Abstract
The process by which committed precursors mature into cardiomyocytes is poorly understood. We found that TLR3 inhibition blocked cardiomyocyte maturation; precursor cells committed to the cardiomyocyte lineage failed to express maturation genes and sarcomeres did not develop. Using various approaches we found that the effects of TLR3 upon cardiomyocyte maturation were dependent upon the RelA subunit of NFκB. Importantly, under conditions that promote the development of mature cardiomyocytes NFκB became significantly enriched at the promoters of cardiomyocyte maturation genes. Furthermore, activation of the TLR3-NFκB pathway enhanced cardiomyocyte maturation. This study therefore demonstrates that the TLR3-NFκB pathway is necessary for the maturation of committed precursors into mature cardiomyocytes.
Keywords: cardiomyocyte maturation, cardiomyocyte development, cardiac reprogramming, innate immunity, TLR3, microRNAs, NFκB
Graphical abstract
Activation of TLR3 enhances direct cardiac reprogramming
Introduction
Studying cardiomyocyte development presents unique challenges when compared to other cell types. Cardiomyocytes are an essential component of the heart. Genetic manipulation can cause abnormal cardiac morphogenesis; typically leading to embryonic lethality. This embryonic lethality is difficult to diagnose prenatally and limits our understanding of the process by which precursors commit to the cardiac lineage and mature into fully functional cardiomyocytes. Due to this hurdle, many researchers have taken to replicating cardiomyocyte development in the culture dish. Various methods have been employed including reprogramming strategies [1–8] [9–14].
In vitro modelling suggests that cardiomyocyte development has two phases; Initiation and Maturation [15–18]. In the Initiation phase of cardiomyocyte development, the precursor cell initially expresses a number of so-called pioneer transcription factors. These pioneer transcription factors induce significant epigenetic remodeling; the precursor phenotype is silenced and various genes that are necessary for commitment to the cardiomyocyte lineage are activated. The pioneer transcription factors have been identified: combined expression of Gata4, Tbx5, Mef2C and Hand1 is necessary for the initial commitment to the cardiomyocyte lineage [16]. Recently, we and others have shown that the key epigenetic mechanism in the Initiation phase of cardiomyocyte development is histone methylation. H3K4 becomes methylated [19] whereas H3K27 is de-methylated [14]. These epigenetic modifications work in concert to activate and repress numerous genes that are necessary to stabilize commitment to the cardiomyocyte lineage. Whereas the steps in the Initiation phase of cardiomyocyte development are well known, the process of cardiomyocyte maturation has not been studied in detail.
Two recent studies have shown that TLR3 is important for reprogramming fibroblasts to iPS [20] and endothelial cells [21]. Specifically, activation of TLR3 causes global changes in the expression and activity of epigenetic modifiers that favor DNA accessibility, and phenotypic fluidity. Interestingly, TLR3 plays a role in the inflammatory response and it is known that inflammation plays a major role in the cardiac response to injury [22, 23]. Consequently, we asked ourselves if TLR3 played a hitherto unknown role in cardiac reprogramming. In this study we show that cardiomyocyte maturation requires TLR3 activated NFκB. Precursor cells that had committed to the cardiomyocyte lineage were prevented from maturing into cardiomyocytes by TLR3 inhibitors or TLR3 knockdown. Further experiments demonstrated that TLR3 controlled cardiomyocyte maturation via NFκB. Pharmacological inhibition of NFκB, as well as knockdown of Ikbkb (Inhibitor of Nuclear Factor Kappa B Kinase) which activates NFκB, prevented cardiomyocyte maturation. Moreover, conditions that induce cardiomyocyte formation induced NFκB binding to the promoters of cardiomyocyte maturation genes. Moreover, we found that microRNAs activate TLR3.
Materials & Methods
Chemicals
TLR agonists were purchased from Invivogen (Mouse TLR1-9 agonist kit, tlrl-kit1mw). The TLR3 antagonist CU-CPT-4a, NFκB antagonist Bay 11-7085 and the AP1 antagonist SR 11302 were purchased from Tocris.
MicroRNA transfection
Mouse (C57BL/6) neonatal cardiac fibroblasts were isolated from 2 day old mouse neonates according to the method outlined in Jayawardena et al [10]. Following isolation fibroblasts were cultured in growth media containing DMEM (ATCC, Catalogue number 30-2002) supplemented with 15%v/v FBS (Thermo Scientific Hyclone Fetal bovine serum, Catalogue number SH30071.03, Lot number AXK49952) and 1%v/v penicillin/streptomycin (Gibco, Catalogue number 15140-122, 100units Penicillin, 100ug/ml Streptomycin). Fibroblasts were passaged once the cells had reached 70–80% confluence using 0.05% w/v trypsin (Gibco, Catalogue number 25300-054). Freshly isolated fibroblasts were labelled as Passage 0. Experiments were conducted with cells at passage 2. For all experiments, cells were seeded at 5000 cells/cm2 in growth media. After 24 hours, the cells were transfected with transfection reagent alone (Dharmafect-I, ThermoScientific), with transfection reagent plus non-targeting microRNAs (negmiR), or with transfection reagent plus our previously reported combination of cardiac reprogramming microRNAs[9] (miR combo, miR-1, miR-133, miR-208, miR-499).
qPCR
Total RNA was extracted using Quick-RNA MiniPrep Kit according to the manufacturer’s instructions (Zymo Research). Total RNA (50ng-100ng) was converted to cDNA using a high capacity cDNA reverse transcription kit (Applied Biosystems). cDNA was used in a standard qPCR reaction involving FAM conjugated gene specific primers and TaqMan Gene Expression Master Mix (Applied Biosystems). The following primers were used for qPCR: Gapdh (Mm99999915_m1), Tnni3 (Mm00437164_m1), Actn2 (Mm00473657_m1), Myh6 (Mm00440359_m1), Cacna1c (Mm00437917_m1), Mef2C (Mm01340482_m1), Tbx5 (Mm00803518_m1), Gata4 (Mm00484689_m1) and Hand2 (Mm00439247_m1).
Immunofluorescence
Cells were fixed with 2%v/v paraformaldehyde (EMS) as described previously [24]. Fixed cells were blocked in antibody buffer (5%w/v BSA, 0.1%v/v Tween-20, in PBS) for 1 hr at room temperature. Following blocking, cells were incubated overnight at 4°C with α-sarcomeric actinin antibody (Sigma, A7811, 1:100) in antibody buffer. After the overnight incubation, cells were washed three times in antibody buffer. Following washing, cells were incubated with Alexa-Fluor conjugated secondary antibodies (Invitrogen, Goat Anti-mouse 594nm) at a 1:500 dilution in antibody buffer for 1hr at room temperature. Nuclei were stained by DAPI at 1μg/ml for 30 minutes at room temperature in antibody buffer. Following washing in PBS to remove unbound complexes, immunofluorescence was measured using a Zeiss Axiovert 200 inverted microscope.
siRNA knockdown
siRNA pools (four siRNAs targeting the gene) and a negative control siRNA were purchased from Dharmacon. siRNAs were made to 20μM in nuclease free water, aliquoted, and stored -80°C until use. Fibroblasts were seeded into 12 well plates at 20,000 cells per well one day prior to transfection. On the day of transfection siRNAs were diluted to 5μM in nuclease free water. For each well, 5μl of the working siRNA solution was diluted with 95μl Optimem-Serum Free media. In a separate tube 5μl of Dharmafect-I (Dharmacon) was diluted with 95μl Optimem-Serum Free media. After 5 minute incubation the two solutions were combined. After 20 minutes complete media lacking antibiotics was added (800μl) and the transfection complexes added to the cells.
