With the dissemination of carbapenemase-producing Enterobacteriaceae (CPE) strains worldwide, carbapenem-hydrolyzing enzymes are increasingly reported among isolates of Escherichia coli, the first hospital and community-acquired opportunistic pathogen. Here, we have performed an epidemiological survey of carbapenemase-producing E. coli (CP-Ec) isolates received at the French National Reference Centre (F-NRC) in 2012 and 2013.
KEYWORDS: MLST, rep-PCR, OXA-48, NDM, VIM, KPC, epidemiology, carbapenemase, molecular epidemiology
ABSTRACT
With the dissemination of carbapenemase-producing Enterobacteriaceae (CPE) strains worldwide, carbapenem-hydrolyzing enzymes are increasingly reported among isolates of Escherichia coli, the first hospital and community-acquired opportunistic pathogen. Here, we have performed an epidemiological survey of carbapenemase-producing E. coli (CP-Ec) isolates received at the French National Reference Centre (F-NRC) in 2012 and 2013. Antimicrobial susceptibilities for last-resort antibiotics and antimicrobial compounds commonly used to treat urinary tract infections were determined by broth microdilution. Clonal relationship was assessed using repetitive sequence-based PCR (rep-PCR) and multilocus sequence typing (MLST). From this collection of 140 carbapenemase-producing E. coli isolates, 74% produced an OXA-48-like carbapenemase and 21% produced an NDM carbapenemase. A link with a foreign country was suspected for 37% of infected/colonized patients. Most of the isolates were from screening (56%) and from urine samples (26%). Colistin, fosfomycin, and nitrofurantoin possessed the most consistent activity, with 100%, 95%, and 96% isolates susceptible, respectively. A wide diversity of carbapenemase-producing E. coli isolates has been found (50 different sequence types [STs]). The most prevalent clones were (i) E. coli sequence type 38 (ST38) producing OXA-48 (n = 21), a clone linked to Turkey and North African countries, (ii) E. coli ST-90 producing OXA-204 (n = 9), which was responsible for an outbreak related to a contaminated duodenoscope, and (iii) E. coli ST-410 producing OXA-181 (n = 5), which was recovered from patients of different geographical origins. These specific clones might be considered high-risk clones for the dissemination of carbapenemases in E. coli. The wide diversity of STs, combined with the increasing number of CP-Ec isolates received by the F-NRC, suggests a likely dissemination of CP-Ec isolates in the community.
INTRODUCTION
Antimicrobial resistance has become a major challenge for public health worldwide. One of the most worrying threats is the emergence and rapid dissemination of carbapenem-resistant Gram-negative bacteria, which is mainly caused by the spread of carbapenemase-producing Enterobacteriaceae (CPE). These CPE strains are often multidrug resistant, if not pan-drug resistant, leaving only very few therapeutic options for treating serious infections.
Carbapenem-hydrolyzing β-lactamases (i.e., carbapenemases) encountered in Enterobacteriaceae belong to either (i) Ambler class A, including KPC, IMI, and GES enzymes, (ii) Ambler class B metallo-β-lactamases (MBLs) of the NDM, VIM, and IMP types, or (iii) Ambler class D enzymes, including OXA-48 and its variants (mostly OXA-181, OXA-204, and OXA-232) (1). Even though France is not considered a country where CPE are endemic (2), local, regional, and interregional outbreaks are increasingly reported (2, 3). These epidemic disseminations most often involve Klebsiella pneumoniae isolates (4, 5). However, Escherichia coli is the second species in terms of isolation frequency among CPE isolates, and its frequency rose from 15.2% in 2012 to 23.8% in 2014 (6). The emergence of carbapenemase-producing E. coli (CP-Ec) is a matter of concern, because unlike K. pneumoniae, which is primarily a nosocomial pathogen, E. coli isolates are responsible for both community- and hospital-acquired infections, thus raising the fear of CPE dissemination in the community. Here, we performed an epidemiological survey of CP-Ec isolates received at the French National Reference Centre (F-NRC) in 2012 and 2013 to identify the emergence of potential high-risk clones and analyze their antimicrobial susceptibility profiles.
RESULTS
Sources of CP-Ec isolates.
A large majority of CP-Ec isolates (n = 140) have been isolated in hospitals (n = 112; 80%). These isolates were recovered from clinical specimens (n = 53; 38%) and screening samples (n = 79; 56%) (detailed information could not be obtained for 8 samples). The distribution of clinical samples was as follows: urine samples (n = 36), blood samples (n = 4), bile samples (n = 4), wound samples (n = 3), vaginal swabs (n = 3), respiratory specimen (n = 1), hip pus sample (n = 1), and gastric fluid of a newborn (n = 1).
Among the 140 studied isolates, 104 (74.3%) produced an OXA-48-like carbapenemase. The other isolates produced NDM-type (20.7%), VIM-type (3.6%), and KPC-3 (0.7%) carbapenemases (see Table S1 in the supplemental material). One isolate coproduced OXA-232 and NDM-1 (0.7%).
