Abstract
Purpose
Genome-wide association studies and targeted sequencing studies of candidate genes have identified common and rare variants that are associated with age-related macular degeneration (AMD). Whole-exome sequencing (WES) studies allow a more comprehensive analysis of rare coding variants across all genes of the genome and will contribute to a better understanding of the underlying disease mechanisms. To date, the number of WES studies in AMD case-control cohorts remains scarce and sample sizes are limited. To scrutinize the role of rare protein-altering variants in AMD cause, we performed the largest WES study in AMD to date in a large European cohort consisting of 1125 AMD patients and 1361 control participants.
Design
Genome-wide case-control association study of WES data.
Participants
One thousand one hundred twenty-five AMD patients and 1361 control participants.
Methods
A single variant association test of WES data was performed to detect variants that are associated individually with AMD. The cumulative effect of multiple rare variants with 1 gene was analyzed using a gene-based CMC burden test. Immunohistochemistry was performed to determine the localization of the Col8a1 protein in mouse eyes.
Main Outcome Measures
Genetic variants associated with AMD.
Results
We detected significantly more rare protein-altering variants in the COL8A1 gene in patients (22/2250 alleles [1.0%]) than in control participants (11/2722 alleles [0.4%]; P = 7.07×10–5). The association of rare variants in the COL8A1 gene is independent of the common intergenic variant (rs140647181) near the COL8A1 gene previously associated with AMD. We demonstrated that the Col8a1 protein localizes at Bruch’s membrane.
Conclusions
This study supported a role for protein-altering variants in the COL8A1 gene in AMD pathogenesis. We demonstrated the presence of Col8a1 in Bruch’s membrane, further supporting the role of COL8A1 variants in AMD pathogenesis. Protein-altering variants in COL8A1 may alter the integrity of Bruch’s membrane, contributing to the accumulation of drusen and the development of AMD.
Abbreviations and Acronyms: AMD, age-related macular degeneration; ARMS2, Age-Related Maculopathy Susceptibility 2; ANNOVAR, Annotate Variation; CADD, Combined Annotation Dependent Depletion; CFH, complement factor H; CFI, complement factor I; CMC, Combined Multivariate and Collapsing; CNV, choroidal neovascularization; COL8A1, Collagen Type VIII Alpha 1 Chain; GA, geographic atrophy; GATK, Genome Analysis ToolKit; GWAS, genome-wide association study; MAF, minor allele frequency; NC1, non-collagenous domain 1; NC2, non-collagenous domain 2; PBS, phosphate-buffered saline; SLC16A8, Solute Carrier Family 16 Member 8; SIFT, Sorting Tolerant From Intolerant; SNP, single nucleotide polymorphism; TIMP3, Tissue Inhibitor of Metalloproteinases 3; UBE3D, Ubiquitin Protein Ligase E3D; VQSLOD, Variant Quality Score Log-ODds; WES, whole-exome sequencing
Age-related macular degeneration (AMD) is the leading cause of irreversible vision loss among persons older than 50 years in the developed world.1, 2 The disease is characterized by progressive damage to the retinal pigment epithelium and photoreceptors in the macula, ultimately leading to visual impairment and blindness. In the early stages of AMD, a spectrum of changes occur, including hypopigmentations and hyperpigmentations of the retina and the formation of extracellular deposits (drusen) in Bruch’s membrane.2 These drusen increase in size and number during the intermediate stages. Two types of AMD can develop in the end stage of the disease. Geographic atrophy (GA), also referred to as the dry form of AMD, is characterized by retinal pigment epithelium cell atrophy, causing photoreceptor cell death. Choroidal neovascularization (CNV), also called the wet form of AMD, is characterized by the formation of new blood vessels, leading to leakage, hemorrhages, and sudden loss of vision.
Age-related macular degeneration is a multifactorial disease influenced by a variety of environmental factors, including age, smoking history, and sunlight exposure during working life.3, 4 There is a large genetic component to the cause of AMD, with an estimated heritability between 46% to 71%.5 Initially, genetic studies in AMD mainly focused on common variants in the population through genome-wide association studies (GWAS) using single nucleotide polymorphism (SNP) microarrays.6, 7, 8, 9 These studies identified genetic variants in or near genes belonging to 4 main pathways, including the complement system, lipoprotein metabolism, angiogenesis, and extracellular matrix remodeling. However, most common genetic variants identified by GWAS are located in noncoding or intergenic regions, and subsequently it is not always apparent which gene near the top-associated SNP is the causative gene.
Involvement of genes in a disease can be established further by identification of protein-altering variants in the coding regions, which often are rare in the population.8 Thus, several studies focused on the discovery of rare variants by sequencing genes in AMD loci. In these studies, rare variants were identified in complement factor H (CFH), complement factor I (CFI), complement C3, and complement C910, 11, 12, 13, 14 that are associated individually with AMD. Recently, a GWAS performed by the International AMD Genomics Consortium using an exome array enriched with rare variants identified 52 AMD-associated variants at 34 genomic loci. Of these 52 variants, 7 variants were rare and 45 variants were common.8
Testing the association of individual rare variants can be challenging, because very large sample sizes are needed to obtain sufficient power.15 Instead of testing each variant individually, gene-based burden tests can evaluate the cumulative effects of multiple genetic variants within a gene, leading to an increased study power.16 Sequence analysis of the coding regions of 681 genes within AMD-associated loci in 1676 AMD patients and 745 control participants identified a higher burden of rare variants in CFI in patients (7.8%) than in control participants (2.3%).12, 17, 18 Furthermore, evaluation of the cumulative effect of rare protein-altering variants, using exome array data by the International AMD Genomics Consortium, identified a significant burden in 4 AMD-associated genes: CFH, CFI, tissue inhibitor of metalloproteinases 3 (TIMP3), and solute carrier family 16 member 8 (SLC16A8).8 A limitation of these studies is that either rare variants in a limited set of genes12 or a limited number of rare variants across the genome8 were tested.
Whole-exome sequencing (WES) studies allow a more comprehensive analysis of rare protein-altering variants across all genes of the genome.19 To date, the number of WES studies in AMD case-control cohorts remain few and sample sizes are limited. Whole-exome sequencing of 213 neovascular AMD patients and 1553 healthy control participants from East Asian populations showed association of a variant in ubiquitin protein ligase E3D (UBE3D) with AMD.20 More recently, WES of 39 individuals with bilateral CNV with low genetic risk scores and 36 unaffected control participants with high genetic risk did not detect any genes that reached genome-wide significance.21
The main goal of the present study was the identification of rare protein-altering variants that are associated with AMD. To achieve this goal, we performed WES in a large European cohort consisting of 1125 patients and 1361 control participants to scrutinize the role of coding variants across the human genome in the cause of AMD.
Methods
Study Population
A cohort of 2516 individuals of European ancestry (1493 women and 1023 men with a mean age of 79 years) was recruited from the European Genetic Database (www.eugenda.org; n = 799) and the Rotterdam Study (n = 1717). From the European Genetic Database, 667 AMD patients (488 patients with late AMD) and 132 healthy control participants were evaluated for this study. Inclusion of individuals took place between December 2005 and June 2014. All participants underwent clinical evaluation by a retinal specialist and were graded for AMD according to the Cologne Image Reading Center protocol.22 Fundus photographs and spectral-domain OCT images were used to classify AMD by the presence of pigmentary changes together with at least 10 small drusen (<63-μm diameter) or the presence of intermediate drusen (63–124-μm diameter) or large drusen (≥125-μm diameter) in the Early Treatment Diabetic Retinopathy Study grid. Furthermore, late AMD was defined as either AMD with subfoveal GA or CNV in at least 1 eye. Control individuals were included in the study when they exhibited no signs of AMD in either eye and were at least 65 years of age at inclusion.
