Abstract
How graphene influences the behavior of living cells or tissues remains a critical issue for its application in biomedical studies, despite the general acceptance that graphene is biocompatible. While direct contact between cells and graphene is not a requirement for all biomedical applications, it is often mandatory for biosensing. Therefore, it is important to clarify whether graphene impedes the ability of cells to interact with biological elements in their environment. Here, a systematic study is reported to determine whether applying graphene on top of matrix substrates masks interactions between these substrates and retinal ganglion cells (RGCs). Six different platforms are tested for primary RGC cultures with three platforms comprised of matrix substrates compatible with these neurons, and another three having a layer of graphene placed on top of the matrix substrates. The results demonstrate that graphene does not impede interactions between RGCs and underlying substrate matrix, such that their positive or negative effects on neuron viability and vitality are retained. However, direct contact between RGCs and graphene reduces the number, but increases basal activity, of functional cation channels. The data indicate that, when proper baselines are established, graphene is a promising biosensing material for in vitro applications in neuroscience.
Keywords: cell survival, extracellular matrices, graphene, ion channels, primary neuron cultures
1. Introduction
Graphene has garnered immense attention ever since its first successful exfoliation in 2004.[1–4] Tremendous efforts have been devoted to demonstrating and understanding the superior mechanical, electrical and thermal properties of graphene, as well as exploring its applications in electronics, energy, water desalination, functional composites, etc.[5–17] Graphene also has the potential to make transformative impacts in the field of medicine. Significant research endeavors are ongoing to explore its potential applications for imaging, biosensing, drug delivery, and tissue engineering.[18–22] While great progresses have been made on almost all fronts, many fundamental questions pertaining to biomedical applications of graphene still remain unanswered.
One critical issue for biomedical studies is how graphene influences the behavior of living cells or tissues, which is still not well understood despite the general acceptance that graphene is biocompatible.[23–30] For example, based on its transparent nature and super-high electron mobility, graphene has been proposed as a sensing element that could enable many novel assays in electrophysiology.[31,32] To function optimally in this capacity, direct contact between cellular processes and graphene is required and this contact should minimally impact interactions between cells and their environment.
In biomedical studies, in vitro model systems utilizing cultured primary cells and cell lines are essential for delineating cellular mechanisms that cannot be readily examined in whole organisms. This includes neurobiological studies, for which graphene is of particular interest as a biosensor for electro-physiological assays.[18–22] Over the century long in vitro cell culture practice, many standard protocols have been developed for optimal cell culture. For neuronal cultures, the culture substrate is of primary importance for cell survival, differentiation and functionality; and the needs of a specific neuronal subtype or cell line dictate the optimal culture substrate.[33–38] Common matrix substrates for neuronal cultures include poly-l-lysine, poly-d-lysine (PDL), and laminin, which are coated on glass prior to cell plating. To account for the influence of matrix substrates on cell behavior, culture studies sometimes utilize control samples in which neurons are cultured on bare glass or silicon.[24–27,29]
Envisioning the great potential of graphene for sensing in neurobiological applications, a few groups have conducted studies to examine in vitro viability of primary neuronal cultures or human neuroblastoma cell lines on graphene substrates.[23–25,27–30] To date, both direct culture on bare graphene that is adhered to thin glass slides[24–26,28,29] and on graphene coated with popular matrix substrates[23,30] have been reported (see Table S1, Supporting Information). The comparison of outcomes of these studies is done while keeping in mind the fact that different cell types were used (i.e., neuroblastoma cell lines, and primary cells from different developmental stages and areas of the nervous system) and preparation of graphene substrates may also vary in different assays. Important features that may differ between studies are noted in Table S1 in the Supporting Information. Cell culture on bare graphene allows for direct interactions between the cells and graphene. However, graphene on bare glass substrates omits the supportive properties of matrix substrates, which increase in importance from neuroblastoma cell lines to primary neuronal cultures. The resultant studies suggest that neurons can be grown on bare graphene adhered to glass, but are conflicted on whether graphene influences cell viability and vitality.[10,23,25,26,28] Studies that combine graphene and matrix substrates utilize platforms in which graphene is sandwiched between a thin glass slide and a layer of matrix substrate, with cells plated on top of the matrix substrate. While this approach does achieve a combinatorial substrate of graphene and organic matrix, direct contact between cellular processes and graphene is prevented. Although the outcomes of these studies are also conflicting, most suggest that the presence of graphene underneath organic matrix substrates does not adversely impact cell adhesion, viability or vitality.[23,30]
Direct contact between cells and graphene is not a requirement for some neurobiological applications such as neuroprosthetics, in which graphene serves only as a flexible substrate to promote neural repair or regeneration.[27] However, direct contact between cells and graphene is often mandatory for biosensing applications. Therefore, it is important to clarify whether graphene impedes the ability of neurons to interact with biological elements in their environment that exist prior to graphene deposition. Here, we report on a systematic study to determine whether applying graphene on top of organic matrix substrates masks interactions between these matrix substrates and primary cultures of purified neurons. We fabricated six different platforms for primary cultures of retinal ganglion cells (RGCs) that were comprised of matrix substrates known to have low, medium and high efficacy for these neurons. Three of the culture platforms included a layer of graphene placed on top of the matrix substrate. Using these platforms, we determined whether graphene overlay and the resultant direct contact between RGCs and the graphene layer alters substrate efficacy, as measured by several important indices of cell viability and vitality, including receptor-mediated endocytosis and neurite outgrowth. We further assessed the specific potential for graphene to serve as a biosensor in electrophysiological assays by measuring substrate and graphene effects on cation channel activity. Results indicate that graphene overlay does not impede interactions between RGCs and underlying substrate matrix, such that the positive or negative effects of culture substrates are retained. This likely arises from the ability of graphene, with its atomic thickness, to conform to the structure of each substrate matrix. However, direct contact between RGCs results in increased cation channel activity, regardless of substrate. This is accompanied by a reduction in the number of functional voltage-gated sodium and potassium channels. Increased activity appears to serve as a compensatory mechanism for the change in ion channel representation, resulting in similar biophysical properties to RGCs cultured on substrate only platforms. Overall, our data indicate that, when the proper baselines are established, graphene is a promising biosensing material for in vitro applications in neuroscience, such as electrophysiological assays.
2. Results and Discussion
To test whether graphene overlay impedes interactions of neurons with underlying biological elements, we performed our studies with primary cultures of RGCs. RGCs are projection neurons, whose axons form the optic nerve. Like other projection neurons, establishing and maintaining RGCs as primary cell cultures can be challenging. Optimal establishment of these cultures requires precise culture conditions,[39] including application of a substrate matrix and media enrichment with supplemental amino acids and an extensive panel of neurotrophic and growth factors.[40–42] The sensitive nature of these neurons in vitro improves applicability of our findings to other neuronal subtypes, many with less stringent culture requirements.