ChIP assays
ChIP assays were performed according the manufacturer’s instructions (Cell Signaling, SimpleChIP Enzymatic Chromatin IP kit #9003). Neonatal cardiac fibroblast nuclei were digested with 0.1ul Micrococcal nuclease per 4×106 cells (amount of Micrococcal nuclease was empirically determined according the manufacturer’s instructions). Immunoprecipitation was performed with ChIP validated antibodies: (1) rabbit IgG control (Cell Signaling, #2729); (2) Histone H3 (Cell Signaling, #4620); and (3) RelA (Cell Signaling, #8242). Immunoprecipitated DNA was quantified by qPCR (ThermoFisher, Power SYBR Green PCR Master Mix, #4367659) with primers for the promoters of Myh6 (Qiagen, EpiTect ChIP qPCR Primer Assay For Mouse Myh6, NM_010856.3 (-)08Kb #GPM1045733(-)08A and EpiTect ChIP qPCR Primer Assay For Mouse Myh6, NM_010856.3 (-)01Kb #GPM1045733(-)01A), Actn2 (Qiagen, EpiTect ChIP qPCR Primer Assay For Mouse Actn2, NM_033268.3 (-)01Kb, #GPM1044781(-)01A) and Tnni3 (Qiagen, EpiTect ChIP qPCR Primer Assay For Mouse Tnni3, NM_009406.3 (-)01Kb, #GPM1052593(-)01A). PCR reactions included the positive control Histone H3 sample and the negative control rabbit IgG sample. A serial dilution of the 2% input chromatin DNA (undiluted, 1:5, 1:25, 1:125) was used to create a standard curve and determine efficiency of amplification. Percent input was calculated and negative control IgG values subtracted. Data is presented as the fold change of percent input between miR combo and negmiR treated samples.
IL6 ELISA
IL6 ELISA kits were from R&D Systems. Fresh media (1ml) was added to the cells one day prior to assaying for IL6. Per manufacturer’s instructions 50ul of media was assayed and the amount of IL6 in pg/ml in the culture media was determined via a standard curve. The IL6 pg/ml value was then adjusted for the total volume of the media (1ml) and the total cellular protein in each well to correct for differences in cell number[25].
Generating beating reprogrammed cardiomyocytes
Isolated mouse (C57BL/6) neonatal cardiac fibroblasts (passage 2) were seeded into 12-well dishes at 15000 cells/cm2 in growth media. Twenty-four hours later growth media was removed and the cells transfected with negmiR or miR combo as described above. One day later, the transfection complexes were removed and media was replaced with a chemically defined reprogramming media[12] that contained 1ug/ml Poly(I:C) (LMW). For the next four days, cells received fresh chemically defined reprogramming media [12] containing 1ug/ml Poly(I:C) (LMW) daily. After this period, the cells received chemically defined reprogramming media [12] without Poly(I:C) (LMW) for a further 10 days. Media was replaced every other day. Beating colonies were identified with a Zeiss Axiovert 200 inverted microscope.
Images
Images were processed with CorelDraw and Zeiss software (Axiovision Rel4.8 and Zen Blue).
Statistics
All statistical analysis was performed using GraphPad. Experiments containing two conditions a t-test was performed. ANOVA was used for experiments with three or more conditions followed by Bonferroni post-hoc tests for comparisons between individual groups. A P-value of less than 0.05 was considered significant.
Results
TLR3 inhibition blocks the maturation phase of cardiac reprogramming
The mechanisms by which committed cells mature into cardiomyocytes are unclear. Two recent studies have shown that TLR3 is important for reprogramming fibroblasts to iPS [20] and endothelial cells [21]. Moreover, TLR3 induces inflammation and inflammation is known to be important in injury. Consequently, we asked ourselves if TLR3 played a hitherto unknown role in the development of mature cardiomyocytes. We were interested in TLRs as these receptors are key mediators of the inflammatory responses in the heart.
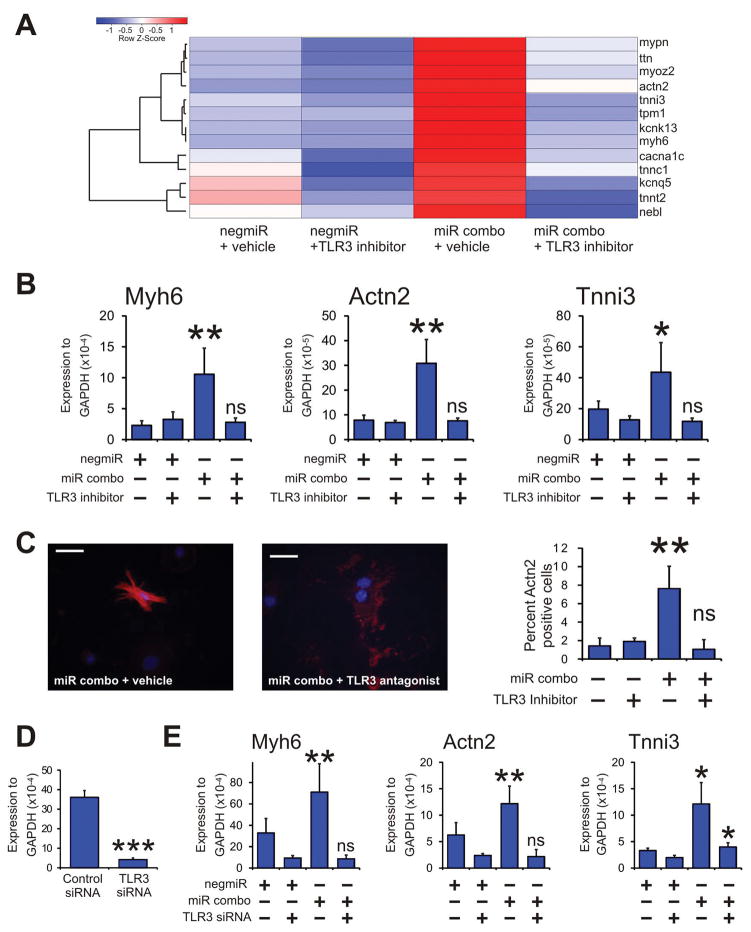
In the first instance, we tested our hypothesis with the specific TLR3 pharmacological inhibitor CU-CPT-4a [26, 27]. We were specifically interested in the maturation phase of cardiac reprogramming. To that end, we carried out an initial screen for the mRNA levels for components of that are involved in cardiomyocyte sarcomere function. We used our previously described miR combo to induce cardiac reprogramming. MiR combo is a combination of four microRNAs (miR-1, -133, -208, -499) that robustly induces cardiac reprogramming both in vitro and in vivo[9, 11–14]. As shown in Figure 1 miR combo significantly induced the expression of 13 components of the cardiomyocyte sarcomere. The effect of miR combo upon cardiomyocyte sarcomere gene expression was completely abolished by the TLR3 pharmacological inhibitor CU-CPT-4a (Figure 1; quantification supplied in Supplementary Figure 1).
Figure 1. TLR3 inhibition inhibits maturation of reprogrammed fibroblasts into cardiomyocytes.
Neonatal cardiac fibroblasts were transfected with negative control miR (negmiR) or miR combo. The day after transfection media was replaced and the cells incubated with either vehicle or the TLR3 pharmacological inhibitor CU-CPT-4a (10uM) for a further 4 days. After incubation with the TLR3 pharmacological inhibitor, cells were cultured in normal growth media for a further 6 days. Quantitative PCR was used to analyze mRNA levels of 13 components of the cardiomyocyte sarcomere.