The design of the Rotterdam Study has been described previously in detail.23, 24 This prospective, population-based follow-up study started in 1990 and has follow-up visits every 5 years. For this analysis, we included a total of 466 AMD patients (74 patients with late AMD) and 1269 control participants from the Rotterdam Study I subcohort 55 years of age and older with WES data. All participants underwent fundus photography of the macula using a 35° film fundus camera (Topcon TRV-50VT; Topcon Global Gateway, Tokyo, Japan) after pupillary dilation. For the last 2 follow-up visits, a Topcon digital 35° color fundus camera (Topcon TRC 50EX; with a Sony DXC-950P 0.44 megapixel digital camera; Sony Corporation, Minato, Japan) was used. Fundus photographs were graded according to the Rotterdam Classification, which is based on the Wisconsin Age-Related Maculopathy Grading System25 and the modified International Classification System.26 Patients were participants with early or late AMD, which is at least soft distinct drusen (≥63-μm diameter), in combination with hypopigmentary or hyperpigmentary changes or soft indistinct drusen (≥125-μm diameter) or reticular drusen. Control participants were those older than 65 years with no signs of AMD or those older than 75 years of age with hard or soft distinct drusen (≥63-μm diameter) or pigmentary abnormalities.
In both cohorts, both eyes of all participants were graded separately by experienced graders (T.S.), who were under the supervision of senior retinal specialists (P.T.V.M.dJ., J.R.V., C.C.W.K., and S.F.). The worst affected eye was used to classify the individual. Written informed consent was obtained from all participants. The study was approved by the local ethics committees on research involving human subjects of the participating centers, and all procedures were conducted according to the tenets of the Declaration of Helsinki. The Rotterdam Study was approved by the Medical Ethics Committee of the Erasmus Medical Center and by the Ministry of Health, Welfare and Sport of The Netherlands, implementing the Wet Bevolkingsonderzoek: ERGO (Population Studies Act: Rotterdam Study).
Whole-Exome Sequencing Capture and Variant Calling
Genomic DNA of all participants was isolated from blood samples according to standard procedures. DNA was fragmented into 200- to 400-bp fragments, and the exome library was prepared on a Caliper Sciclone NGS workstation (Caliper Life Science, Hopkinton, MA). The exome was captured with the Nimblegen SeqCap EZ Exome version 2.0 44-Mb kit (Roche Nimblegen, Inc, Madison, WI), covering 329 028 exons and 710 miRNAs. Paired-end sequencing was performed on 2 Illumina HiSeq2000 sequencers using Illumina TruSeq V3 chemistry (Illumina, Inc, San Diego, CA). High-quality reads were mapped to the UCSC hg19 reference genome using the Burrows-Wheeler alignment tool.27 Variant calling was performed by Genome Analysis ToolKit (GATK) HaplotypeCaller, following the GATK best practice guidelines (available at https://software.broadinstitute.org/gatk; accessed July 2016). Single nucleotide variants and indels were filtered separately using GATKs Variant-Quality Score Recalibration module. Variants with a variant quality score log-odds (VQSLOD) score lower than –7.2 were removed. Variant annotation was done using annotate variation (ANNOVAR)28 and an in-house pipeline developed by the Department of Human Genetics of the Radboud University Medical Center.29 Functional effects of variants were predicted by 3 different prediction algorithms: Sorting Tolerant From Intolerant (SIFT),30 PolyPhen-2,31 and Combined Annotation Dependent Depletion (CADD)32 (threshold of deleteriousness for CADD, ≥20). In addition, conservation of candidate variants was estimated by PhyloP (threshold for deleteriousness, ≥2.7) and Grantham (threshold for deleteriousness, ≥80).
Data Quality Control
Stringent quality control steps were performed with PLINK version 1.0733 to exclude those positions that had high chances of being false positive results. Variants were removed according to the following criteria: (1) genotypes with a missing rate of more than 5% of individuals and (2) common variants (minor allele frequency, >0.05) that were not in Hardy-Weinberg equilibrium in control participants. After these quality control steps, a total of 744 022 variants were available for analysis. Subject-level quality control was carried out, excluding individuals with a call rate of less than 95% or an extreme inbreeding coefficient (cutoff, ±0.12).34 Pairwise identity by descent was calculated to confirm the lack of relatedness among all samples (PI-HAT, <0.25). A multidimensional scaling was performed with PLINK version 1.07 to obtain the principal components, which were used to confirm that all individuals were clustered as European samples and to correct for population stratification (Fig S1, available at www.aaojournal.org). After all quality controls, a cohort of 1125 AMD patients and 1361 control participants was selected for association analyses.
Statistical Analyses
A single variant association test was carried out with RAREMETALWORKER (available at http://genome.sph.umich.edu/wiki/RAREMETALWORKER; accessed January 2017) using a linear mixed model. This software performs a score statistics-based rare-variant association analysis, providing single-variant results and a variance–covariance matrix. Linkage disequilibrium relationships between markers within 1 Mb are stored in the covariance matrix to perform the gene-level analyses. Analysis was performed using an additive model controlling for age, gender, clinic, and the first 4 components. Genome-wide significance levels used for single-variant analysis were defined based on Bonferroni correction (P ≤ 5×10–8).
By definition, single-variant analyses have limited power to detect rare variant associations, especially for limited sample sizes. Association power was increased by evaluating the accumulated association of multiple rare exonic variants within each gene.35 Gene-based burden tests were carried out by RAREMETAL36 using the summary statistics and linkage disequilibrium matrices generated in the single-variant analysis. Three different methods were used: the Combined Multivariate and Collapsing (CMC)_counts and Variable Thresholds tests, which are burden tests that assume all alleles to influence the association in the same direction, and the sequence kernel association test (SKAT) test, which evaluates risk and protective alleles to maximize power. A subset of 308 784 rare protein-altering variants (minor allele frequency, <0.05) were used in the analysis to avoid the major presence of non–protein-altering variants (n = 435 238) diluting the burden because of deleterious variants. We selected rare variants that alter amino acid residues (nonsynonymous variants), truncate proteins (nonsense and stop-gain variants), or affect RNA splicing (variants affecting the invariate splice donor and splice acceptor sites).
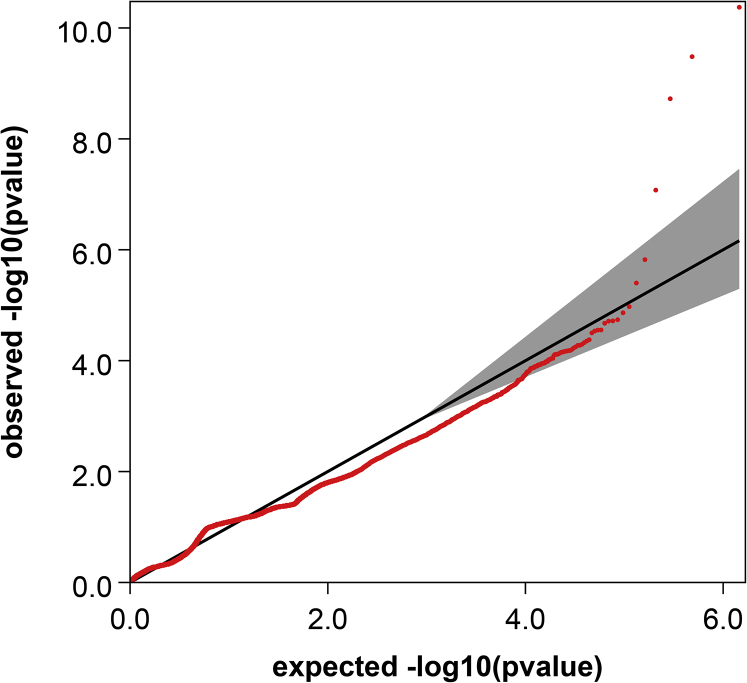
First, we focused on the 34 previously reported AMD loci and we applied a Bonferroni-corrected significance threshold based on the 619 genes located within 500 kb of the top-associated SNP in each of the AMD loci (according to ref.8) and carrying at least 1 rare protein-altering variant (P < 0.05/619 = 8.07×10–5). Haploview37 was used to reconstruct the region of interest to validate that the rare Collagen Type VIII Alpha 1 Chain (COL8A1) variants belong to different haplotype blocks than the common risk variant rs140647181 identified in a previous single-variant test.8 In a secondary analysis, we extended the search of rare variant burden to all genes across the genome, applying a Bonferroni-corrected significance threshold of 0.05/17 596 = 2.84×10–6. Quantile-quantile plots of P values from single-variant analysis and gene-based tests were generated to discard any batch effect or population substructure.
