Abstract
Understanding the risks of biological invasion posed by ballast water—whether in the context of compliance testing, routine monitoring, or basic research—is fundamentally an exercise in biodiversity assessment, and as such should take advantage of the best tools available for tackling that problem. The past several decades have seen growing application of genetic methods for the study of biodiversity, driven in large part by dramatic technological advances in nucleic acids analysis. Monitoring approaches based on such methods have the potential to increase dramatically sampling throughput for biodiversity assessments, and to improve on the sensitivity, specificity, and taxonomic accuracy of traditional approaches. The application of targeted detection tools (largely focused on PCR but increasingly incorporating novel probe-based methodologies) has led to a paradigm shift in rare species monitoring, and such tools have already been applied for early detection in the context of ballast water surveillance. Rapid improvements in community profiling approaches based on high throughput sequencing (HTS) could similarly impact broader efforts to catalogue biodiversity present in ballast tanks, and could provide novel opportunities to better understand the risks of biotic exchange posed by ballast water transport—and the effectiveness of attempts to mitigate those risks. These various approaches still face considerable challenges to effective implementation, depending on particular management or research needs. Compliance testing, for instance, remains dependent on accurate quantification of viable target organisms; while tools based on RNA detection show promise in this context, the demands of such testing require considerable additional investment in methods development. In general surveillance and research contexts, both targeted and community-based approaches are still limited by various factors: quantification remains a challenge (especially for taxa in larger size classes), gaps in nucleic acids reference databases are still considerable, uncertainties in taxonomic assignment methods persist, and many applications have not yet matured sufficiently to offer standardized methods capable of meeting rigorous quality assurance standards. Nevertheless, the potential value of these tools, their growing utilization in biodiversity monitoring, and the rapid methodological advances over the past decade all suggest that they should be seriously considered for inclusion in the ballast water surveillance toolkit.
Keywords: Ballast water, Monitoring, Nucleic acids, PCR, High throughput sequencing, Compliance
1. The importance of ballast water monitoring in research and regulatory contexts
Maritime trade has long been recognized as a major driver of invasive species spread globally, and ballast water continues to serve as a primary vector for non-indigenous marine species introductions (Drake and Lodge, 2004; Keller et al., 2011; Seebens et al., 2013). To address the environmental, economic and public health impacts of ballast water-borne invasive species, a number of government entities have enacted comprehensive regulations involving the use of best management practices and the application of treatment technologies for use on-board commercial vessels (Costello et al., 2007). Ballast water discharges are currently regulated at multiple levels of governance by multiple agencies, including internationally by the International Maritime Organization (IMO) and within the U.S. by the Coast Guard (USCG) and the Environmental Protection Agency (EPA). These policies are the outcome of decades of research and concerted effort by the global scientific and shipping communities to understand the risks posed by ballast water-borne invasions and to determine the management strategies best positioned to mitigate those risks.
In February 2004, the IMO, through its Marine Environmental Protection Committee and participating member nations, enacted the International Convention for the Control and Management of Ship’s Ballast Water and Sediments. More widely known as the BWM Convention (Lloyd’s Register, 2016), it is intended to help reduce the risk of new invasions of non-native species through the implementation of an interim ballast water exchange Standard (D-1) and a more restrictive numerical organism discharge Standard (D-2) for shipboard treatment systems. The technology performance standard targets two size classes of plankton in addition to limits for toxigenic Vibrio cholerae, Escherichia coli and intestinal enterococci. Ship owners required to meet the D-2 standard must have on-board treatment systems retrofitted to existing vessels or designed into the ballasting systems of new build vessels according to a prescribed timeline. On September 8, 2016 the total world shipping tonnage of ratifying states reached the 35% minimum required to trigger the one-year period for the BWM Convention to enter into force on September 8, 2017.
Ballast water discharges in the U.S. are regulated under the Nonindigenous Aquatic Nuisance Prevention and Control Act of 1990 (NANPCA), as amended by the National Invasive Species Act of 1996 (NISA). The USCG is the responsible agency for developing and enforcing regulations under this act. The USCG published a “Notice of Proposed Rule Making” in 2009, and after reviews and public comments, the “Standards for living organisms in ships’ ballast water discharged in U.S. waters; final rule” was published in the Federal Register on March 23, 2012 (USCG, 2012). Both the IMO and USCG discharge standards for treatment systems are identical, as follows:
For organisms greater than or equal to 50 μm in minimum dimension: discharge must contain fewer than 10 live organisms per cubic meter (m3) of ballast water.
For organisms <50 μm and greater than or equal to 10 μm in minimum dimension: discharge must contain <10 living organisms per milliliter (ml) of ballast water.
- Indicator organisms must not exceed:
- For toxigenic V. cholerae (serotypes 01 and 0139): a concentration of <1 colony forming unit (cfu) per 100 ml.
- For Escherichia coli: a concentration of fewer than 250 cfu per 100 ml.
- For intestinal enterococci: a concentration of fewer than 100 cfu per 100 ml.
That document also contains a specific timetable for installation of treatment technology based on the vessel’s construction date and ballast capacity. In December of 2016 USCG issued its first type approval for a ballast water management system (BWMS) to Optimarin AS of Norway. USCG continues to evaluate additional BWMS at several USCG accepted testing facilities.
Since 2008, EPA has also regulated ballast water discharges and other discharges incidental to the normal operation of vessels under the Clean Water Act (CWA) through the Vessel General Permit (VGP) (USEPA, 2013). Sixteen states also have specific ballast water management requirements imposed either through separate regulations or CWA Section 401 Certifications for the VGP Program. EPA had previously exempted these discharges from its regulations, citing the fact that USCG had promulgated and administered ballast water regulations pursuant to its Congressional mandate in NISA. However, a 2005 court decision vacated that exemption, leading to implementation of the first VGP in 2008 (Northwest Environmental Advocates v. EPA, Northern District Court of California, 2005). In March 2013, EPA issued a revised VGP replacing the previous 2008 version, and requiring existing vessels to meet the IMO D-2 and USCG ballast water discharge performance standard. The VGP includes monitoring requirements for installed BWMS that address functionality depending on the technology used by the treatment system, and periodic biological organism monitoring using total heterotrophic bacteria, E. coli, and enterococci as indicators of treatment performance.
Regulatory agencies and ship owners need assurance that treatment technology will perform successfully in a wide range of water quality and under harsh environmental conditions on board ships. Developing appropriate testing procedures to verify biological treatment efficacy at land-based facilities has proven to be a challenging endeavor. Since 2001, USCG and EPA have participated in a joint activity under EPA’s Environmental Technology Verification Program (ETV) to develop performance verification protocols for BWMS. Assisted by stakeholders and technical panel input, a working draft was produced in 2004. Additional research and testing by the USCG at the Naval Research Laboratory resulted in significant improvements to the draft which was eventually released by EPA as the document entitled “Generic protocol for the verification of ballast water treatment technology” (USEPA, 2010). The ETV protocol contains a biological assay for enumerating live organisms based on a combination of two vital fluorescence stains (fluorescein diacetate [FDA] and 5-chloromethylfluorescein diacetate [CMFDA]) with direct microscope observation and probing for motility verification. Although vital fluorescent stains have been demonstrated useful in evaluation the efficacy of BWMS, the ETV protocol recommends testing facilities validate the effectiveness of stains with ambient populations before use.
The USCG is required to periodically monitor the performance of BWMS installed on commercial vessels. It is impractical for inspectors to perform shipboard biological testing to the degree conducted at land-based facilities. Therefore, portable and rapid test kits are being developed by at least seven manufacturers to enable inspectors to determine if a sample of ballast water appears to have been successfully treated, or if a condition of “gross exceedance” exists. The inspector can then arrange for a more detailed biological test if time permits, or notify the ship operator to have the treatment system fully evaluated at the next port of call. The USCG is currently evaluating several candidate test kits based on different assay technologies.
The ETV protocol recognizes that improvements in biological assays and instrumentation for BWMS testing are expected to occur in the coming years. USCG and EPA remain committed to updating and improving the ETV protocol periodically to include such advances in procedures and technology which will enhance the quality and cost effectiveness of the BWMS testing program. In addition, USCG and EPA continue to encourage research aimed at better understanding the relationship between the number of viable propagules discharged with ballast water, and the risk of non-native species establishment—the so-called “risk release” relationship (Carlton et al., 2011). These efforts are rooted in concerns expressed by some that existing discharge limits are not sufficiently protective of water quality, concerns highlighted by recent court ruling challenging currently adopted U.S. standards (Natural Resources Defense Council et al. v. EPA, United States Court of Appeals for the Second Circuit, 2015). USCG and EPA have backed a number of research efforts aimed at developing more robust scientific support for current and future numerical discharge standards (Carlton et al., 2011; Lee et al., 2013; USEPA and ANSTF, 2013).