Most of the samples were isolated in three regions: Paris and suburbs, the northeast of France, and the southeast of France. A probable link with a foreign country was established for 37% (52/140) of the patients infected or colonized with a CPE (e.g., hospitalization abroad within the year preceding the CPE isolation, recent travel abroad, or living outside France) (Table 1). Twelve (40%) of the NDM-producing isolates were linked to the Indian subcontinent, which is advocated to be the main reservoir of this type of metallo-β-lactamase (7). Among the 104 OXA-48-like carbapenemase producers, 35 (34%) were associated with cross-border transfers, especially with North African countries, which is in agreement with the epidemiological data known for OXA-48-like carbapenemases (8–10). For the 69 remaining OXA-48-like carbapenemase cases, no clear link with a foreign country could be established. Finally, only one KPC-3-producing isolate was recovered from a patient having links with Italy, where KPC-producing K. pneumoniae is known to be endemic (2).
TABLE 1.
Carbapenemases and suspected origin
| Country data | Carbapenemase-producing isolate(s) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OXA-48-like |
NDM |
VIM |
KPC-3 | NDM-1 + OXA-232 | |||||||||
| OXA-48 | OXA-204 | OXA-244 | OXA-181 | NDM-1 | NDM-4 | NDM-5 | NDM-6 | NDM-7 | VIM-1 | VIM-4 | |||
| Country (n) | |||||||||||||
| Algeria | 8 | ||||||||||||
| Burma | 1 | ||||||||||||
| Canada | 1 | ||||||||||||
| Chad | 1 | ||||||||||||
| Djibouti | 1 | ||||||||||||
| Egypt | 2 | ||||||||||||
| India | 1 | 4 | 2 | 1 | 1 | 3 | 1 | ||||||
| Italy | 1 | 1 | |||||||||||
| Lebanon | 1 | ||||||||||||
| Libya | 2 | ||||||||||||
| Mali | 1 | ||||||||||||
| Morocco | 9 | ||||||||||||
| Niger | 1 | ||||||||||||
| Portugal | 1 | ||||||||||||
| Republic of Mauritius | 1 | ||||||||||||
| Romania | 1 | ||||||||||||
| Singapore | 1 | ||||||||||||
| Spain | 1 | ||||||||||||
| Turkey | 3 | ||||||||||||
| Vietnam | 1 | ||||||||||||
| Link with foreign country | |||||||||||||
| No. | 30 | 0 | 0 | 5 | 4 | 3 | 3 | 1 | 4 | 0 | 0 | 1 | 1 |
| % | 34 | 0 | 0 | 83 | 24 | 100 | 75 | 100 | 100 | 0 | 0 | 100 | 100 |
| Unknown or no link with foreign country | |||||||||||||
| No. | 58 | 9 | 1 | 1 | 13 | 0 | 1 | 0 | 0 | 3 | 2 | 0 | 0 |
| % | 66 | 100 | 100 | 17 | 76 | 0 | 25 | 0 | 0 | 100 | 100 | 0 | 0 |
| Total (no.) | 88 | 9 | 1 | 6 | 17 | 3 | 4 | 1 | 4 | 3 | 2 | 1 | 1 |
Antimicrobial susceptibility of CP-Ec.
Susceptibility testing was performed for eight antibiotics (ceftazidime, nitrofurantoin, fosfomycin, imipenem, amdinocillin, trimethoprim-sulfamethoxazole, colistin, and temocillin). These molecules were selected because of their potential use in urinary tract infections, as E. coli is the main uropathogen.
All isolates except 3 CP-Ec producers were tested. Colistin, fosfomycin, and nitrofurantoin possessed the most consistent activity, with 100%, 95% (130/137), and 96% (131/137) of isolates susceptible, respectively. Ceftazidime, imipenem, amdinocillin, and trimethoprim-sulfamethoxazole were active only on 41% (56/137), 81% (111/137), 64% (87/137), and 31% (42/137) of the isolates, respectively. Finally, all strains except one were resistant to temocillin. (Table 2).
TABLE 2.
Antibiotic susceptibility of CPE isolatesa
| Antibiotic | Class B (32 isolates) (%) |
Class D (103 isolates) (%) |
All CPEs (137 isolates) (%) |
|||
|---|---|---|---|---|---|---|
| Susceptibility | Resistance | Susceptibility | Resistance | Susceptibility | Resistance | |
| Ceftazidime | 0 | 100 | 54 | 46 | 41 | 59 |
| Colistin | 100 | 0 | 100 | 0 | 100 | 0 |
| Fosfomycin | 81 | 19 | 99 | 1 | 95 | 5 |
| Imipenem | 25 | 75 | 99 | 1 | 81 | 19 |
| Amdinocillin | 28 | 72 | 76 | 24 | 64 | 36 |
| Nitrofurantoin | 84 | 16 | 99 | 1 | 96 | 4 |
| Temocillin | 0 | 100 | 0 | 100 | 1 | 99 |
| Trimethoprim-sulfamethoxazole | 22 | 78 | 32 | 68 | 31 | 69 |
aAccording to Comité de l'antibiogramme de la Société Française de Microbiologie (CA-SFM)-EUCAST breakpoints.
