SUMMARY
Prevalence of diabetes and obesity in Mexican Pima Indians is low, while prevalence is high in Pima Indians in the United States (US); although lifestyle likely accounts for much of the difference, the role of genetic factors is not well-explored. To examine this, we genotyped 359 single nucleotide polymorphisms, including established type 2 diabetes and obesity variants from genome-wide association studies (GWAS) and 96 random markers, in 342 Mexican Pimas. A multimarker risk score of obesity variants was associated with body mass index (BMI; β = 0.81 kg/m2 per SD, P = 0.0066). The mean value of the score was lower in Mexican Pimas than in US Pimas (P = 4.3×10−11), and differences in allele frequencies at established loci could account for ~7% of the population difference in BMI; however, the difference in risk scores was consistent with evolutionary neutrality given genetic distance. To identify loci potentially under recent natural selection, allele frequencies at 283 variants were compared between US and Mexican Pimas, accounting for genetic distance. The largest differences were seen at HLA markers (e.g., rs9271720, difference = 0.75, P = 8.7×10−9); genetic distances at HLA were greater than at random markers (P = 1.6×10−46). Analyses of GWAS data in 937 US Pimas also showed sharing of alleles identical by descent at HLA that exceeds its genomic expectation (P = 7.0×10−10). These results suggest that, in addition to the widely-recognized balancing selection at HLA, recent directional selection may also occur, resulting in marked allelic differentiation between closely related populations.
Keywords: Type 2 Diabetes Mellitus, Obesity, HLA, American Indians
INTRODUCTION
Diabetes mellitus and obesity are common metabolic disorders in human populations. These disorders are related, as obesity is a strong risk factor for type 2 diabetes, which is by far the most common form of diabetes. Both conditions are strongly heritable (Willemsen et al., 2015, Elks et al., 2012), and their prevalence differs widely among human populations (Knowler et al., 1978, Kelly et al., 2008, King et al., 1998, Hanson et al., 1995). There has, thus, been a great deal of speculation about how evolutionary factors may have influenced these traits. In recent years, genome-wide association studies (GWAS) have identified a number of loci at which specific alleles are reproducibly associated with type 2 diabetes or obesity in humans (Zeggini et al., 2008, Saxena et al., 2012, Morris et al., 2012, Kooner et al., 2011, Tsai et al., 2010, Williams et al., 2014, Thorleifsson et al., 2009, Speliotes et al., 2010). Prevalence of type 2 diabetes and obesity in the Pima Indians of Arizona, USA, is extraordinarily high (Knowler et al., 1978, Hanson et al., 2015, Hanson et al., 1995), while their prevalence is much lower in Pima Indians from the village of Maycoba in Sonora, Mexico (Schulz et al., 2006, Esparza-Romero et al., 2015). The genetic distance between the populations is relatively small (Schulz et al., 2006, Tishkoff and Kidd, 2004), and, although the historical divergence time is uncertain, linguistic analysis suggests the populations diverged ~750 years ago (Hale, 1958). Lifestyle in the US Pimas is more “modern”, with greater access to technology and processed foods, while that in the Mexican Pimas is more “traditional”, with greater reliance on manual labor and locally-produced food. It is likely that these lifestyle differences account for much of the difference in prevalence of obesity and type 2 diabetes, but the contribution of genetic factors to these population differences has remained largely unexplored. In the present study, we analyze established and putative susceptibility variants for type 2 diabetes and obesity in Mexican Pimas and compare allele frequencies with those in US Pimas to determine the extent to which these established loci can account for the differences in disease prevalence. We also compare differences in frequency at these variants with those at randomly selected variants, as allele frequency differences that are greater than the genomic expectation between closely related populations can be an indication of recent natural selection (Price et al., 2009, Bhatia et al., 2011). The HLA locus is of particular interest for diabetes studies, as HLA variants are associated with type 2 diabetes in both Europeans and US Pimas (Saxena et al., 2012, Williams et al., 2011), and strongly associated with type 1 diabetes (Hu et al., 2015). As HLA is also a strong candidate for natural selection, we investigated this locus in more detail.
MATERIALS and METHODS
Participants
The study included participants in the Maycoba Project (Urquidez-Romero et al., 2014, Esparza-Romero et al., 2015), a survey to examine diabetes and obesity in the residents of Maycoba, a village in Sonora, Mexico, and the surrounding area; this area includes many Pima Indians, as well as Mexicans who are not Pimas. Surveys of individuals who were ≥ 18 years old were conducted in 1995 and 2010. For the present study, all participants in the 2010 survey with available DNA were selected for genotyping. This included 176 individuals whose heritage was full Pima by self-report (Pima-MX), 166 with partial Pima heritage (PrtPima-MX, defined as reporting at least one parent with Pima heritage, but not full heritage Pima), and 251 with no Indian heritage (NonInd-MX). For comparison with US Pimas, we also selected a random sample of 402 participants in a longitudinal study in Arizona who were full Pima heritage by self-report (Pima-US) and were ≥ 18 years old with available DNA (Knowler et al., 1978). For further comparison, an additional 212 participants in this Arizona longitudinal study were included whose heritage was full American Indian, but who reported no Pima heritage (AmInd-US); these were largely from other tribes in the southwestern US, and they were included to represent a “general” Amerindian population, across diverse tribal groups. Body mass index (BMI, kg/m2) was measured and diabetes was diagnosed with an oral glucose tolerance test. For additional details see Supplemental Methods.
Genotypes
Single nucleotide polymorphisms (SNPs) were genotyped by BeadXpress (Illumina, San Diego, CA) according to manufacturer’s instructions. We genotyped 47 “established” type 2 diabetes and 37 “established” BMI associated variants, identified as having associations at genome-wide statistical significance (P < 5.0×10−8) in “early” GWAS (largely before 2013) (Zeggini et al., 2008, Morris et al., 2012, Saxena et al., 2012, Kooner et al., 2011, Tsai et al., 2010, Williams et al., 2014, Thorleifsson et al., 2009, Speliotes et al., 2010). We also genotyped 48 “putative” Pima type 2 diabetes and 57 “putative” Pima BMI variants, which achieved suggestive significance (though generally not genome-wide significance) in our mapping studies in US Pimas (Hanson et al., 2007, Hanson et al., 2014, Muller et al., 2013, Bian et al., 2010, Malhotra et al., 2011, Bian et al., 2013, Traurig et al., 2009, Traurig et al., 2012). SNPs in HLA and TREH were included among variants with “putative” associations with type 2 diabetes in US Pimas. However, additional SNPs at these two loci, which have previously been studied in detail in US Pimas (Muller et al., 2013, Williams et al., 2011), were genotyped so that we could capture information about classical HLA alleles and capture haplotypes that predict plasma trehalase activity. We also typed 49 ancestry informative markers with large allele frequency differences between American Indians and Europeans (Tian et al., 2007). For comparative purposes, we typed 96 markers randomly selected from among those successfully genotyped in our GWAS (Malhotra et al., 2011). See Table S1 for a list of all markers.
Genetic Associations with Diabetes and Obesity
The associations between genotypes and type 2 diabetes and BMI were analyzed in the 342 Mexican Pimas who were either of full or partial Pima heritage. The association between diabetes and the number of “risk” alleles at each marker was analyzed using logistic regression, while association with BMI was similarly analyzed with linear regression. Additional details are given in Supplemental Methods.
Genetic Risk Scores
To test aggregate associations of “established” type 2 diabetes and obesity variants, multiallelic genetic risk scores were constructed. The scores were constructed by selecting one independent SNP for each locus, and this resulted in 42 SNPs for the diabetes score and 29 SNPs for the BMI score. The genetic risk score (GRS) over g established variants was calculated as:
Where βi is the effect size at the ith SNP, and li is the number of risk alleles carried by the individual at the ith SNP. Effect sizes for type 2 diabetes were taken as the logarithms of the odds ratios from large meta-analyses (Hanson et al., 2015), while those for BMI were taken as the standardized regression coefficients in the GIANT meta-analysis (Locke et al., 2015). A t-test was used to compare mean values of genetic risk scores between populations; an empirical bootstrap method was used to account for genetic distances. Additional details are given in Supplemental Methods.
Comparisons across Major Continental Populations
To place comparisons in the context of those among other populations constituting major continental groups, we obtained genotypic data for the same markers genotyped in Mexican Pimas for several populations from the HapMap Project (The International HapMap Consortium. 2005). These populations included Europeans from the Centre d’Etude du Polymorphism Humain families in Utah (CEU), East Asians from Han Chinese in Beijing (CHB), Africans from the Yoruba in Ibadan, Nigeria (YRI), and individuals of Mexican ancestry from Los Angeles (MEX). Genotypic data were obtained from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/) or, if not available from HapMap, from the 1000 Genomes Project (http://www.1000genomes.org/).