(A) Heat-map overview of the qPCR analysis. Expression values were normalized to the average expression of the negmiR vehicle samples and then averaged (N=3 technical replicates (fibroblasts were derived from single litter and seeded into 3 individual wells). Averages for each gene were then converted to Z-scores. Centroid linkage and Euclidean methods were employed for clustering and distance measurements respectively.
(B & C) Neonatal cardiac fibroblasts were transfected with negative control miR (negmiR) or miR combo. The day after transfection media was replaced and the cells incubated with either vehicle or the TLR3 pharmacological inhibitor CU-CPT-4a (10uM) for a further 4 days. After incubation with the TLR3 pharmacological inhibitor, cells were cultured in normal growth media for a further 10 days.
(B) RNA levels of the cardiomyocyte sarcomere components Myh6 (αmyosin heavy chain), Actn2 (αsarcomeric actinin) and Tnni3 (cardiac troponin-I) was determined by qPCR. N=4 independent experiments.
(C) Left: Cells were fixed and stained with anti-Actn2 antibodies (red). Nuclei were stained with DAPI (blue). Scale bar 50 microns. Right: Quantification of immunostaining shown in B. Cells expressing Actn2 were counted and expressed as a percentage of the total cell population. N=6 independent experiments.
(D & E) Neonatal cardiac fibroblasts were first transfected with either a control siRNA or a siRNA that targeted TLR3. Two days later, the cells were transfected again with either the negative control miR negmiR or miR combo. The day after transfection with miRNAs the media was replaced and the cells cultured in normal growth media for 14 days.
(D) Quantification of TLR3 knockdown by qPCR. N=3 independent experiments.
(E) RNA levels of the cardiomyocyte structural proteins Myh6 (αmyosin heavy chain), Actn2 (αsarcomeric actinin) and Tnni3 (cardiac troponin-I) was determined by qPCR. N=4 independent experiments. Data represented as Mean ± SEM. ***P<0.001, **P<0.01, *P<0.05, ns: not significant. For A, B and D comparisons are made between miR combo and negmiR for each group. For C, comparison is made between control siRNA and siRNA targeting TLR3.
We verified our initial screen by measuring the mRNA levels of three components of the cardiomyocyte sarcomere: Myh6 (αmyosin heavy chain), Actn2 (αsarcomeric actinin) and Tnni3 (cardiac troponin-I). As we observed in our initial screen, pharmacological inhibition of TLR3 completely inhibited miR combo reprogramming with respect to the expression of Myh6, Actn2, and Tnni3 (Figure 1B). We then assessed the effects of TLR3 inhibition upon the maturation of reprogrammed fibroblasts at the cellular level. The ability of miR combo to generate mature cardiomyocytes with organized sarcomeres was completely inhibited by CU-CPT-4a (Figure 1C). These results were then verified by siRNA mediated knockdown of TLR3. Knockdown of TLR3 by siRNA was robust (Figure 1D) and completely abrogated miR combo reprogramming with respect to the expression of Myh6, Actn2, and Tnni3 (Figure 1E).
Neither TLR3 inhibition nor TLR3 activation affects the initiation phase of cardiac reprogramming
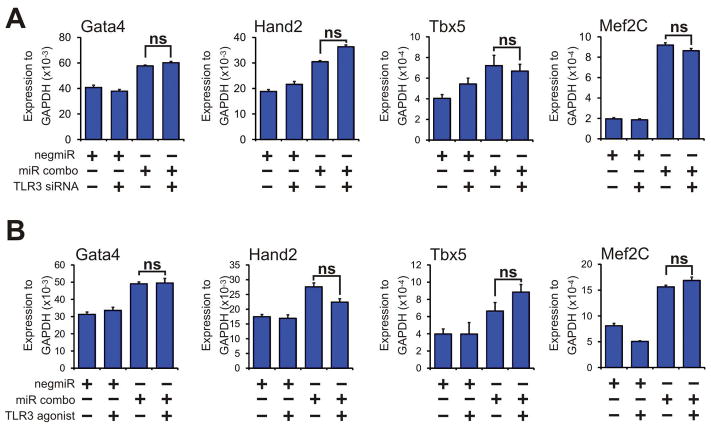
Following these results, we wanted to investigate the mechanism by which TLR3 influenced the maturation of reprogrammed cells in more detail. During heart development, in the initial phase of differentiation of precursors into cardiomyocytes epigenetic processes act to turn on expression of the cardiomyocyte-lineage commitment factors Gata4, Hand2, Tbx5 and Mef2C are expressed[28]. Similarly, increased expression of these cardiomyocyte-lineage commitment factors in fibroblasts represents the initial phase of cardiac reprogramming [1–6, 9, 12, 14]. We found that the expression of the cardiomyocyte-lineage commitment factors Gata4, Hand2, Tbx5 and Mef2C, that was induced by miR combo, was not affected by either TLR3 knockdown (Figure 2A) or by TLR3 activation (Figure 2B). This data indicates that the effects of TLR3 upon the cardiac reprogramming were not due to changes in the initiation phase of cardiac reprogramming.
Figure 2. Neither TLR3 inhibition nor TLR3 activation affects early stage cardiac reprogramming.
(A) Neonatal cardiac fibroblasts were first transfected with either a control siRNA or a siRNA that targeted TLR3. Two days later, the cells were transfected again with either the negative control miR negmiR or miR combo. The day after transfection with miRNAs, the media was replaced and the cells cultured in normal growth media for 3 days. RNA levels of the cardiomyocyte-lineage commitment factors Gata4, Hand2, Tbx5 and Mef2C was determined by qPCR. N=9 independent experiments. Comparisons are made between miR combo + control siRNA and miR combo + TLR3 siRNA, ns: not significant. Data represented as Mean ± SEM.
(B) Neonatal cardiac fibroblasts were transfected with negative control miR (negmiR) or miR combo. The day after transfection media was replaced and the cells incubated with vehicle or the TLR3 agonist Poly(I:C) LMW (low molecular weight Poly(I:C)) for a further 3 days. RNA levels of the cardiomyocyte-lineage commitment factors Gata4, Hand2, Tbx5 and Mef2C was determined by qPCR. N=6 independent experiments. Comparisons are made between miR combo + vehicle and miR combo + TLR3 agonist, ns: not significant. Data represented as Mean ± SEM.