As demonstrated by significant previous literature,[33–38] laminin is the preferred substrate for primary RGC cultures. Anecdotally, bare glass and PDL are generally considered sub-optimal, with PDL being preferable to bare glass. While these substrates have not been formally compared for efficacy, field standards for primary RGC cultures indicate that these three substrates represent a continuum of efficacy for the establishment of viable and vital RGCs. As such, we designed six different culture schemes, using these three substrates. We produced three substrate-only platforms. The first was a bare glass coverslip with no matrix substrate. The second and third platforms were glass coverslips coated with either laminin or PDL. We then adopted three graphene-integrated versions of the substrate-only platforms. For one platform, we placed graphene on bare glass (Figure 1A, left-hand panel). For the remaining two platforms, we coated glass coverslips with either laminin or PDL followed by graphene overlay (Figure 1A, middle and right-hand panels). As depicted in Figure 1A, RGCs were in direct contact with graphene in the graphene-integrated platforms. All cultures, regardless of platform, were maintained in optimal growth media for RGCs, as previously described.[42,43]
Figure 1.
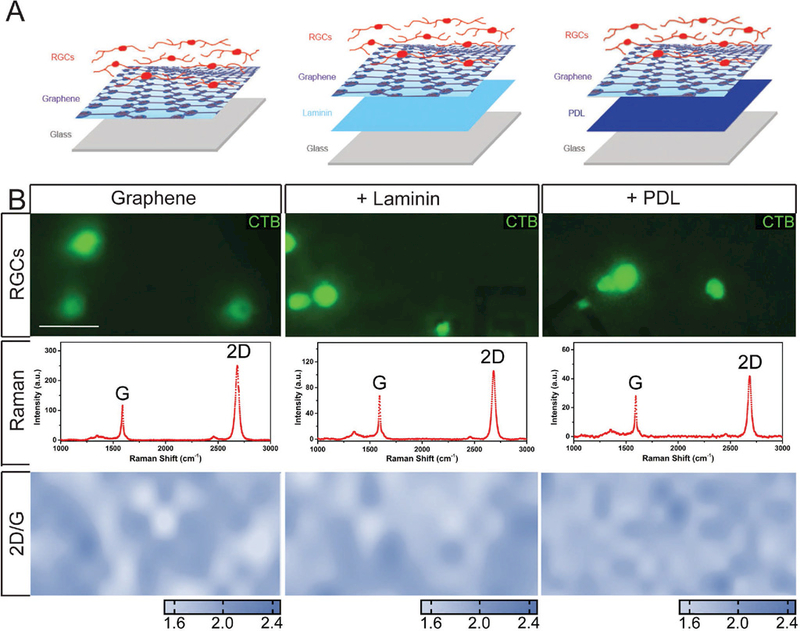
Matrix substrates and standard culture conditions do not affect the quality of graphene. A) Assembly schematics for graphene-integrated devices. Glass coverslips were coated with laminin or PDL. Graphene was then placed on top of laminin or PDL coating. RGCs were plated directly on top of graphene. B) (top panel) Representative fluorescent micrographs of RGCs cultured graphene alone (left), laminin (middle), or PDL (right) with graphene overlay. RGCs were labeled with CTB conjugated to Alexa Fluor-488 (green). (B) (second panel) Quality of the graphene alone, or on laminin or PDL, was assessed by Raman spectra and (B) (third panel) intensity ratio mapping of 2D and G peaks. Scale bar = 10 μm; Images taken at 40×.
To view cells in our culture, RGCs were labeled with the neural tracer cholera toxin beta subunit (CTB) conjugated to Alexa 488 fluorophore (Figure 1B; top panel). To make sure that these cells were cultured on graphene, we examined the graphene using Raman spectroscopy following cell plating and exposure to standard culture conditions for one week. As shown in the second panel of Figure 1B, the 2D (~2681 cm−1) peak exhibited a symmetric shape and the 2D-to-G (~1583 cm−1) intensity ratio was about 2, which indicates that the graphene had a monolayer structure. To characterize the continuity of the graphene, we extracted and plotted intensity ratio mapping of the characteristic graphene Raman 2D-G intensity ratio. This spatial mapping revealed continuous distribution of graphene underneath RGCs with glass, laminin, and PDL substrates (Figure 1B; third panel). These data indicate that: 1) successful transfer of graphene on laminin and PDL matrix is feasible, 2) graphene remains intact following cell plating and after exposure to the environmental conditions of cell culture (i.e., 37 °C and 5% CO2) for at least one week, and 3) direct contact with neurons does not disrupt the graphene layer, regardless of underlying substrate.
Viability, the ability of a cell to survive or live in its conditions successfully, is a key parameter for in vitro cell culture. Therefore, we measured RGC survival as a function of culture substrate and determined the impact of graphene overlay on the survival baselines for each of these substrates. RGC survival was determined by plasma membrane integrity and the presence of intracellular enzyme activity, using a Viability/Cytotoxicity assay (LIVE/DEAD Kit, Thermo Fisher). We labeled RGC cultures one week after plating with calcein-AM (green) and ethidium homodimer-1 (EthD-1, red) to measure intracellular enzyme activity and plasma membrane integrity, respectively. Calcein-AM is a cell-permeable dye that enters cells through passive diffusion. Once inside the cell, esterase enzymes remove the acetomethoxy (AM) group to produce a cell-impermeant, polar molecule that is highly fluorescent.[44] EthD-1 is a DNA/RNA stain that is impermeable to cells with an intact plasma membrane.[45] Cells that label calcein+ only are designated as “live” or “viable” cells, whereas cells that label EthD-1+ only are designated as “dead” cells.[46,47] Cells that label positive for both calcein and EthD-1 are designated as “compromised” due to a disrupted plasma membrane that allowed EthD-1 to enter, but still retain active intracellular enzyme activity to convert calcein-AM to calcein.
For cultures plated on bare glass as well as both matrix substrates a majority of RGCs were calcein+ (green) with minimal colabeling with EthD-1 (yellow), as shown in broad-field fluoro-micrographs (Figure 2A). This baseline level of cell compromise was consistent with previous studies.[43] EthD-1+ only cells (red) were essentially not detected at this one-week time point in any of our culture platforms (Figure 2A). Quantification of calcein and EthD-1 labeling revealed that graphene overlay did not alter RGC density, as compared to their respective substrate-only platforms (p > 0.05; Figure 2B). Importantly, comparison between platforms with and without graphene overlay indicates that graphene does not alter cell viability and the overall health of the cells in these two groups is similar. While it is not our main concern here, we found that the density of RGCs was 31% and 41% lower in the PDL platform, as compared to bare glass and laminin platforms, respectively (p < 0.05 for both; Figure 2B). The reduced RGC density noted in the PDL and graphene-PDL platforms could be due to either a reduction in cell adherence at the time of plating or to increased cell attrition prior to the one-week time point.
Figure 2.