Genetic Attributable Fraction
To estimate the extent to which differences in allele frequencies at established obesity or type 2 diabetes variants may explain the population difference in mean BMI, or diabetes prevalence, the genetic attributable fraction (GAF) was calculated (Hanson et al., 2015). We define the GAF as the proportion of the difference in mean BMI (or in diabetes prevalence) between a high-risk “target” population (US Pimas) and a lower risk “reference” population (Mexican Pimas) that can be explained by differences in allele frequencies across established loci. Full details are given in Supplemental Methods.
Analyses of Allele Frequency Differences
As identification of variants for which the difference in allele frequency between closely related populations exceeds that expected under evolutionary neutrality can provide a powerful test for selection (Price et al., 2009, Bhatia et al., 2011), we analyzed allele frequency differences between full-heritage Mexican and US Pimas (Pima-MX and Pima-US) for each of the 283 SNPs with minor allele frequency > 0.05 (excluding admixture markers). We arbitrarily selected one allele at each SNP and calculated, by allele counting, its frequency in Mexican Pimas (fMX), frequency in US Pimas (fUS) and frequency in both populations combined (fT); we also calculated the absolute value of the allele frequency difference between populations (|δ|). The test for statistical significance of the allele frequency difference for each SNP was taken as:
Where the summation is over the g randomly selected markers (Price et al., 2009). This quantity follows a χ2 distribution on 1 degree of freedom that is subject to a genomic control procedure which accounts for the genetic distance between populations, as well was for stratification due to admixture or the presence of related individuals (Price et al., 2009). The false discovery rate (FDR) procedure was used to assess statistical significance, with control for multiple statistical tests (Benjamini and Hochberg, 1995). The genomic control procedure performs optimally when the genetic distance between populations is relatively close (e.g., FST<0.01), and with larger genetic distances, variation in allele frequency differences can be larger than expected under genomic control. To account for this, we also calculated the P-value empirically by simulation for the SNPs with large |δ| values. Full details are given in Supplemental Methods.
To further examine the observed allele frequency differences in a genomic context, we obtained GWAS data from the Human Genome Diversity Project (HGDP) (Li et al., 2008)). This constitutes data on 660,918 SNPs typed on the Illumina 650Y array on 1043 individuals from 51 populations around the world. We selected 27 populations with ≥ 15 genotyped individuals, and calculated FST across 2637 markers selected randomly from 2 Mb segments assigned across all autosomes (resulting in ~1 Mb between markers). Allele frequency differences were calculated across all autosomal SNPs with an average minor allele frequency >0.05 for each pair of populations for which FST was 0.0296–0.0425 (FST ± one standard error between Mexican and US Pimas); this resulted in 21,830,844 comparisons across 40 pairs of populations. The proportion of comparisons with |δ| ≥ the observed value between Mexican and US Pimas was taken as a measure of the genomic expectation for populations at comparable genetic distance.
Genetic Distances
To summarize allele frequencies differences across multiple markers, the co-ancestry coefficient (FST) was calculated as a measure of genetic distance between populations. FST represents the proportion of variance in allele frequency in the combined population explained by membership in the subpopulations, and it was calculated by the method of Hudson (Hudson et al., 1992), as this method provides valid evolutionary inferences when sample sizes differ between populations (Bhatia et al., 2013). We compared FST calculated across diabetes, obesity or HLA markers with that calculated across the randomly selected markers. For statistical significance tests, the standard error of the difference between FST across the markers of interest and FST across random markers was calculated by a bootstrap procedure. Individuals from each population were resampled, with replacement, in each iteration to construct the studied sample size; to account for variation in marker selection, a new set of random markers was also selected by resampling the same number of random markers in each iteration. A value of FST which is significantly higher than that at random markers is consistent with differential directional selection, while an FST significantly lower than that at random markers is consistent with balancing selection, or with concurrent directional selection across populations (Suzuki, 2010).
Excess Sharing of Alleles Identical by Descent
Directional selection results in excess sharing of alleles IBD, particularly among distantly related individuals. Thus, identification of regions where the mean proportion of alleles shared IBD among pairs of individuals significantly exceeds its genomic average can provide a powerful test for selection (Albrechtsen et al., 2010, Han and Abney, 2013). We, therefore, analyzed locus-wise IBD sharing among 937 full-heritage US Pimas who had participated in a GWAS and thereby had suitable data for estimation of IBD (Malhotra et al., 2011). Genotypic data for 398,430 autosomal SNPs, generated on the Affymetrix Genome-wide Human SNP Array 6.0 (Affymetrix, Santa Clara, CA) were analyzed. For each pair of individuals (n=437,691, excluding first degree relatives), the proportion of alleles shared IBD was estimated with BEAGLE as described previously (Browning and Browning, 2010, Hsueh et al., 2017). We compared the mean IBD observed at each genomic location with its genome-wide average. Details are given in Supplemental Methods. To further investigate natural selection at particular SNPs, extended haplotype homozygosity (EHH) scores were calculated in these US Pimas (n=506 after exclusion of first degree relatives) using SELSCAN (Sabeti et al., 2002, Szpiech and Hernandez, 2014). Details are given in Supplemental Methods.
RESULTS and DISCUSSION
Diabetes and Obesity Variants in Mexican Pima Indians
We analyzed associations with diabetes and BMI in Mexican Pima Indians, including 176 full-heritage Pimas and 166 partial-heritage Pimas (see Table S2 for characteristics of participants). Results for nominally statistically significant (P<0.05) associations are shown in Table 1, and results for all markers in Tables S3 and S4. The established type 2 diabetes variant at CDKAL1 was significantly associated with diabetes in Mexican Pimas in a direction consistent with the established association, while obesity-susceptibility variants in NEGR1, BDNF and FAIM2 were similarly associated with BMI. This suggests that these diabetes and obesity variants may be particularly important in Mexican Pimas. However, the effect sizes of all the variants tested were modest in the original GWAS in which they were identified, and the current sample size of Mexican Pimas is small, so power to detect statistically significant associations with individual markers is low. We, thus, proceeded to analyze multiallelic genetic risk scores for type 2 diabetes and BMI.
Table 1.
SNPS with statistically significant (P<0.05) association with diabetes or BMI in Mexican Pima Indians
| Associations with Diabetes | Proportion with Diabetes (N) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Class | Chr | Mb | Gene | SNP | R/N | Freq PimaUS | Freq PimaMX | RR | NR | NN | OR (95% CI) | P-val |
| Established | 6 | 20.68 | CDKAL1 | rs7756992 | C/T | 0.32 | 0.58 | 0.13(82) | 0.08 (149) | 0.03 (92) | 2.10 (1.05, 4.17) | 0.0347 |
| Putative | 14 | 69.07 | RAD51L1 | rs4902613 | G/A | 0.50 | 0.42 | 0.13(52) | 0.09 (169) | 0.03 (102) | 2.41 (1.17, 4.98) | 0.0174 |
| Putative | 14 | 69.12 | RAD51L1 | rs4899250 | T/C | 0.50 | 0.43 | 0.16(50) | 0.09 (176) | 0.03 (94) | 2.60 (1.24, 5.44) | 0.0112 |
| Associations with BMI | Mean BMI in kg/m2 (N) | |||||||||||
| Class | Chr | Mb | Gene | SNP | R/N | Freq PimaUS | Freq PimaMX | RR | NR | NN | β (95% CI) | P-val |
| Established | 1 | 72.81 | NEGR1 | rs2815752 | A/G | 0.92 | 0.67 | 27.72(181) | 26.66 (115) | 26.3 (30) | 1.08 (0.25, 1.90) | 0.0103 |
| Putative | 2 | 210.24 | MAP2 | rs12475149 | G/T | 0.83 | 0.85 | 27.69(190) | 26.27 (118) | 27.0 (12) | 1.43 (0.42, 2.43) | 0.0053 |
| Established | 11 | 27.67 | BDNF | rs925946 | T/G | 0.19 | 0.23 | 29.01(11) | 27.78 (134) | 26.7 (180) | 1.06 (0.10, 2.02) | 0.0312 |
| Established | 12 | 50.25 | FAIM2 | rs7138803 | T/C | 0.12 | 0.24 | 30.05(20) | 27.65 (117) | 26.6 (188) | 1.23 (0.33, 2.13) | 0.0076 |
Chr is chromosome, Mb is the physical position in megabases (build37), R/N represent the two alleles with the risk allele listed first. Frequencies are calculated for the risk allele. RR, NR and NN represent the number of individuals homozygous for the risk allele, heterozygous and homozygous for the low risk allele respectively. OR is the odds ratio (per copy of the risk allele) and β is the regression coefficient (kg/m2 per copy of the risk allele). Results are adjusted for age, sex and Amerindian admixture.