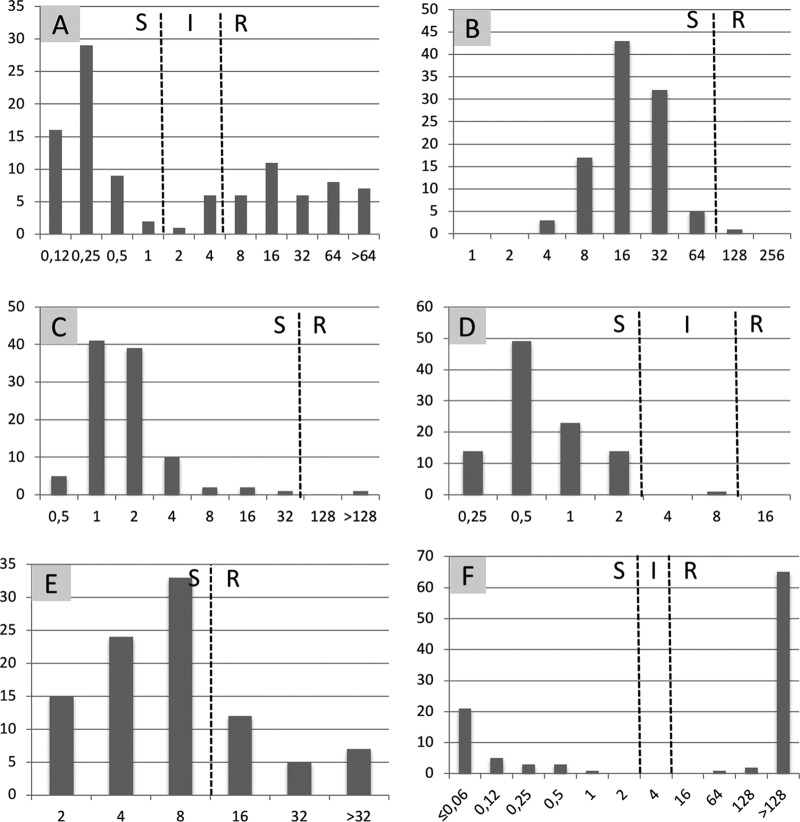
When considering susceptibility testing on the 103 OXA-48-like producers, colistin, nitrofurantoin, and fosfomycin retain excellent in vitro activity toward OXA-48 like producers, with 100%, 99%, and 99% of isolates susceptible, respectively (Fig. 1, Table 2). All strains were susceptible to colistin, with MICs of 0.25 mg/liter (85.5%), 0.5 mg/liter (12.5%), and 1 mg/liter (2%). Although these isolates produce a carbapenemase, all strains except one may be categorized as susceptible to imipenem, according to the Comité de l'antibiogramme de la Société Française de Microbiologie (CA-SFM)-EUCAST guidelines (11). Indeed, OXA-48 enzymes are known to exhibit only low hydrolytic activity toward carbapenems (12). Ceftazidime remained active toward 54% (n = 56) of the isolates. Amdinocillin was active on most of the isolates (n = 78; 76%), but with high MICs ranging from 2 mg/liter to 8 mg/liter, whereas wild-type strains have MICs between 0.064 and 0.5 mg/liter. Finally, in most of the cases, isolates were resistant to trimethoprim-sulfamethoxazole (n = 70; 68%). As expected, all strains were resistant to temocillin (MICs ranging from 64 to >256 mg/liter), which is currently considered a marker for suspected OXA-48 production.
FIG 1.
Antimicrobial susceptibility of class-D-carbapenemase-producing E. coli (n = 103). x axis, MIC in mg/liter; y axis, number of isolates. A, ceftazidime; B, nitrofurantoin; C, fosfomycin; D, imipenem; E, amdinocillin; F, trimethoprim-sulfamethoxazole; S, susceptible; I, intermediate; R, resistant.
When considering susceptibility testing on the 32 MBL producers (27 NDM and 5 VIM), all were resistant to ceftazidime (MICs > 64 mg/liter) and temocillin (MICs from 16 to >256 mg/liter). Only 7 isolates (22%) were susceptible to trimethoprim-sulfamethoxazole. In contrast, colistin was fully active toward all tested strains (MICs of 0.25 or 0.5 mg/liter). Fosfomycin and nitrofurantoin remained active in 81% (26/32) and 84% (27/32) of the MBL isolates, respectively. For 28% (9/32) and 25% (8/32) of MBL-producing E. coli isolates, respectively, amdinocillin and imipenem were categorized as susceptible.