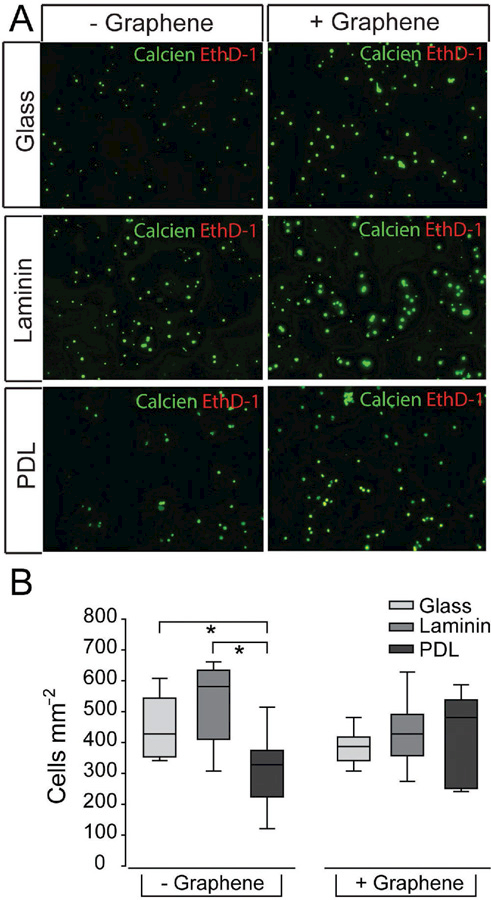
Matrix substrate, but not graphene overlay, impacts RGC density. A) Representative fluorescent micrographs of RGCs cultured on glass (top), laminin (middle), or PDL (bottom) with (+) or without (−) graphene overlay. RGCs were labeled with calcein (green) and ethidium homodimer-1 (red). Scale bar = 100 μm. B) Box plot of total cell density (y-axis; cells mm−2) in each culture platform. Asterisks indicate p < 0.05.
In addition to cell viability, it is of critical importance that graphene overlay does not impede the ability of matrix substrate to improve RGC vitality, or activity. To determine whether graphene overlay altered the ability of RGCs to execute a more complex cellular function, we first assessed receptor-mediated endocytosis across our culture platforms, using the active uptake, active transport tracer CTB. This tracer is commonly used to measure axon transport and connectivity in RGCs.[48–52] CTB binds to the GM1 ganglioside receptor and enters RGCs via caveolin-1-mediated endocytosis.[51,52] Following endocytosis, CTB is trafficked to RGC terminals via the microtubule network.[48,50] The activity required for uptake and transport of CTB makes it an ideal marker of not only viability, but also cell vitality. It is important to distinguish between a cell that is technically “viable” as it is able to survive in its conditions, and a cell that is maintaining vital, activity-dependent intracellular processes.[47]
In our studies, one week after plating in our six culture platforms, RGCs were treated with CTB conjugated to Alexa Fluor-594 (red) for 24 h. CTB uptake and transport was visualized by live cell fluorescent imaging. All culture platforms contained CTB + RGCs, which exhibited CTB in both the soma and along neurites (Figure 3A). Quantification of CTB + RGC density shows that the density of CTB + RGCs in graphene-integrated platforms did not differ statistically from their respective substrate-only platforms (p > 0.05 for all; Figure 3B). As such, we conclude that RGCs in the graphene-integrated platforms exhibited similar vitality to those in their respective substrate-only platforms, indicating that graphene overlay did not alter the measured efficacy of glass, laminin or PDL substrates.
Figure 3.
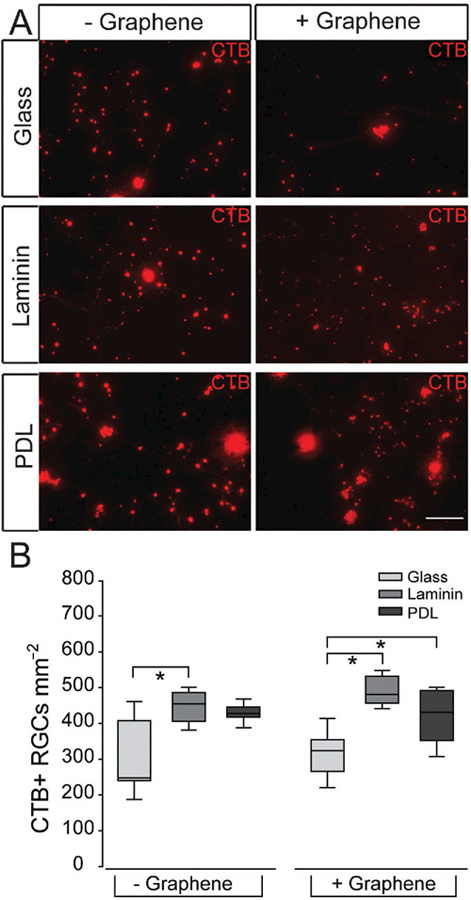
Receptor-mediated endocytosis of CTB in RGCs on culture substrates with graphene overlay. A) Representative fluorescent micrographs of RGCs cultured on glass (top), laminin (middle), or PDL (bottom) with (+) or without (−) graphene overlay. RGCs were labeled with CTB conjugated to Alexa Fluor-594 (red). Scale bar = 100 μm. B) Box plot of CTB+ cell density (y-axis; RGCs mm−2) in each culture platform. Asterisks indicate p < 0.05.
As to the effects of different matrix substrates, we found that the laminin-only platform contained 32% more CTB + RGCs than the bare glass platform (p < 0.05; Figure 3B). The PDL- only platform contained a median density of CTB + RGCs that was between bare glass and laminin platforms (p > 0.05; Figure 3B). For the graphene-integrated platforms, graphene overlay on bare glass contained 35% and 25% lower density of CTB + RGCs than graphene overlay on laminin and PDL, respectively (p < 0.05 for both; Figure 3B). These data suggest that while PDL substrate yields the lowest absolute density of RGCs (Figure 2), the vitality of these RGCs, as measured by receptor-mediated endocytosis, is higher than that achieved with a bare glass substrate.
In addition to receptor-mediated endocytosis, we adopted neurite outgrowth as another important indicator of neuronal vitality in vitro to assess whether graphene overlay affects the cell function. To measure neurite outgrowth in our six culture platforms, we quantified the complexity of RGC neurites with live cell imaging in cultures labeled with either calcein-AM or CTB (Figure 4A). Neurite complexity was measured by counting the number of times calcein+/CTB+ neurites intersected the lines of a 25 gm × 25 gm grid mask placed on each fluoromicrograph. This method of quantification accounts for changes in both the length and complexity of RGC neurites. The complexity of axonal branching is particularly important in establishing neural circuits; one of the primary reasons this method of quantification was chosen.[53] The traditional method of measuring neurite length was also performed and yielded the same findings as our combined analysis of length and complexity (Figure S1, Supporting Information).
Figure 4.
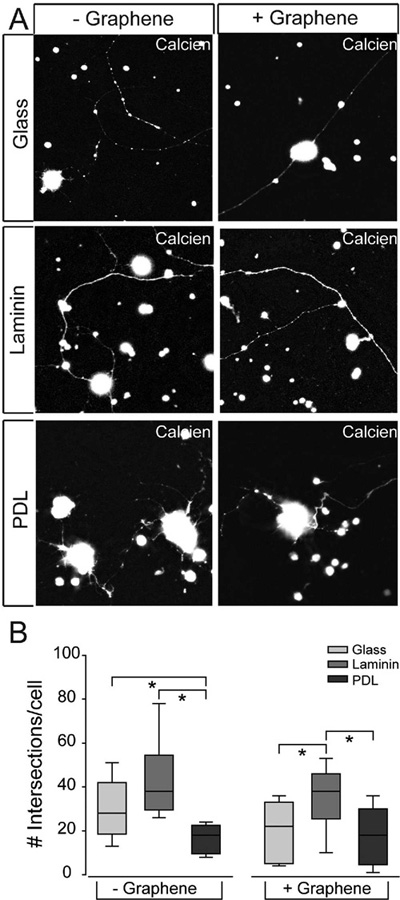
RGC neurite outgrowth on culture substrates with graphene overlay. A) Representative fluorescent micrographs of RGCs cultured on glass (top), laminin (middle), or PDL (bottom) with (+) or without (−) graphene overlay. RGCs were labeled with calcein (white). Scale bar = 100 μm. B) Box plot of the number of intersections per cell (y-axis) in each culture platform. Intersections were counted as the number of times any neurite crossed a line in the 25 × 25 μm grid that was overlaid on fluoromicrographs of 20× magnification. Asterisks indicate p < 0.05.