Characterization of Phenotypic Features of COL8A1 Variant Carriers
Phenotypic characterization was performed including participants from the Rotterdam Study. Age-related macular degeneration features were based on the eye with the most severe phenotype. Glaucoma-related features were the mean of both eyes at the last visit during follow-up. Refraction was based on the mean spherical equivalent of both eyes at the last visit during follow-up, or the last visit before cataract extraction. Statistical significance was tested with an independent sample t test for continuous variables, a chi-square or Fisher exact test for dichotomous variables, and a Mann–Whitney U test for drusen area because of its nonnormal distribution. All tests performed were 2-sided.
Mouse Retina Staining
Eyes from P60 C57BL/6J wild-type mice were enucleated and embedded in Tissue-Tek O.C.T. Compound (4583, Sakura Finetek, Alphen aan den Rijn, the Netherlands). Seven-micrometer sections were dried for 1 hour at room temperature. Using the hydrophobic PAP pen (Z377821-1EA, Sigma-Aldrich, St. Louis, MO), a circle was drawn surrounding the sections. Retinas then were incubated for 20 minutes in phosphate-buffered saline (PBS) 0.05% Tween (8.22184.0500, Merk millipore, Burlington, MA) and 0.05% Triton X-100 (9002-93-1, Sigma-Aldrich) at room temperature. After blocking in 0.1% ovo albumin (A4344,0250, AppliChem GmbH, Darmstadt, Germany), 0.5% fish gelatin (G7041-100G, Sigma-Aldrich), and 5% bovine serum albumin (A7906-100G, Sigma-Aldrich) in PBS for 30 minutes, primary antibodies were added and incubated overnight at 4° C. Primary antibodies used included rabbit polyclonal anticollagen type VIII α 1 (1:50; HPA053107, Sigma-Aldrich) and rat monoclonal Laminin β-1 (1:50; MA5-14657, ThermoFisher Scientific, Waltham, MA). Retinas were washed 4×5 minutes in PBS and incubated with the goat antirabbit Alexa 568 (1:500; A11006, Life Technologies, Carlsbad, CA) and goat antirat Alexa 488 secondary antibody (1:500; A11006, Life Technologies) for 45 minutes at room temperature (dilution, 1:500 in blocking solution). Nuclei staining with 4,′6-diamidino-2-phenylindole (1:8000; 0100-20, I.T.K Diagnostics B.V, Uithoorn, the Netherlands) was combined with the secondary antibody incubation. Sections then were washed 4×5 minutes in PBS, rinsed in MilliQ-purified water, and mounted in Prolong Gold antifade reagent (P36930, Life Technologies). Imaging was performed using a Zeiss Z1 Imager. All images were obtained at the same intensity. An image with ZEN software was created to obtain TIFF or JPEG files.
Results
Whole-Exome Sequencing
We performed WES on 2516 unrelated individuals (1125 patients and 1361 control participants), obtaining an average of 2.8 billion bases per individual and a mean coverage of ×63. After variant calling and recalibration, a total of 759 450 variants were identified, being 754 503 single nucleotide variants and 4947 insertions or deletions (indels). Of the complete set of variants, 7.6% (n = 57 571) were common variants, and the remaining 92.4% (n = 701 879) were classified as rare variants with a minor allele frequency of less than 0.05. Genotype data obtained from WES were checked for concordance with the genotype data of a customized Illumina exome array,8 available for a subset of the study population (n = 1330). Variants genotyped by both WES and exome array (n = 80 779) had a concordance rate of more than 99%, demonstrating the high quality of our sequencing data and the high accuracy of our genotype calling.
Single Variant and Gene-Based Association Analyses
We first performed a genome-wide single-variant association analysis for individual common and rare variants using the WES data of 1125 AMD patients and 1361 control participants of European ancestry. Results confirmed association of variants in the CFH and Age-Related Maculopathy Susceptibility 2 (ARMS2) genes with AMD in this cohort.8 Two common coding variants in CFH (rs1061170 [P = 4.24×10–11] and rs1061147 [P = 3.30×10–10]) and 1 common variant in ARMS2 (rs10490924 [P = 1.89×10–9]) were associated with AMD above the threshold of genome-wide significance (P ≤ 5×10–8; see Figs S2 and S3, available at www.aaojournal.org).
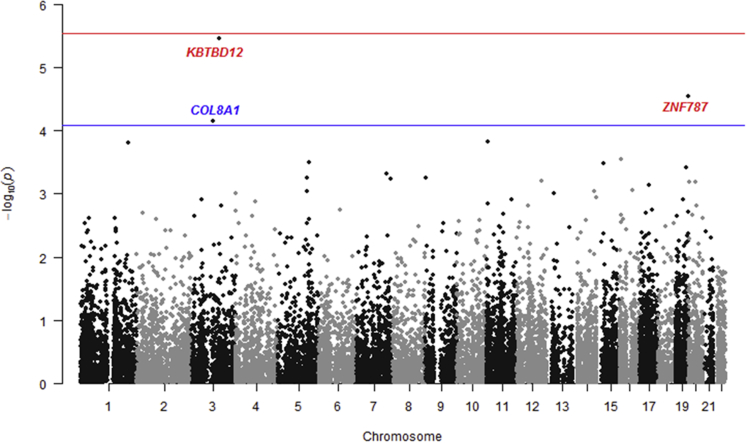
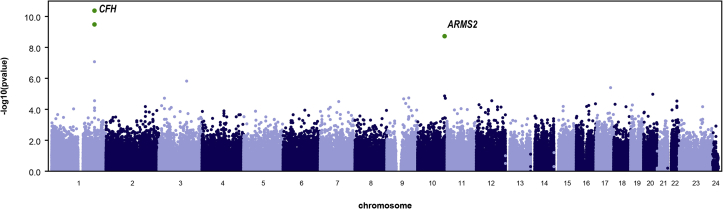
Subsequently, we evaluated the burden of rare protein-altering variants in genes at previously identified AMD loci using gene-based burden tests. For this analysis, 619 genes were selected that are within 500 kb of the top-associated SNP at 34 AMD loci identified in a recent GWAS8 (Table S1, available at www.aaojournal.org). A CMC burden test (applying genomic control λ = 0.940) showed a significant burden of rare variants in the COL8A1 gene (P = 7.07×10–5; Fig 1).
Figure 1.