While regulation continues to be the driving motivation behind much ballast water monitoring, the task of understanding risks associated with ballast water-borne invasions is also important in a variety of non-regulatory contexts (Barry et al., 2008). Ballast water likely continues to facilitate the spread of a host of high profile invasive species, and determining the patterns and mechanisms of that spread remains an important goal. Detection of target species in ballast water provides immediate information relevant to risk of invasion to recipient ports and also aids in reconstructing invasion pathways and better understanding invasion histories. In addition, fundamental questions remain regarding the overall risk posed by ballast water transport and the degree to which recent changes in management policy have reduced that risk. Research efforts aimed at answering those questions and others require substantial investment in ballast water monitoring and application of a broad analytical toolkit capable of characterizing ballast water in the context of multiple research and management needs (USEPA and ANSTF, 2013).
2. Advantages of nucleic acids-based monitoring tools
Ballast water monitoring is fundamentally a task in understanding the biodiversity present in ballast tanks—which species are present, how many individual propagules there are, and the capacity of those propagules to establish new populations upon release (i.e. condition or viability). Over the past decade a dramatic shift has occurred in the development of tools for biodiversity monitoring, with growing recognition that nucleic acids-based technologies may offer unprecedented advances in sensitivity, specificity, and cost-effectiveness. Specimen identification based on morphological taxonomy continues to be invaluable, and to provide the ultimate foundation for all biodiversity assessment. However, these traditional approaches are burdened with widely recognized inadequacies that may limit their utility for certain conservation and natural resource applications. Morphology-based bio-diversity assessments tend to be slow and costly and rely on professional taxonomic expertise that may itself be endangered (Pfrender et al., 2010). They are also limited generally to identifying whole adult specimens (Besansky et al., 2003; Mallet and Willmott, 2003) and are challenged by the fact that a great many taxa continue to elude comprehensive taxonomic assessment, including a number of speciose and ecologically important groups such as eukaryotic meiofauna (Creer et al., 2010; Bik et al., 2012).
In response to these challenges, molecular ecologists have touted the potential value of nucleic acids-based technologies for biodiversity monitoring (Lodge et al., 2012; Taberlet et al., 2012; Ji et al., 2013; Cristescu, 2014), including applications specific to biological invasions (Darling and Blum, 2007; Bott et al., 2010; Blanchet, 2012). These methods arguably exhibit advantages over traditional morphological approaches (Table 1). Not all of these benefits have yet been fully realized. Nevertheless, available evidence suggests that continued development of molecular tools will only further solidify many of these advantages. Nucleic acids-based technologies have already proven capable of detecting diversity that is overlooked in traditional approaches, rendering them particularly relevant for assessments of those species that are present at extremely low abundances and thus likely to evade detection (Lindeque et al., 2013; Zhou et al., 2013). Other studies have demonstrated that molecular biodiversity monitoring can be less expensive and more responsive to immediate regulatory and management needs than morphological approaches, and can generate standardized data more amenable to audit and less vulnerable to variability in taxonomic expertise across studies (Pfrender et al., 2010; Ji et al., 2013; Stein et al., 2014a; Stein et al., 2014b).
Table 1.
Relative merits of genetic and traditional methods. Criteria are worded so that desirable outcomes are always reflected in a “high” rating. Advantages of genetic methods should be taken with some caution, as many nucleic acids tools are still in development phases and have not yet fully achieved their expected potential. For instance, the affordability of genetic tools currently depends on availability of appropriate in-house or extramural molecular support, which varies widely across users, and also on sample throughput.
| Criterion | Genetic methods | Traditional morphological methods |
|---|---|---|
| Sensitivity | HIGH | low |
| Specificity | HIGH | low |
| Ability to identify sub-adult or partial specimens | HIGH | low |
| Ability to identify cryptic taxa | HIGH | low |
| Quantification | low | HIGH |
| Opportunity for passive surveillance | HIGH | low |
| Affordability of up-front costs | low | HIGH |
| Affordability per sample | HIGH* | low |
| Speed of analytical turnaround | HIGH | low |
| False negative avoidance | HIGH | low |
| False positive avoidance | low | HIGH |
Affordability of genetic methods is highly scale-dependent, and per sample costs are strongly negatively correlated with sample throughput.
All of these considerations suggest that nucleic acids-based approaches could provide critically valuable tools for answering the most pressing questions associated with ballast water monitoring (Table 2). Indeed, many of the most important criteria for ballast water monitoring—high sensitivity, species-specificity, and applicability to a broad taxonomic range of potential targets—may be better met by molecular tools than by standard morphological methods. However, several crucially important conditions have not yet been met by available molecular tools; most notably, the ability to assess viability and to quantify abundance remain extremely limited. These caveats will clearly constrain the applicability of molecular tools for answering certain questions at the center of managers’ concerns, such as whether or not ballast samples can be said to meet compliance standards (see Section 3.3 below). Nevertheless, we emphasize that the role of ballast water monitoring is broad, and extends from immediate management interests such as compliance to long-term research questions such as the relationship between ballast water-borne propagule pressure and invasion risk.
Table 2.
Questions associated with ballast water monitoring, and applicable genetic tools. Different nucleic acids-based detection methods satisfy different criteria and are associated with different challenges to technology development and deployment.
| Management/science question | Criteria that must be satisfied to answer question | Possible genetic tools | Most significant challenges |
|---|---|---|---|
| Single species approaches | |||
| Does the sample contain target species X? | Target specificity, sensitivity | PCR/qPCR or other probe-based detection methods | Managing false positive and negative errors |
| What is the abundance of target species X in the sample? | Target specificity, sensitivity, quantification | qPCR or other probe-based detection method calibrated for quantification of target | Managing false positive and negative errors, plus calibration for robust quantification of target |
| Does the sample comply with a standard? | Target specificity, sensitivity, quantification, viability | qPCR targeting transcripts associated with viability | Managing false positive and negative errors, calibration for robust quantification of target, plus identification of targets tightly associated with viability and possibly additional costs associated with handling RNA targets |
| Community approaches | |||
| What species are in the sample? | Broad community profiling, sensitivity | HTS metabarcoding | Gaps in reference databases, difficulty interpreting data from rare species, not amenable to fast turnaround |
| What is the overall biodiversity (species richness and abundance) in the sample? | Broad community profiling, sensitivity, quantification | HTS metabarcoding, calibration of sequence frequency data to relative abundance | Gaps in reference databases, difficulty interpreting data from rare species, not amenable to fast turnaround, plus calibration for robust quantification |
3. Examples of nucleic acids-based tools
In the following we discuss in detail three fundamental approaches to ballast water monitoring, the potentially relevant nucleic acids-based detection technologies, and their associated benefits and limitations.
3.1. Targeted detection
Perhaps the most straightforward task in ballast water monitoring is determining whether a particular species is present in a sample. Nucleic acids-based tools have already shown considerable promise for targeted detection by relying on the fact that very short segments of an organism’s genome can be utilized to identify that organism as a member of a particular species. Although this idea has been widely recognized by molecular ecologists for decades, only in recent years has it become more broadly popularized among natural resource managers through association with the concept of DNA barcoding (Hebert et al., 2003b; Hebert et al., 2003a). Despite some concerns regarding applicability of the approach—most importantly, limitation of existing reference sequence databases (see below; Page et al., 2005; Trebitz et al., 2015) and questions regarding the true universality of barcode loci (Zhan et al., 2014b)—DNA barcoding has been widely adopted to improve species-level identifications and to highlight possible cases of previously undescribed diversity, in both research and environmental management contexts.
DNA barcoding entails the generation of sequence from a barcode locus and comparison of that sequence to reference databases that associate sequences with known species (Hebert et al., 2003b). Targeted detection typically takes a more indirect route, applying the fundamental concept of barcoding (short sequences diagnostic for species identification) to more rapid techniques for determining the presence of a target species. Generally, these techniques involve the development of species-specific nucleic acid “probes.” When these probes are introduced into bulk nucleic acids extracted from environmental samples, they bind only to complementary DNA or RNA released from individuals of the target species. Binding of probe to target nucleic acid thus provides a signal of target species presence. Probe-based targeted detection approaches differ in the methods they adopt to detect this signal, and they vary in availability, sensitivity, analytical turnaround time, and their capacity to quantify target nucleic acid concentration.