TLR3 controls the maturation phase of cardiac reprogramming via the RelA subunit of NFκB
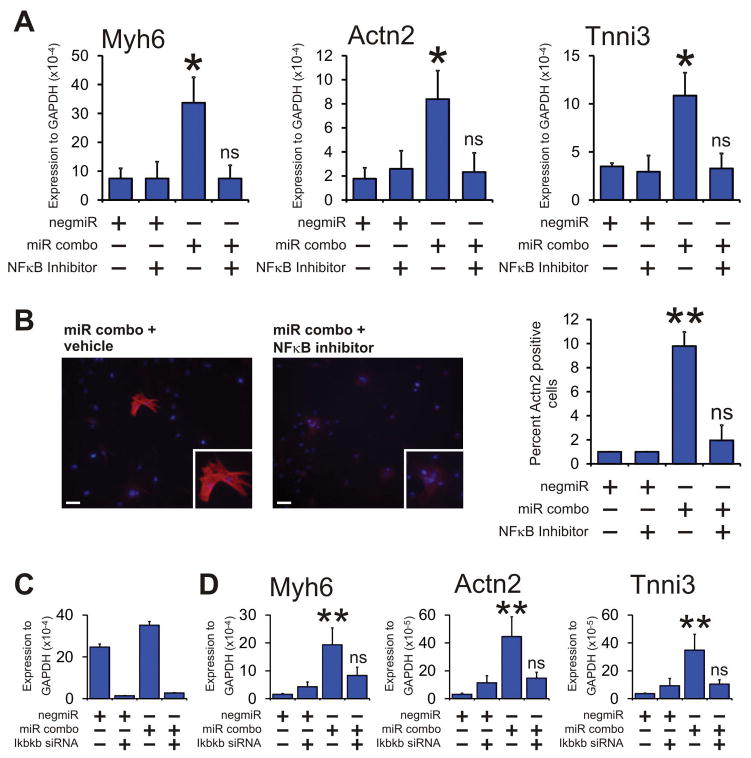
TLR3 mediates the activation of a number of transcription factors[22]. Of these transcription factors, two mediate the vast majority of the effects of TLR3: AP1 and NFκB[22]. Consequently, we hypothesized that TLR3 would influence maturation of reprogrammed cells via AP1 and/or NFκB. Pharmacological inhibition of AP1 had no effect on the ability of miR combo to reprogram fibroblasts (Supplementary Figure 2). In contrast, the pharmacological inhibition of NFκB completely inhibited miR combo reprogramming at both the RNA (Figure 3A) and protein (Figure 3B) level.
Figure 3. NFκB is important for miR combo reprogramming.
(A & B) Neonatal cardiac fibroblasts were transfected with negative control miR (negmiR) or miR combo. The day after transfection media was replaced and the cells incubated with vehicle or the NFκB antagonist Bay 11-7085. After one day of treatment, the media was replaced with normal growth media and cells cultured for a further 12 days.
(A) RNA levels of the cardiomyocyte structural proteins Myh6 (αmyosin heavy chain), Actn2 (αsarcomeric actinin) and Tnni3 (cardiac troponin-I) following treatment with the NFκB antagonist Bay 11-7085 was determined by qPCR. N=3 independent experiments.
(B) Left: Cells were fixed and stained with anti-Actn2 antibodies (red). Nuclei were stained with DAPI (blue). Scale bar 100 microns. Inset pictures are at 5x magnification. Right: Quantification of immunostaining shown in C. Cells expressing Actn2 were counted and expressed as a percentage of the total cell population. N=3 independent experiments.
(C & D) Neonatal cardiac fibroblasts were transfected with microRNAs (negmiR or miR combo) and siRNA (control siRNA or a siRNA that targeted Ikbkb). The day after transfection with miRNAs the media was replaced and the cells cultured in normal growth media for either 4 days (to assess knockdown efficiency) or 14 days (to assess RNA levels of cardiomyocyte structural proteins).
(C) Quantification of Ikbkb knockdown by qPCR. N=3 independent experiments.
(D) RNA levels of the cardiomyocyte structural proteins Myh6 (αmyosin heavy chain), Actn2 (αsarcomeric actinin) and Tnni3 (cardiac troponin-I) was determined by qPCR. N=3 independent experiments.
Data represented as Mean ± SEM. Comparisons are made between miR combo and negmiR for each group, **P<0.01, *P<0.05, ns: not significant.
We further verified a role for NFκB in the maturation of reprogrammed cells by targeting Ikbkb; a kinase that is necessary for NFκB activation[29]. Knockdown of Ikbkb was robust (Figure 3C). Importantly, knockdown of Ikbkb completely inhibited miR combo reprogramming with respect to the expression of the cardiomyocyte maturation markers Myh6, Actn2, and Tnni3 (Figure 3D). In agreement with the studies described above, Ikbkb knockdown did not influence the initiation phase of cardiac reprogramming (Supplementary Figure 3). Moreover; Ikbkb knockdown did not affect miR combo mediated suppression of endodermal, ectodermal and vascular markers (Supplementary Figure 4).
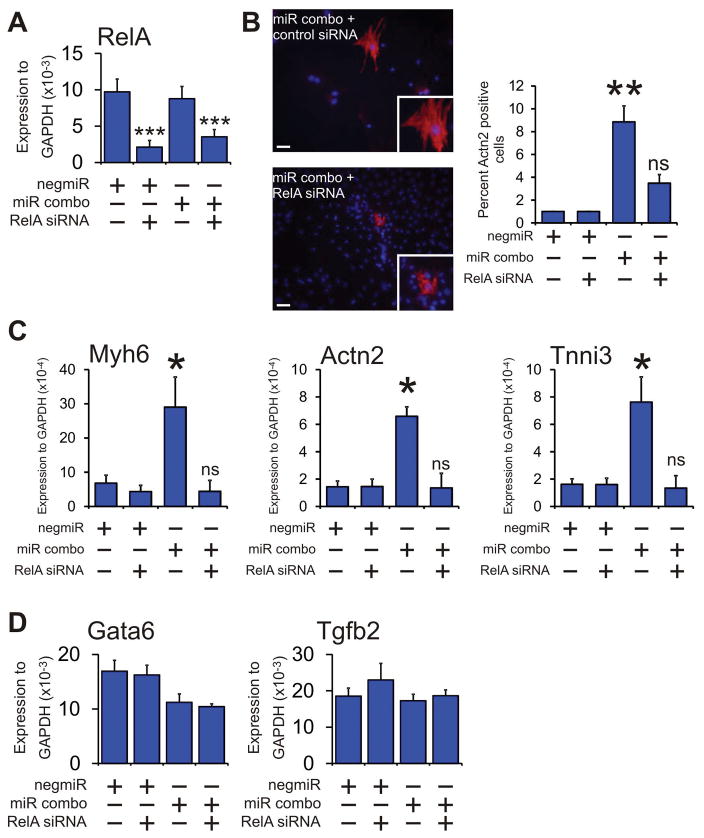
There are five NFκB proteins: NF-κB1 (p105/p50); NF-κB2 (p100/p52); RelA (p65); RelB; and c-Rel. Only RelA, RelB and c-Rel induce transcription. We focused on RelA as it is the most highly expressed Rel protein. Knockdown of RelA, which was found to be robust (Figure 4A), prevented the appearance of Actn2(+) cells in miR combo transfected fibroblasts (Figure 4B). Effects at the protein level were also observed at the mRNA level; targeting RelA with siRNA completely inhibited miR combo reprogramming with respect to sarcomere-related gene expression (Figure 4C). We also noted that RelA knockdown had no effect on the expression of the endodermal marker Gata6 or the general differentiation marker Tgfb2 (Figure 4D).
Figure 4. RelA mediates the effects of NFκB.
Neonatal cardiac fibroblasts were first transfected with either a control siRNA or a siRNA that targeted the NFκB subunit RelA. Two days later, the cells were transfected again with either the negative control miR negmiR or miR combo. The day after transfection with miRNAs, the media was replaced and the cells cultured in normal growth media for 13 days.
(A) Quantification of RelA knockdown by qPCR.
(B) Left: Cells were fixed and stained with anti-Actn2 antibodies (red). Nuclei were stained with DAPI (blue). Scale bar 100 microns. Inset pictures are at 5x magnification. Right: Quantification of immunostaining shown in B. Cells expressing Actn2 were counted and expressed as a percentage of the total cell population. N=3 independent experiments.
(C) RNA levels of the cardiomyocyte structural proteins Myh6 (αmyosin heavy chain), Actn2 (αsarcomeric actinin) and Tnni3 (cardiac troponin-I) was determined by qPCR. N=3 independent experiments.