Gene-based burden test for rare variants using whole-exome sequencing data of 1125 age-related macular degeneration (AMD) patients and 1361 control participants of European ancestry. The blue line indicates the significance threshold (P < 0.05/619 = 8.07×10–5) for testing 619 genes located in or near AMD-associated loci. The COL8A1 gene reaches the significance threshold and is depicted in blue. The red line indicates the genome-wide significant threshold (P < 0.05/17 596 = 2.84×10–6) for genes outside the AMD-associated loci. The KBTBD12 and ZNF787 genes do not reach genome-wide significance and are depicted in red. Bonferroni correction was applied to both significance thresholds.
We then expanded the burden analysis to protein-altering variants across the genome. The CMC burden test (applying genomic control λ = 1.057) showed a suggestive association in the KBTBD12 (P = 3.50×10–6) and ZNF787 (P = 2.89×10–5) genes, but these associations did not reach the genome-wide significance level (Fig 1). The signal at the KBTBD12 gene did reach the genome-wide significance threshold when the SKAT test was applied (P = 4.45×10–7). In both tests, the association signal was driven mainly by the effect of 1 rare variant (rs148151101; P = 1.52×10–6). However, this particular variant in KBTBD12 was not associated with AMD in an exome array analysis in a cohort of 16 144 AMD patients and 17 832 control participants of European ancestry by the International AMD Genomics Consortium (P = 0.387).8
Rare Variant Burden in the COL8A1 Gene
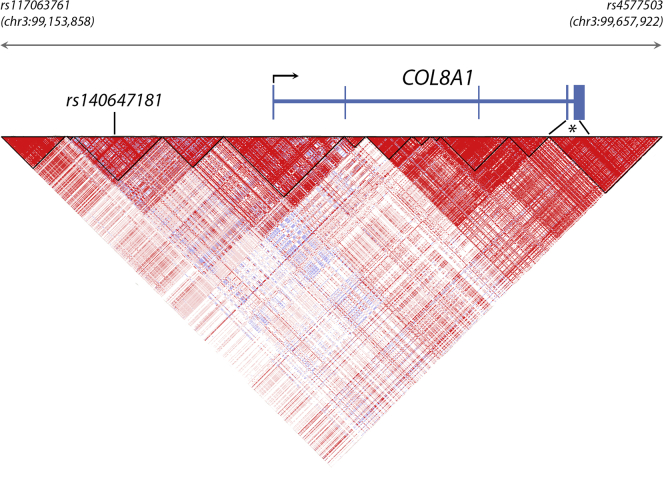
We next determined whether the rare variant burden in COL8A1 is independent of the previously identified AMD-associated common variant (rs140647181) near the COL8A1 gene.8 This common variant is intergenic, located 560 kb downstream of DCBLD2 and 177 kb upstream of COL8A1. To evaluate the independence between the rare protein-altering variants and the common intergenic variant rs140647181, we reconstructed the haplotype block structure at the COL8A1 locus to visualize which regions of the gene are linked closely and are inherited together. Several recombination events between the rare protein-altering variants in the COL8A1 gene and rs140647181 were observed, meaning that the region containing the rare variants is not inherited together with the region containing the common intergenic variant (Fig 2). These results support that the rare variant burden observed in this study is independent of the common intergenic variant previously associated with AMD.
Figure 2.
Haploblock structure of the genomic region encompassing the COL8A1 gene and the age-related macular degeneration-associated common intergenic variant rs140647181. A Haploview plot was generated based on common single nucleotide variants extracted from the 1000 Genomes phase 3 dataset. Red triangles marked with black lines represent genomic regions that are closely linked and are inherited together. This haplotype block distribution shows that the rare protein-altering variants identified in the COL8A1 gene (indicated with an asterisk) are not located in the same haplotype block as rs140647181, meaning that the rare variants are not inherited together with the common intergenic variant. This supports that the rare variant burden in COL8A1 is independent of the common intergenic variant rs140647181.
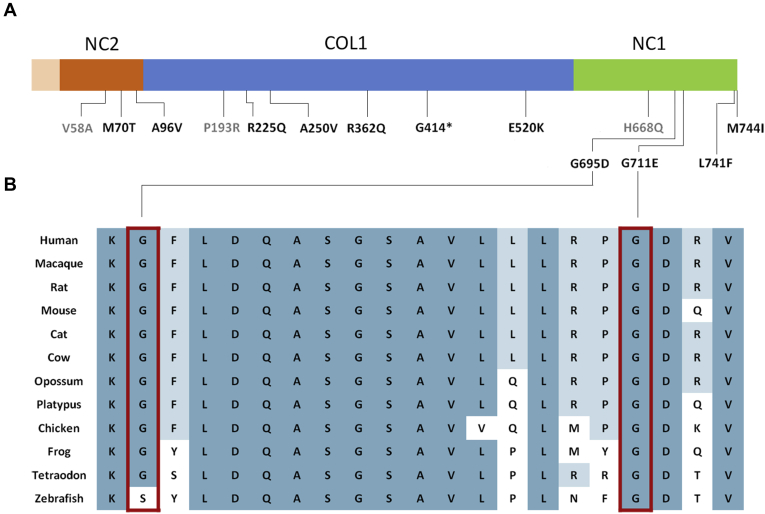
The COL8A1 burden is explained by 14 rare protein-altering variants spread across the protein (Fig 3), which are found more often in patients (22/2250 alleles [1.0%]) than in control participants (11/2722 alleles [0.4%]; Table 1; Table S2, available at www.aaojournal.org). Six variants, including 1 nonsense variant (p.G414*) and 5 missense variants (p.M70T, p.A96V, p.E520K, p.G711E, and p.M744I), were identified only in patients, but not in control participants, and 5 additional variants (p.R225Q, p.A250V, p.R362Q, p.G695D, and p.L741F) were found at a higher frequency in patients than in control participants. The nonsense variant p.G414* is predicted to lead to a premature termination in the COL1 domain, or may cause nonsense-mediated decay of the COL8A1 mRNA. Two missense variants (p.G695D and p.G711E) are predicted to be deleterious with all conservation and pathogenicity tests used and have a CADD score of 20 or more, which classifies them among the top 0.75% most deleterious mutations that are found in the human genome (Table 1). The 2 missense variants p.G695D and p.G711E affect 2 highly conserved amino acid residues in the noncollagenous 1 domain (Fig 3).
Figure 3.
Location and conservation of protein-coding variants in COL8A1. A, Location of rare protein-altering variants identified in age-related macular degeneration (AMD) patients and control participants in the different COL8A1 domains: triple-helical region (COL1), noncollagenous domain 1 (NC1), and noncollagenous domain 2 (NC2). Variants detected only in control individuals are depicted in gray. B, Alignment of COL8A1 protein sequences of different species. Boxed missense variants identified in AMD patients, predicted to be deleterious in all conservation and pathogenicity tests (Table 1), affect highly conserved glycine residues in the NC1 domain.
Table 1.