By far the most common method is amplification of bound target via the polymerase chain reaction (PCR). PCR-based detection has become a mainstay of targeted monitoring efforts, in large part due to the broad accessibility of the technology. The application of these methods has extended to ballast water monitoring for target species across multiple regulatory size classes. A number of studies have demonstrated the utility of PCR-based detection for determining the presence of pathogenic bacteria (Aridgides et al., 2004; Mimura et al., 2005; Emami et al., 2012) and harmful algae (Doblin et al., 2004; Drake et al., 2005; Patil et al., 2005; Burkholder et al., 2007; Doblin et al., 2007) in ballast tanks, and some have extended the methodology to detect zooplankton species (Harvey et al., 2009; Egan et al., 2015). While most applications of PCR-based monitoring involve straightforward presence/absence detection, some studies have taken advantage of quantitative PCR (qPCR) methods to estimate abundance of target species. For instance, Doblin et al. adopted a qPCR approach based on amplification of the mitochondrial 16S locus to determine cell concentrations of toxic Microcystis and Anabaena algal species in ballast on ships transiting the Great Lakes (Doblin et al., 2007).
A variety of other methods have been adopted for probe-based targeted detection in ballast water samples, including Southern hybridization (Aridgides et al., 2004), fluorescence in situ hybridization (FISH; Joachimsthal et al., 2004), and nucleic acid sequence based amplification (NASBA, an isothermal method for rapid RNA amplification; Fykse et al., 2012), though given its ubiquity PCR is very likely to remain the most commonly applied technology. More recent studies have attempted to extend the utility of probe-based monitoring by leveraging novel engineering solutions to create platforms with potential for future field deployment (Mahon et al., 2011; Egan et al., 2013; Mahon et al., 2013). For instance, Egan et al. (2015) were able to detect Dreissena polymorpha (zebra mussel) and D. bugensis (quagga mussel) propagules at very low concentrations in ballast water samples using Laser Transmission Spectroscopy (LTS), a technology that enables sensitive detection in seconds of probe bound to extracted target DNA, with a physical footprint small enough to be carried onto ships. Such technologies, if coupled with protocols for rapid nucleic acid extraction (Tomlinson et al., 2010), could enable analytical turnaround times on the order of minutes, essentially providing point-of-testing diagnostics.
These probe-based approaches are capable of detecting targets including eggs, larvae, and other life stages lacking diagnostic morphological characteristics (Pfrender et al., 2010); cryptic taxa or taxa that cannot be resolved morphologically at species level (Bucklin et al., 2010); and viable but non-culturable (VBNC) microorganisms (Li et al., 2014). For example, in one recent study Stehouwer et al. found that genetic methods could identify phytoplankton species present in ballast tanks that could not be identified to species level by either microscopy or flow cytometry (Stehouwer et al., 2012). A number of recent studies have also demonstrated that in many cases physical capture of the target organism may be unnecessary for positive detection. Probe-based monitoring based on extraction of environmental DNA (eDNA, or DNA collected without direct attempt to sample target organisms) has shown that even DNA disassociated from the target organism can be detected in aquatic samples (Lodge et al., 2012; Bohmann et al., 2014; Barnes and Turner, 2015). Furthermore, targeted probe-based methods can be extremely sensitive, in some cases out-performing traditional detection tools in active field monitoring efforts (Jerde et al., 2011). Applications to ballast water monitoring suggest that sensitivity of PCR-based methods is sufficient to address management-relevant questions; for instance, Fyske et al. demonstrated that two different amplification-based approaches (qPCR and NASBA) were capable of detecting V. cholerae at levels far below the IMO standard of <1 cfu per 100 ml of ballast water (Fykse et al., 2012).
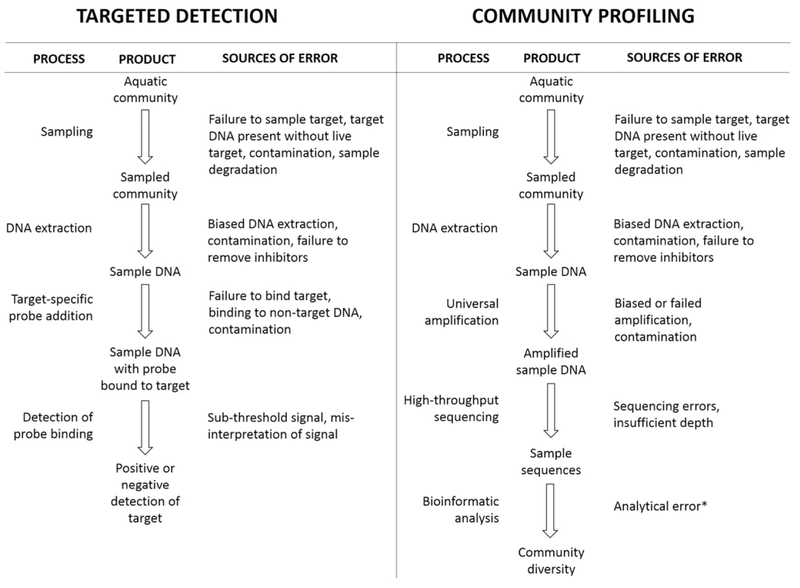
Despite these advantages, certain limitations continue to limit application of nucleic acids-based tools for targeted detection. Quantification, for instance, remains problematic in most cases. While a number of methodologies, most notably qPCR, are capable of precisely determining concentrations of target nucleic acid in extracted samples, translating those values into organism counts is a non-trivial matter. The solution is most straightforward in the case of single celled target species, for which relatively tight correlations between cell count and nucleic acid concentration can be derived (Doblin et al., 2007; Fykse et al., 2012). For larger, more complex organisms, such as zooplankton, individual variation in body mass can render these inferences more difficult and prone to error (Darling and Blum, 2007). Also of concern in many monitoring contexts is the general inability of probe-based detection to discern viability of target organisms. Finally, while the persistence of DNA outside of the living organism may allow for broader monitoring efforts, it also raises the specter of false positive detections in which DNA is present despite the absence of a living individual. Solutions to this problem may involve adopting novel nucleic acids targets more tightly linked to viability (Section 3.3). False positive detections are a general concern for targeted detection (see Fig. 1), as the extremely high sensitivity afforded by probe-based technologies means that very low levels of contamination or mis-probing (binding of probe to non-target sequences) can potentially result in positive detections. These concerns are best addressed by thorough testing of probe-based methods prior to field application, including rigorous determination of error rates and demonstration of species specificity in relevant sampling contexts (Darling and Mahon, 2011). Despite these limitations, probe-based monitoring has already been deployed in broad invasive species surveillance programs (Jerde et al., 2013). While such deployments demonstrate the value of rigorously vetted tools for informing management decisions, they do not necessarily imply fitness of those tools for diagnostic applications in regulatory contexts—specifically, compliance with discharge standards. Such fitness can only be assured through appropriate method validation, as discussed in Section 3.3 below.
Fig. 1.
Workflow for targeted probe-based detection (left) and HTS-based community profiling (right), including potential sources of error introduced at each process step. False positive and false negative errors can derive from multiple steps in each process. *Encompasses a broad range of error sources including, but not limited to, poor filtering of reads, failure to remove chimeras, and inaccuracies in taxonomic assignment.
3.2. Community profiling
A number of important monitoring questions require more comprehensive understanding of ballast water-borne biodiversity than can be provided by targeted approaches (Table 2). For example, systematic assessments of invasion risk require detection not only of particular high risk species, but of all potentially invasive propagules introduced into recipient ports. Only recently have advances in nucleic acids analysis enabled this more exhaustive characterization of biological communities. Specifically, the advent of high throughput sequencing (HTS, also referred to as next- or second-generation sequencing) is opening up unprecedented opportunities for biodiversity assessment (Bik et al., 2012; Taberlet et al., 2012; Bohmann et al., 2014). HTS allows for the expansion of DNA barcoding into DNA metabarcoding (Cristescu, 2014); while the former entails generating barcode sequence from a single organism, the latter enables generation of an entire population of barcode sequences representing all of the organisms in a complex environmental sample.
This simplistic representation obviously belies the true complexity of the metabarcoding approach. Nevertheless, metabarcoding in principle provides a means of describing in great detail diverse biotic assemblages. Recent studies have indicated that the approach can deliver results comparable to morphological taxonomic surveys, with similar management implications, at considerably lower cost (Ji et al., 2013). Given the current trajectory of technological advance, it is reasonable to expect that the ability to generate sequence data will only become less expensive and more accessible in the future (Shokralla et al., 2012). Metabarcoding approaches also have the advantage of extremely high sensitivity, and introduce the possibility of broadening biodiversity assessments to include taxa typically excluded from traditional surveys (Creer et al., 2010; Medinger et al., 2010; Lindeque et al., 2013; Pagenkopp Lohan et al., 2016). These benefits have led some researchers to explore the utility of HTS-based approaches in various environmental management contexts, primarily focusing on water quality monitoring (Chariton et al., 2010; Hajibabaei et al., 2011; Kermarrec et al., 2014).
