, Arnér ES. Selenocysteine insertion at a predefined UAG codon in a release factor 1 (RF1)-depleted Escherichia coli host strain bypasses species barriers in recombinant selenoprotein translation. J Biol Chem. 2017;292:5476–87. doi: 10.1074/jbc.M117.776310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Selmer M, Su XD. Crystal structure of an mRNA-binding fragment of Moorella thermoacetica elongation factor SelB. EMBO J. 2002;21:4145–53. doi: 10.1093/emboj/cdf408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Itoh Y, Sekine S, Yokoyama S. Crystal structure of the full-length bacterial selenocysteine-specific elongation factor SelB. Nucleic Acids Res. 2015;43:9028–38. doi: 10.1093/nar/gkv833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Berchtold H, Reshetnikova L, Reiser CO, et al.. Crystal structure of active elongation factor Tu reveals major domain rearrangements. Nature. 1993;365:126–32. doi: 10.1038/365126a0 [DOI] [PubMed] [Google Scholar]
- [91].Li C, Reches M, Engelberg-Kulka H. The bulged nucleotide in the Escherichia coli minimal selenocysteine insertion sequence participates in interaction with SelB: a genetic approach. J Bacteriol. 2000;182:6302–7. doi: 10.1128/JB.182.22.6302-6307.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Liu Z, Reches M, Groisman I, et al.. The nature of the minimal 'selenocysteine insertion sequence' (SECIS) in Escherichia coli. Nucleic Acids Res. 1998;26:896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Yoshizawa S, Rasubala L, Ose T, et al.. Structural basis for mRNA recognition by elongation factor SelB. Nat Struct Mol Biol. 2005;12:198–203. doi: 10.1038/nsmb890 [DOI] [PubMed] [Google Scholar]
- [94].Soler N, Fourmy D, Yoshizawa S. Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB. J Mol Biol. 2007;370:728–41. doi: 10.1016/j.jmb.2007.05.001 [DOI] [PubMed] [Google Scholar]
- [95].Rodnina MV, Fischer N, Maracci C, et al.. Ribosome dynamics during decoding. Philos Trans R Soc Lond B Biol Sci. 2017;372:doi: 10.1098/rstb.2016.0182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–4. doi: 10.1038/334362a0 [DOI] [PubMed] [Google Scholar]
- [97].Ogle JM, Murphy FV, Tarry MJ, et al.. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell. 2002;111:721–32. doi: 10.1016/S0092-8674(02)01086-3 [DOI] [PubMed] [Google Scholar]
- [98].Schmeing TM, Voorhees RM, Kelley AC, et al.. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–94. doi: 10.1126/science.1179700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Helgstrand M, Mandava CS, Mulder FA, et al.. The ribosomal stalk binds to translation factors IF2, EF-Tu, EF-G and RF3 via a conserved region of the L12 C-terminal domain. J Mol Biol. 2007;365:468–79. doi: 10.1016/j.jmb.2006.10.025 [DOI] [PubMed] [Google Scholar]
- [100].Mohr D, Wintermeyer W, Rodnina MV. GTPase activation of elongation factors Tu and G on the ribosome. Biochemistry. 2002;41:12520–8. doi: 10.1021/bi026301y [DOI] [PubMed] [Google Scholar]
- [101].Kothe U, Wieden HJ, Mohr D, et al.. Interaction of helix D of elongation factor Tu with helices 4 and 5 of protein L7/12 on the ribosome. J Mol Biol. 2004;336:1011–21. doi: 10.1016/j.jmb.2003.12.080 [DOI] [PubMed] [Google Scholar]
- [102].Diaconu M, Kothe U, Schlünzen F, et al.. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015 [DOI] [PubMed] [Google Scholar]