Analysis of Genetic Risk Scores for Diabetes and BMI
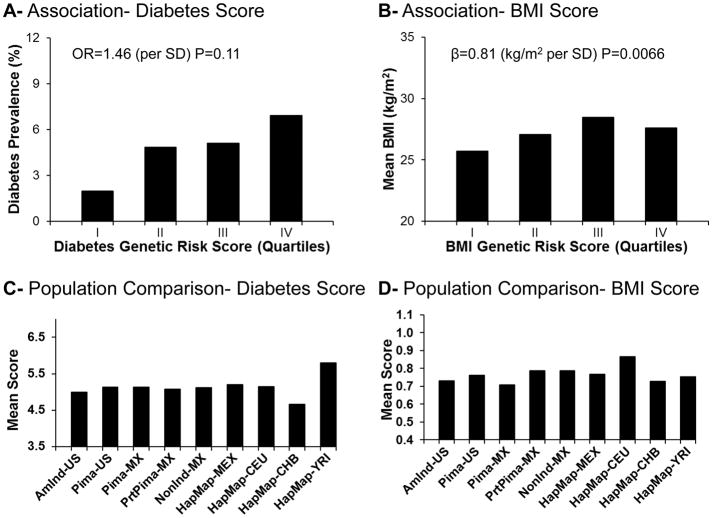
We constructed multiallelic genetic risk scores, weighted by the published effect size for each locus, across all established type 2 diabetes and obesity variants, and we analyzed these for association with diabetes and BMI in Mexican Pimas. The diabetes genetic risk score was associated with higher diabetes prevalence (odds ratio [OR]=1.46 per SD, 95% confidence interval [CI], 0.94–2.26), but this association was not statistically significant (P=0.11, Figure 1A). On the other hand, a higher BMI genetic risk score was significantly associated with BMI with an effect of 0.81 kg/m2 per SD (95% CI 0.27–1.34, P=0.0066, Figure 1B). These analyses suggest that established obesity alleles in aggregate influence BMI even in the context of a “traditional” lifestyle in a population with a mean BMI of 27.2 kg/m2.
Figure 1.
A: Association between type 2 diabetes genetic risk score and diabetes prevalence in Mexican Pima Indians. Prevalence of diabetes, adjusted for age, sex and European admixture, is shown by quartile of the genetic risk score. B: Association between obesity genetic risk score and BMI in Mexican Pima Indians. Mean BMI, adjusted for age, sex and European admixture, is shown by quartile of the genetic risk score. C: Mean values of the type 2 diabetes genetic risk score by population. D: Mean values of the obesity genetic risk score by population.
To evaluate the extent of divergence between populations in genetic risk for type 2 diabetes and BMI, we compared the mean value of the genetic risk score across populations. Mean genetic risk scores for type 2 diabetes were comparable between Mexican and US Pimas (Figure 1C, Online Supplement Table S5), while the highest values were observed in Africans. The mean obesity genetic risk score was significantly lower in Mexican Pimas than in US Pimas (P=4.3×10−11 by t-test); the highest values for the BMI genetic risk score were observed in Europeans (Figure 1D). To limit the potential influence of unidentified European admixture, we repeated the analysis with exclusion of those with >10% European ancestry according to genetic estimates, and the differences between Mexican and US Pimas remained highly significant (P=9.2×10−9).
To evaluate the extent to which differences in allele frequencies at established susceptibility variants between populations could explain differences in diabetes or obesity risk, we estimated the genetic attributable fraction (GAF), or the proportion of the difference in population prevalence for type 2 diabetes between US and Mexican Pimas, attributable to differences in allele frequencies (Hanson et al., 2015). The age-sex adjusted prevalence of diabetes in Mexican Pimas was 9.4%, while that in US Pimas was 52.9% (OR=10.8, P=3.0×10−13); the GAF for the difference between US and Mexican Pimas was 0.2% (P=0.94), and this suggests that allele frequencies across these established type 2 diabetes susceptibility variants do not account for a significant portion of the population difference in diabetes prevalence. The age-sex adjusted mean BMI was 26.9 kg/m2 in Mexican Pimas and 35.2 kg/m2 in US Pimas (P=2.5×10−24); the GAF for the difference in mean BMI is 7.3% (P=1.1×10−6), and this suggests that differences between US Pimas and Mexican Pimas in frequencies of established obesity variants can account for a modest but significant portion, about 7% (0.6 kg/m2), of the difference in mean BMI between populations. Thus, differences in obesity between the populations may not be wholly attributable to the well-documented lifestyle differences (Schulz et al., 2006). For these analyses, we weighted alleles according to effect sizes observed in European populations, but the optimal weights for Amerindian populations are not known. These analyses are also based on diabetes and BMI variants identified in the first wave of GWAS. Additional variants have been identified for both traits (Mahajan et al., 2014, Locke et al., 2015), and it is likely that many more remain unidentified; it is uncertain if the same GAF results would be obtained with inclusion of additional variants. However, these first wave variants have the strongest effect sizes in Europeans, and these effect sizes are often comparable across diverse populations (Hanson et al., 2015, Carlson et al., 2013), thus, these variants have large potential individual contributions to population differences in risk.
The variance estimate used in the standard t-test for differences in means between populations does not take genetic distance into account; therefore, while a significant result reflects differences in the mean values of the risk scores, such differences may arise on the basis of the genetic distance between populations, and thus, may be consistent with the effects of neutral variation across markers rather than selection. To account for this, we constructed an empirical expectation for the differences in genetic risk scores using a bootstrap procedure. When P-values were thus calculated empirically, none of the differences between populations for either the diabetes or BMI risk scores achieved statistical significance after correction for the number of pairwise tests (P<0.0014, given 36 pairwise tests for each score). This indicates that the extent of the genetic differences in diabetes or obesity risk among populations for these markers is consistent with evolutionary neutrality, given the genetic distances. The strongest difference was observed in diabetes risk between East Asian and African populations (empirical P=0.0069). Previous studies using a smaller set of diabetes variants and a larger number of populations reported a similar, but significant, gradient in type 2 diabetes genetic risk from African to Asian (and Amerindian) populations, and suggested that this reflects the effects of differential selection (Klimentidis et al., 2011, Corona et al., 2013). On the other hand, analyses of homozygosity and the extent of linkage disequilibrium across established type 2 diabetes loci have not generally suggested selection at these loci (Ayub et al., 2014).
Allele Frequency Differences between Mexican and US Pimas
While the analyses of directional allelic differentiation suggest that differences between Mexican and US Pimas across type 2 diabetes and BMI variants are generally consistent with neutrality, individual loci may have been subject to selection. To assess allelic differentiation at individual variants, we compared allele frequencies between Mexican and US Pimas, using a “genomic control” procedure based on the 96 randomly selected markers to account for genetic distance. With adjustment for the number of markers analyzed (n = 283) by the FDR procedure, five variants had significant (FDR < 0.05) differences between Mexican and US Pimas (Table 2A); results for all variants are shown in Table S6. Four of the significantly differentiated variants (rs9271720, rs9272219, rs9268858, rs502771) were in the HLA-DR/DQ region, while one (rs117619140) was in TREH. While the FDR procedure evaluates experiment-wise statistical significance, some of the HLA variants achieved genome-wide significance [P<5×10−7, based on the number of effectively independent variants estimated from GWAS data in US Pimas (Malhotra et al., 2011)]. Although the genomic control procedure is generally expected to account for admixture and other demographic factors, allele frequency differences can still be subject to residual confounding. To assess robustness of the genomic control procedure, we also calculated P-values empirically from data simulated under a model of neutral genetic “drift”. These 5 markers showed empirical P-values comparable to those obtained with genomic control. In our primary analyses we classified individuals according to self-reported ethnicity, but similar results were obtained when analyses were restricted to those whose genetic ancestry estimate was >90% Amerindian (Table S7). This suggests the results are unlikely to be confounded by European admixture, but the present data do not allow estimation of admixture from other Amerindian groups. While our results suggest that the degree of allelic differentiation observed at these loci is highly unlikely under a simple model of genetic “drift”, a demographic explanation cannot be entirely excluded.