These potential advantages of metabarcoding have not been lost on molecular ecologists interested in understanding ballast water-borne biodiversity. Recent applications to ballast water monitoring include studies aimed at cataloguing biodiversity across multiple taxonomic groups and regulatory size classes, ranging from viruses to zooplankton (Table 3). Several recent studies have taken advantage of the capacity of HTS to assess richness of taxonomic groups poorly amenable to traditional morphological characterization. Ng et al. (2015) applied metabarcoding of the mitochondrial 16S locus to explore relationships between microbial communities in ballast water and recipient harbor waters in the tropical port of Singapore. That study revealed expected declines in phototrophic taxa in ballast samples taken after lengthy voyages (107 days in one case). HTS was also able to detect indicator organisms (V. cholerae and E. coli) in ballast at levels lower than IMO numerical discharge standards. Metabarcoding has also been employed to investigate the impacts of ballast water treatment on microbial communities, revealing that certain genera may exhibit resistance to particular treatment systems (Fujimoto et al., 2014). Even viral communities have proven accessible to metabarcoding; Kim et al. (2015) explored viral community (“virome”) diversity among ballast water samples on ships transiting the Great Lakes, revealing distinct virome structure among the Lakes and detecting a number of viral fish and shrimp pathogens being transported in ballast. Other studies have investigated eukaryotic zoo- and phytoplankton. Three other recent publications summarizing HTS-based assessment of ballast water samples from the R/V Polarstern (Ardura et al., 2015; Zaiko et al., 2015b; Zaiko et al., 2015a) revealed decreases in community DNA quality and overall sequence richness, consistent with DNA degradation associated with high in situ mortality, despite increases in frequency of sequences assigned to potentially invasive taxa, suggesting that they may remain viable throughout lengthy voyages and pose high risks of invasion upon discharge. These studies and others also underscore the possibility of leveraging the high sensitivity and taxonomic resolution of metabarcoding approaches to conduct even targeted monitoring, while simultaneously generating data suitable for passive surveillance of non-targets (e.g. Simmons et al., 2016).
Table 3.
Published direct applications of molecular genetic tools for ballast water monitoring and/or research. Note that in a number of these publications nucleic acids-based methods are not adopted as the primary detection technology. Abbreviations for methodologies are defined in the text.
| Authors | Journal | Target organisms | Target loci | Methodology | |
|---|---|---|---|---|---|
| 1. | Aridgides et al., 2004 | Marine Pollution Bulletin | Vibrio cholerae, Aureococcus anophageferens | 18S,ORF1, tcpA | PCR, qPCR |
| 2. | Doblin et al., 2004 | Harmful Algae | Pfiesteria | 18S | qPCR |
| 3. | Joachimsthal et al., 2004 | Marine Pollution Bulletin | Total bacteria, enterobacteria, Vibrio cholerae and E. coli | Fluorescence In situ Hybridization (FISH) | |
| 4. | Drake et al., 2005 | Biological Invasions | Pfiesteria | 18S | qPCR |
| 5. | Mimura et al., 2005 | Marine Pollution Bulletin | Vibrio cholera | 16S | Sanger sequencing, barcoding |
| 6. | Patil et al., 2005 | Biological Invasions | Cymnodinium catenatum | 18S | PCR |
| 7. | Burkholder et al., 2007 | Harmful Algae | Harmful algae | 18S, 16S | PCR |
| 8. | Doblin et al., 2007 | Harmful Algae | Microcystis | mcyE | PCR and qPCR |
| 9. | Harvey et al., 2009 | Journal of Experimental Marine Biology and Ecology | multiple | ITS1, ITS2, 5.8S | PCR, Sanger sequencing |
| 10. | Ma et al., 2009 | Progress in Natural Science | Planktonic bacterial community | 16S | Restriction fragment length polymorphisms (RFLP) |
| 11. | Hess-Erga et al., 2010 | Water Research | microbial community | 16S | Denaturing gradient gel electrophoresis (DGGE) |
| 12. | Briski et al., 2010 | Biological Invasions | Diapausing eggs | 16S | Sanger sequencing, barcoding |
| 13. | Xu et al., 2011 | International Journal of Systematic and Evolutionary Microbiology | Kordiimonas lacus | 16S | Sanger sequencing, barcoding |
| 14. | Emami et al., 2012 | PLoS One | Vibrio cholerae and Pseudomonas | 16S | PCR |
| 15. | Fykse et al., 2012 | Marine Pollution Bulletin | Vibrio cholera | groEL, tcpA | qPCR, NASBA |
| 16. | Stehouwer et al., 2012 | Journal of Applied Phycology | Phytoplankton | 16S | DGGE |
| 17. | Fujimoto et al., 2014 | PLoS One | Microbial community | 16S | HTS with propidium mono-azide (PMA) cross-linking |
| 18. | Steichen et al., 2014 | Marine Pollution Bulletin | Eukaryotic community | 18S | DGGE |
| 19. | Ardura et al., 2015 | Journal ofMolluscan Studies | Peringia ulvae | COI | HTS |
| 20. | Egan et al., 2015 | Environmental Science and Technology | Dreissena polymorpha and D. bugensis | CO1, species Specific | PCR and LTS |
| 21. | Kim et al., 2015 | Environmental Science and Technology | Viral community | 16S | HTS |
| 22. | Ng et al., 2015 | PLoS One | Microbial community | 16S | HTS and qPCR |
| 23. | Steichen and Quigg, 2015 | Marine Pollution Bulletin | Phytoplankton | 18S | Sanger sequencing, barcoding |
| 24. | Zaiko et al., 2015a | Marine Pollution Bulletin | Multiple | COI, RBC | HTS |
| 25. | Zaiko et al., 2015b | Marine Environmental Research | Multiple | COI | HTS |
| 26. | Pagenkopp Lohan et al., 2016 | Microbial Ecology | Microbial eukaryotes | SSU RNA | HTS |
HTS is a relatively young technology, and metabarcoding is a relatively new application with attendant limitations (Zaiko et al., 2015a; Viard et al., 2016). For one thing, turnaround time for analytical results is, as expected, considerably longer than for probe-based targeted detection. Although rapid improvements have been made in accessibility of HTS technology, metabarcoding is a technically demanding process and weeks to months may be required for generation of sequence data and appropriate analyses. As noted above in the case of targeted detection, metabarcoding approaches are also generally insensitive to organism viability (though see Section 3.3 below). Furthermore, while they are clearly capable of highly sensitive detection of rare taxa, there remains some question regarding the robustness of such detections. A number of studies have now demonstrated that metabarcoding can provide more comprehensive estimates of sample biodiversity than traditional approaches (Ji et al., 2013; Lindeque et al., 2013), and recent experimental work has further shown that they are capable of detecting rare taxa (Pochon et al., 2013; Zhan et al., 2013). However, other studies have found that sequencing errors can lead to overestimation of rare species richness in the absence of stringent filtering protocols (Kunin et al., 2010), protocols which unfortunately may negatively impact the ability to detect rare species present in the sample (Zhan et al., 2014a). This awkward catch-22 clearly suggests the need for novel solutions to the problem of low-abundance species, including not only more robust error detection algorithms but also strategies that quantify the effects of artifact removal on the ability to detect rare species (Zhan et al., 2014a). Also problematic are current limitations to existing reference sequence databases. Recently published assessments confirm the patchy taxonomic coverage of available reference databases (Trebitz et al., 2015; Briski et al., 2016), and these limitations have impacted the ability to fully catalogue diversity in ballast water samples.
Additionally, quantification of organism abundance remains a significant challenge for metabarcoding approaches. Although some studies indicate the possibility of approximate abundance estimates (Kelly et al., 2014; Elbrecht and Leese, 2015), a number of attributes of HTS-based approaches render such estimates extremely challenging. Since target locus copy number can vary dramatically even across individual cells, it is exceedingly difficult to accurately correlate organism number to number of generated sequences; this problem is obviously only exacerbated in the case of multicellular diversity (Elbrecht and Leese, 2015). Although this problem exists for targeted detection, as well (see Section 3.1, above), at least in that case it is possible to experimentally determine calibration curves for the target species; that task is generally unrealistic for comprehensive community diversity assessments. Furthermore, bias in both DNA extraction and amplification of target DNA using “universal” barcoding primers is inevitable, resulting ultimately in over-representation of certain taxa. Strategies for overcoming this limitation may include comparison of metabarcoding results across multiple loci (Cristescu, 2014), development of amplification-free workflows (Zhou et al., 2013), or assessments of risk that rely on patterns of positive/negative detections across samples rather than abundance estimates within any one sample (Jerde et al., 2011; Takahara et al., 2012).