Clonal relationship.
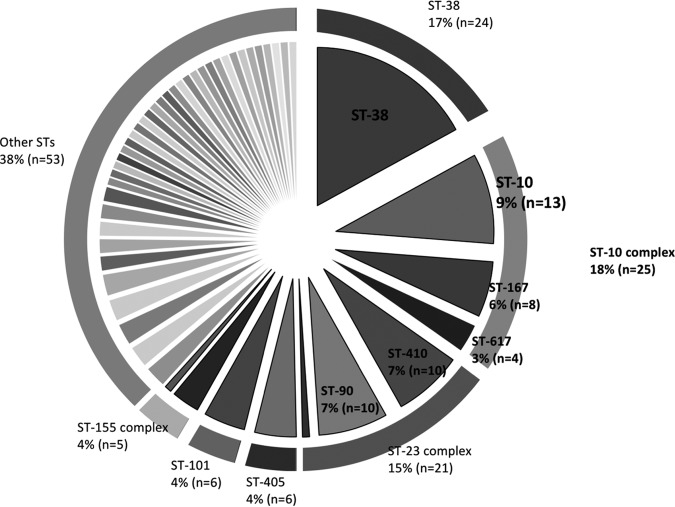
The distribution of the sequence types among the E. coli isolates is represented in Fig. 2 and shows a high diversity; 50 different sequence types (STs) were identified (including 2 new STs). ST38, clonal complex 10 (CC10, which includes ST10, ST167, ST617, and ST48), and clonal complex 23 (CC23, which includes ST88, ST90, and ST410) were the most commonly identified, representing 17.1% (n = 24), 17.8% (n = 25), and 15.0% (n = 21), respectively, of 140 carbapenemase-producing E. coli isolates. Six isolates (4.3%) belonged to ST405 and ST101 and five isolates (3.6%) belonged to clonal complex 155 (CC155, which includes ST155 and ST58). The other isolates (n = 53; 37.8%) belonged to single diverse STs (Fig. 2). Plasmid DNA was extracted from all OXA-48-producing ST38 isolates (n = 21). A large majority (n = 17; 81%) of the strains has no plasmid (data not shown). Furthermore, all electrotransformation attempts were negative, strongly suggesting that blaOXA-48 gene is inserted into the chromosome of this particular clone, as previously suggested (13).
FIG 2.
Diversity of sequence types (ST) among carbapenemase-producing E. coli isolates. Major STs are indicated in %; n, total number of isolates.
Clonal relationships were further analyzed using repetitive sequence-based PCR (rep-PCR). Results were then compared to those obtained by multilocus sequence typing (MLST) (Fig. 3). Among 9 OXA-204 producers, which displayed similar rep-PCR patterns, 8 originated from the same hospital and were thus suspected to be part of an outbreak. This outbreak was further investigated and could be linked to a contaminated endoscope (14). Of note, poor similarity was found for most of the isolates from CC10, suggesting the absence of localized ongoing outbreak and the strong ability of this particular clonal background to acquire plasmids harboring carbapenemase genes. The same kind of result was also observed for isolates of ST38, which could be divided into 4 branches according to the rep-PCR results (Fig. 3). However, among all ST38 isolates (n = 24), 21 possessed the same rep-PCR pattern (including 20 OXA-48 producers and 1 NDM-1 producer), suggesting clonal diffusion of a particular ST38 strain. Finally, one ST767 isolate expressing VIM-1 shared a similar rep-PCR pattern to those of the four ST58 isolates expressing OXA-48. Finally, isolates of ST101 are quite diverse in terms of Diversilab results (3 different patterns) and in terms of produced carbapenemases.
FIG 3.
Comparisons of E. coli isolates by MLST and Diversilab methods. Gray boxes indicate clonal complexes. Number of isolates with an identical Diversilab pattern is indicated. Expressed carbapenemases are indicated for each Diversilab pattern. Numbers in parentheses correspond to the number of isolates with a given carbapenemase or multilocus sequence type.
DISCUSSION
Before 2011, in France the spread of carbapenemase-producing Enterobacteriaceae was mainly associated with hospital dissemination (15). However, identification of CP-Ec strains strongly suggest a dissemination of these strains outside the hospital. This is further supported by the increasing number of CP-Ec isolates received by the F-NRC despite the lack of hospital outbreaks (6). Only one hospital outbreak was clearly established in our study, an endoscopy-associated transmission of a carbapenem-resistant ST90 and OXA-204-producing E. coli strain (14).
Epidemiological analysis of the 140 CP-Ec recovered in France in 2012 and 2013 showed a high diversity of circulating clones, in a similar manner to that previously reported between 2001 to 2012 (15–17). However, in the present study, which combined MLST and rep-PCR typing results, 6 clonal complexes, namely, CC10 (especially ST10 and ST167), ST38, CC23 (especially ST410 and ST90), CC155, ST101 and ST405 are clearly overrepresented.