Table 2.
Allele frequency differences between Mexican and US Pima Indians for statistically significant (FDR<0.05) SNPs and for haplotypes tagging low-resolution HLA-DRB1 alleles.
| A. Allele frequency differences for SNPs with statistically significant differences between populations | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Mb | Gene | SNP | Alleles | fMX | fUS | |δ| | P-val* | FDR | Empirical P-val† |
| 6 | 32.59 | HLA | rs9271720 | T/C | 0.15 | 0.90 | 0.75 | 8.7×10−9 | 0.000033 | <4.0×10−8 |
| 6 | 32.60 | HLA | rs9272219 | T/G | 0.36 | 0.93 | 0.58 | 1.2×10−6 | 0.000691 | 1.4×10−5 |
| 6 | 32.43 | HLA | rs9268858 | T/C | 0.38 | 0.94 | 0.55 | 2.4×10−6 | 0.000769 | 7.4×10−6 |
| 6 | 32.58 | HLA | rs502771 | T/C | 0.76 | 0.17 | 0.59 | 7.9×10−6 | 0.001521 | 6.0×10−6 |
| 11 | 118.53 | TREH | rs117619140 | A/C | 0.47 | 0.07 | 0.40 | 2.3×10−4 | 0.022371 | 3.7×10−4 |
| B. Frequency differences for low-resolution HLA-DRB1 alleles inferred by haplotypes | ||||||||||
| Chr | Mb | Gene | Haplotype(s) | Allele | fMX | fUS | |δ| | P-val* | ||
| 6 | 32.55 | HLA | CTCT/CTCG | DRB1*04 | 0.53 | 0.06 | 0.47 | 2.8×10−5 | ||
| 6 | 32.55 | HLA | TTCT | DRB1*08 | 0.20 | 0.03 | 0.17 | 3.2×10−2 | ||
| 6 | 32.55 | HLA | TCTT | DRB1*14 | 0.14 | 0.83 | 0.69 | 3.8×10−7 | ||
| 6 | 32.55 | HLA | TTTT/TTCG | DRB1*16 | 0.04 | 0.07 | 0.03 | 6.0×10−1 | ||
Chr is chromosome, Mb is the physical position in megabases (build37), For table 2A, “Alleles” represent the two bases, frequencies are calculated for the allele listed first. For table 2B, “Allele” represents the low-resolution allele inferred by the designated haplotype(s); bases for the haplotype are given in the following order: rs9268858, rs502771, rs9271720, rs9272219.
P-value calculated with genomic control procedure.
P-value calculated empirically by simulation.
Based on previous HLA typing conducted in US Pimas (Williams et al., 2009) the four HLA SNPs with significant frequency differences between Mexican and US Pimas tag all of the observed common classic “low-resolution” HLA-DRB1 alleles. Frequency differences between Mexican and US Pimas for these low-resolution alleles, inferred from haplotypes, are shown in Table 2B. The frequency differences across the individual SNPs largely reflect differences at HLA-DRB1*14. This allele is relatively common in Amerindian populations, but uncommon in other populations, and, as described previously, has an extraordinarily high frequency (0.83) in US Pimas (Williams et al., 2009); we find its frequency is much lower (0.14) in Mexican Pimas, among whom the most common allele is HLA-DRB1*04.
Many studies have suggested that recent natural selection has occurred at HLA in human populations (Black and Hedrick, 1997, Hedrick, 1998, Meyer and Thomson, 2001, Solberg et al., 2008, Meyer et al., 2018). Variants in HLA have been associated with numerous autoimmune diseases, including type 1 diabetes; there are associations with type 2 diabetes as well in both Europeans and in US Pimas (Saxena et al., 2012, Williams et al., 2011). The variant associated with type 2 diabetes, however, did not differ in frequency between the Mexican and US Pimas (rs9268852, |δ|=0.05, P=0.41).
In contrast to HLA, to our knowledge previous studies in humans have not implicated natural selection at TREH, which encodes for trehalase, an enzyme that digests trehalose, a sugar present in some foods including desert plants and mushrooms. Variants in TREH are strongly associated with plasma trehalase activity and modestly associated with type 2 diabetes in US Pimas (Muller et al., 2013). The type 2 diabetes-associated variant did not differ significantly between US and Mexican Pimas (rs558907, P=0.71), but rs117619140 did differ significantly between populations. The A allele, which is more frequent in Mexican than in US Pimas, is associated with much higher plasma trehalase activity (Muller et al., 2013). Differential selection on trehalase activity is a potential explanation for the allele frequency differences we observe between US and Mexican Pimas.
Comparisons with HDGP Data
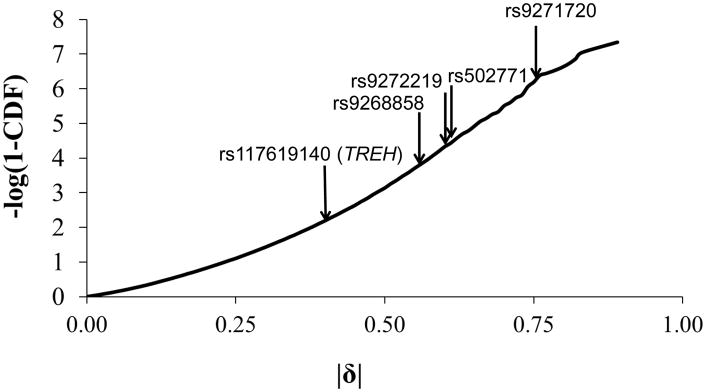
Dense genotypic genome-wide data are not available for the Mexican Pimas. Therefore, to obtain a global genomic context for these allele frequency differences, we compared them with differences observed between populations at comparable genetic distances in GWAS data from the HDGP. The distribution of |δ|, calculated across 21,830,844 SNP-wise comparisons for 40 pairs of populations is shown in Figure 2. On a genome-wide basis, it is very unusual to observe allele frequency differences of the magnitude we observed between US and Mexican Pimas at the HLA-DR/DQ markers between other populations at comparable genetic distances. For rs9271720, for which |δ|=0.75 between Mexican and US Pimas, the proportion of SNPs at which differences of this magnitude were observed between HGDP populations was 5.9×10−7. On the other hand, allele frequency differences as great or greater than those observed between Mexican and US Pimas at the TREH SNP rs117619140 (|δ|=0.40) were more common, occurring at a proportion of 0.003. Thus, in a genomic context, the evidence for differential natural selection between Mexican and US Pimas at HLA is particularly strong. The evidence at TREH is weaker, and further studies are required to establish selection at TREH with greater confidence.
Figure 2.
Cumulative distribution function (CDF) of allele frequency differences between Human Genome Diversity Project populations with FST 0.0296–0.0425. The negative base 10 logarithm of 1-CDF is shown; higher values indicate more unusual allele frequency differences. Arrows indicate allele frequency differences between Mexican and US Pimas observed for the SNP of interest.
Analyses of Genetic Distances
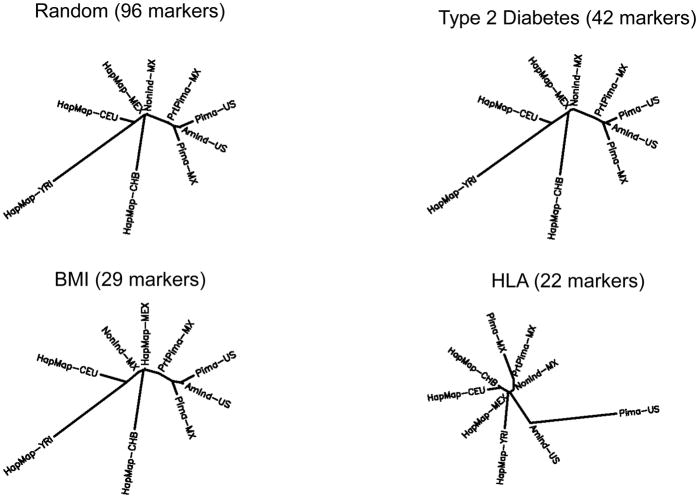
To further assess allelic differentiation across multiple genetic markers, we analyzed FST across BMI, diabetes and HLA markers. Results of these analyses are shown in Figure 3 and Table S8. None of the differences between FST values calculated across the BMI markers and FST values across random markers achieved statistical significance after correction for the number of pairwise comparisons (P<0.0014), while for the type 2 diabetes markers, the only significant difference was seen between the US Indians who were not Pimas and Africans (YRI). The Mexican and US Pimas, however, were much more highly divergent at the HLA markers than at the random markers (FST=0.229 versus FST=0.036, P=1.6×10−46). The distance between US Pimas at the HLA markers was also significantly greater than that at random markers for several other populations, and the distance between Mexican Pimas and US Indians who were not Pimas at HLA markers was also greater than at random markers (FST=0.086 versus 0.034, P=7.3×10−6). Differences in genetic distances that are greater than expected are often reflective of differential directional selection between populations and the tests for allele frequency differences between closely related populations presented above are also designed to detect differential directional selection (Price et al., 2009, Suzuki, 2010, Bhatia et al., 2011). Directional selection occurs when one allele is favored (or disfavored) such that allelic diversity is lost at a faster rate than under neutrality. The prevailing theory among many population geneticists, however, is that HLA has been subject to balancing selection, as this can account for the high degree of heterozygosity, the large number of common alleles observed, and the similarity of many allele frequencies across populations (Black and Hedrick, 1997, Hedrick, 1998, Meyer and Thomson, 2001, Solberg et al., 2008, Meyer et al., 2018). Balancing selection is a type of natural selection in which allelic diversity is maintained for a longer time than expected under neutral genetic “drift” (e.g., if there is a heterozygote advantage). For several global populations, we observed that FST values were significantly smaller at the HLA markers than at random markers (e.g, CEU and CHB, YRI and CEU); this is consistent with long-term balancing selection at HLA. In this context, the large differences we observe between Mexican and US Pimas, seem particularly striking.