The quantification results revealed that the number of intersections between the grid mask and RGC neurites was twofold higher in the laminin platform and 1.5-fold higher in bare glass platform than in the PDL platform (p < 0.05 for both; Figure 4B). Importantly, RGCs cultured on the graphene-integrated laminin platform also exhibited the most neurite outgrowth, with ~1.5-fold more intersections than graphene-integrated glass and PDL platforms (p < 0.05 for both; Figure 4B). Overall, there is no significant difference in neurite outgrowth, as measured by the number of intersections, between graphene-integrated platforms and their respective substrate-only platforms (p > 0.05; Figure 4B), indicating that graphene overlay does not alter efficacy of glass, laminin, or PDL substrates.
As to the effects of different matrix substrates, Figure 4B suggests that PDL matrix impedes neurite outgrowth in RGCs, while laminin matrix promotes neurite outgrowth. Bare glass exhibits median efficacy as a substrate for neurite outgrowth that is similar to laminin in substrate-only devices and similar to PDL in graphene-integrated devices.
Ion channel activity is essential to neuronal function, particularly neurotransmission, and is the basis for electro-physiological assays. To more specifically assess the potential application of graphene as a biosensor for electrophysiological assays, which require direct contact between neuronal processes and graphene, we measured the effect of graphene overlay on cation channel activity in RGCs across our three culture substrates.
One week after plating cells in our six culture platforms, we performed real-time, thallium flux imaging.[54] In this assay, thallium acts as a surrogate for cations and a fluorescent signal is generated by thallium binding to a cell-permeable Thallosdye. Similar to traditional calcium imaging, increased fluorescent signal indicates opening of cation channels, which are promiscuously permeable to thallium. The representative images in Figure 5A depict baseline fluorescence of Thallos dye (left-hand panels) and fluorescence after the addition of thallium (right-hand panels). For illustration purposes, heat maps of the fluorescence signal for individual cells are depicted in panel insets (Figure 5A). For statistical comparison, we quantified cation channel activity as the change in fluorescent intensity of thallium between baseline and peak measurements (peak/baseline intensity) for individual RGCs (Figure 5B). RGCs cultured on bare glass and laminin matrix platforms exhibited similar levels of cation channel activity, as indicated by thallium flux (p > 0.05; Figure 5B). In contrast, RGCs cultured on the PDL matrix platform exhibited approximately 19–23% less cation channel activity than both glass and laminin substrates (p < 0.05; Figure 5B). Similarly, RGCs cultured on the graphene-integrated PDL platform exhibited 24% and 42% less thallium flux than RGCs cultured on graphene-integrated glass and laminin platforms, respectively (p < 0.05; Figure 5B). Like the substrate-only platforms, the graphene-integrated glass and laminin platforms exhibited similar levels of cation channel activity (p > 0.05; Figure 5B).
Figure 5.
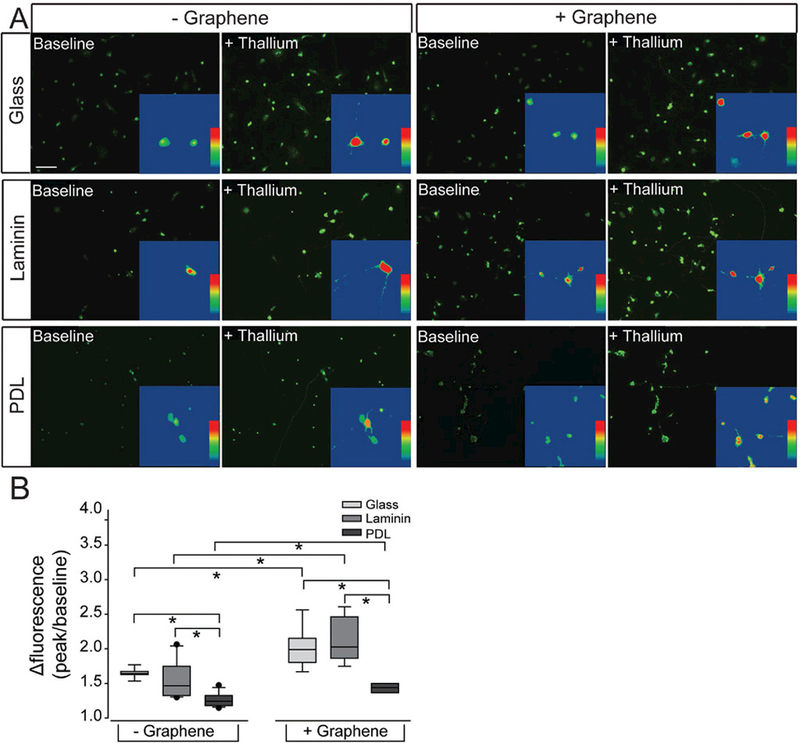
Cation channel activity in RGCs on culture substrates with graphene overlay. A) Representative fluorescent micrographs of RGCs cultured on glass (top), laminin (middle), or PDL (bottom) platforms with (+) or without (−) graphene overlay. RGCs were loaded with the cell-permeable dye Thallos (green). Images were taken at baseline and after addition of thallium, which binds to and increases the fluorescent intensity of Thallos dye. Scale bar = 100 μm. Insert: zoom of an individual cell within the larger image analyzed with a heat map showing the fluorescent signal of Thallos dye. B) Box plot of the change in the fluorescent intensity of each cell (peak intensity/baseline intensity).
Importantly, RGCs cultured on each of the graphene-integrated platforms exhibited greater cation channel activity than their respective substrate-only platforms (p < 0.05 for all; Figure 5B). This increase in thallium flux ranged from 6% to 37% (Figure 5B). These data indicate that: 1) PDL matrix impairs cation channel activity in RGCs, 2) graphene overlay increases overall cation channel activity, regardless of substrate, and 3) graphene overlay does not change the relative efficacy of glass, laminin, and PDL, which suggests that despite overall enhancement of cation channel activity, graphene overlay does not obscure the cation channel phenotype induced by the culture substrate.
To further inform the changes in ion channel activity seen from the thallium flux studies, we performed whole-cell, patch-clamp recordings on RGCs on laminin and graphene-integrated laminin platforms to measure inward and outward current activities of the cells. Since the effect of graphene overlay on cation channel activity was similar regardless of substrate and cell integrity is paramount for whole-cell patch clamp, we chose to only culture and record from cells on the preferred cultured substrate for RGCs, laminin. RGCs cultured on both laminin and graphene-integrated laminin platforms showed large somas with neurites extending from the cell body (Figure 6A, B). RGCs were further identified physiologically by applying brief, 40 ms, depolarizing test potentials from −80 to 30 mV in 10 mV increments.