Rare Protein-Altering Variants Identified in the COL8A1 Gene in 1125 Age-Related Macular Degeneration Patients and 1361 Controls
| Protein Change | cDNA Change | Domain | PhyloP∗ | Grantham∗ | SIFT (Score)∗ | PolyPhen2 (Score)∗ | CADD∗ | Counts Patients (n = 2250) | Counts Controls (n = 2722) | Single-Variant P Value | Single-Variant Odds Ratio (95% Confidence Interval) | Burden Test P Value | Burden Test Odds Ratio (95% Confidence Interval) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| V58A | 173T→C | NC2 | 4.317 | 64 | Damaging (0.014) | Poss. damaging (0.646) | 19.7 | 0 | 1 | 0.56 | 0.78232 (0.34–1.79) | 7.07 × 10–5 | 1.34153 (1.16–1.55) |
| M70T | 209T→C | NC2 | 4.216 | 81 | Damaging (0.004) | Benign (0.001) | 22.5 | 1 | 0 | 0.11 | 1.9552 (0.85–4.47) | ||
| A96V | 287C→T | NC2 | 1.266 | 64 | Tolerated (1) | Benign (0) | 5.6 | 1 | 0 | 0.06 | 2.18674 (0.96–5) | ||
| P193R | 578C→G | COL1 | 4.028 | 103 | Damaging (0.02) | Poss. damaging (0.463) | 22.1 | 0 | 1 | 0.61 | 0.80691 (0.35–1.84) | ||
| R225Q | 674G→A | COL1 | 6.782 | 43 | Tolerated (0.328) | Benign (0.008) | 22.9 | 1 | 1 | 0.48 | 1.23339 (0.69–2.21) | ||
| A250V | 749C→T | COL1 | 2.257 | 64 | Tolerated (0.338) | Benign (0) | 2.5 | 1 | 1 | 0.83 | 0.93868 (0.52–1.68) | ||
| R362Q | 1085G→A | COL1 | 3.041 | 43 | Tolerated (0.105) | Benign (0.071) | 20.5 | 6 | 3 | 0.06 | 1.3026 (0.99–1.72) | ||
| G414∗ | 1240G→T | COL1 | 9.803 | NA | NA | NA | 39 | 2 | 0 | 0.01 | 2.14633 (1.20–3.85) | ||
| E520K | 1558G→A | COL1 | 9.828 | 56 | Tolerated (0.296) | Poss. damaging (0.945) | 21.6 | 1 | 0 | 0.82 | 1.1015 (0.48–2.52) | ||
| H668Q | 2004C→G | NC1 | 2.728 | 24 | Damaging (0.004) | Prob. damaging (0.989) | 25.9 | 0 | 1 | 0.45 | 0.72856 (0.32–1.66) | ||
| G695D | 2084G→A | NC1 | 9.873 | 94 | Damaging (0.005) | Prob. damaging (0.977) | 26.7 | 4 | 2 | 0.08 | 1.3489 (0.96–1.89) | ||
| G711E | 2132G→A | NC1 | 9.873 | 98 | Damaging (0.014) | Prob. damaging (1) | 26.7 | 1 | 0 | 0.08 | 2.1159 (0.92–4.84) | ||
| L741F | 2223G→T | NC1 | 0.615 | 22 | Tolerated (0.084) | Benign (0.366) | 22.2 | 2 | 1 | 0.11 | 1.47754 (0.92–2.38) | ||
| M744I | 2232G→C | NC1 | 9.477 | 10 | Tolerated (0.186) | Benign (0.001) | 24.8 | 2 | 0 | 0.13 | 1.56374 (0.87–2.81) |
COL1 = triple-helical region; CADD = Combined Annotation Dependent Depletion; NA = not applicable; NC1 = noncollagenous domain 1; NC2 = noncollagenous domain 2; Poss. = possibly; Prob. = probably; Sift = Sorting Tolerant From Intolerant.
Thresholds for deleteriousness: PhyloP ≥ 2.7, Grantham ≥ 80, SIFT ≤ 0.1, PolyPhen ≥ 0.4, and CADD ≥ 20.
Phenotypic Features of COL8A1 Variant Carriers
We examined the effect of the COL8A1 variants on the AMD phenotype in participants from the Rotterdam Study only, because it is a population-based cohort study without prior selection on phenotype. This group consists of 16 AMD patients carrying a COL8A1 variant, 11 individuals carrying a COL8A1 variant without AMD, and 450 AMD patients without a COL8A1 variant (Table 2). Features of early AMD were not significantly different between COL8A1 carriers and noncarriers with AMD, although COL8A1 carriers had a somewhat higher proportion of hyperpigmentary changes (P = 0.062). No statistically significant differences were found for glaucoma-related features such as intraocular pressure and vertical cup-to-disc ratio. The groups differed significantly in spherical equivalent, with COL8A1 carriers being more myopic (P = 0.005). However, there was no significant difference in the proportion of participants with mild and severe myopia.
Table 2.
Comparison of Phenotypic Features between Carriers and Noncarriers of COL8A1 Variants in the Rotterdam Study
| Protein Change | COL8A1 Variant and Age-Related Macular Degeneration (n = 16) | COL8A1 Variant, No Age-Related Macular Degeneration (n = 11) | No COL8A1 Variant and Age-Related Macular Degeneration (n = 450) |
|---|---|---|---|
| Age at last visit (yrs) | 79.6 (SD, 6.3) | 82.5 (SD, 7.9) | 80.0 (SD, 6.5) |
| Spherical equivalent | –0.37 (SD, 1.86)∗ | 1.14 (SD, 1.83) | 1.26 (SD, 2.29)∗ |
| Mild myopia (–3 to –6 D; %) | 3/16 (19) | 1/11 (9) | 23/437 (5) |
| Severe myopia (≤ –6 D; %) | 0/16 (0) | 0/11 (0) | 2/437 (0) |
| Corneal curvature (mm) | 7.72 (SD, 0.32) | 7.58 (SD, 0.26) | 7.70 (SD, 0.26) |
| IOP (mmHg) | 13.8 (SD, 3.0) | 14.3 (SD, 2.8) | 13.9 (SD, 3.3) |
| VCDR | 0.36 (SD, 0.18) | 0.37 (SD, 0.24) | 0.32 (SD, 0.18) |
| Subtype of AMD (no.) | 3 GA, 0 CNV, 0 mixed†, 13 early | — | 29 GA, 21 CNV, 21 mixed†, 379 early |
| Drusen area >10% (%) | 4/16 (25) | 0/11 (0) | 88/450 (20) |
| Presence of hyperpigmentation (%) | 14/16 (88) | 1/11 (9) | 287/450 (64) |
| Presence of reticular drusen (%) | 0/16 (0) | 0/11 (0) | 27/450 (6) |
| Presence of drusen outside grid (%) | 10/16 (63) | 7/11 (64) | Not available |
AMD = age-related macular degeneration; CNV = choroidal neovascularization (wet AMD); D = diopters; GA = geographic atrophy (dry AMD); IOP = intraocular pressure; SD = standard deviation; VCDR = vertical cup-to-disc ratio.
P = 0.005 independent samples t test (2-tailed) between AMD patients carrying a COL8A1 variant (n = 16) and AMD patients without variants in COL8A1 (n = 437), t = 2.81, degrees of freedom, 451.
Geographic atrophy and CNV.