Five OXA-181-producing E. coli ST410 isolates were recovered from patients with no epidemiological link. One of these isolates was identified by a community-serving diagnostic laboratory from a patient with no history of recent travel. This clone might be an emergent clone, since ST410 has already been described as a reservoir of resistance genes, particularly of the blaCTX-M-15 gene (18–20). Interestingly, ST410 was found several times in epidemiological studies of multidrug-resistant (MDR) E. coli strains in animals, suggesting a dissemination of this clone through the food chain (18–20). Since 2012, the proportion of the OXA-181 variant is increasing in the class D carbapenemases, from less than 1% in 2012 to 8% in 2015 (however, it is far behind OXA-48, which was found in ∼90% of the class D carbapenemases in 2015; L. Gauthier, personal data). Furthermore, this variant may be underestimated since carbapenem hydrolytic activity is very low, with sometimes false-negative results obtained with phenotypic methods such as the Carba NP test (21).
Other identified STs or CCs were already described as multidrug resistant, particularly as extended-spectrum β-lactamase (ESBL) producers, namely CC10 complex (22) and ST405 (23). Their resistance profiles, geographical origins, and carbapenemase types varied, suggesting a common origin but with further independent evolution. Nevertheless, among the 8 E. coli ST167 (CC10) isolates, 5 were positive for NDM and 3 were linked to India. In addition, among 4 E. coli ST-617 isolates, 3 had a link with Morocco. Susceptibility to non-β-lactam antibiotics varied among them, but a link with a foreign country could be established (2 with Algeria, one with Morocco, one with Vietnam), suggesting the spread of this clone in North African countries and may be in South East Asia. Among the 6 E. coli ST101 isolates, 4 produced an NDM-like carbapenemase, and three of them had a link with the Indian subcontinent, a result that is in accordance with another study (24).
Surprisingly, only one CP-Ec isolate belonged to ST131, although this globally disseminated and multidrug-resistant clone is notably responsible for the worldwide dissemination of the ESBL CTX-M-15. Of note, carbapenemase-producing E. coli ST131 isolates have been previously identified (25, 26), but most of them were positive for KPC or VIM carbapenemases and were reported from Italy, China, and the United States, which does not correspond to the French epidemiology (mostly OXA-48 producers).
Regarding antimicrobial susceptibilities, colistin (100%), fosfomycin (95%), and nitrofurantoin (96%) remain the most active drugs against CP-Ec isolates. These results are of great importance, since fosfomycin and nitrofurantoin are frequently prescribed for urinary tract infections in the community, even without microbiological documentation. These results are in accordance with those of previous studies (27, 28), except for those for nitrofurantoin, for which a susceptibility of 40% was found for 7 E. coli isolates tested (27). Almost all OXA-48-like carbapenemase producers were categorized as susceptible to imipenem according to EUCAST breakpoints. However, it is always controversial to propose imipenem for the treatment of infections due to OXA-48-producing bacteria, as the results obtained in vitro cannot be translated to the in vivo setting (29, 30). Ceftazidime remains active against half of the OXA-48-like producing E. coli isolates that do not coexpress an ESBL. Accordingly, it can be a reliable molecule for the treatment of infections caused by OXA-48-producing E. coli strains that do not produce an ESBL or a plasmid-mediated AmpC cephalosporinase, since MICs are in that case comparable to those of wild-type isolates. In the case of coproduction of an enzyme (ESBL or plasmid-mediated AmpC cephalosporinase) that is able to hydrolyze expanded-spectrum cephalosporins, ceftazidime-avibactam therapy was shown to be a relevant therapeutic option. A majority of OXA-48-producing E. coli isolates are multisusceptible (e.g., 50/87 do not coexpress an ESBL). Indeed, the IncL/M type plasmid that carries the blaOXA-48 gene does not carry other resistance genes. In 39% (34/87) of the CP-Ec cases, an initial OXA-48-producing K. pneumoniae isolate was found, suggesting an in vivo transfer within the patient's gut.
The isolates in this study were isolated 5 years ago, but their features are quite similar to those of more recent years. While the number of CP-Ec cases has increased (488 isolates received at the F-NRC in 2016), E. coli remains the second most common species among CPE isolates (31.5%) behind K. pneumoniae (38.5%) and far ahead of E. cloacae (12%) (T. Naas, personal communication). OXA-48-like carbapenemases remain the main carbapenemase in France (82% of the CP-Ec isolates in 2016), with still a large majority of OXA-48 variants (>90%). A significant difference is the increasing number of isolates addressed to the F-NRC by community-serving (nonhospital) laboratories, which further suggests a dissemination of CPEs in the community.
MATERIALS AND METHODS
Bacterial isolates.