Figure 6.
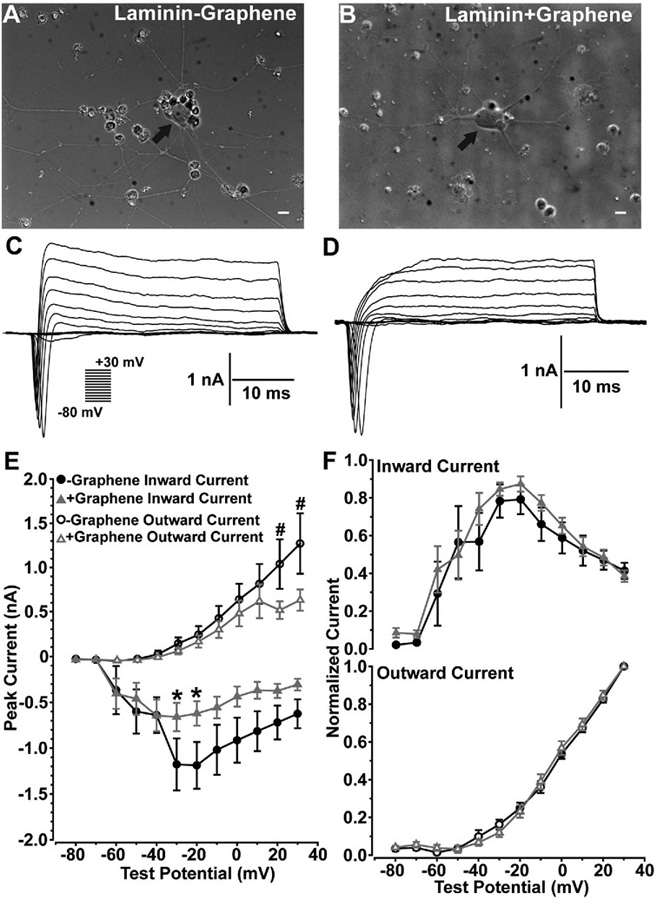
Graphene overlay does not alter the biophysical interaction between voltage-gated sodium and potassium channels in cultured RGCs. RGCs cultured on laminin platforms without (−), A) or with (+), B) graphene overlay showed large cell bodies (arrows) with widespread neurite processes. Scale bars = 20 μm. RGCs cultured on laminin platforms without (−), C) or with (+), D) graphene overlay produced transient inward currents followed by prolonged outward currents to depolarizing test potentials. E) RGCs cultured on graphene-integrated platforms showed significantly reduced inward (*) and outward (#) currents. F) However, the reduction in inward and outward currents does not alter the biophysical interaction between inward and outward currents. (n = 7, -Graphene; n = 10, +Graphene; *p < 0.025; #p < 0.015).
RGCs cultured on both laminin and graphene-integrated laminin platforms produced transient inward currents followed by prolonged outward currents to depolarizing test potentials (Figure 6C, D). RGC responses were quantified by measuring the peak of each transient inward and outward currents produced by each test potential. RGCs cultured on both laminin and graphene-integrated laminin platforms activated inward current statistically different from zero at −60 mV (Figure 6E). RGCs cultured on laminin platforms showed increased inward currents up to −20 mV followed by decreased inward currents from −10 to 30 mV, which is likely due to an interaction between inactivating voltage-gated sodium channels and activation of voltage-gated potassium channels at these higher test potentials. RGCs cultured on both laminin and graphene-integrated laminin platforms showed increased inward currents from −60 to −30 mV followed by a decrease in inward current for remaining test potentials. RGCs cultured on graphene-integrated laminin platforms showed a modest but statistically significant decrease in inward currents for test potentials between −30 and −20 mV as compared to laminin only substrate (*, p < 0.025). Both RGCs cultured on laminin and graphene-integrated laminin platforms showed outward current activation at −30 mV, and outward current generally increased as test potential increased. However, RGCs cultured on graphene-integrated laminin platforms showed a statistically significant decrease in peak outward current between 10 and 30 mV (#, p < 0.015). Reduced inward and outward currents for RGCs cultured on graphene-integrated platforms suggest a decrease in the number of functional voltage-gated sodium and potassium channels.
To understand if graphene impacts the biophysical characteristics of these channels mediating inward and outward currents, we normalized peak inward and outward current values produced at each test potential by the maximum inward or outward current value for each cell. Here, we found no significant 0.5 difference between inward and outward current profiles (Figure 6F) of RGCs cultured on laminin or graphene-integrated laminin platforms, suggesting the reduced number of voltage-gated sodium and potassium channels in RGCs plated on graphene does not influence the biophysical interaction between voltage-gated sodium and potassium channels governing action potential initiation and membrane repolarization.
3. Conclusion
These studies address one critical issue for neurobiological applications of graphene, that is, how graphene influences the behavior of living neurons, which is still not well understood despite the general acceptance that graphene is biocompatible.[23–30] Through systematic studies, we examine the effect of graphene through comparing the outcomes of viability, vitality, and electrophysiological function in primary cultures of RGCs on each of three common substrates (glass, laminin, and PDL) with and without graphene overlay. Our results confirm that culture substrate influences the health of primary neurons in culture. When all outcomes are considered, laminin substrate yielded the most robust RGC cultures, as anticipated. Based on anecdotal evidence, we expected PDL matrix to yield more robust RGC cultures than bare glass. However, our data suggest that the efficacy of bare glass and PDL matrix as substrates for RGC cultures depends on the outcome examined. For three of the four outcomes we examined, the bare glass platform yielded more robust RGC cultures than the PDL matrix platform, which suggests that interactions between RGCs and PDL matrix could negatively influence RGC viability, vitality, and function.
For all viability and vitality indices examined, graphene-integrated platforms exhibited the same pattern of RGC phenotypes as the substrate-only platforms. This indicates that direct contact between RGCs and graphene, a monolayer atomic structure, does not impede interactions between RGCs and underlying substrate matrix, such that the positive or negative effects of culture substrates are retained. It is likely that the atomically thin graphene conforms to the structure of underlying matrix. Mechanical interaction between cells and extracellular matrix and matrix structure is a key factor in this interaction.
Interestingly, we did observe that graphene enhances cation channel activity, as illustrated by an increase in the magnitude of thallium influx. Other studies examining the physiology of neurons cultured on graphene have found a potentiation of neurotransmission through increased presynaptic vesicle number, release probability, and turnover rate.[55] Graphene producing an increase in presynaptic neurotransmission could lead to increased postsynaptic cation channel activity, which is consistent with our thallium flux results on graphene-integrated platforms.
Electrophysiological recordings showed reduced inward and outward currents for RGCs cultured on graphene-integrated platforms, which indicates a decrease in the number of functional voltage-gated sodium and potassium channels. This reduction in cation channel expression could arise from a myriad of alterations, including transcription, translation, protein trafficking, and membrane structure.[55] Despite differences in ion channel representation, analysis of overall inward and outward current profiles revealed no significant difference between RGCs cultured on substrate-only or graphene-integrated platforms, indicating that graphene overlay does not alter the overall biophysical properties of RGCs. This is supported by previous studies showing that neurons cultured on graphene substrates do not show significantly altered electro-physiological properties, compared to neurons cultured on traditional culture substrates.[26] Increased cation channel activity, like that noted in our thallium flux assay, could serve as compensatory mechanism to maintain the biophysical properties of RGCs, despite graphene-mediated reductions in the number of functional voltage-gated sodium and potassium channels.