Figure 3.
Dendrograms summarizing genetic distances between populations for random markers, type 2 diabetes variants, obesity variants and HLA variants. FST values were taken as genetic distances and dendrograms were generated with PHYLIP.
Analyses of Identity-by-Descent in US Pimas
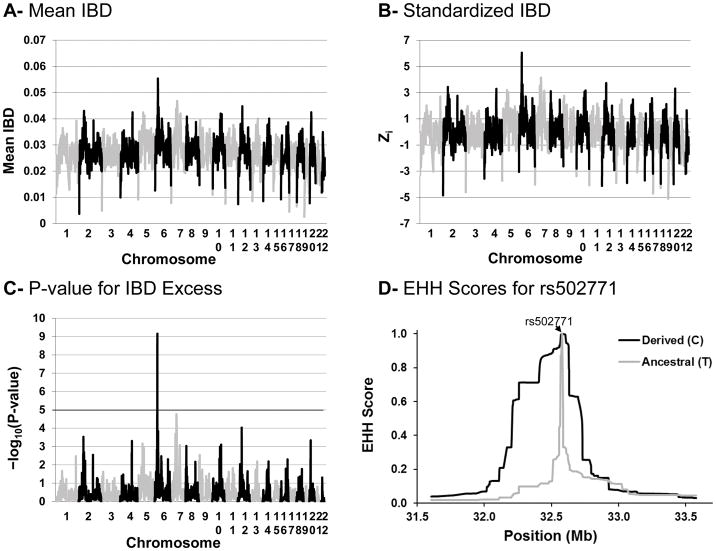
Identification of regions at which the proportion of alleles shared identical by descent (IBD) between pairs of individuals in a population greatly exceeds IBD at other regions of the genome can also provide a powerful test for directional selection (Albrechtsen et al., 2010, Han and Abney, 2013). Like tests of allelic differentiation, but in contrast to many other methods, analysis of IBD can detect selection when it occurs on standing variation, rather than on a new mutation or previously rare variant (Albrechtsen et al., 2010); however, since comparison is made with genomic IBD sharing within a population, this method is more robust to demographic factors that differ across populations than tests of allele frequency differences. Since GWAS data suitable for calculation of IBD based on phased haplotypes were available in a separate sample constituting 937 full-heritage US Pimas (Malhotra et al., 2011), we analyzed these GWAS data to determine if IBD at the HLA region was increased relative to the rest of the genome. As shown in Figure 4, the highest mean IBD across the genome was observed on chromosome 6p in the HLA region (30.18 Mb); the mean IBD at this region was 0.055, whereas the genomic average was 0.027 (standardized Z=6.06, P=7.0×10−10). With correction for the number of independent regions tested, the HLA region was the only one showing a statistically significant increase in IBD (P<1.5×10−5).
Figure 4.
Genome-wide analysis of IBD sharing in 937 US Pimas. A: Mean value of the proportion of alleles shared IBD by chromosomal location in all 437,691 pairs of individuals. B: Allele sharing score (Zi), standardized for the expected sharing within pairs and for the genomic expectation across all loci. C. P-value for the null hypothesis that the proportion of alleles shared IBD is within its genomic expectation against the alternative that it exceeds its expectation at each genomic location. Results are plotted by physical location (build 37) on each chromosome. D. Extended haplotype homozygosity (EHH) scores for alleles at rs502771. EHH scores represent the probability that two haplotypes carrying a given allele (derived or ancestral) at rs502771, selected randomly from the population, are homozygous at a given location and at all intervening SNPs (and thus inherited IBD).
To further explore the possibility for natural selection at individual markers, we analyzed EHH scores. We found that linkage disequilibrium with the derived allele at rs502771 (C, frequency=0.83 in US Pimas), which is highly concordant with HLA-DRB1*14, occurs over a longer range than with the ancestral allele (Figure 4D). This combination of high allele frequency and extended long-range haplotypes [higher EHH scores across greater distances (with a difference between scores for derived and ancestral alleles of 0.75 at distances >100 kb)] is consistent with recent directional selection around HLA-DRB1*14. This is an unusual pattern in US Pimas- of 1168 chromosome 6 SNPs outside the HLA region with comparable derived allele frequency (0.80–0.86) only 1.4% have EHH score differences ≥ 0.75 extending > 100 kb. Given this pattern of EHH scores, the excess of IBD sharing and the allele frequency differences between US and Mexican Pimas that are much greater than expected given the genetic distance, recent directional selection at HLA seems the most likely explanation for these findings.
Implications
The present study demonstrates significant differences in the frequencies of established obesity variants between Mexican and US Pimas. Although our analyses suggest that these differences are consistent with neutral genetic “drift”, or demographic factors, they nonetheless illustrate the importance of measuring genetic risk even when there are large environmental differences between groups. We also demonstrate marked allele frequency differences at HLA between Mexican and US Pimas, which are consistent with recent differential directional selection. Although balancing selection has been widely observed at HLA, recently, some studies have suggested that directional selection has occurred as well (Bhatia et al., 2011, Kawashima et al., 2012). The magnitude of allele frequency differences observed in earlier studies (|δ|≈0.3) is smaller than that observed between Mexican and US Pimas. Thus, the present study provides further evidence that directional selection has also shaped the genetic landscape at HLA, alongside balancing selection. This directional selection may result in marked allelic differentiation between closely related population, and is perhaps illustrative of the powerful and diverse influence of natural selection at HLA. One caveat is that, although “overdominance” (i.e., a heterozygote advantage) has often been considered the most likely mechanism by which balancing selection at HLA occurs, “frequency-based” models, (i.e., whether an allele is favored depends on its frequency, such as when it is favored when rare, but disfavored once it becomes very common) provide an equally good fit to the data (Meyer and Thomson, 2001, Takahata and Nei, 1990). Since “frequency-based” balancing selection operates as directional selection over the short-term, it can be difficult to distinguish between these possibilities. In addition, the highly differentiated HLA variants are not those previously associated with diabetes, and it is not clear whether natural selection at HLA has resulted in differences in risk for diabetes, obesity or other diseases between populations. Given the phenotypic differences and the extensive genetic differences between Mexican and US Pimas at HLA, further genetic and phenotypic studies, potentially including sequencing, are warranted to investigate the role of natural selection at HLA in metabolic and immunologic traits in these populations.
Supplementary Material
Acknowledgments
The authors thank the participants who volunteered for these studies. The Maycoba Project was funded by the National Institutes of Health in the United States. (Project Grant # 1R01DK082568-01A1 to LOS). This work was also supported in part by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases. This work was presented in part at the annual meeting of the American Society of Human Genetics, Boston, MA, October 22–26, 2013.
Footnotes
Conflict of Interest Statement:
The authors have no conflicts of interest to disclose.