Metabarcoding workflows are complex, with a large number of processing steps standing between an environmental sample and information that may be useful to decision-makers. The options available at each processing step can result in substantial variation between metabarcoding studies, as each study tailors its workflow to the needs of a particular application (Cristescu, 2014). Unfortunately, this variation frequently leads to analytical results that are not directly comparable across studies. Recent work has demonstrated that choices in HTS data processing can dramatically influence analytical outcomes, and numerous studies have revealed the dependence of diversity assessments on the choice of barcode locus (Meusnier et al., 2008; Zhan et al., 2014b; Zhan et al., 2014a; Flynn et al., 2015). These results highlight the diversity of options available to develop metabarcoding tools appropriate for specific ballast water monitoring tasks, but also emphasize the challenges associated with developing standardized protocols that are transferrable across studies and readily auditable by third parties, considerations that may prove important in developing comprehensive ballast water surveillance programs.
3.3. Compliance testing
Of all the applications reviewed here, compliance testing is the only one that carries the weight of enforcement, and thus the one most burdened by specific criteria guiding development of testing protocols. As noted above, such development is a complex undertaking. At this time the ETV protocol for approval of BWMS in the US relies on biological as-says based on vital staining alongside direct microscopic visualization and motility testing for enumerating live organisms (USCG, 2012). However, these standard approaches are not without their shortcomings. While staining with FDA and CMFDA has been shown to reliably reflect viability in marine protists, accuracy can vary depending on taxonomic composition of the ballast water community (Steinberg et al., 2011) and in the case of many taxa considerable overlap exists in the frequency distribution of fluorescence intensity between living and dead cells, leading inevitably to errors in viability assessment (Adams et al., 2014; MacIntyre and Cullen, 2016). USCG, EPA, and other regulatory agencies outside of the U.S. thus continue to seek novel technologies that would improve the quality and cost effectiveness of compliance testing protocols.
3.3.1. Assessing viability
Viability is, of course, the central challenge of compliance tools. As we have already seen, this is generally problematic for nucleic acids-based technologies. However, there is good reason to believe that identification of appropriate analytical targets may ultimately overcome this limitation. Recent development of methods based on adenosine tri-phosphate (ATP) quantification indicate substantial promise as future compliance testing tools (van Slooten et al., 2015; Wright et al., 2015). Those methods take advantage of the universal role of ATP in cellular metabolism, adopting the molecule as a suitable and measurable indicator of organismal viability. Similar reasoning can be applied to nucleic acids-based methods for biodiversity monitoring by identifying nucleic acid targets that should be closely associated with viability. This tactic has already been adopted in other environmental management contexts. Water quality monitoring frequently relies on assessment of microbiological indicators (primarily E. coli, but also other species), a task hampered by the slow response time of culture-based methods as well as the potential importance of VBNC strains. Nucleic acids-based tools have been explored extensively as possible solutions to these problems and have even been adopted by regulatory agencies to test for the presence of water-borne pathogens, attesting to their potential applicability in enforcement contexts (Keer and Birch, 2003; Gonzalez and Noble, 2014; Mendes Silva and Domingues, 2015). Generally, two approaches have been adopted: 1) identify targets for amplification (by PCR, qPCR, or NASBA) that are tightly coupled with viability due both to their metabolic roles and their lability, such as ribosomal RNAs or longer messenger RNAs associated with important cellular functions (e.g. heat-shock proteins or even proteins involved in pathogenicity); 2) prevent detection of dead cells by treatment with propidium monoazide (PMA), a dye that only enters cells lacking an intact membrane and that binds DNA, precluding amplification. Interestingly, both of these approaches have been implemented in the context of ballast water monitoring, although not in compliance settings. In one study, targeted detection of the harmful cyanobacteria Microcystis in ships’ ballast transiting the Great Lakes was conducted using qPCR amplification of a variety of nucleic acids targets, allowing quantitative assessment at multiple levels (Doblin et al., 2007): 16S DNA was adopted to determine overall concentration of Microcystis; the toxin-producing microcystin synthetase gene (mcyE) to estimate the proportion of toxic Microcystin species; 16S rRNA to assess the proportion of metabolically active cells; and mcyE RNA to determine the level of active toxin production. This approach powerfully illustrates the value of pursuing different loci for generating detailed information on the presence, abundance, and potential viability of target species of interest, although it is important to note that assays employing RNA loci introduce additional complications and expense. Another study coupled HTS with PMA cross-linking to test the anti-microbial efficacy of an alkali ballast water treatment system (Fujimoto et al., 2014). In addition to confirming dramatic decreases in viable microbial diversity associated with treatment, results of PMA processing suggested that non-viable bacteria did not appreciably contribute to the sequence pool after 3-day alkali treatments, indicating that nucleic acids from dead bacteria had been largely removed through degradation.
The above considerations adopt a straightforward interpretation of the term “viable” to mean “living,” consistent with current USCG regulations (USCG, 2012). In other words, the metabolic targets mentioned above generally enable determination of whether a target organism is alive or dead. It is important to note that a broader interpretation of viability allows that some organisms may be alive and yet incapable of future survival and/or reproduction, and thus pose no risk of invasion. Organisms that are “inviable” in this sense may nevertheless register as living in many of the described tests. Navigating this distinction will prove challenging not only for regulators but for those tasked with developing appropriate compliance testing methodologies. In July 2016 the USCG denied the appeals of four manufacturers to accept Type Approval test data supported by a proposed alternative biological method using the Most Probable Number (MPN) assay to determine organism viability associated with treatment systems based on ultraviolet (UV) irradiation. Although MPN is a recognized procedure for single species with well-characterized growth media requirements, its more novel application to enumerate mixed assemblages found in ballast water has not yet been validated. USCG cited additional concerns with organisms rendered nonviable after treatment but capable of repairing damage caused by UV radiation, thus restoring reproductive capacity and viability. The USCG also stated that acceptance of the alternative method would effectively require a change to the discharge standard, since it measures a different end point and is therefore not equivalent to the standard specified in the regulatory requirements. Nucleic acid-based detection methods could conceivably provide a means to side-step some of the problematic issues inherent in the use of culture dilution methods, offering a more palatable alternative for regulatory consideration. Analysis of molecular targets has, in fact, enabled assessment of potentially sub-lethal and reparable UV damage to target organisms in other water treatment contexts, revealing the possibility of VBNC states in both bacterial and viral pathogens (Eischeid et al., 2009; Zhang et al., 2015). However, to our knowledge there have not yet been attempts to identify specific molecular targets capable of identifying organisms that may be alive and yet non-reproductive or otherwise unable to successfully establish new invasive populations.
3.3.2. Estimating abundance
Together, these studies suggest that assessment of viability is not an unreasonable future goal for nucleic acids-based ballast water monitoring, and that applications in the context of compliance testing are a distinct possibility. Since compliance testing requires not just recognizing viable targets but also counting them, the promise of nucleic acids-based tools is likely much higher for microbial indicators given the greater amenability of those targets to quantification. In fact, currently available technologies may already provide adequate means of quantifying viable cell counts for IMO-defined indicator species (Mehrabadi et al., 2012; Mendes Silva and Domingues, 2015), and application to ballast water compliance testing may be a matter of establishing effective protocols that meet the necessary challenge criteria. High sensitivity of qPCR and other probe-based technologies would very likely enable detection at thresholds well below desired discharge limits, a wide variety of potential nucleic acid targets provide options for measuring molecular activity tightly coupled to metabolic functions of interest, and well established protocols exist for forensic-level quality assurance of molecular genetic workflows. Such approaches could plausibly be expanded beyond the currently recognized indicator taxa, perhaps to consider other pathogenic species that pose considerable risk of introduction via ballast water (e.g. toxic phytoplankton or parasitic protozoa).
Even in the case of indicator taxa, nucleic acids-based detection methods must rely on indirect measures of target organism abundance. As noted above, in many cases this is not particularly problematic; single celled targets, for instance, are amenable to relatively precise calibration of nucleic acid concentration and cell counts. Such calibration becomes more difficult in the case of larger size classes; the >50 μm size class, in particular, represents a complex assemblage of diverse taxa, with associated variation in individual body mass, cell count, and metabolic activity (Darling and Blum, 2007). Although individual-based abundance counts remain the gold standard, it is worth noting that there is clear precedent for development of tools that only indirectly quantify organism abundance. In addition to the microbial methods discussed above, the ATP-based methods currently being explored in the context of ballast water compliance testing similarly depend on inference of abundance independent of direct organism counts, by determining correlation of the measured metabolite with target cell viability in standardized experiments (van Slooten et al., 2015). Conceivably, similar calibrations could be utilized to associate total viable organism counts of any size class with the activity of some universal metabolite—for instance, various “housekeeping” genes in the case of nucleic acids targets. Only extensive experimental study will determine whether such methods can be applied universally to samples that differ substantially not only in terms of abiotic variables (i.e. various challenge conditions) but also in terms of taxonomic diversity.