Overall, our data suggest that while graphene does not alter the biophysical phenotype of RGCs, it does alter the way in which this phenotype is achieved. Although further studies are required to elucidate the cause of changes in ion channel expression and the underlying mechanism for increased cation channel activity, the identification of these graphene-dependent changes is important for interpretation of electrophysiological assays utilizing graphene as the biosensor. Our data indicate that, when the proper baselines are established, graphene is a promising biosensing material for in vitro applications in neuroscience, such as electrophysiological assays.
4. Experimental Section
Graphene Synthesis:
Copper foils (Alfa Aesar 0.025 mm, 99.8% pure) with a grain size of =100 μm were cut into strips of 8 mm in width and then a 5% nitric acid (10 min) bath was used to eliminate contamination. A copper strip was loaded onto a quartz boat, which was transferred to a horizontal furnace system. After the system was pumped down to 10 mTorr, the copper strip was annealed at 1000 °C with 100sccm H2 for 1 h. A mixed gas of 80sccm H2 and 20sccm CH4 was then introduced into the furnace for the growth of graphene at 1000 °C for 30 min.
Substrate Preparation:
The metal markers were patterned on coverslips via photolithography, and deposited with 5 nm Cr and 40 nm Au by thermal evaporation. 8 × 8 mm squares were then cut from a graphene/copper strip. After spin coating with an ultrathin polymethyl methacrylate (PMMA) layer, a wet etching process was performed to remove the copper foil by copper etchant. The PMMA/graphene film was then transferred to the coverslips that were pre-coated with laminin or PDL. After waiting for the film dry overnight, acetone, isopropyl alcohol and deionized water baths were used to remove the PMMA layer and clean the substrates. Finally, Raman spectroscopy was utilized to identify the thickness and quality of graphene with a 532 nm excitation laser. A typical optical image of graphene on laminin-coated coverslips is shown in Figure S2 in the Supporting Information. The uniformity of graphene sheet and graphene edge can be identified via Raman mapping of the 2D peak intensity of graphene. The continuity of graphene can be further confirmed by optical images of graphene on SiO2/Si substrate with higher contrast ratio. To test the substrate quality, the laminin-coated substrates with anti-laminin antibodies before and after graphene transfer process were loaded. As shown in Figure S3 in the Supporting Information, laminin coating is present and consistent before and after transfer process, indicating that graphene transfer does not damage the laminin coating.
Purified Retinal Ganglion Cell Primary Cultures:
Primary cultures of purified RGCs were prepared as previously described.[39,40,42,43] Briefly, RGCs were purified by immunomagnetic separation, using mouse anti-rat Thy-1.1/Cd90 IgG antibody (5 μg mL−1, Cat# 554 895, BD Biosciences, San Jose, CA) and metallic microbeads conjugated to anti-mouse IgG secondary antibody (Cat# 130–047-102, Miltenyi Biotec, Auburn, CA). RGCs were plated in 24-well plates on glass coverslips (12 mm circle, Cat# 89 015–724, VWR, Radnor, PA) with the following coatings: glass alone, graphene on glass, laminin (0.01 mg mL−1; Cat# L6274, Sigma), graphene on laminin, PDL (0.1 mg mL−1, Cat# P6407, Sigma, Saint Louis, MO), or graphene on PDL (see Figure 1A). RGCs were grown in serum-free, Neurobasal A media (Cat# 21 103 049, Gibco, Carlsbad, CA), containing the following supplements: 2% B27 (Cat# 17 504 044, Gibco), 1% N2 (Cat# 17 502 048, Gibco), 2 × 10−3 mL-glutamine (Cat# G7513, Sigma), 100 × 10−6 m inosine (Cat# 58–63-9, Sigma), 0.1% gentamycin (Cat# 15 710–064, Gibco), 50 ng mL−1 brain-derived nerve growth factor (Cat# PHC7074, Gibco), 20 ng mL−1 ciliary neurotrophic factor (Cat# PRC7015, Gibco), and 10 ng mL−1 basic fibroblast growth factor (Cat# 13 256–029, Gibco). Experiments were performed on RGCs approximately 1 week after plating.
Cell Survival Assay:
To determine the extent of cell death in RGC cultures plated on different substrates, with or without graphene overlay, a LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells was utilized (Cat# L3224, ThermoFisher). Calcein-AM was used to label cells with active intracellular enzyme activity and ethidium homodimer-1 (EthD-1) was used to label cells with a compromised plasma membrane. RGCs were incubated in a phosphate-buffered saline solution containing 6 × 10−6 m calcein-AM and 4 × 10−6 m EthD-1 for 15 min at 37 °C. Fluorescent microscopy was used to image live cells following calcein-AM and EthD-1 staining. In subsequent fluoromicrographs, calcein+, EthD-1+, and calcein+/EthD-1+ RGCs were counted and the density (cells mm−2) of calcein+ and EthD-1+ RGCs (n = 13, glass; 10–11, laminin; 5–9, PDL) was calculated and compared between the three substrates with and without graphene.
Receptor-Mediated Endocytosis Assay:
To measure receptor-mediated endocytosis in RGC cultures plated on different substrates, with or without graphene overlay, the neural tracer cholera toxin β (CTB; μg μL−1 CTB-594, Cat# C34777; 1 μg μL−1 CTB-488, Cat# C34775, ThermoFisher) was utilized. 2 μL of CTB was added to each well (24-well plate, =800 μL media per well) and incubated for approximately 24 h at 37 °C. Live cell fluorescent microscopy was used to image uptake and transport of CTB in RGCs. In subsequent fluoromicrographs, CTB+ RGCs were counted and the cell density was quantified by calculating the number of cells per mm2. The quantification of CTB+ cell density (n = 7, glass; 9–12, laminin; 5–10, PDL) was compared between the three substrates with and without graphene.
Neurite Outgrowth Assay:
RGC cultures were labeled with calcein, as described above. Neurite complexity was measured in calcein + RGCs by overlaying fluoromicrographs with a 25 × 25 μm grid mask and counting the number of times neurites intersected with the grid. The quantification of the number of intersections between calcein+ (n = 5–6, glass; 5–6, laminin; 6–18, PDL) neurites and the grid mask was compared between the three substrates with and without graphene.
Ion Channel Activity Assay:
Thallium flux imaging was utilized to assession channel activity in RGC cultures plated on different substrates, with or without graphene overlay. Cells were loaded with Thallos-AM dye (0.5 μg μL−1, Cat# 0902, TEFlabs, Austin, TX), generously provided by Dave Weaver ofVanderbilt University, by incubating with the dye for 30 min at 37 °C. Following dye loading, cells were washed with fresh media. Using live cell fluorescence microscopy, the baseline fluorescence of the Thallos dye was recorded for continuously 30 s (images taken every 10 s). 2 × 10−3 m thallium (TI+) solution was then added to cell culture media and live imaging was performed continuously for the 5 min (images taken every 10 s). Fluorescence intensity was calculated for each image using Nikon NIS-Elements software. To determine the change in fluorescence intensity of the thallos dye after TI+ was added, the peak intensity after TI+ was divided by the baseline intensity to generate Δfluorescence. Δfluorescence was compared between the three substrates with and without graphene (n = 6–9, glass; 5, laminin; 4–7, PDL).