(D) RNA levels of the endodermal marker Gata6 and the general marker of differentiation Tgfb2 were determined by qPCR. N=3 independent experiments.
Data represented as Mean ± SEM. Comparisons are made between miR combo and negative control miR (negmiR) for each group, **P<0.01, *P<0.05, ns: not significant.
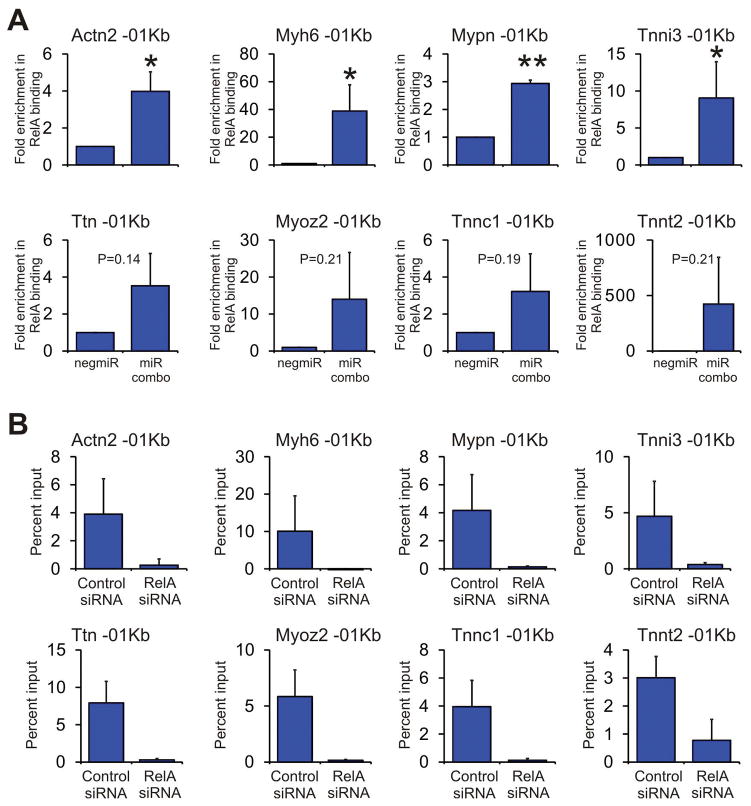
Finally, we wanted to determine how RelA controlled the expression of cardiomyocyte maturation genes. Consequently, we used ChIP assays to determine if miR combo induced RelA binding to the promoters of components of the cardiomyocyte sarcomere. Significant enrichment of RelA was observed at the Actn2, Myh6, Mypn and Tnni3 promoters (Figure 5A) following miR combo treatment. Similar enrichment was also observed for the Ttn, Myoz2, Tnnc1 and Tnnt2 promoters; however, this failed to reach P<0.05 significance (Figure 5A). There was no enrichment in the unrelated gene RPL30 (Supplementary Figure 5). Targeted knockdown of RelA completely removed the ChIP signal. This result verified that RelA was indeed binding to the promoters of the cardiomyocyte sarcomere genes (Figure 5B).
Figure 5. The NFκB subunit RelA binds to the promoters of cardiomyocyte maturation genes.
(A) Neonatal cardiac fibroblasts were transfected with negmiR or miR combo. After 7 days, chromatin DNA was subjected to ChIP analysis. Primers were designed to target the first 1Kb of the indicated cardiomyocyte sarcomere genes (represented by -01Kb). Results are presented as the fold enrichment in RelA binding where percent input of the negmiR control was taken to be 1. N=3 independent experiments. Data represented as Mean ± SEM. Comparisons are made between miR combo and negative control miR (negmiR), **P<0.01, *P<0.05, ns: not significant.
(B) Neonatal cardiac fibroblasts were transfected with miR combo and either a control siRNA or a siRNA that targeted RelA. After 7 days, chromatin DNA was subjected to ChIP analysis. Primers were designed to target the first 1Kb of the indicated cardiomyocyte sarcomere genes (represented by -01Kb). Results are presented as the percentage of chromatin input. N=3 independent experiments. Data represented as Mean ± SEM.
MicroRNAs activate TLR3
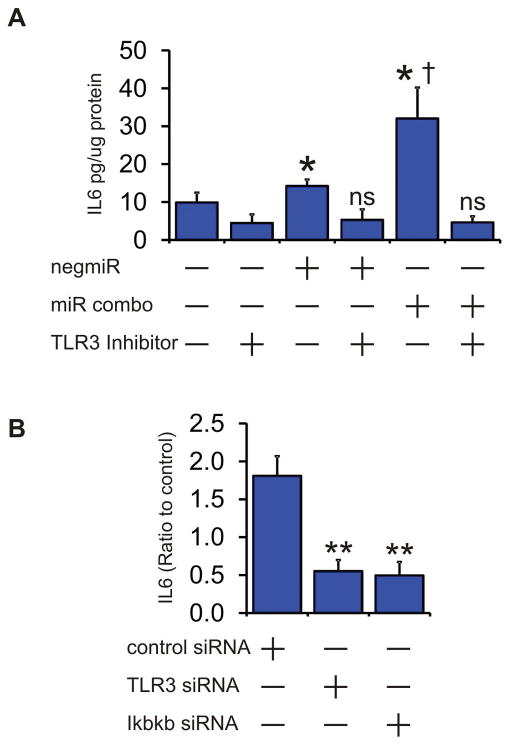
The pharmacological inhibitor and siRNA mediated knockdown experiments suggested that miR combo activated TLR3. To test this further we transfected cells with microRNAs and assessed TLR3 activity by measuring IL6 secretion into the media. IL6 secretion is an accepted measurement of the activity of TLRs, including TLR3 [30–37]. When compared to mock transfected fibroblasts both the control non-targeting miRNA (negmiR) and miR combo significantly induced IL6 secretion (Figure 6A). Comparisons between negmiR and miR combo indicated that miR combo had the stronger effect. The induction of IL6 secretion by microRNAs was TLR3 dependent; the addition of the TLR3 inhibitor CU-CPT-4a, which inhibits interaction between RNA and TLR3, completely ablated the effect of negmiR and miR combo upon IL6 secretion (Figure 6A). Targeted knockdown of TLR3 or Ikbkb inhibited miR combo induced IL6 secretion; further validating that miR combo activated the TLR3-NFκB pathway (Figure 6B).
Figure 6. microRNAs activate TLR3.
(A) Neonatal cardiac fibroblasts were transfected with negmiR or miR combo. A mock transfection where lipid reagent alone was added to the cells was also used. The TLR3 inhibitor CU-CPT-4a, which interferes with RNA binding to TLR3, was added one day post-transfection. IL6 concentration in the media was assessed 4 days post-transfection and values expressed as pg IL6 per ug of total protein. N=4 independent experiments. Data represented as Mean ± SEM. Comparisons are made to the respective mock transfected group (*P<0.05, ns: not significant) and between negmiR and miR combo (†P<0.05).
(B) Neonatal cardiac fibroblasts were transfected with microRNAs (mock, miR combo) and siRNA (non-targeting control, TLR3, Ikbkb). IL6 concentration in the media was assessed 4 days post-transfection and values expressed as a ratio between miR combo and mock transfected cells. N=3 independent experiments. Data represented as Mean ± SEM. Comparisons are made to miR combo plus non-targeting control siRNA group, **P<0.01, *P<0.05.