References
- Albrechtsen A, Moltke I, Nielsen R. Natural selection and the distribution of identity-by-descent in the human genome. Genetics. 2010;186:295–308. doi: 10.1534/genetics.110.113977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayub Q, Moutsianas L, Chen Y, Panoutsopoulou K, Colonna V, Pagani L, Prokopenko I, Ritchie GR, Tyler-Smith C, Mccarthy MI, Zeggini E, Xue Y. Revisiting the thrifty gene hypothesis via 65 loci associated with susceptibility to type 2 diabetes. Am J Hum Genet. 2014;94:176–185. doi: 10.1016/j.ajhg.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:12. [Google Scholar]
- Bhatia G, Patterson N, Pasaniuc B, Zaitlen N, Genovese G, Pollack S, Mallick S, Myers S, Tandon A, Spencer C, Palmer CD, Adeyemo AA, Akylbekova EL, Cupples LA, Divers J, Fornage M, Kao WH, Lange L, Li M, Musani S, Mychaleckyj JC, Ogunniyi A, Papanicolaou G, Rotimi CN, Rotter JI, Ruczinski I, Salako B, Siscovick DS, Tayo BO, Yang Q, Mccarroll S, Sabeti P, Lettre G, De Jager P, Hirschhorn J, Zhu X, Cooper R, Reich D, Wilson JG, Price AL. Genome-wide comparison of African-ancestry populations from CARe and other cohorts reveals signals of natural selection. Am J Hum Genet. 2011;89:368–381. doi: 10.1016/j.ajhg.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatia G, Patterson N, Sankararaman S, Price AL. Estimating and interpreting FST: the impact of rare variants. Genome Res. 2013;23:1514–1521. doi: 10.1101/gr.154831.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian L, Hanson RL, Ossowski V, Wiedrich K, Mason CC, Traurig M, Muller YL, Kobes S, Knowler WC, Baier LJ, Bogardus C. Variants in ASK1 are associated with skeletal muscle ASK1 expression, in vivo insulin resistance, and type 2 diabetes in Pima Indians. Diabetes. 2010;59:1276–1282. doi: 10.2337/db09-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian L, Traurig M, Hanson RL, Marinelarena A, Kobes S, Muller YL, Malhotra A, Huang K, Perez J, Gale A, Knowler WC, Bogardus C, Baier LJ. MAP2K3 is associated with body mass index in American Indians and Caucasians and may mediate hypothalamic inflammation. Hum Mol Genet. 2013;22:4438–4449. doi: 10.1093/hmg/ddt291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black FL, Hedrick PW. Strong balancing selection at HLA loci: evidence from segregation in South Amerindian families. Proc Natl Acad Sci U S A. 1997;94:12452–12456. doi: 10.1073/pnas.94.23.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. High-resolution detection of identity by descent in unrelated individuals. Am J Hum Genet. 2010;86:526–539. doi: 10.1016/j.ajhg.2010.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CS, Matise TC, North KE, Haiman CA, Fesinmeyer MD, Buyske S, Schumacher FR, Peters U, Franceschini N, Ritchie MD, Duggan DJ, Spencer KL, Dumitrescu L, Eaton CB, Thomas F, Young A, Carty C, Heiss G, Le Marchand L, Crawford DC, Hindorff LA, Kooperberg CL. Generalization and dilution of association results from European GWAS in populations of non-European ancestry: the PAGE study. PLoS Biol. 2013;11:e1001661. doi: 10.1371/journal.pbio.1001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona E, Chen R, Sikora M, Morgan AA, Patel CJ, Ramesh A, Bustamante CD, Butte AJ. Analysis of the genetic basis of disease in the context of worldwide human relationships and migration. PLoS Genet. 2013;9:e1003447. doi: 10.1371/journal.pgen.1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elks CE, Den Hoed M, Zhao JH, Sharp SJ, Wareham NJ, Loos RJ, Ong KK. Variability in the heritability of body mass index: a systematic review and meta-regression. Front Endocrinol (Lausanne) 2012;3:29. doi: 10.3389/fendo.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparza-Romero J, Valencia ME, Urquidez-Romero R, Chaudhari LS, Hanson RL, Knowler WC, Ravussin E, Bennett PH, Schulz LO. Environmentally Driven Increases in Type 2 Diabetes and Obesity in Pima Indians and Non-Pimas in Mexico Over a 15-Year Period: The Maycoba Project. Diabetes Care. 2015;38:2075–2082. doi: 10.2337/dc15-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale K. Internal diversity in Uto-Aztecan: I. International Journal of American Linguistics. 1958;24:101–107. [Google Scholar]
- Han L, Abney M. Using identity by descent estimation with dense genotype data to detect positive selection. Eur J Hum Genet. 2013;21:205–211. doi: 10.1038/ejhg.2012.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RL, Bogardus C, Duggan D, Kobes S, Knowlton M, Infante AM, Marovich L, Benitez D, Baier LJ, Knowler WC. A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes. 2007;56:3045–3052. doi: 10.2337/db07-0462. [DOI] [PubMed] [Google Scholar]
- Hanson RL, Mccance DR, Jacobsson LT, Narayan KM, Nelson RG, Pettitt DJ, Bennett PH, Knowler WC. The U-shaped association between body mass index and mortality: relationship with weight gain in a Native American population. J Clin Epidemiol. 1995;48:903–916. doi: 10.1016/0895-4356(94)00217-e. [DOI] [PubMed] [Google Scholar]
- Hanson RL, Muller YL, Kobes S, Guo T, Bian L, Ossowski V, Wiedrich K, Sutherland J, Wiedrich C, Mahkee D, Huang K, Abdussamad M, Traurig M, Weil EJ, Nelson RG, Bennett PH, Knowler WC, Bogardus C, Baier LJ. A genome-wide association study in American Indians implicates DNER as a susceptibility locus for type 2 diabetes. Diabetes. 2014;63:369–376. doi: 10.2337/db13-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RL, Rong R, Kobes S, Muller YL, Weil EJ, Curtis JM, Nelson RG, Baier LJ. Role of established type 2 diabetes-susceptibility genetic variants in a high prevalence American Indian population. Diabetes. 2015;64:2646–2657. doi: 10.2337/db14-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. Balancing selection and MHC. Genetica. 1998;104:207–214. doi: 10.1023/a:1026494212540. [DOI] [PubMed] [Google Scholar]
- Hsueh WC, Nair AK, Kobes S, Chen P, Goring HHH, Pollin TI, Malhotra A, Knowler WC, Baier LJ, Hanson RL. Identity-by-Descent Mapping Identifies Major Locus for Serum Triglycerides in Amerindians Largely Explained by an APOC3 Founder Mutation. Circ Cardiovasc Genet. 2017:10. doi: 10.1161/CIRCGENETICS.117.001809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Deutsch AJ, Lenz TL, Onengut-Gumuscu S, Han B, Chen WM, Howson JM, Todd JA, De Bakker PI, Rich SS, Raychaudhuri S. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Genet. 2015;47:898–905. doi: 10.1038/ng.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima M, Ohashi J, Nishida N, Tokunaga K. Evolutionary analysis of classical HLA class I and II genes suggests that recent positive selection acted on DPB1*04:01 in Japanese population. PLoS One. 2012;7:e46806. doi: 10.1371/journal.pone.0046806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T, Yang W, Chen CS, Reynolds K, He J. Global burden of obesity in 2005 and projections to 2030. Int J Obes (Lond) 2008;32:1431–1437. doi: 10.1038/ijo.2008.102. [DOI] [PubMed] [Google Scholar]
- King H, Aubert RE, Herman WH. Global burden of diabetes, 1995–2025: prevalence, numerical estimates, and projections. Diabetes Care. 1998;21:1414–1431. doi: 10.2337/diacare.21.9.1414. [DOI] [PubMed] [Google Scholar]