Electrophysiological Recordings:
Cultured RGCs, which showed circular somas with neurite processes extending from the cell body, were targeted for intracellular recordings. The culture medium was exchanged with an extracellular solution containing (in mM) 140 NaCl, 5 KCl, 2 CaCl2, 2 MgCl2, 10 Hepes, 10 dextrose adjusted to a pH of 7.4 with NaOH (290 osm). Intracellular recording pipettes were pulled from thick-walled borosilicate glass and filled with (in mM) 130 KCl, 10 NaCl, 0.5 CaCl2, 2 MgCl2, 5 EGTA, 10 Hepes, 3 Na2-ATP adjusted to pH 7.2 with KOH (280 osm). Recording pipettes had a resistance between 8 and 11 MΩ. External and internal solutions were the same as used by Ruiz-Ederra et al.[56] to investigate retinal cultured Muller cell physiology. Electrophysiological recordings were performed at room temperature (23 °C). RGCs were viewed using an upright microscope (Olympus BX51, 40x) under differential interference contrast, using a CCD (charge- coupled device) camera (Andor). The recording pipette was positioned near an RGC, using a micromanipulator, and the potential difference between intracellular and extracellular solutions was subtracted online. The recording pipette was then positioned onto an RGC, forming a tight seal (>1 GΩ) between the recording pipette and cultured RGC. Then, brief negative pressure pulses were applied to break into cells to form a whole-cell configuration (access resistance typically <50 MΩ) and voltage clamped at −80 mV. To assess basic electrical activity from cultured RGCs, currents produced by brief, 40 ms, voltage steps from −80 to +30 mV in +10 mV increments were measured. Changes in input current were subtracted from the response to the test potential by averaging six hyperpolarizing pulses after the test potential (P/6 protocol). Analog electrical activity was obtained (Multiclamp 700B, Molecular Devices),digitized (sampling rate 50 kHz, Digidata 1550A, Molecular Devices), and analyzed using Clampfit 10.6 (Molecular Devices). Here, peak inward and outward currents generated during each test potential were measured. Inward and outward currents were respectively normalized by dividing each response by the maximum response of each cell (n = 7, -Graphene; n = 10, +Graphene). Two-way analysis of variance [ANOVAs, one between subjects (culture substrate) and one within subjects (test potential)] was used to compare inward and outward currents produced by each test potential between RGCs cultured on laminin platforms or graphene-integrated laminin platforms; following ANOVAs, Tukey post-hoc tests to assess multiple comparisons were used. Statistical significance was defined as P < 0.05. Sigma Plot (Version 12.5, Systat Software Inc.) to perform statistical analyses was used.
Statistical Analysis:
Statistical analysis was conducted with SigmaPlot Version 13 (Systat Software Inc, San Jose, CA). For all assays, data were assessed for normality by Shapiro-Wilkes test and for equal variance by Brown-Forsythe test. All data passed normality test (p > 0.05 for all). For comparisons within substrate-only and graphene-integrated platforms that passed equal variance testing (p > 0.05), statistical significance was assessed by a one-way ANOVA with post-hoc correction and pairwise comparison by Holm-Sidak Method. For comparisons within substrate-only and graphene-integrated platforms that failed equal variance testing (p < 0.05), statistical significance was assessed by a Kruskal-Wallis one-way ANOVA on ranks with post-hoc correction and pairwise comparison by Dunn’s Method. Statistical comparisons between graphene-integrated platforms and their respective substrate-only platforms were conducted with a T-test. For all statistical comparisons, p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgements
R.A.F. and Y.Z. contributed equally to this work. These studies were supported with funding from National Eye Institute: P30EY008126 (RMS; VVRC), R01EY027729 (DL,YX,RMS), R21EY026176 (DL,YX,RMS), T32EY007135 (RAS;VVRC); Unrestricted Departmental Award, Research to Prevent Blindness, Inc. (VEI).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
References
- [1].Novoselov KS, Geim AK, Morozov SV, Jiang D, Zhang Y, Dubonos SV, Grigorieval IV, Firsov AA, Science 2004, 306, 666. [DOI] [PubMed] [Google Scholar]
- [2].Castro Neto AH, Guinea F, Peres NMR, Novoselov KS, Geim AK, Rev. Mod. Phys 2009, 81, 109. [Google Scholar]
- [3].Mueller T, Xia F, Avouris P, Nat. Photonics 2010, 4, 297. [Google Scholar]
- [4].Engel M, Steiner M, Lombardo A, Ferrari AC, Lohneysen HV, Avouris P, Krupke R, Nat. Commun 2012, 3, 906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang Y, Shao Y, Matson DW, Li J, Lin Y, ACS Nano 2010, 4, 1790. [DOI] [PubMed] [Google Scholar]
- [6].Yang Y, Yang X, Zou X, Wu S, Wan D, Cao A, Liao L, Yuan Q, Duan X, Adv. Funct. Mater 2017, 27, 1604096. [Google Scholar]
- [7].Liu S, Guo X, NPG Asia Mater 2012, 4, e23. [Google Scholar]
- [8].Gao N, Gao T, Yang X, Dai X, Zhou W, Zhang A, Lieber CM, Proc. Natl. Acad. Sci. USA 2016, 113, 14633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kireev D, Seyock S, Lewen J, Maybeck V, Wolfrum B, Offenhausser A, Adv. Healthc. Mater 2017, 6. [DOI] [PubMed] [Google Scholar]
- [10].Park DW, Brodnick SK, Ness JP, Atry F, Krugner-Higby L, Sandberg A, Mikael S, Richner TJ, Novello J, Kim H, Baek D-H, Bong J, Frye ST, Thongpang S, Swanson KI, Lake W, Pashaie R, Williams JC, Ma Z, Nat. Protoc 2016, 11, 2201. [DOI] [PubMed] [Google Scholar]
- [11].Marchesan S, B L, Prato M, Science 2017, 356, 1010. [DOI] [PubMed] [Google Scholar]
- [12].Koerbitzer B, Krauss P, Nick C, Yadav S, Schneider JJ, Thielemann C, 2D Mater 2016, 3, 024004. [Google Scholar]
- [13].Liu C, Yu Z, Neff D, Zhamu A, Jang BZ, Nano Lett 2010, 10, 4863. [DOI] [PubMed] [Google Scholar]