Between January 2012 and December 2013, 140 CP-Ec isolates received by the F-NRC were investigated. These isolates were identified at the species level using MALDI-TOF mass spectrometry (MALDI Biotyper system; Brucker Daltonics, Wissembourg, France).
Susceptibility testing.
Antimicrobial susceptibility testing was performed by the disk diffusion method on Mueller-Hinton (MH) agar (Bio-Rad, Marnes-la-Coquette, France) and interpreted according to updated 2018 EUCAST breakpoint tables for interpretation of MICs and zone diameters, version 8.0 (11). To determine susceptibility to temocillin, breakpoints from CA-SFM/EUCAST 2018 were used (31). MICs were determined for ceftazidime, imipenem, colistin, fosfomycin, amdinocillin, temocillin, trimethoprim-sulfamethoxazole, and nitrofurantoin using broth microdilution (Sensititre; Thermo Fisher Scientific, Paris, France). The production of extended-spectrum β-lactamases (ESBLs) was evidenced by a double-disk synergy test using cefepime, ceftazidime, and ticarcillin-clavulanic acid disks.
PCR and sequencing of β-lactamase-encoding genes.
Whole-cell DNA was extracted using the QIAmp DNA minikit following the manufacturer's recommendations (Qiagen, Courtaboeuf, France). All isolates were screened by PCR for the carbapenemase-encoding genes blaKPC, blaOXA-48-like, blaVIM, blaNDM, and blaIMP, as previously described (32). ESBL-producing strains were screened for the genes blaCTX-M, blaTEM, and blaSHV. In the case of a positive signal, the entire genes were amplified and subsequently sequenced using an automated sequencer (3130 Genetic Analyzer; Applied Biosystems, Les Ullis, France). The nucleotide and deduced protein sequences were analyzed using the BLAST module of the Beta-Lactamase DataBase (BLDB) website (33).
Clonal relationship.
Multilocus sequence typing (MLST) with seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) was performed for E. coli isolates according to Wirth et al. (34). Allelic and ST numbers were determined using the Warwick scheme (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli). The genetic relationship between the E. coli isolates was studied using Diversilab, a semiautomated typing system based on repetitive sequence-based PCR (rep-PCR), following the manufacturer's instructions (bioMérieux, La Balme les Grottes, France). A 95% cutoff value of similarity was used to define a cluster, as recommended by the manufacturer.
Plasmid DNA analysis and transformation assays.
Plasmid DNA was extracted from the isolates using the Kieser technique, as previously described (35). E. coli NCTC50192, harboring four plasmids of 154, 66, 48, and 7 kb, was used as a plasmid size marker (34). Plasmid DNAs were analyzed by agarose gel electrophoresis. Direct transfer of OXA-48 resistance markers was attempted by electrotransformation of purified plasmid DNA, using E. coli TOP10 as the recipient (36). Selection was performed on Trypticase soy agar plates supplemented with temocillin (16 μg/ml).
ACKNOWLEDGMENTS
We are very grateful to Diana Chin and Lea Le Fahler for technical assistance.
This work was partially funded by a grant from the Université Paris-Sud, France, Assistance Publique—Hôpitaux de Paris, French Ministry of Health, by the French Public Health Agency (Santé Publique France), and by the Laboratory of Excellence LERMIT, supported by a grant from ANR (ANR-10-LABX-33) and by the Joint Programming Initiative on Antimicrobial Resistance (JPIAMR) DesInMBL (grant ANR-14-JAMR-002).
Laurent Dortet is coinventor of the Carba NP test, the patent of which has been licensed to bioMérieux (La Balmes les Grottes, France).