Pharmacological activation of TLR3 enhances maturation of reprogrammed fibroblasts
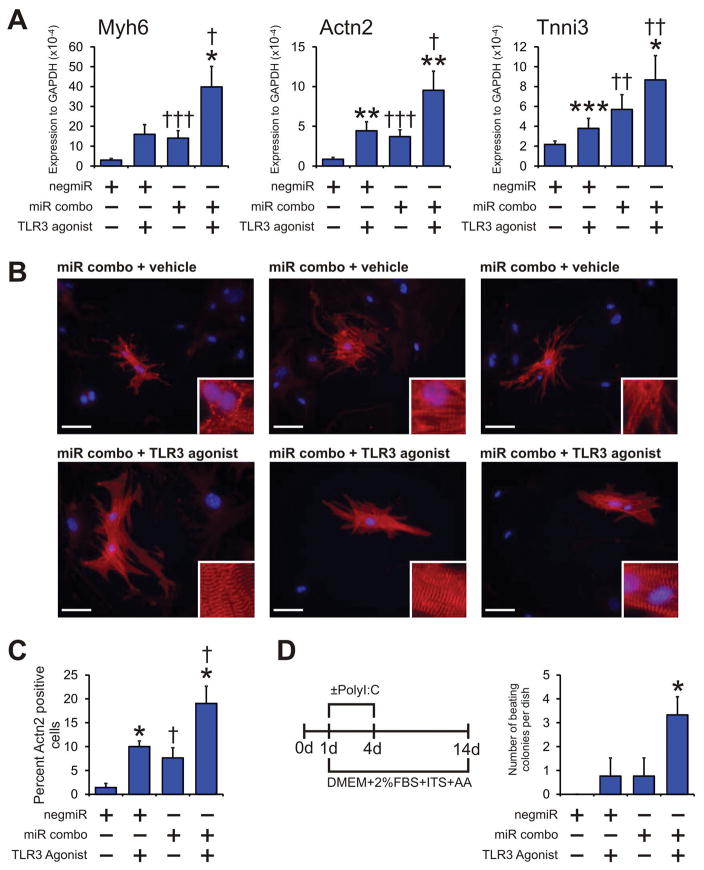
Following the identification of the mechanism by which TLR3 controlled miR combo reprogramming, we next examined if stimulation of TLR3 could enhance the efficiency of miR combo. As expected, miR combo increased RNA levels of Myh6, Actn2 and Tnni3 (Figure 7A). The effect of miR combo upon Myh6, Actn2 and Tnni3 expression was significantly enhanced by the addition of the TLR3 agonist Poly(I:C) (Figure 7A). Intriguingly, Poly(I:C) also induced expression of Myh6, Actn2 and Tnni3 in the control negmiR samples (Figure 7A). This effect, considering that Poly(I:C) had no effect on the expression of cardiomyocyte-commitment factors, is further evidence that the TLR3 pathway controls the maturation phase of cardiac reprogramming. We then performed immunostaining to determine if the effects at the RNA level were also observed at the protein level. Indeed, we found that the number of Actn2(+) cells that formed in response to miR combo treatment was increased by the TLR3 agonist Poly(I:C) (Figure 7B, with quantification provided in Figure 7C). We also noted that TLR3 activation enhanced sarcomere maturation (see figure inserts in Figure 7B).
Figure 7. TLR3 agonists enhance maturation of miR combo reprogrammed cardiomyocytes.
(A–C) Neonatal cardiac fibroblasts were transfected with negative control miR (negmiR) or miR combo. The day after transfection media was replaced and the cells incubated with vehicle or the TLR3 agonist Poly(I:C) LMW (low molecular weight Poly(I:C)) for a further 4 days. After incubation with the TLR3 agonist, cells were cultured in normal growth media for a further 10 days.
(A) RNA levels of the cardiomyocyte structural proteins Myh6 (αmyosin heavy chain), Actn2 (αsarcomeric actinin) and Tnni3 (cardiac troponin-I) was determined by qPCR. N=5–14.
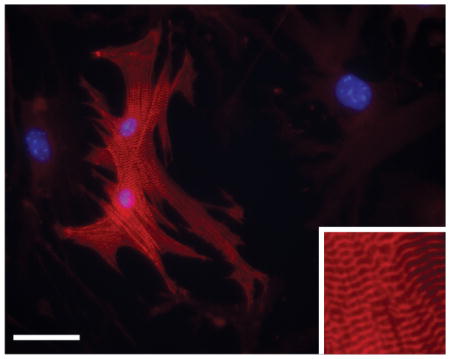
(B) Cells were fixed and stained with anti-Actn2 antibodies (red). Nuclei were stained with DAPI (blue). N=6 independent experiments. Scale bar 50 microns. Inset pictures are at 5x magnification to show sarcomere structure.
(C) Quantification of immunostaining shown in B. Cells expressing Actn2 were counted and expressed as a percentage of the total cell population.
(D) Neonatal cardiac fibroblasts were transfected with negative control miR (negmiR) or miR combo. The day after transfection media was replaced and the cells incubated with differentiation media (DMEM + 2%FBS + ITS + AA) and the TLR3 agonist Poly(I:C) LMW for the indicated times. Fourteen days after the transfection, the numbers of beating colonies were counted. N=4 independent experiments. Data represented as Mean ± SEM. *Comparisons made between vehicle and TLR3 agonist for each group ***P<0.001, **P<0.01, *P<0.05. †Comparisons made between miR combo and negmiR for each group †††P<0.001, ††P<0.01, †P<0.05.
In accordance with our previous study [9], we found that transfecting fibroblasts with miR combo led to the appearance of spontaneously beating colonies (Figure 7D). The ability of miR combo to form spontaneously beating colonies was increased 3-fold by the addition of the TLR3 agonist Poly(I:C) (Figure 7D). Importantly, mature beating colonies were observed within one week of transfection (Supplementary Movie 1).
Discussion
In this study we demonstrate that TLR3 activated NFκB is an important mechanism for the maturation of committed precursors into cardiomyocytes.
Our study clearly identified a role for TLR3 activated NFκB specifically in the maturation phase of cardiac reprogramming. This differs from previous studies which have linked TLR3 activated NKκB to the reprogramming to iPS [20] or endothelial cells [21]. These previous studies demonstrate that TLR3-NFκB causes global changes in the expression and activity of epigenetic modifiers that favors increased DNA accessibility. In this open chromatin configuration, the activation of the pluripotency program by the Yamanaka factors[20], or the induction of endothelial lineage by trans-differentiation factors[21], is facilitated. In these studies, the epigenetic plasticity that is induced by TLR3 activation is largely mediated by NFκB, as shown using pharmacological or molecular antagonists of NFκB.
We have extended this work by examining the role of TLR in maturation of cardiomyocyte precursors. We found that TLR3 activation increased the binding of NFκB directly to cardiomyocyte sarcomere genes. By contrast, we found that TLR3 played no role in the commitment of precursors into the cardiomyocyte lineage. TLR3 inhibition or knockdown did not influence the expression of various transcription factors that are necessary for commitment into the cardiomyocyte lineage.
Our findings may explain why cardiac reprogramming strategies are more efficient in the injured heart. After cardiac injury, dying cells in the injured zone release molecules which are recognized by innate immunity receptors such as TLRs [38, 39]. Moreover, various immune cells that invade the dead tissue secrete pro-inflammatory cytokines. The net result is an inflammatory environment in the post-MI heart that activates the RelA subunit of NFκB [40–42]. Indeed, our NFκB model for cardiomyocyte maturation may also explain why we have previously seen a degree of in vivo fibroblast conversion into mature cardiomyocytes even in the absence of reprogramming factors [11].