- Klimentidis YC, Abrams M, Wang J, Fernandez JR, Allison DB. Natural selection at genomic regions associated with obesity and type-2 diabetes: East Asians and sub-Saharan Africans exhibit high levels of differentiation at type-2 diabetes regions. Hum Genet. 2011;129:407–418. doi: 10.1007/s00439-010-0935-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowler WC, Bennett PH, Hamman RF, Miller M. Diabetes incidence and prevalence in Pima Indians: a 19-fold greater incidence than in Rochester, Minnesota. Am J Epidemiol. 1978;108:497–505. doi: 10.1093/oxfordjournals.aje.a112648. [DOI] [PubMed] [Google Scholar]
- Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P, Been LF, Chia KS, Dimas AS, Hassanali N, Jafar T, Jowett JB, Li X, Radha V, Rees SD, Takeuchi F, Young R, Aung T, Basit A, Chidambaram M, Das D, Grundberg E, Hedman AK, Hydrie ZI, Islam M, Khor CC, Kowlessur S, Kristensen MM, Liju S, Lim WY, Matthews DR, Liu J, Morris AP, Nica AC, Pinidiyapathirage JM, Prokopenko I, Rasheed A, Samuel M, Shah N, Shera AS, Small KS, Suo C, Wickremasinghe AR, Wong TY, Yang M, Zhang F, Abecasis GR, Barnett AH, Caulfield M, Deloukas P, Frayling TM, Froguel P, Kato N, Katulanda P, Kelly MA, Liang J, Mohan V, Sanghera DK, Scott J, Seielstad M, Zimmet PZ, Elliott P, Teo YY, Mccarthy MI, Danesh J, Tai ES, Chambers JC. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43:984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL, Myers RM. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, Powell C, Vedantam S, Buchkovich ML, Yang J, Croteau-Chonka DC, Esko T, Fall T, Ferreira T, Gustafsson S, Kutalik Z, Luan J, Magi R, Randall JC, Winkler TW, Wood AR, Workalemahu T, Faul JD, Smith JA, Hua Zhao J, Zhao W, Chen J, Fehrmann R, Hedman AK, Karjalainen J, Schmidt EM, Absher D, Amin N, Anderson D, Beekman M, Bolton JL, Bragg-Gresham JL, Buyske S, Demirkan A, Deng G, Ehret GB, Feenstra B, Feitosa MF, Fischer K, Goel A, Gong J, Jackson AU, Kanoni S, Kleber ME, Kristiansson K, Lim U, Lotay V, Mangino M, Mateo Leach I, Medina-Gomez C, Medland SE, Nalls MA, Palmer CD, Pasko D, Pechlivanis S, Peters MJ, Prokopenko I, Shungin D, Stancakova A, Strawbridge RJ, Ju Sung Y, Tanaka T, Teumer A, Trompet S, Van Der Laan SW, Van Setten J, Van Vliet-Ostaptchouk JV, Wang Z, Yengo L, Zhang W, Isaacs A, Albrecht E, Arnlov J, Arscott GM, Attwood AP, Bandinelli S, Barrett A, Bas IN, Bellis C, Bennett AJ, Berne C, Blagieva R, Bluher M, Bohringer S, Bonnycastle LL, Bottcher Y, Boyd HA, Bruinenberg M, Caspersen IH, Ida Chen YD, Clarke R, Daw EW, De Craen AJ, Delgado G, Dimitriou M, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I, Saleheen D, Wang X, Zeggini E, Abecasis GR, Adair LS, Almgren P, Atalay M, Aung T, Baldassarre D, Balkau B, Bao Y, Barnett AH, Barroso I, Basit A, Been LF, Beilby J, Bell GI, Benediktsson R, Bergman RN, Boehm BO, Boerwinkle E, Bonnycastle LL, Burtt N, Cai Q, Campbell H, Carey J, Cauchi S, Caulfield M, Chan JC, Chang LC, Chang TJ, Chang YC, Charpentier G, Chen CH, Chen H, Chen YT, Chia KS, Chidambaram M, Chines PS, Cho NH, Cho YM, Chuang LM, Collins FS, Cornelis MC, Couper DJ, Crenshaw AT, Van Dam RM, Danesh J, Das D, De Faire U, Dedoussis G, Deloukas P, Dimas AS, Dina C, Doney AS, Donnelly PJ, Dorkhan M, Van Duijn C, Dupuis J, Edkins S, Elliott P, Emilsson V, Erbel R, Eriksson JG, Escobedo J, Esko T, Eury E, Florez JC, Fontanillas P, Forouhi NG, Forsen T, Fox C, Fraser RM, Frayling TM, Froguel P, Frossard P, Gao Y, Gertow K, Gieger C, Gigante B, Grallert H, Grant GB, Grrop LC, Groves CJ, Grundberg E, Guiducci C, Hamsten A, Han BG, Hara K, Hassanali N, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Kobes S, Knowler WC, Baier LJ, Bogardus C, Hanson RL. A genome-wide association study of BMI in American Indians. Obesity (Silver Spring) 2011;19:2102–2106. doi: 10.1038/oby.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Aguiar VRC, Bitarello BD, Brandt DYC, Nunes K. A genomic perspective on HLA evolution. Immunogenetics. 2018;70:5–27. doi: 10.1007/s00251-017-1017-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Thomson G. How selection shapes variation of the human major histocompatibility complex: a review. Ann Hum Genet. 2001;65:1–26. doi: 10.1046/j.1469-1809.2001.6510001.x. [DOI] [PubMed] [Google Scholar]
- Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A, Prokopenko I, Kang HM, Dina C, Esko T, Fraser RM, Kanoni S, Kumar A, Lagou V, Langenberg C, Luan J, Lindgren CM, Muller-Nurasyid M, Pechlivanis S, Rayner NW, Scott LJ, Wiltshire S, Yengo L, Kinnunen L, Rossin EJ, Raychaudhuri S, Johnson AD, Dimas AS, Loos RJ, Vedantam S, Chen H, Florez JC, Fox C, Liu CT, Rybin D, Couper DJ, Kao WH, Li M, Cornelis MC, Kraft P, Sun Q, Van Dam RM, Stringham HM, Chines PS, Fischer K, Fontanillas P, Holmen OL, Hunt SE, Jackson AU, Kong A, Lawrence R, Meyer J, Perry JR, Platou CG, Potter S, Rehnberg E, Robertson N, Sivapalaratnam S, Stancakova A, Stirrups K, Thorleifsson G, Tikkanen E, Wood AR, Almgren P, Atalay M, Benediktsson R, Bonnycastle LL, Burtt N, Carey J, Charpentier G, Crenshaw AT, Doney AS, Dorkhan M, Edkins S, Emilsson V, Eury E, Forsen T, Gertow K, Gigante B, Grant GB, Groves CJ, Guiducci C, Herder C, Hreidarsson AB, Hui J, James A, Jonsson A, Rathmann W, Klopp N, Kravic J, Krjutskov K, Langford C, Leander K, Lindholm E, Lobbens S, Mannisto S, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller YL, Hanson RL, Knowler WC, Fleming J, Goswami J, Huang K, Traurig M, Sutherland J, Wiedrich C, Wiedrich K, Mahkee D, Ossowski V, Kobes S, Bogardus C, Baier LJ. Identification of genetic variation that determines human trehalase activity and its association with type 2 diabetes. Hum Genet. 2013;132:697–707. doi: 10.1007/s00439-013-1278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Helgason A, Palsson S, Stefansson H, St Clair D, Andreassen OA, Reich D, Kong A, Stefansson K. The impact of divergence time on the nature of population structure: an example from Iceland. PLoS Genet. 2009;5:e1000505. doi: 10.1371/journal.pgen.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, Mcdonald GJ, Ackerman HC, Campbell SJ, Altshuler D, Cooper R, Kwiatkowski D, Ward R, Lander ES. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Saxena R, Elbers CC, Guo Y, Peter I, Gaunt TR, Mega JL, Lanktree MB, Tare A, Castillo BA, Li YR, Johnson T, Bruinenberg M, Gilbert-Diamond D, Rajagopalan R, Voight BF, Balasubramanyam A, Barnard J, Bauer F, Baumert J, Bhangale T, Bohm BO, Braund PS, Burton PR, Chandrupatla HR, Clarke R, Cooper-Dehoff RM, Crook ED, Davey-Smith G, Day IN, De Boer A, De Groot MC, Drenos F, Ferguson J, Fox CS, Furlong CE, Gibson Q, Gieger C, Gilhuijs-Pederson LA, Glessner JT, Goel A, Gong Y, Grant SF, Grobbee DE, Hastie C, Humphries SE, Kim CE, Kivimaki M, Kleber M, Meisinger C, Kumari M, Langaee TY, Lawlor DA, Li M, Lobmeyer MT, Maitland-Van Der Zee AH, Meijs MF, Molony CM, Morrow DA, Murugesan G, Musani SK, Nelson CP, Newhouse SJ, O’connell JR, Padmanabhan S, Palmen J, Patel SR, Pepine CJ, Pettinger M, Price TS, Rafelt S, Ranchalis J, Rasheed A, Rosenthal E, Ruczinski I, Shah S, Shen H, Silbernagel G, Smith EN, Spijkerman AW, Stanton A, Steffes MW, Thorand B, Trip M, Van Der Harst P, Van Der AD, Van Iperen EP, Van Setten J, Van Vliet-Ostaptchouk JV, Verweij N, Wolffenbuttel BH, Young T, Zafarmand MH, Zmuda JM, Boehnke M, Altshuler D, Mccarthy M, Kao WH, Pankow JS, Cappola TP, Sever P, et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90:410–425. doi: 10.1016/j.ajhg.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz LO, Bennett PH, Ravussin E, Kidd JR, Kidd KK, Esparza J, Valencia ME. Effects of traditional and western environments on prevalence of type 2 diabetes in Pima Indians in Mexico and the U.S. Diabetes Care. 2006;29:1866–1871. doi: 10.2337/dc06-0138. [DOI] [PubMed] [Google Scholar]