REFERENCES
- 1.Patel G, Bonomo RA. 2013. ‘Stormy waters ahead’: Global emergence of carbapenemases. Front Microbiol 4:48. doi: 10.3389/fmicb.2013.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albiger B, Glasner C, Struelens MJ, Grundmann H, Monnet DL, European Survey of Carbapenemase-Producing Enterobacteriaceae working group . 2015. Carbapenemase-producing Enterobacteriaceae in Europe: assessment by national experts from 38 countries, 2015. Euro Surveill 20 (45):pii=30062 https://www.eurosurveillance.org/content/10.2807/1560-7917.ES.2015.20.45.30062. [DOI] [PubMed] [Google Scholar]
- 3.Grundmann H, Glasner C, Albiger B, Aanensen DM, Tomlinson CT, Andrasević AT, Cantón R, Carmeli Y, Friedrich AW, Giske CG, Glupczynski Y, Gniadkowski M, Livermore DM, Nordmann P, Poirel L, Rossolini GM, Seifert H, Vatopoulos A, Walsh T, Woodford N, Monnet DL; European Survey of Carbapenemase-Producing Enterobacteriaceae (EuSCAPE) Working Group. 2017. Occurrence of carbapenemase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis 17:153–163. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 4.Semin-Pelletier B, Cazet L, Bourigault C, Juvin ME, Boutoille D, Raffi F, Hourmant M, Blancho G, Agard C, Connault J, Corvec S, Caillon J, Batard E, Lepelletier D. 2015. Challenges of controlling a large outbreak of OXA-48 carbapenemase-producing Klebsiella pneumoniae in a French university hospital. J Hosp Infect 89:248–253. doi: 10.1016/j.jhin.2014.11.018. [DOI] [PubMed] [Google Scholar]
- 5.Lee C-R, Lee JH, Park KS, Kim YB, Jeong BC, Lee SH. 2016. Global dissemination of carbapenemase-producing Klebsiella pneumoniae: epidemiology, genetic context, treatment options, and detection methods. Front Microbiol 7:895. doi: 10.3389/fmicb.2016.00895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dortet L, Cuzon G, Ponties V, Nordmann P. 2017. Trends in carbapenemase-producing Enterobacteriaceae, France, 2012 to 2014. Euro Surveill 22:30461. doi: 10.2807/1560-7917.ES.2017.22.6.30461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson AP, Woodford N. 2013. Global spread of antibiotic resistance: the example of New Delhi metallo-β-lactamase (NDM)-mediated carbapenem resistance. J Med Microbiol 62:499–513. doi: 10.1099/jmm.0.052555-0. [DOI] [PubMed] [Google Scholar]
- 8.van Duin D, Doi Y. 2017. The global epidemiology of carbapenemase-producing Enterobacteriaceae. Virulence 8:460–469. doi: 10.1080/21505594.2016.1222343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poirel L, Potron A, Nordmann P. 2012. OXA-48-like carbapenemases: the phantom menace. J Antimicrob Chemother 67:1597–1606. doi: 10.1093/jac/dks121. [DOI] [PubMed] [Google Scholar]
- 10.Cuzon G, Bentchouala C, Vogel A, Héry M, Lezzar A, Smati F, Dortet L, Naas T. 2015. First outbreak of OXA-48-positive carbapenem-resistant Klebsiella pneumoniae isolates in Constantine, Algeria. Int J Antimicrob Agents 46:725–727. doi: 10.1016/j.ijantimicag.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 11.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). 2018. Breakpoint tables for interpretation of MICs and zone diameters. Version 8.0, valid from 2018-01-01. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_8.0_Breakpoint_Tables.pdf.
- 12.Oueslati S, Nordmann P, Poirel L. 2015. Heterogeneous hydrolytic features for OXA-48-like β-lactamases. J Antimicrob Chemother 70:1059–1063. doi: 10.1093/jac/dku524. [DOI] [PubMed] [Google Scholar]
- 13.Poirel L, Bernabeu S, Fortineau N, Podglajen I, Lawrence C, Nordmann P. 2011. Emergence of OXA-48-producing Escherichia coli clone ST38 in France. Antimicrob Agents Chemother 55:4937–4938. doi: 10.1128/AAC.00413-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potron A, Bernabeu S, Cuzon G, Pontiès V, Blanchard H, Seringe E, Naas T, Nordmann P, Dortet L. Analysis of OXA-204 carbapenemase-producing Enterobacteriaceae reveals possible endoscopy-associated transmission, France, 2012 to 2014. Euro Surveill 2018 22(49):pii=17-00048. doi: 10.2807/1560-7917.ES.2017.22.49.17-00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Potron A, Poirel L, Rondinaud E, Nordmann P. 2013. Intercontinental spread of OXA-48 beta-lactamase-producing Enterobacteriaceae over a 11-year period, 2001 to 2011. Euro Surveill 18(31):pii=20549 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20549. [DOI] [PubMed] [Google Scholar]
- 16.Dortet L, Naas T, Boytchev I, Fortineau N. 2015. Endoscopy-associated transmission of carbapenemase-producing Enterobacteriaceae: return of 5 years' experience. Endoscopy 47:561. doi: 10.1055/s-0034-1392098. [DOI] [PubMed] [Google Scholar]
- 17.Pantel A, Boutet-Dubois A, Jean-Pierre H, Marchandin H, Sotto A, Lavigne J-P, CARB-LR group . 2014. French regional surveillance program of carbapenemase-producing Gram-negative bacilli: results from a 2-year period. Eur J Clin Microbiol Infect Dis 33:2285–2292. doi: 10.1007/s10096-014-2189-5. [DOI] [PubMed] [Google Scholar]