Localization of COL8A1 to Bruch’s Membrane
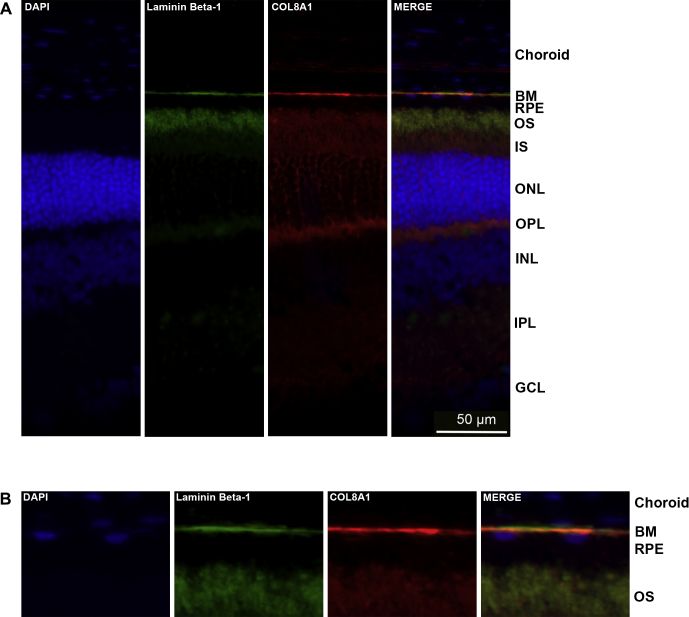
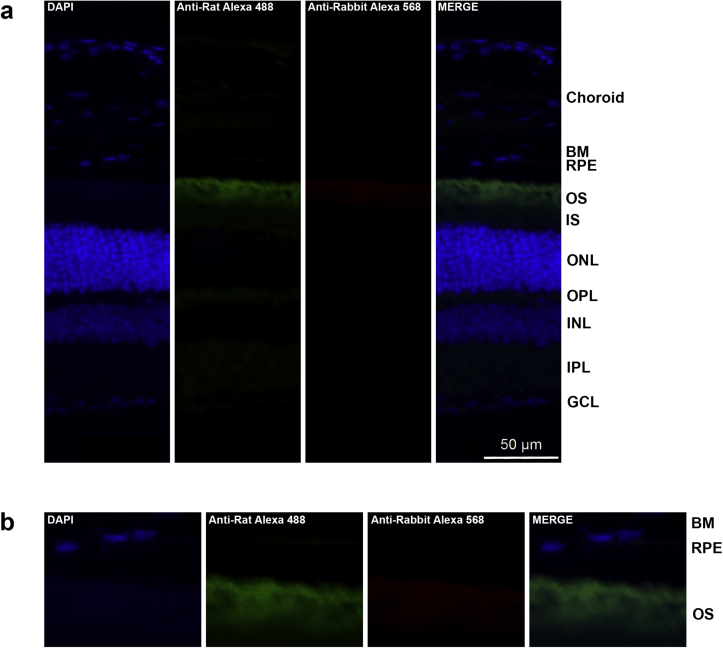
Localization of COL8A1 in the retina has not yet been described in the literature. To assess whether COL8A1 is localized at Bruch’s membrane, the main AMD disease site, we performed immunohistochemistry on retinas of wild-type C57BL/6J adult mice. Mice were selected for these experiments because mouse and human retinas exhibit a common basic architecture38 and often are used to model human retinal disease, although mice lack a macula and have a higher photoreceptor cell density and a relatively thicker Bruch’s membrane in the central retina.39 Laminin β-1 was used as a marker for Bruch’s membrane.40 The coimmunostaining of Laminin β-1 and Col8a1 robustly demonstrated that both proteins localize at Bruch’s membrane (Fig 4). In addition, Col8a1 showed some expression in the photoreceptor layer, being most evident at the outer plexiform layer, the synaptic region between the photoreceptor cells, and the inner nuclear layer cells. To exclude that the staining was the result of background staining derived from the use of secondary antibodies, we performed the same procedure without adding primary antibody (Fig S4, available at www.aaojournal.org). This confirmed that the Col8a1 and Laminin β-1 staining observed at Bruch’s membrane is the result of the primary antibody.
Figure 4.
Localization of Col8a1 in mouse retinas. A, The localization of Col8a1 (in red) was studied on P90 retinas derived from wild-type C57BL/6J mice. Laminin β-1 (Lamb1; green) was used as a Bruch’s membrane marker. Col8a1 colocalizes with Lamb1 at Bruch’s membrane. Col8a1 staining also showed a weaker signal in other layers of the retina. B, Magnifications of the outer region of the retina, where the colocalization between Lamb1 and Col8a1 can be appreciated. DAPI (4′,6-diamidino-2-phenylindole) (blue) was used to stain cell nuclei. BM = Bruch’s membrane; GCL = ganglion cell layer; INL = inner nuclear layer; IPL = inner plexiform layer; IS = inner segment; ONL = outer nuclear layer; OPL = outer plexiform layer; OS = outer segment; RPE = retinal pigment epithelium.
Discussion
In this study, we aimed to scrutinize the role of rare protein-altering variants in the cause of AMD using WES. By focusing on rare protein-altering variants in the coding regions, we sought to determine the causality of genes in the disease. Because most top SNPs identified in GWAS studies for AMD are in noncoding or intergenic regions,8 it is not always apparent which gene near the top-associated SNP is the causative gene. In this study, WES analysis in 1125 AMD patients and 1361 control participants revealed a significant burden of rare protein-altering variants in the COL8A1 gene in AMD. The COL8A1 burden is explained by 14 rare protein-altering variants spread across the protein, which are found more often in patients (22/2250 alleles [1.0%]) than in control participants (11/2722 alleles [0.4%]). The association of rare variants in the COL8A1 gene is independent of the common AMD-associated intergenic variant rs140647181, located 560 kb downstream of DCBLD2 and 177 kb upstream of COL8A1.8 No rare-variant burden was observed in the DCBLD2 gene, nor in other genes at the same AMD locus. Taken together, these findings support that the previously observed association of the common intergenic variant rs140647181 is driven by effects on COL8A1 rather than by other genes at the locus.
COL8A1 encodes 1 of the 2 α chains of collagen type VIII, which is a major component of ocular basement membranes.41 Several studies have investigated the association between alterations in genes encoding the 2 subunits of collagen VIII (COL8A1 and COL8A2) and ocular abnormalities such as myopic CNV, anterior segment dysgenesis, and thin corneal stroma.42, 43, 44, 45 Although several studies postulated a role for COL8A1 in ocular basement membranes, so far no published data confirm the localization of COL8A1 in Bruch’s membrane. There are several lines of evidence to support that Bruch’s membrane plays a crucial role in AMD. Because of its location, Bruch’s membrane is involved intensively in the exchange of numerous biomolecules, nutrients, and waste products between the retinal pigment epithelium and the choroidal capillary bed.46 A disturbed integrity or stability of Bruch’s membrane can lead to accumulation of these products in drusen or can weaken the physical barrier against the invasion of new blood vessels into the retina.47 In this study, we demonstrate the presence of Col8a1 in Bruch’s membrane, further supporting the role of COL8A1 variants in AMD pathogenesis.48
Protein-altering variants in COL8A1 may lead to structural alterations in Bruch’s membrane, which can be responsible for the development of AMD.43 Interestingly, we describe 14 rare protein-altering variants in COL8A1, including 1 nonsense variant (p.G414*) and 2 deleterious missense variants (p.G695D and p.G711E) that affect highly conserved residues in the C-terminal noncollagenous 1 domain. The noncollagenous 1 domain mediates proper folding of the protein and the assembly of collagen VIII and X into polygonal lattices.49, 50, 51 Therefore, these COL8A1 variants may lead to an aberrantly folded protein, impairing transport of the protein to Bruch’s membrane or altering Bruch’s membrane integrity or stability. Consequently, this may contribute to the development of early AMD. In our study, we observed a higher, albeit nonsignificant, proportion of hyperpigmentary changes in AMD patients carrying COL8A1 variants. Larger patient populations are needed to validate this finding. Previous studies have implicated COL8A1 in retinal angiogenesis by mediating proliferation and migration of endothelial cells,43 suggesting that COL8A1 variants could contribute to the development of neovascularization in late AMD. In the Rotterdam Study, we identified 3 COL8A1 carriers with GA, but no carriers who demonstrated CNV (Table 2). However, in the European Genetic Database cohort, we identified 1 carrier with GA, 3 carriers with CNV, and 2 carriers with the mixed type of AMD with GA and CNV (data not shown). Therefore, we cannot conclude that there is an overrepresentation of CNV in carriers of COL8A1 variants. Interestingly, COL8A1 variants seem to contribute to refractive error, although the contribution to severe myopic errors was insignificant. In the Rotterdam Study, the refractive error is, on average, emmetropic in AMD patients carrying COL8A1 variants. Therefore, it is unlikely that myopic thinning of Bruch’s membrane contributed to the development of AMD in these carriers.