4. Conclusions
Ballast water monitoring comprises a suite of related tasks, including direct assessment of compliance with regulatory standards, surveillance for incursions of known high profile invasive species, and general research aimed at understanding the risks of invasion posed by ballast water-borne organisms. Each of these tasks poses particular questions, and each of those questions introduces criteria that must be met by the analytical tools employed to answer them (Table 2). Given the rapidly increasing application of nucleic acids-based methods for biodiversity assessment in a broad array of environmental management disciplines, it stands to reason that these tools represent a potential resource for increasing the effectiveness of ballast water monitoring efforts. While a number of these methods have already been utilized in the context of ballast water research and/or surveillance (Table 3), further refinement is required not only to better tailor genetic tools for ballast water applications (e.g. by addressing the particular challenge conditions unique to ballast water monitoring) but also to surmount existing technological shortcomings. A number of those shortcomings could be addressed with concerted research efforts aimed at modifying existing methods for use in ballast water settings—for instance, through development of probe-based methods for detecting specific target species likely to be transferred in ballast, or through rigorous calibration aimed at quantifying indicator organism abundance using qPCR or related approaches. Other limitations, such as gaps in existing reference sequence databases, will likely take longer to overcome and will be better addressed as part of broader research and development efforts. Nucleic acids-based tools for biodiversity assessment are only likely to increase in utility as technologies grow more accessible, and as molecular and bioinformatics workflows become more efficient and more standardized. Researchers and managers concerned with understanding ballast water biodiversity are thus presented with an opportunity to leverage these broader efforts to develop tools that enable more accurate, sensitive, and cost-effective means of understanding and mitigating invasion risks posed by ballast water.
Acknowledgements
The authors wish to thank members of the ICES Working Group on Ballast and Other Ship Vectors (WGBOSV) for constructive discussion of an earlier version of this work; specifically, we thank Sarah Bailey for the invitation to present to WGBOSV and to develop the current version of the paper for this special issue. The U.S. Environmental Protection Agency, through its Office of Research and Development, supported the work described here. Though it has been subjected to Agency administrative review and approved for publication, its content does not necessarily reflect official Agency policy.
References
- Adams J, Briski E, Ram J, Bailey S, 2014. Evaluating the response of freshwater organisms to vital staining. Manag. Biol. Invasions 5, 197–208. [Google Scholar]
- Ardura A, Zaiko A, Martinez JL, Samuiloviene A, Borrell Y, Garcia-Vazquez E, 2015. Environmental DNA evidence of transfer of North Sea molluscs across tropical waters through ballast water. J. Molluscan Stud. 81, 495–501. [Google Scholar]
- Aridgides LJ, Doblin MA, Berke T, Dobbs FC, Matson DO, Drake LA, 2004. Multiplex PCR allows simultaneous detection of pathogens in ships’ ballast water. Mar. Pollut. Bull. 48, 1096–1101. [DOI] [PubMed] [Google Scholar]
- Barnes MA, Turner CR, 2015. The ecology of environmental DNA and implications for conservation genetics. Conserv. Genet. 17, 1–17. [Google Scholar]
- Barry SC, Hayes KR, Hewitt CL, Behrens HL, Dragsund E, Bakke SM, 2008. Ballast water risk assessment: principles, processes, and methods. ICES J. Mar. Sci. 65, 121–131. [Google Scholar]
- Besansky NJ, Severson DW, Ferdig MT, 2003. DNA barcoding of parasites and invertebrate disease vectors: what you don’t know can hurt you. Trends Parasitol. 19, 545–546. [DOI] [PubMed] [Google Scholar]
- Bik HM, Porazinska DL, Creer S, Caporaso JG, Knight R, Thomas WK, 2012. Sequencing our way towards understanding global eukaryotic biodiversity. Trends Ecol. Evol. 27, 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchet S, 2012. The use of molecular tools in invasion biology: an emphasis on freshwater ecosystems. Fish. Manag. Ecol. 19, 120–132. [Google Scholar]
- Bohmann K, Evans A, Gilbert MTP, Carvalho GR, Creer S, Knapp M, Yu DW, de Bruyn M, 2014. Environmental DNA for wildlife biology and biodiversity monitoring. Trends Ecol. Evol. 29, 358–367. [DOI] [PubMed] [Google Scholar]
- Bott NJ, Ophel-Keller KM, Sierp MT, Herdina, Rowling KP, McKay AC, Loo MGK, Tanner JE, Deveney MR, 2010. Toward routine, DNA-based detection methods for marine pests. Biotechnol. Adv. 28, 706–714. [DOI] [PubMed] [Google Scholar]
- Briski E, Cristescu ME, Bailey SA, MacIsaac HJ, 2010. Use of DNA barcoding to detect invertebrate invasive species from diapausing eggs. Biol. Invasions 13, 1325–1340. [Google Scholar]
- Briski E, Ghabooli S, Bailey SA, MacIsaac HJ, 2016. Are genetic databases sufficiently populated to detect non-indigenous species? Biol. Invasions 18, 1911–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucklin A, Ortman BD, Jennings RM, Nigro LM, Sweetman CJ, Copley NJ, Sutton T, Wiebe PH, 2010. A “Rosetta Stone” for metazoan zooplankton: DNA barcode analysis of species diversity of the Sargasso Sea (Northwest Atlantic Ocean). Deep-Sea Res. II Top. Stud. Oceanogr. 57, 2234–2247. [Google Scholar]
- Burkholder JM, Hallegraeff GM, Melia G, Cohen A, Bowers HA, Oldach DW, Parrow MW, Sullivan MJ, Zimba PV, Allen EH, Kinder CA, Mallin MA, 2007. Phytoplankton and bacterial assemblages in ballast water of U.S. military ships as a function of port of origin, voyage time, and ocean exchange practices. Harmful Algae 6, 486–518. [Google Scholar]
- Carlton JT, Ruiz GM, Byers JE, Cangelosi A, Dobbs FC, Grosholz ED, Leung B, MacIsaac HJ, Wonham MJ, 2011. Assessing the Relationship between Propagule Pressure and Invasion Risk in Ballast Water. National Research Council.
- Chariton AA, Court LN, Hartley DM, Colloff MJ, Hardy CM, 2010. Ecological assessment of estuarine sediments by pyrosequencing eukaryotic ribosomal DNA. Front. Ecol. Environ. 8, 233–238. [Google Scholar]
- Costello C, Drake JM, Lodge DM, 2007. Evaluating an invasive species policy: ballast water exchange in the Great Lakes. Ecol. Appl. 17, 655–662. [DOI] [PubMed] [Google Scholar]
- Creer S, Fonseca VG, Porazinska DL, Giblin-Davis RM, Sung W, Power DM, Packer M, Carvalho GR, Blaxter ML, Lambshead PJ, Thomas WK, 2010. Ultrasequencing of the meiofaunal biosphere: practice, pitfalls and promises. Mol. Ecol. 19 (Suppl. 1), 4–20. [DOI] [PubMed] [Google Scholar]
- Cristescu ME, 2014. From barcoding single individuals to metabarcoding biological communities: towards an integrative approach to the study of global biodiversity. Trends Ecol. Evol. 29, 566–571. [DOI] [PubMed] [Google Scholar]
- Darling JA, Blum MJ, 2007. DNA-based methods for monitoring invasive species: a review and prospectus. Biol. Invasions 9, 751–765. [Google Scholar]
- Darling JA, Mahon AR, 2011. From molecules to management: adopting DNA-based methods for monitoring biological invasions in aquatic environments. Environ. Res. 111, 978–988. [DOI] [PubMed] [Google Scholar]
- Doblin MA, Drake LA, Coyne KJ, Rublee PA, Dobbs FC, 2004. Pfiesteria species identified in ships’ ballast water and residuals: a possible vector for introductions to coastal areas. In: Steidinger KA, Landsberg JH, Tomas CR, Vargo GA (Eds.), Harmful Algae 2002. Florida Fish and Wildlife Conservation Commission. [Google Scholar]
- Doblin MA, Coyne KJ, Rinta-Kanto JM, Wilhelm SW, Dobbs FC, 2007. Dynamics and short-term survival of toxic cyanobacteria species in ballast water from NOBOB vessels transiting the Great Lakes—implications for HAB invasions. Harmful Algae 6, 519–530. [Google Scholar]
- Drake JM, Lodge DM, 2004. Global hot spots of biological invasions: evaluating options for ballast-water management. Proc. R. Soc. Lond. B Biol. Sci. 271, 575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake LA, Meyer AE, Forsberg RL, Baier RE, Doblin MA, Heinemann S, Johnson WP, Koch M, Rublee PA, Dobbs FC, 2005. Potential invasion of microorganisms and pathogens via ‘interior hull fouling’: biofilms inside ballast water tanks. Biol. Invasions 7, 969–982. [Google Scholar]