- Solberg OD, Mack SJ, Lancaster AK, Single RM, Tsai Y, Sanchez-Mazas A, Thomson G. Balancing selection and heterogeneity across the classical human leukocyte antigen loci: a meta-analytic review of 497 population studies. Hum Immunol. 2008;69:443–464. doi: 10.1016/j.humimm.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, Lango Allen H, Lindgren CM, Luan J, Magi R, Randall JC, Vedantam S, Winkler TW, Qi L, Workalemahu T, Heid IM, Steinthorsdottir V, Stringham HM, Weedon MN, Wheeler E, Wood AR, Ferreira T, Weyant RJ, Segre AV, Estrada K, Liang L, Nemesh J, Park JH, Gustafsson S, Kilpelainen TO, Yang J, Bouatia-Naji N, Esko T, Feitosa MF, Kutalik Z, Mangino M, Raychaudhuri S, Scherag A, Smith AV, Welch R, Zhao JH, Aben KK, Absher DM, Amin N, Dixon AL, Fisher E, Glazer NL, Goddard ME, Heard-Costa NL, Hoesel V, Hottenga JJ, Johansson A, Johnson T, Ketkar S, Lamina C, Li S, Moffatt MF, Myers RH, Narisu N, Perry JR, Peters MJ, Preuss M, Ripatti S, Rivadeneira F, Sandholt C, Scott LJ, Timpson NJ, Tyrer JP, Van Wingerden S, Watanabe RM, White CC, Wiklund F, Barlassina C, Chasman DI, Cooper MN, Jansson JO, Lawrence RW, Pellikka N, Prokopenko I, Shi J, Thiering E, Alavere H, Alibrandi MT, Almgren P, Arnold AM, Aspelund T, Atwood LD, Balkau B, Balmforth AJ, Bennett AJ, Ben-Shlomo Y, Bergman RN, Bergmann S, Biebermann H, Blakemore AI, Boes T, Bonnycastle LL, Bornstein SR, Brown MJ, Buchanan TA, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. Statistical methods for detecting natural selection from genomic data. Genes Genet Syst. 2010;85:359–376. doi: 10.1266/ggs.85.359. [DOI] [PubMed] [Google Scholar]
- Szpiech ZA, Hernandez RD. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol Biol Evol. 2014;31:2824–2827. doi: 10.1093/molbev/msu211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, Jonsdottir T, Olafsdottir EJ, Olafsdottir GH, Jonsson T, Jonsson F, Borch-Johnsen K, Hansen T, Andersen G, Jorgensen T, Lauritzen T, Aben KK, Verbeek AL, Roeleveld N, Kampman E, Yanek LR, Becker LC, Tryggvadottir L, Rafnar T, Becker DM, Gulcher J, Kiemeney LA, Pedersen O, Kong A, Thorsteinsdottir U, Stefansson K. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Tian C, Hinds DA, Shigeta R, Adler SG, Lee A, Pahl MV, Silva G, Belmont JW, Hanson RL, Knowler WC, Gregersen PK, Ballinger DG, Seldin MF. A genomewide single-nucleotide-polymorphism panel for Mexican American admixture mapping. Am J Hum Genet. 2007;80:1014–1023. doi: 10.1086/513522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Kidd KK. Implications of biogeography of human populations for ‘race’ and medicine. Nat Genet. 2004;36:S21–27. doi: 10.1038/ng1438. [DOI] [PubMed] [Google Scholar]
- Traurig M, Mack J, Hanson RL, Ghoussaini M, Meyre D, Knowler WC, Kobes S, Froguel P, Bogardus C, Baier LJ. Common variation in SIM1 is reproducibly associated with BMI in Pima Indians. Diabetes. 2009;58:1682–1689. doi: 10.2337/db09-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traurig MT, Perez JM, Ma L, Bian L, Kobes S, Hanson RL, Knowler WC, Krakoff JA, Bogardus C, Baier LJ. Variants in the LEPR gene are nominally associated with higher BMI and lower 24-h energy expenditure in Pima Indians. Obesity (Silver Spring) 2012;20:2426–2430. doi: 10.1038/oby.2012.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM, Chang CC, Chen P, Chen CH, Fann CS, Chen YT, Wu JY. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. PLoS Genet. 2010;6:e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urquidez-Romero R, Esparza-Romero J, Chaudhari LS, Begay RC, Giraldo M, Ravussin E, Knowler WC, Hanson RL, Bennett PH, Schulz LO, Valencia ME. Study design of the Maycoba Project: obesity and diabetes in Mexican Pimas. Am J Health Behav. 2014;38:370–378. doi: 10.5993/AJHB.38.3.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen G, Ward KJ, Bell CG, Christensen K, Bowden J, Dalgard C, Harris JR, Kaprio J, Lyle R, Magnusson PK, Mather KA, Ordonana JR, Perez-Riquelme F, Pedersen NL, Pietilainen KH, Sachdev PS, Boomsma DI, Spector T. The concordance and heritability of type 2 diabetes in 34,166 twin pairs From international twin registers: The Discordant Twin (DISCOTWIN) Consortium. Twin Res Hum Genet. 2015;18:762–771. doi: 10.1017/thg.2015.83. [DOI] [PubMed] [Google Scholar]
- Williams AL, Jacobs SB, Moreno-Macias H, Huerta-Chagoya A, Churchhouse C, Marquez-Luna C, Garcia-Ortiz H, Gomez-Vazquez MJ, Burtt NP, Aguilar-Salinas CA, Gonzalez-Villalpando C, Florez JC, Orozco L, Haiman CA, Tusie-Luna T, Altshuler D. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams R, Chen YF, Endres R, Middleton D, Trucco M, Williams JD, Knowler W. Molecular variation at the HLA-A, B, C, DRB1, DQA1, and DQB1 loci in full heritage American Indians in Arizona: private haplotypes and their evolution. Tissue Antigens. 2009;74:520–533. doi: 10.1111/j.1399-0039.2009.01381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RC, Muller YL, Hanson RL, Knowler WC, Mason CC, Bian L, Ossowski V, Wiedrich K, Chen YF, Marcovina S, Hahnke J, Nelson RG, Baier LJ, Bogardus C. HLA-DRB1 reduces the risk of type 2 diabetes mellitus by increased insulin secretion. Diabetologia. 2011;54:1684–1692. doi: 10.1007/s00125-011-2122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, De Bakker PI, Abecasis GR, Almgren P, Andersen G, Ardlie K, Bostrom KB, Bergman RN, Bonnycastle LL, Borch-Johnsen K, Burtt NP, Chen H, Chines PS, Daly MJ, Deodhar P, Ding CJ, Doney AS, Duren WL, Elliott KS, Erdos MR, Frayling TM, Freathy RM, Gianniny L, Grallert H, Grarup N, Groves CJ, Guiducci C, Hansen T, Herder C, Hitman GA, Hughes TE, Isomaa B, Jackson AU, Jorgensen T, Kong A, Kubalanza K, Kuruvilla FG, Kuusisto J, Langenberg C, Lango H, Lauritzen T, Li Y, Lindgren CM, Lyssenko V, Marvelle AF, Meisinger C, Midthjell K, Mohlke KL, Morken MA, Morris AD, Narisu N, Nilsson P, Owen KR, Palmer CN, Payne F, Perry JR, Pettersen E, Platou C, Prokopenko I, Qi L, Qin L, Rayner NW, Rees M, Roix JJ, Sandbaek A, Shields B, Sjogren M, Steinthorsdottir V, Stringham HM, Swift AJ, Thorleifsson G, Thorsteinsdottir U, Timpson NJ, Tuomi T, Tuomilehto J, Walker M, Watanabe RM, Weedon MN, Willer CJ, Illig T, Hveem K, Hu FB, Laakso M, Stefansson K, Pedersen O, Wareham NJ, Barroso I, Hattersley AT, Collins FS, Groop L, Mccarthy MI, Boehnke M, Altshuler D. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.