As mentioned above, we found that miR combo induced RelA binding to the promoters of various components of the cardiomyocyte sarcomere. Canonical RelA binding sites are present in the Myh6 promoter but are absent in the promoters of the Actn2, Mypn and Tnni3 genes. Non-canonical RelA binding sites have been identified in other genes [43], and they are present in in the Actn2, Mypn and Tnni3 gene promoters. However, it is also possible that RelA influences cardiac gene expression through an indirect mechanism. RelA, and NFκB, have been shown to modulate gene expression through binding to other proteins [44–46] as well as by modulating the activity of the epigenetic machinery. It is possible that the RelA subunit of NFκB plays a similar role in cardiomyocyte maturation.
Our study suggests that microRNAs directly activate TLR3. Several TLRs are known to bind to nucleic acids: TLR3; TLR7; TLR8; and TLR9[47]. TLR3 recognizes double-stranded (ds) RNA; whereas TLR7 and TLR8 bind to single-stranded RNA. In contrast, TLR9 is activated by unmethylated CpG sequences in DNA molecules[47]. Only a limited number of reports have demonstrated that microRNAs bind to TLRs. Even though microRNAs are dsRNA molecules, the microRNAs miR-21, miR-29a, and Let-7b bind, and activate, TLR7 and TLR8[48, 49]. With respect to TLR3, it was originally suggested that microRNAs might be too small to induce efficient dimerization, and thus activation, of TLR3 [50]. However, this assumption is likely to need revision both in light of our results as well as the recent report that the plant derived microRNA FνmiR168 binds to dendritic cell TLR3[51]. We found that miR combo more strongly induced TLR3 than the negative control microRNA used in our studies. This may suggest that TLR3 activation by microRNAs is sequence dependent. In support of this notion, siRNA mediated activation of TLRs has been shown to be sequence dependent[52].
Finally, our study has clinical implications for the therapeutic use of cardiac reprogramming. Based on the findings of our study, the addition of TLR3 ligands to reprogramming cocktails would be expected to enhance reprogramming efficiency; specifically by increasing the number of mature cardiomyocytes and thereby increasing the level of functional recovery that occurs when reprogramming agents are delivered into the infarcted heart. Systemic administration of TLR ligands can lead to septic shock [22, 23]; however, synthetic TLR3 ligands that induce little or no inflammatory response have recently been identified [53]. As such these synthetic ligands may be suitable for in vivo studies. We are currently investigating this possibility in conjunction with fibroblast lineage-tracing models in vivo with a view to determining if in vivo TLR3 activation enhances the functional improvement of direct reprogramming and, as such, be useful as an adjunctive therapy for cardiac reprogramming.
Conclusion
Cardiomyocyte maturation requires TLR3 and NFκB.
Supplementary Material
Significance Statement.
The process by which precursor cells mature into cardiomyocytes is poorly understood. In this study, we show that cardiomyocyte maturation is critically dependent upon TLR3 and NFκB.
Acknowledgments
Research conducted in these studies was supported by a NIH grant as as the Edna and Fred L. Mandel, Jr. Foundation.
Research conducted in these studies was supported by National Heart, Lung, and Blood Institute grant R01 HL131814-01A1 as well as the Edna and Fred L. Mandel, Jr. Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest
JC is a patent holder with Cooke Consulting LLC. All other authors decalred no conflicts of interest with respect to this study.
Author contributions: Conrad P Hodgkinson: Conception & Design, Collection and/or assembly of data, Data analysis & interpretation, Manuscript writing, Final approval of manuscript. Richard E Pratt: Conception & Design, Data analysis & interpretation, Manuscript writing, Final approval of manuscript. Imke Kirste: Conception & Design, Collection and/or assembly of data. Sophie Dal-Pra: Collection and/or assembly of data. John P Cooke: Conception & Design, Data analysis & interpretation, Final approval of manuscript. Victor J Dzau: Conception & Design, Financial support, Data analysis & interpretation, Manuscript writing, Final approval of manuscript.
References
- 1.Qian L, Huang Y, Spencer CI, et al. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fu JD, Stone NR, Liu L, et al. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Reports. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nam YJ, Song K, Luo X, et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci U S A. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Song K, Nam YJ, Luo X, et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nam YJ, Lubczyk C, Bhakta M, et al. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 2014;141:4267–4278. doi: 10.1242/dev.114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ifkovits JL, Addis RC, Epstein JA, et al. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One. 2014;9:e89678. doi: 10.1371/journal.pone.0089678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narita H, Shima F, Yokoyama J, et al. Engraftment and morphological development of vascularized human iPS cell-derived 3D-cardiomyocyte tissue after xenotransplantation. Sci Rep. 2017;7:13708. doi: 10.1038/s41598-017-14053-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamakawa H, Ieda M. Strategies for heart regeneration: approaches ranging from induced pluripotent stem cells to direct cardiac reprogramming. Int Heart J. 2015;56:1–5. doi: 10.1536/ihj.14-344. [DOI] [PubMed] [Google Scholar]
- 9.Jayawardena TM, Egemnazarov B, Finch EA, et al. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ Res. 2012;110:1465–1473. doi: 10.1161/CIRCRESAHA.112.269035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jayawardena T, Mirotsou M, Dzau VJ. Direct reprogramming of cardiac fibroblasts to cardiomyocytes using microRNAs. Methods Mol Biol. 2014;1150:263–272. doi: 10.1007/978-1-4939-0512-6_18. [DOI] [PubMed] [Google Scholar]
- 11.Jayawardena TM, Finch EA, Zhang L, et al. MicroRNA induced cardiac reprogramming in vivo: evidence for mature cardiac myocytes and improved cardiac function. Circ Res. 2015;116:418–424. doi: 10.1161/CIRCRESAHA.116.304510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang X, Hodgkinson CP, Lu K, et al. Selenium Augments microRNA Directed Reprogramming of Fibroblasts to Cardiomyocytes via Nanog. Sci Rep. 2016;6:23017. doi: 10.1038/srep23017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Dal-Pra S, Mirotsou M, et al. Tissue-engineered 3-dimensional (3D) microenvironment enhances the direct reprogramming of fibroblasts into cardiomyocytes by microRNAs. Sci Rep. 2016;6:38815. doi: 10.1038/srep38815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dal-Pra S, Hodgkinson CP, Mirotsou M, et al. Demethylation of H3K27 Is Essential for the Induction of Direct Cardiac Reprogramming by miR Combo. Circ Res. 2017 doi: 10.1161/CIRCRESAHA.116.308741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kathiriya IS, Nora EP, Bruneau BG. Investigating the transcriptional control of cardiovascular development. Circ Res. 2015;116:700–714. doi: 10.1161/CIRCRESAHA.116.302832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol. 2013;5:a008292. doi: 10.1101/cshperspect.a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.David L, Polo JM. Phases of reprogramming. Stem Cell Res. 2014;12:754–761. doi: 10.1016/j.scr.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet. 2013;14:427–439. doi: 10.1038/nrg3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Z, Chen O, Zheng M, et al. Re-patterning of H3K27me3, H3K4me3 and DNA methylation during fibroblast conversion into induced cardiomyocytes. Stem Cell Res. 2016;16:507–518. doi: 10.1016/j.scr.2016.