- Egan SP, Barnes MA, Hwang C-T, Mahon AR, Feder JL, Ruggiero ST, Tanner CE, Lodge DM, 2013. Rapid invasive species detection by combining environmental DNA with light transmission spectroscopy. Conserv. Lett. 6, 402–409. [Google Scholar]
- Egan SP, Grey E, Olds B, Feder JL, Ruggiero ST, Tanner CE, Lodge DM, 2015. Rapid molecular detection of invasive species in ballast and harbor water by integrating environmental DNA and light transmission spectroscopy. Environ. Sci. Technol. 49, 4113–4121. [DOI] [PubMed] [Google Scholar]
- Eischeid AC, Meyer JN, Linden KG, 2009. UV disinfection of adenoviruses: molecular indications of DNA damage efficiency. Appl. Environ. Microbiol. 75, 23–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbrecht V, Leese F, 2015. Can DNA-based ecosystem assessments quantify species abundance? Testing primer bias and biomass-sequence relationships with an innovative metabarcoding protocol. PLoS One 10, e0130324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami K, Askari V, Ullrich M, Mohinudeen K, Anil AC, Khandeparker L, Burgess JG, Mesbahi E, 2012. Characterization of bacteria in ballast water using MALDI-TOF mass spectrometry. PLoS One 7, e38515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn JM, Brown EA, Chain FJ, MacIsaac HJ, Cristescu ME, 2015. Toward accurate molecular identification of species in complex environmental samples: testing the performance of sequence filtering and clustering methods. Ecol. Evol. 5, 2252–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto M, Moyerbrailean GA, Noman S, Gizicki JP, Ram ML, Green PA, Ram JL, 2014. Application of ion torrent sequencing to the assessment of the effect of alkali ballast water treatment on microbial community diversity. PLoS One 9, e107534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fykse EM, Nilsen T, Nielsen AD, Tryland I, Delacroix S, Blatny JM, 2012. Real-time PCR and NASBA for rapid and sensitive detection of Vibrio cholerae in ballast water. Mar. Pollut. Bull. 64, 200–206. [DOI] [PubMed] [Google Scholar]
- Gonzalez RA, Noble RT, 2014. Comparisons of statistical models to predict fecal indicator bacteria concentrations enumerated by qPCR- and culture-based methods. Water Res. 48, 296–305. [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Shokralla S, Zhou X, Singer GA, Baird DJ, 2011. Environmental barcoding: a next-generation sequencing approach for biomonitoring applications using river benthos. PLoS One 6, e17497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey JBJ, Hoy MS, Rodriguez RJ, 2009. Molecular detection of native and invasive marine invertebrate larvae present in ballast and open water environmental samples collected in Puget Sound. J. Exp. Mar. Biol. Ecol. 369, 93–99. [Google Scholar]
- Hebert PDN, Ratnasingham S, deWaard JR, 2003a. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B Biol. Sci. 270, S96–S99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Cywinska A, Ball SL, DeWaard JR, 2003b. Biological identifications through DNA barcodes. Proc. R. Soc. Lond. B Biol. Sci. 270, 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess-Erga OK, Blomvagnes-Bakke B, Vadstein O, 2010. Recolonization by heterotrophic bacteria after UV irradiation or ozonation of seawater; a simulation of ballast water treatment. Water Res. 44, 5439–5449. [DOI] [PubMed] [Google Scholar]
- Jerde CL, Mahon AR, Chadderton WL, Lodge DM, 2011. “Sight-unseen” detection of rare aquatic species using environmental DNA. Conserv. Lett. 4, 150–157. [Google Scholar]
- Jerde CL, Chadderton WL, Mahon AR, Renshaw MA, Corush J, Budny ML, Mysorekar S, Lodge DM, 2013. Detection of Asian carp DNA as part of a Great Lakes basin-wide surveillance program. Can. J. Fish. Aquat. Sci. 70, 522–526. [Google Scholar]
- Ji Y, Ashton L, Pedley SM, Edwards DP, Tang Y, Nakamura A, Kitching R, Dolman PM, Woodcock P, Edwards FA, Larsen TH, Hsu WW, Benedick S, Hamer KC, Wilcove DS, Bruce C, Wang X, Levi T, Lott M, Emerson BC, Yu DW, 2013. Reliable, verifiable and efficient monitoring of biodiversity via metabarcoding. Ecol. Lett. 16, 1245–1257. [DOI] [PubMed] [Google Scholar]
- Joachimsthal EL, Ivanov V, Tay ST, Tay JH, 2004. Bacteriological examination of ballast water in Singapore Harbour by flow cytometry with FISH. Mar. Pollut. Bull. 49, 334–343. [DOI] [PubMed] [Google Scholar]
- Keer JT, Birch L, 2003. Molecular methods for the assessment of bacterial viability. J. Microbiol. Methods 53, 175–183. [DOI] [PubMed] [Google Scholar]
- Keller RP, Drake JM, Drew MB, Lodge DM, 2011. Linking environmental conditions and ship movements to estimate invasive species transport across the global shipping network. Divers. Distrib. 17, 93–102. [Google Scholar]
- Kelly RP, Port JA, Yamahara KM, Crowder LB, 2014. Using environmental DNA to census marine fishes in a large mesocosm. PLoS One 9, e86175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermarrec L, Franc A, Rimet F, Chaumeil P, Frigerio J-M, Humbert J-F, Bouchez A, 2014. A next-generation sequencing approach to river biomonitoring using benthic diatoms. Freshwat. Sci. 33, 349–363. [Google Scholar]
- Kim Y, Aw TG, Teal TK, Rose JB, 2015. Metagenomic investigation of viral communities in ballast water. Environ. Sci. Technol. 49, 8396–8407. [DOI] [PubMed] [Google Scholar]
- Kunin V, Engelbrektson A, Ochman H, Hugenholtz P, 2010. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ. Microbiol. 12, 118–123. [DOI] [PubMed] [Google Scholar]
- Lee H, Reusser DA, Frazier M, 2013. Approaches to setting organism-based ballast water discharge standards. Ecol. Appl. 23, 301–310. [DOI] [PubMed] [Google Scholar]
- Li L, Mendis N, Trigui H, Oliver JD, Faucher SP, 2014. The importance of the viable but non-culturable state in human bacterial pathogens. Front. Microbiol. 5, 258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeque PK, Parry HE, Harmer RA, Somerfield PJ, Atkinson A, 2013. Next generation sequencing reveals the hidden diversity of zooplankton assemblages. PLoS One 8, e81327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge D, Turner CR, Jerde CL, Barnes MA, Chadderton L, Egan SP, Feder JL, Mahon AR, Pfrender ME, 2012. Conservation in a cup of water: estimating biodiversity and population abundance from environmental DNA. Mol. Ecol. 21, 2555–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Xiong H, Tang S, Yang Q, Li M, 2009. Comparison of the community structure of planktonic bacteria in ballast water from entry ships and local sea water in Xiamen Port. Prog. Nat. Sci. 19, 947–953. [Google Scholar]
- MacIntyre HL, Cullen JJ, 2016. Classification of phytoplankton cells as live or dead using the vital stains fluorescein diacetate and 5-chloromethylfluorescein diacetate. J. Phycol. 52, 572–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon AR, Barnes MA, Senapati S, Feder JL, Darling JA, Chang HC, Lodge D, 2011. Molecular detection of invasive species in heterogeneous mixtures using a microfluidic carbon nanotube platform. PLoS One 6, e17280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahon AR, Barnes MA, Li F, Egan SP, Tanner CE, Ruggiero ST, Feder JL, Lodge DM, 2013. DNA-based species detection capabilities using laser transmission spectroscopy. J. R. Soc. Interface 10, 20120637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallet J, Willmott K, 2003. Taxonomy: renaissance or Tower of Babel? Trends Ecol. Evol. 18, 57–59. [Google Scholar]
- Medinger R, Nolte V, Pandey RV, Jost S, Ottenwalder B, Schlotterer C, Boenigk J, 2010. Diversity in a hidden world: potential and limitation of next-generation sequencing for surveys of molecular diversity of eukaryotic microorganisms. Mol. Ecol. 19 (Suppl. 1), 32–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabadi JF, Morsali P, Nejad HR, Imani Fooladi AA, 2012. Detection of toxigenic Vibrio cholerae with new multiplex PCR. J. Infect. Public Health 5, 263–267. [DOI] [PubMed] [Google Scholar]