The findings described herein need to be interpreted in light of several strengths and limitations. We demonstrated that WES with relatively large cohorts is an efficient strategy to detect rare variants in AMD-associated genes. Previous studies that detected rare variants in AMD were focused on predefined gene sets using targeted sequencing12 or predefined variant sets using exome arrays,8 whereas our study performed a comprehensive exome-wide search for rare variants using WES. The main advantage of performing WES is that it enables the identification of all rare variants present in coding regions across the genome, allowing a more comprehensive evaluation of rare variants than other approaches based on a limited set of genes or variants. A burden of rare variants has been described previously in CFH, CFI, TIMP3, and SLC16A8,8, 12 but these findings were not confirmed in our study. This may be because although we had a relatively large cohort, our study may not have had sufficient power to detect these associations. In the study by Fritsche et al,8 a larger cohort was used, consisting of 16 144 AMD patients and 17 832 control participants. However, most of the COL8A1 variants (11/14) identified by WES in our study were not present on the exome array that was used by Fritsche et al, which may explain why a burden of rare COL8A1 variants was not observed in that study.8 In addition, differences in study designs and populations, case definition, geographical origin, statistical tests used, or correction for confounding factors may explain the different results observed among these studies.
In conclusion, we performed an exome-wide sequence analysis of rare protein-altering variants in AMD and we detected a burden of rare variants in the COL8A1 gene. A common intergenic variant near this gene was associated previously with AMD risk,7, 8 but no protein-altering variants within the gene have been described in AMD so far. This work supports a role for protein-altering variants in the COL8A1 gene in AMD pathogenesis and suggests that the previously observed association of the common intergenic variant is driven by effects on COL8A1. In this study, we demonstrate the presence of Col8a1 in Bruch’s membrane, further supporting the role of COL8A1 variants in AMD pathogenesis. Protein-altering variants in COL8A1 may alter the integrity of Bruch’s membrane, contributing to the accumulation of drusen and the development of AMD. This study showed that WES provides a fruitful approach for gene and variant identification in complex disorders such as AMD. Collaborative efforts among the scientific community are needed to perform even larger exome- or genome-wide sequencing studies52 that will increase our understanding of the genetic architecture and disease mechanisms of AMD further.
Acknowledgments
The authors thank Pascal Arp, Mila Jhamai, André Uitterlinden, and Marijn Verkerk for their help in creating the Rotterdam Study Exome Sequencing dataset.
Manuscript no. 2017-2357.
Footnotes
Financial Disclosure(s): The author(s) have made the following disclosure(s): T.S.: Lecturer − Bayer Health Care.
S.F.: Employee − Roche.
C.C.W.K.: Consultant − Bayer, Novartis, Thea Pharma; Lecturer − Thea Pharma.
A.I.dH.: Consultant − Ionis Pharmaceuticals.
Supported by the European Research Council under the European Union’s Seventh Framework Programme (FP/2007–2013)/ERC grant agreement no. 310644 (MACULA). This project has received funding from the European Union’s Horizon 2020 research and innovation programme under grant agreement no. 634479 (EYE-RISK). Financial support for the study was provided by the Oogfonds; MaculaFonds; Landelijke Stiching voor Blinden en Slechtzienden; and Novartis Fonds (Uitzicht grant no.: 2015-36). The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University, Rotterdam, The Netherlands; Netherlands Organization for the Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly (RIDE); the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); and the Municipality of Rotterdam, Rotterdam, The Netherlands. The exome sequencing data set of the Rotterdam Study was funded by the Netherlands Genomics Initiative (NGI)/Netherlands Organisation for Scientific Research (NWO) sponsored Netherlands Consortium for Healthy Aging (NCHA; project no. 050-060-810), by the Genetic Laboratory of the Department of Internal Medicine, Erasmus MC, and by the Complementation Project of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL; www.bbmri.nl; project number CP2010-41). The sponsors and funding organization had no role in the design or conduct of this research.
HUMAN SUBJECTS: Both human and animal subjects were used in this study. DNA samples of human subjects were used for whole exome sequencing and retina sections of wild-type adult mice were used for immunohistochemistry. The Medical Ethics Committee of the Erasmus Medical Center and the Ministry of Health, Welfare and Sport of The Netherlands, implementing the Wet Bevolkingsonderzoek: ERGO (Population Studies Act: Rotterdam Study), approved the Rotterdam Study and the study was approved by the local ethics committees on research involving human subjects of the participating centers. Informed consent to participate in the study was obtained from all participants. The study was performed in accordance with the tenets of the Declaration of Helsinki.
Author Contributions:
Conception and design: Corominas, Colijn, Geerlings, Klaver, den Hollander
Analysis and interpretation: Corominas, Colijn, Pauper, Amin, Lores Motta, Verlouw, van Rooij, Kraaij, E.K.de Jong, van Duijn, Klaver, den Hollander
Data collection: Corominas, Colijn, Geerlings, Pauper, Bakker, Amin, Kersten, Garanto, Verlouw, van Rooij, Kraaij, P.T.V.M.de Jong, Hofman, Vingerling, Schick, Fauser, van Duijn, Hoyng, Klaver, den Hollander
Obtained funding: None
Overall responsibility: Corominas, Colijn, Geerlings, E.K.de Jong, Klaver, den Hollander
Supplemental material available at www.aaojournal.org.
Supplementary Data
Figure S1.
Figure S2.
Figure S3.
Figure S4.