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J, Sayed N, Hunter A, et al. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayed N, Wong WT, Ospino F, et al. Transdifferentiation of human fibroblasts to endothelial cells: role of innate immunity. Circulation. 2015;131:300–309. doi: 10.1161/CIRCULATIONAHA.113.007394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hodgkinson CP, Ye S. Toll-like receptors, their ligands, and atherosclerosis. ScientificWorldJournal. 2011;11:437–453. doi: 10.1100/tsw.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin YT, Verma A, Hodgkinson CP. Toll-like receptors and human disease: lessons from single nucleotide polymorphisms. Curr Genomics. 2012;13:633–645. doi: 10.2174/138920212803759712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hodgkinson CP, Naidoo V, Patti KG, et al. Abi3bp is a multifunctional autocrine/paracrine factor that regulates mesenchymal stem cell biology. Stem Cells. 2013;31:1669–1682. doi: 10.1002/stem.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hodgkinson CP, Laxton RC, Patel K, et al. Advanced glycation end-product of low density lipoprotein activates the toll-like 4 receptor pathway implications for diabetic atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:2275–2281. doi: 10.1161/ATVBAHA.108.175992. [DOI] [PubMed] [Google Scholar]
- 26.Cheng K, Wang X, Yin H. Small-molecule inhibitors of the TLR3/dsRNA complex. J Am Chem Soc. 2011;133:3764–3767. doi: 10.1021/ja111312h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faksh A, Britt RD, Jr, Vogel ER, et al. TLR3 activation increases chemokine expression in human fetal airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2016;310:L202–211. doi: 10.1152/ajplung.00151.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruneau BG. Transcriptional regulation of vertebrate cardiac morphogenesis. Circ Res. 2002;90:509–519. doi: 10.1161/01.res.0000013072.51957.b7. [DOI] [PubMed] [Google Scholar]
- 29.Banerjee A, Gerondakis S. Coordinating TLR-activated signaling pathways in cells of the immune system. Immunol Cell Biol. 2007;85:420–424. doi: 10.1038/sj.icb.7100098. [DOI] [PubMed] [Google Scholar]
- 30.Hahnlein JS, Ramwadhdoebe TH, Semmelink JF, et al. Distinctive expression of T cell guiding molecules in human autoimmune lymph node stromal cells upon TLR3 triggering. Sci Rep. 2018;8:1736. doi: 10.1038/s41598-018-19951-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bernard JJ, Cowing-Zitron C, Nakatsuji T, et al. Ultraviolet radiation damages self noncoding RNA and is detected by TLR3. Nat Med. 2012;18:1286–1290. doi: 10.1038/nm.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaefer TM, Fahey JV, Wright JA, et al. Innate immunity in the human female reproductive tract: antiviral response of uterine epithelial cells to the TLR3 agonist poly(I:C) J Immunol. 2005;174:992–1002. doi: 10.4049/jimmunol.174.2.992. [DOI] [PubMed] [Google Scholar]
- 33.Town T, Jeng D, Alexopoulou L, et al. Microglia recognize double-stranded RNA via TLR3. J Immunol. 2006;176:3804–3812. doi: 10.4049/jimmunol.176.6.3804. [DOI] [PubMed] [Google Scholar]
- 34.Kumar A, Zhang J, Yu FS. Toll-like receptor 3 agonist poly(I:C)-induced antiviral response in human corneal epithelial cells. Immunology. 2006;117:11–21. doi: 10.1111/j.1365-2567.2005.02258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lan T, Wang D, Bhagat L, et al. Design of synthetic oligoribonucleotide-based agonists of Toll-like receptor 3 and their immune response profiles in vitro and in vivo. Org Biomol Chem. 2013;11:1049–1058. doi: 10.1039/c2ob26946e. [DOI] [PubMed] [Google Scholar]
- 36.Mastri M, Shah Z, McLaughlin T, et al. Activation of Toll-like receptor 3 amplifies mesenchymal stem cell trophic factors and enhances therapeutic potency. Am J Physiol Cell Physiol. 2012;303:C1021–1033. doi: 10.1152/ajpcell.00191.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sironi M, Biasin M, Cagliani R, et al. A common polymorphism in TLR3 confers natural resistance to HIV-1 infection. J Immunol. 2012;188:818–823. doi: 10.4049/jimmunol.1102179. [DOI] [PubMed] [Google Scholar]
- 38.Epelman S, Liu PP, Mann DL. Role of innate and adaptive immune mechanisms in cardiac injury and repair. Nat Rev Immunol. 2015;15:117–129. doi: 10.1038/nri3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nahrendorf M, Swirski FK. Innate immune cells in ischaemic heart disease: does myocardial infarction beget myocardial infarction? Eur Heart J. 2016;37:868–872. doi: 10.1093/eurheartj/ehv453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kis A, Yellon DM, Baxter GF. Role of nuclear factor-kappa B activation in acute ischaemia-reperfusion injury in myocardium. Br J Pharmacol. 2003;138:894–900. doi: 10.1038/sj.bjp.0705108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mann DL. The emerging role of innate immunity in the heart and vascular system: for whom the cell tolls. Circ Res. 2011;108:1133–1145. doi: 10.1161/CIRCRESAHA.110.226936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Newton K, Dixit VM. Signaling in innate immunity and inflammation. Cold Spring Harb Perspect Biol. 2012:4. doi: 10.1101/cshperspect.a006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wong D, Teixeira A, Oikonomopoulos S, et al. Extensive characterization of NF-kappaB binding uncovers non-canonical motifs and advances the interpretation of genetic functional traits. Genome Biol. 2011;12:R70. doi: 10.1186/gb-2011-12-7-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashburner BP, Westerheide SD, Baldwin AS., Jr The p65 (RelA) subunit of NF-kappaB interacts with the histone deacetylase (HDAC) corepressors HDAC1 and HDAC2 to negatively regulate gene expression. Mol Cell Biol. 2001;21:7065–7077. doi: 10.1128/MCB.21.20.7065-7077.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamit-Hezi A, Nir S, Wolstein O, et al. Interaction of TAFII105 with selected p65/RelA dimers is associated with activation of subset of NF-kappa B genes. J Biol Chem. 2000;275:18180–18187. doi: 10.1074/jbc.275.24.18180. [DOI] [PubMed] [Google Scholar]
- 46.Islam KN, Mendelson CR. Potential role of nuclear factor kappaB and reactive oxygen species in cAMP and cytokine regulation of surfactant protein-A gene expression in lung type II cells. Mol Endocrinol. 2002;16:1428–1440. doi: 10.1210/mend.16.6.0856. [DOI] [PubMed] [Google Scholar]
- 47.Chen X, Liang H, Zhang J, et al. microRNAs are ligands of Toll-like receptors. RNA. 2013;19:737–739. doi: 10.1261/rna.036319.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fabbri M, Paone A, Calore F, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehmann SM, Kruger C, Park B, et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat Neurosci. 2012;15:827–835. doi: 10.1038/nn.3113. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Liu L, Davies DR, et al. Dimerization of Toll-like receptor 3 (TLR3) is required for ligand binding. J Biol Chem. 2010;285:36836–36841. doi: 10.1074/jbc.M110.167973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cavalieri D, Rizzetto L, Tocci N, et al. Plant microRNAs as novel immunomodulatory agents. Sci Rep. 2016;6:25761. doi: 10.1038/srep25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Judge AD, Sood V, Shaw JR, et al. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- 53.Matsumoto M, Tatematsu M, Nishikawa F, et al. Defined TLR3-specific adjuvant that induces NK and CTL activation without significant cytokine production in vivo. Nat Commun. 2015;6:6280. doi: 10.1038/ncomms7280. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.