- Mendes Silva D, Domingues L, 2015. On the track for an efficient detection of Escherichia coli in water: a review on PCR-based methods. Ecotoxicol. Environ. Saf. 113, 400–411. [DOI] [PubMed] [Google Scholar]
- Meusnier I, Singer GA, Landry JF, Hickey DA, Hebert PD, Hajibabaei M, 2008. A universal DNA mini-barcode for biodiversity analysis. BMC Genomics 9, 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimura H, Katakura R, Ishida H, 2005. Changes of microbial populations in a ship’s ballast water and sediments on a voyage from Japan to Qatar. Mar. Pollut. Bull. 50, 751–757. [DOI] [PubMed] [Google Scholar]
- Ng C, Le TH, Goh SG, Liang L, Kim Y, Rose JB, Yew-Hoong KG, 2015. A comparison of microbial water quality and diversity for ballast and tropical harbor waters. PLoS One 10, e0143123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page TJ, Choy SC, Hughes JM, 2005. The taxonomic feedback loop: symbiosis of morphology and molecules. Biol. Lett. 1, 139–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagenkopp Lohan KM, Fleischer RC, Carney KJ, Holzer KK, Ruiz GM, 2016. Amplicon-based pyrosequencing reveals high diversity of protistan parasites in ships’ ballast water: implications for biogeography and infectious diseases. Microb. Ecol. 71, 530–542. [DOI] [PubMed] [Google Scholar]
- Patil JG, Gunasekera RM, Deagle BE, Bax NJ, Blackburn SI, 2005. Development and evaluation of a PCR based assay for detection of the toxic dinoflagellate, Gymnodinium catenatum (Graham) in ballast water and environmental samples. Biol. Invasions 7, 983–994. [Google Scholar]
- Pfrender ME, Hawkins CP, Bagley MJ, Courtney GW, Creutzburg BR, Epler JH, Fend S, Ferrington LC, Hartwell PL, Jackson S, Larsen DP, Levesque CA, Morse JC, Petersen MG, Ruiter D, 2010. Assessing macroinvertebrate biodiversity in freshwater ecosystems: advances and challenges in DNA-based approaches. Q. Rev. Biol. 85. [DOI] [PubMed] [Google Scholar]
- Pochon X, Bott NJ, Smith KF, Wood SA, 2013. Evaluating detection limits of next-generation sequencing for the surveillance and monitoring of international marine pests. PLoS One 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd’s Register, 2016. Understanding Ballast Water Management: Guidance for Ship Owners and Operators. third ed Lloyd’s Register Group, Ltd. [Google Scholar]
- Seebens H, Gastner MT, Blasius B, 2013. The risk of marine bioinvasion caused by global shipping. Ecol. Lett. 16, 782–790. [DOI] [PubMed] [Google Scholar]
- Shokralla S, Spall JL, Gibson JF, Hajibabaei M, 2012. Next-generation sequencing technologies for environmental DNA research. Mol. Ecol. 21, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Simmons M, Tucker A, Chadderton WL, Jerde CL, Mahon AR, Taylor E, 2016. Active and passive environmental DNA surveillance of aquatic invasive species. Can. J. Fish. Aquat. Sci. 73, 76–83. [Google Scholar]
- Stehouwer PP, Liebich V, Peperzak L, 2012. Flow cytometry, microscopy, and DNA analysis as complementary phytoplankton screening methods in ballast water treatment studies. J. Appl. Phycol. 25, 1047–1053. [Google Scholar]
- Steichen JL, Quigg A, 2015. Assessing the viability of microorganisms in the ballast water of vessels transiting the North Atlantic Ocean. Mar. Pollut. Bull. 101, 258–266. [DOI] [PubMed] [Google Scholar]
- Steichen JL, Schulze A, Brinkmeyer R, Quigg A, 2014. All aboard! A biological survey of ballast water onboard vessels spanning the North Atlantic Ocean. Mar. Pollut. Bull. 87, 201–210. [DOI] [PubMed] [Google Scholar]
- Stein ED, Martinez MC, Stiles S, Miller PE, Zakharov EV, 2014a. Is DNA barcoding actually cheaper and faster than traditional morphological methods: results from a survey of freshwater bioassessment efforts in the United States? PLoS One 9, e95525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein ED, White BP, Mazor RD, Jackson JK, Battle JM, Miller PE, Pilgrim EM, Sweeney BW, 2014. b. Does DNA barcoding improve performance of traditional stream bioassessment metrics? Freshwat. Sci 33, 302–311. [Google Scholar]
- Steinberg MK, Lemieux EJ, Drake LA, 2011. Determining the viability of marine protists using a combination of vital, fluorescent stains. Mar. Biol. 158, 1431–1437. [Google Scholar]
- Taberlet P, Coissac E, Pompanon F, Brochmann C, Willerslev E, 2012. Toward next-generation biodiversity assessment using DNA metabarcoding. Mol. Ecol. 21, 2045–2050. [DOI] [PubMed] [Google Scholar]
- Takahara T, Minamoto T, Yamanaka H, Doi H, Kawabata Z, 2012. Estimation of fish biomass using environmental DNA. PLoS One 7, e35868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JA, Dickinson MJ, Boonham N, 2010. Rapid detection of Phytophthora ramorum and P. kernoviae by two-minute DNA extraction followed by isothermal amplification and amplicon detection by generic lateral flow device. Phytopathology 100, 143–149. [DOI] [PubMed] [Google Scholar]
- Trebitz AS, Hoffman JC, Grant GW, Billehus TM, Pilgrim EM, 2015. Potential for DNA-based identification of Great Lakes fauna: match and mismatch between taxa inventories and DNA barcode libraries. Sci. Rep. 5, 12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USCG, 2012. Standards for living organisms in ships’ ballast water discharged in U. S. waters, final rule. Fed. Regist. 77, 17254–17320. [Google Scholar]
- USEPA, 2010. Generic protocol for the verification of ballast water treatment technology. EPA/600/R-1-/146.
- USEPA, 2013. 2013 Final issuance of National Pollution Discharge Elimination System (NPDES) Vessel General Permit (VGP) for discharges incidental to the normal operation of vessels.
- USEPA, ANSTF, 2013. Developing a comprehensive long-term research strategy to support determination of protective ballast water discharge standards.
- van Slooten C, Wijers T, Buma AGJ, Peperzak L, 2015. Development and testing of a rapid, sensitive ATP assay to detect living organisms in ballast water. J. Appl. Phycol. 27, 2299–2312. [Google Scholar]
- Viard F, David P, Darling JA, 2016. Marine invasions enter the genomic era: three lessons from the past, and the way forward. Curr. Zool. 62, 629–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright DA, Welschmeyer NA, Peperzak L, 2015. Alternative, indirect measures of ballast water treatment efficacy during a shipboard trial: a case study. J. Mar. Eng. Technol. 14, 1–8. [Google Scholar]
- Xu XW, Huo YY, Bai XD, Wang CS, Oren A, Li SY, Wu M, 2011. Kordiimonas lacus sp. nov., isolated from a ballast water tank, and emended description of the genus Kordiimonas. Int. J. Syst. Evol. Microbiol. 61, 422–426. [DOI] [PubMed] [Google Scholar]
- Zaiko A, Martinez JL, Schmidt-Petersen J, Ribicic D, Samuiloviene A, Garcia-Vazquez E, 2015a. Metabarcoding approach for the ballast water surveillance - an advantageous solution or an awkward challenge? Mar. Pollut. Bull. 92, 25–34. [DOI] [PubMed] [Google Scholar]
- Zaiko A, Martinez JL, Ardura A, Clusa L, Borrell YJ, Samuiloviene A, Roca A, Garcia-Vazquez E, 2015b. Detecting nuisance species using NGST: methodology shortcomings and possible application in ballast water monitoring. Mar. Environ. Res. 112, 64–72. [DOI] [PubMed] [Google Scholar]
- Zhan AB, Hulak M, Sylvester F, Huang X, Adebayo AA, Abbott CL, Adamowicz SJ, Heath DD, Cristescu ME, MacIsaac HJ, 2013. High sensitivity of 454 pyrosequencing for detection of rare species in aquatic communities. Methods Ecol. Evol. 4, 558–565. [Google Scholar]
- Zhan A, Xiong W, He S, Macisaac HJ, 2014a. Influence of artifact removal on rare species recovery in natural complex communities using high-throughput sequencing. PLoS One 9, e96928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan A, Bailey SA, Heath DD, Macisaac HJ, 2014b. Performance comparison of genetic markers for high-throughput sequencing-based biodiversity assessment in complex communities. Mol. Ecol. Resour. 14, 1049–1059. [DOI] [PubMed] [Google Scholar]
- Zhang S, Ye C, Lin H, Lv L, Yu X, 2015. UV disinfection induces a VBNC state in Escherichia coli and Pseudomonas aeruginosa. Environ. Sci. Technol. 49, 1721–1728. [DOI] [PubMed] [Google Scholar]
- Zhou X, Li Y, Liu S, Yang Q, Zhou L, Tang M, Fu R, Li J, Huang Q, 2013. Ultra-deep sequencing enables high-fidelity recovery of biodiversity for bulk arthropod samples without PCR amplification. Gigascience 2, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]