- [14].Dean CR, Young AF, Meric I, Lee C, Wang L, Sorgenfrei S, Watanabe K, Taniguchi T, Kim P, Shepard KL, Hone J, Nat Nanotechnol 2010, 5, 722. [DOI] [PubMed] [Google Scholar]
- [15].Park S, Mohanty N, Suk JW, Nagaraja A, An J, Piner RD, Cai W, Dreyer DR, Berry V, Ruoff RS, Adv. Mater 2010, 22, 1736. [DOI] [PubMed] [Google Scholar]
- [16].Surwade SP, Smirnov SN, Vlassiouk IV, Unocic RR, Veith GM, Dai S, Mahurin SM, Nat. Nanotechnol 2015, 10, 459. [DOI] [PubMed] [Google Scholar]
- [17].Kim KH, Oh Y, Islam MF, Nat. Nanotechnol 2012, 7, 562. [DOI] [PubMed] [Google Scholar]
- [18].Zhang Y, Dodson KH, Fischer R, Wang R, Li D, Sappington RM, Xu Y, Nanoscale 2016, 8, 19043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Liu Q, Guo B, Rao Z, Zhang B, Gong JR, Nano Lett 2013, 13, 2436. [DOI] [PubMed] [Google Scholar]
- [20].Cohen-Karni T, Qing Q, Li Q, Fang Y, Lieber CM, Nano Lett 2010, 10, 1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Weaver CL, LaRosa JM, Luo X, Cui XT, ACS Nano 2014, 8, 1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sayyar S, Murray E, Thompson B, Chung J, Officer DL,J. Mater. Chem. 2015, 3, 481. [DOI] [PubMed] [Google Scholar]
- [23].Veliev F, Briancon-Marjollet A, Bouchiat V, Delacour C, Biomaterials 2016, 86, 33. [DOI] [PubMed] [Google Scholar]
- [24].He Z, Zhang S, Song Q, Li W, Liu D, Li H, Tang M, Chai R, Colloids Surf. B Biointerfaces 2016, 146, 442. [DOI] [PubMed] [Google Scholar]
- [25].Lee JS, Lipatov A, Ha L, Shekhirev M, Andalib MN, Sinitskii A, Lim JY, Biochem. Biophys. Res. Commun 2015, 460, 267. [DOI] [PubMed] [Google Scholar]
- [26].Fabbro A, Scaini D, Leon V, Vazquez E, Cellot G, Privitera G,Lombardi L, Torrisi F, Tomarchio Fl., Bonaccorso F, Bosi S, Ferrari AC, Ballerini L, Prato M, ACS Nano 2016, 10, 615. [DOI] [PubMed] [Google Scholar]
- [27].Bendali A, Hess LH, Seifert M, Forster V, Stephan AF, Garrido JA, Picaud S, Adv. Healthc. Mater 2013, 2, 929. [DOI] [PubMed] [Google Scholar]
- [28].Sahni D, Jea A, Mata JA, Marcano DC, Sivaganesan A, Berlin JM, Tatsui CE, Sun Z, Luerssen TG, Meng S, Kent TA, Tour JM, J. Neurosurg. Pediatr 2013, 11, 575. [DOI] [PubMed] [Google Scholar]
- [29].Park H-B,J. Microbiol. Biotechnol. 2013, 23, 274. [DOI] [PubMed] [Google Scholar]
- [30].Li N, Zhang X, Song Q, Su R, Zhang Q, Kong T, Liu L, Jin G, Tang M, Cheng G, Biomaterials 2011, 32, 9374. [DOI] [PubMed] [Google Scholar]
- [31].Kuzum D, Takano H, Shim E, Reed JC, Juul H, Richardson AG, de Vries J, Bink H, Dichter MA, Lucas TH, Coulter DA, Cubukcu E, Litt B, Nat. Commun 2014, 5, 5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Park DW, Schendel AA, Mikael S, Brodnick SK, Richner TJ, Ness JP, Hayat MR, Atry F, Frye ST, Pashaie R, Thongpang S, Ma Z, Williams JC, Nat. Commun 2014, 5, 5258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Meyer-Franke A, Kaplan MR, Pfreiger FW, Barres BA, Neuro-endocrinology 1995, 15, 805. [DOI] [PubMed] [Google Scholar]
- [34].Chen Q, Kinch MS, Lin TH, Burridge K, Juliano RL, J. Biol. Chem 1994, 269, 26602. [PubMed] [Google Scholar]
- [35].Kuhn TB, Brown MD, Bamburg JR, J. Neurobiol 1998, 37, 524. [DOI] [PubMed] [Google Scholar]
- [36].Smalheiser NR, Crain SM, Reid LM, Dev. Brain Res 1984, 12, 136. [DOI] [PubMed] [Google Scholar]
- [37].Cohen J, Burne JF, McKinlay C, Winter J, Dev. Biol 1987, 122, 407. [DOI] [PubMed] [Google Scholar]
- [38].Cohen J, Johnson AR, J. Cell Sci 1991, 15, 1. [DOI] [PubMed] [Google Scholar]
- [39].Sappington RM, Sidorova T, Ward NJ, Chakravarthy R, Ho KW, Calkins DJ, Channels (Austin) 2015, 9, 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Lee SJ, Duncan DS, Echevarria FD, McLaughlin WM, Hatcher JB, Sappington RM, J. Clin. Exp. Ophthalmol 2015, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sappington RM, Calkins DJ, IOVS 2006, 47, 3860. [DOI] [PubMed] [Google Scholar]
- [42].Sappington RM, Sidorova T, Long DJ, Calkins DJ, Invest. Ophthalmol. Vis. Sci 2009, 50, 717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sappington RM, Chan M, Calkins DJ, Invest. Ophthalmol. Vis. Sci 2006, 47, 2932. [DOI] [PubMed] [Google Scholar]
- [44].Legrand O, Simonin G, Perrot J-Y, Zittoun R, Blood 1998, 91, 4480. [PubMed] [Google Scholar]
- [45].Poole CA, Brookes NH, Gilbert RT, Beaumont BW, Crowther A, Scott L, Merrilees MJ, Connect. Tissue Res 2009, 33, 233. [DOI] [PubMed] [Google Scholar]
- [46].Chen S, Nilsen J, Brinton RD, Endocrinology 2006, 147, 5303. [DOI] [PubMed] [Google Scholar]
- [47].Grieshaber P, Lagreze WA, Noack C, Boehringer D, Biermann J, J. Neurosci. Methods 2010, 192, 233. [DOI] [PubMed] [Google Scholar]
- [48].Crish SD, Calkins DJ, Neuroscience 2011, 176, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Formichella CR, Abella SK, Sims SM, Cathcart HM, Sappington RM, J. Clin. Cell Immunol 2014, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Crish SD, Dapper JD, MacNamee SE, Balaram P, Sidorova TN, Lambert WS, Calkins DJ, Neuroscience 2013, 229, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Crish SD, Sappington RM, Inman DM, Horner PJ, Calkins DJ, Proc. Natl. Acad. Sci. USA 2010, 107, 5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Echevarria FD, Walker CC, Abella SK, Won M, Sappington RM, J. Clin. Exp. Ophthalmol 2013, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kalil K, Dent EW, Nat. Rev. Neurosci 2014, 15, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Weaver CD, Harden D, Dworetzky SI, Robertson B, Knox RJ, J. Biomol. Screen 2004, 9, 671. [DOI] [PubMed] [Google Scholar]
- [55].Kitko KE, Hong T, Lazarenko RM, Ying D, Xu YQ, Zhang Q, Nat. Commun 2018, 9, 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Ruiz-Ederra J, Zhang H, Verkman AS, J. Biol. Chem 2007, 282,21866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