References
- 1.Smith W., Assink J., Klein R. Risk factors for age-related macular degeneration: pooled findings from three continents. Ophthalmology. 2001;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 2.Chakravarthy U., Evans J., Rosenfeld P.J. Age related macular degeneration. BMJ. 2010;340 doi: 10.1136/bmj.c981. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y., Bedell M., Zhang K. Age-related macular degeneration: genetic and environmental factors of disease. Mol Interv. 2010;10:271–281. doi: 10.1124/mi.10.5.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schick T., Ersoy L., Lechanteur Y.T.E. History of sunlight exposure is a risk factor for age-related macular degeneration. Retina. 2016;36:787–790. doi: 10.1097/IAE.0000000000000756. [DOI] [PubMed] [Google Scholar]
- 5.Seddon J.M., Cote J., Page W.F. The US twin study of age-related macular degeneration: relative roles of genetic and environmental influences. Arch Ophthalmol. 2005;123:321–327. doi: 10.1001/archopht.123.3.321. [DOI] [PubMed] [Google Scholar]
- 6.Cheng C.-Y., Yamashiro K., Chen L.J. New loci and coding variants confer risk for age-related macular degeneration in East Asians. Nat Commun. 2015;6:6063. doi: 10.1038/ncomms7063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritsche L.G., Chen W., Schu M. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433–439. doi: 10.1038/ng.2578. 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fritsche L.G., Igl W., Bailey J.N.C. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48:134–143. doi: 10.1038/ng.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein R.J., Zeiss C., Chew E.Y. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raychaudhuri S., Iartchouk O., Chin K. A rare penetrant mutation in CFH confers high risk of age-related macular degeneration. Nat Genet. 2011;43:1232–1236. doi: 10.1038/ng.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgason H., Sulem P., Duvvari M.R. A rare nonsynonymous sequence variant in C3 is associated with high risk of age-related macular degeneration. Nat Genet. 2013;45:1371–1374. doi: 10.1038/ng.2740. [DOI] [PubMed] [Google Scholar]
- 12.Seddon J.M., Yu Y., Miller E.C. Rare variants in CFI, C3 and C9 are associated with high risk of advanced age-related macular degeneration. Nat Genet. 2013;45:1366–1370. doi: 10.1038/ng.2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan X., Larson D.E., Wang C. Identification of a rare coding variant in complement 3 associated with age-related macular degeneration. Nat Genet. 2013;45:1375–1379. doi: 10.1038/ng.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Ven J.P.H., Nilsson S.C., Tan P.L. A functional variant in the CFI gene confers a high risk of age-related macular degeneration. Nat Genet. 2013;45:813–817. doi: 10.1038/ng.2640. [DOI] [PubMed] [Google Scholar]
- 15.Asimit J., Zeggini E. Rare variant association analysis methods for complex traits. Annu Rev Genet. 2010;44:293–308. doi: 10.1146/annurev-genet-102209-163421. [DOI] [PubMed] [Google Scholar]
- 16.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Triebwasser M.P., Roberson E.D.O., Yu Y. Rare variants in the functional domains of complement factor H are associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2015;56:6873. doi: 10.1167/iovs.15-17432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kavanagh D., Yu Y., Schramm E.C. Rare genetic variants in the CFI gene are associated with advanced age-related macular degeneration and commonly result in reduced serum factor I levels. Hum Mol Genet. 2015;24:3861–3870. doi: 10.1093/hmg/ddv091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiezun A., Garimella K., Do R. Exome sequencing and the genetic basis of complex traits. Nat Genet. 2012;44:623–630. doi: 10.1038/ng.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang L.-Z., Li Y.-J., Xie X.-F. Whole-exome sequencing implicates UBE3D in age-related macular degeneration in East Asian populations. Nat Commun. 2015;6:6687. doi: 10.1038/ncomms7687. [DOI] [PubMed] [Google Scholar]
- 21.Sardell R.J., Bailey J.N.C., Courtenay M.D. Whole exome sequencing of extreme age-related macular degeneration phenotypes. Mol Vis. 2016;22:1062–1076. [PMC free article] [PubMed] [Google Scholar]
- 22.Ristau T., Ersoy L., Lechanteur Y. Allergy is a protective factor against age-related macular degeneration. Invest Opthalmol Vis Sci. 2014;55:210. doi: 10.1167/iovs.13-13248. [DOI] [PubMed] [Google Scholar]
- 23.Hofman A., Murad S.D., van Duijn C.M. The Rotterdam Study: 2014 objectives and design update. Eur J Epidemiol. 2013;28:889–926. doi: 10.1007/s10654-013-9866-z. [DOI] [PubMed] [Google Scholar]
- 24.Hofman A., Brusselle G.G.O., Murad S.D. The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol. 2015;30:661–708. doi: 10.1007/s10654-015-0082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein R., Davis M.D., Magli Y.L. The Wisconsin age-related maculopathy grading system. Ophthalmology. 1991;98:1128–1134. doi: 10.1016/s0161-6420(91)32186-9. [DOI] [PubMed] [Google Scholar]
- 26.Bird A.C., Bressler N.M., Bressler S.B. An international classification and grading system for age-related maculopathy and age-related macular degeneration. The International ARM Epidemiological Study Group. Surv Ophthalmol. 1995;39:367–374. doi: 10.1016/s0039-6257(05)80092-x. [DOI] [PubMed] [Google Scholar]
- 27.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38 doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Ligt J., Willemsen M.H., van Bon B.W.M. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–1929. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 30.Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adzhubei I.A., Schmidt S., Peshkin L. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kircher M., Witten D.M., Jain P. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purcell S., Neale B., Todd-Brown K. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lohmueller K.E., Sparsø T., Li Q. Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. Am J Hum Genet. 2013;93:1072–1086. doi: 10.1016/j.ajhg.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stitziel N.O., Kiezun A., Sunyaev S. Computational and statistical approaches to analyzing variants identified by exome sequencing. Genome Biol. 2011;12:227. doi: 10.1186/gb-2011-12-9-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu D.J., Peloso G.M., Zhan X. Meta-analysis of gene-level tests for rare variant association. Nat Genet. 2013;46:200–204. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 38.Hoon M., Okawa H., Della Santina L., Wong R.O.L. Functional architecture of the retina: development and disease. Prog Retin Eye Res. 2014;42:44–84. doi: 10.1016/j.preteyeres.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Volland S., Esteve-Rudd J., Hoo J. A comparison of some organizational characteristics of the mouse central retina and the human macula. PLoS One. 2015;10 doi: 10.1371/journal.pone.0125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aisenbrey S., Zhang M., Bacher D. Retinal pigment epithelial cells synthesize laminins, including laminin 5, and adhere to them through alpha3- and alpha6-containing integrins. Invest Ophthalmol Vis Sci. 2006;47:5537–5544. doi: 10.1167/iovs.05-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tamura Y., Konomi H., Sawada H. Tissue distribution of type VIII collagen in human adult and fetal eyes. Invest Ophthalmol Vis Sci. 1991;32:2636–2644. [PubMed] [Google Scholar]
- 42.Leveziel N., Yu Y., Reynolds R. Genetic factors for choroidal neovascularization associated with high myopia. Invest Ophthalmol Vis Sci. 2012;53:5004–5009. doi: 10.1167/iovs.12-9538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Velazquez-Villoria A., Recalde S., Anter J. Evaluation of 10 AMD associated polymorphisms as a cause of choroidal neovascularization in highly myopic eyes. PLoS One. 2016;11 doi: 10.1371/journal.pone.0162296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Desronvil T., Logan-Wyatt D., Abdrabou W. Distribution of COL8A2 and COL8A1 gene variants in Caucasian primary open angle glaucoma patients with thin central corneal thickness. Mol Vis. 2010;16:2185–2191. [PMC free article] [PubMed] [Google Scholar]
- 45.Hopfer U., Fukai N., Hopfer H. Targeted disruption of Col8a1 and Col8a2 genes in mice leads to anterior segment abnormalities in the eye. FASEB J. 2005;19:1232–1244. doi: 10.1096/fj.04-3019com. [DOI] [PubMed] [Google Scholar]
- 46.Booij J.C., Baas D.C., Beisekeeva J. The dynamic nature of Bruch’s membrane. Prog Retin Eye Res. 2010;29:1–18. doi: 10.1016/j.preteyeres.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 47.Chong N.H.V., Keonin J., Luthert P.J. Decreased thickness and integrity of the macular elastic layer of Bruch’s membrane correspond to the distribution of lesions associated with age-related macular degeneration. Am J Pathol. 2005;166:241–251. doi: 10.1016/S0002-9440(10)62248-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Jong P.T.V.M. Age-related macular degeneration. N Engl J Med. 2006;355:1474–1485. doi: 10.1056/NEJMra062326. [DOI] [PubMed] [Google Scholar]
- 49.Sawada H., Konomi H., Hirosawa K. Characterization of the collagen in the hexagonal lattice of Descemet’s membrane: its relation to type VIII collagen. J Cell Biol. 1990;110:219–227. doi: 10.1083/jcb.110.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bogin O., Kvansakul M., Rom E. Insight into Schmid metaphyseal chondrodysplasia from the crystal structure of the collagen X NC1 domain trimer. Structure. 2002;10:165–173. doi: 10.1016/s0969-2126(02)00697-4. [DOI] [PubMed] [Google Scholar]
- 51.Kvansakul M., Bogin O., Hohenester E., Yayon A. Crystal structure of the collagen α1(VIII) NC1 trimer. Matrix Biol. 2003;22:145–152. doi: 10.1016/s0945-053x(02)00119-1. [DOI] [PubMed] [Google Scholar]
- 52.Moutsianas L., Agarwala V., Fuchsberger C. The power of gene-based rare variant methods to detect disease-associated variation and test hypotheses about complex disease. PLOS Genet. 2015;11 doi: 10.1371/journal.pgen.1005165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.