- 18.Giufrè M, Graziani C, Accogli M, Luzzi I, Busani L, Cerquetti M, Escherichia coli Study Group . 2012. Escherichia coli of human and avian origin: detection of clonal groups associated with fluoroquinolone and multidrug resistance in Italy. J Antimicrob Chemother 67:860–867. doi: 10.1093/jac/dkr565. [DOI] [PubMed] [Google Scholar]
- 19.Falgenhauer L, Imirzalioglu C, Ghosh H, Gwozdzinski K, Schmiedel J, Gentil K, Bauerfeind R, Kämpfer P, Seifert H, Michael GB, Schwarz S, Pfeifer Y, Werner G, Pietsch M, Roesler U, Guerra B, Fischer J, Sharp H, Käsbohrer A, Goesmann A, Hille K, Kreienbrock L, Chakraborty T. 2016. Circulation of clonal populations of fluoroquinolone-resistant CTX-M-15-producing Escherichia coli ST410 in humans and animals in Germany. Int J Antimicrob Agents 47:457–465. doi: 10.1016/j.ijantimicag.2016.03.019. [DOI] [PubMed] [Google Scholar]
- 20.López-Cerero L, Egea P, Serrano L, Navarro D, Mora A, Blanco J, Doi Y, Paterson DL, Rodríguez-Baño J, Pascual A. 2011. Characterisation of clinical and food animal Escherichia coli isolates producing CTX-M-15 extended-spectrum β-lactamase belonging to ST410 phylogroup A. Int J Antimicrob Agents 37:365–367. doi: 10.1016/j.ijantimicag.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Bakthavatchalam YD, Anandan S, Veeraraghavan B. 2016. Laboratory detection and clinical implication of oxacillinase-48 like carbapenemase: the hidden threat. J Glob Infect Dis 8:41–50. doi: 10.4103/0974-777X.176149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oteo J, Diestra K, Juan C, Bautista V, Novais A, Pérez-Vázquez M, Moyá B, Miró E, Coque TM, Oliver A, Cantón R, Navarro F, Campos J; Spanish Network in Infectious Pathology Project (REIPI). 2009. Extended-spectrum beta-lactamase-producing Escherichia coli in Spain belong to a large variety of multilocus sequence typing types, including ST10 complex/A, ST23 complex/A and ST131/B2. Int J Antimicrob Agents 34:173–176. doi: 10.1016/j.ijantimicag.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 23.Coque TM, Novais A, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum beta-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mushtaq S, Irfan S, Sarma JB, Doumith M, Pike R, Pitout J, Livermore DM, Woodford N. 2011. Phylogenetic diversity of Escherichia coli strains producing NDM-type carbapenemases. J Antimicrob Chemother 66:2002–2005. doi: 10.1093/jac/dkr226. [DOI] [PubMed] [Google Scholar]
- 25.Accogli M, Giani T, Monaco M, Giufrè M, Garcia-Fernandez A, Conte V, D'Ancona F, Pantosti A, Rossolini GM, Cerquetti M. 2014. Emergence of Escherichia coli ST131 sub-clone H30 producing VIM-1 and KPC-3 carbapenemases, Italy. J Antimicrob Chemother 69:2293–2296. doi: 10.1093/jac/dku132. [DOI] [PubMed] [Google Scholar]
- 26.Peirano G, Bradford PA, Kazmierczak KM, Badal RE, Hackel M, Hoban DJ, Pitout JD. 2014. Global incidence of carbapenemase-producing Escherichia coli ST131. Emerg Infect Dis 20:1928–1931. doi: 10.3201/eid2011.141388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livermore DM, Warner M, Mushtaq S, Doumith M, Zhang J, Woodford N. 2011. What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int J Antimicrob Agents 37:415–419. doi: 10.1016/j.ijantimicag.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Kaase M, Szabados F, Anders A, Gatermann SG. 2014. Fosfomycin susceptibility in carbapenem-resistant Enterobacteriaceae from Germany. J Clin Microbiol 52:1893–1897. doi: 10.1128/JCM.03484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mimoz O, Grégoire N, Poirel L, Marliat M, Couet W, Nordmann P. 2012. Broad-spectrum β-lactam antibiotics for treating experimental peritonitis in mice due to Klebsiella pneumoniae producing the carbapenemase OXA-48. Antimicrob Agents Chemother 56:2759–2760. doi: 10.1128/AAC.06069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daikos GL, Markogiannakis A. 2011. Carbapenemase-producing Klebsiella pneumoniae: (when) might we still consider treating with carbapenems? Clin Microbiol Infect 17:1135–1141. doi: 10.1111/j.1469-0691.2011.03553.x. [DOI] [PubMed] [Google Scholar]
- 31.Comité de l'antibiogramme de la Société Française de Microbiologie (CA-SFM). 2018. Comité de l'antibiogramme de la Société Française de Microbiologie (CA-SFM), Recommandations 2018 V.1.0 Février. http://www.sfm-microbiologie.org/UserFiles/files/casfm/CASFM%20V1_0%20FEVRIER%202018.pdf.
- 32.Dortet L, Bréchard L, Cuzon G, Poirel L, Nordmann P. 2014. Strategy for rapid detection of carbapenemase-producing Enterobacteriaceae. Antimicrob Agents Chemother 58:2441–2445. doi: 10.1128/AAC.01239-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, Dortet L, Retailleau P, Iorga BI. 2017. Beta-Lactamase DataBase (BLDB)—structure and function. J Enzyme Inhib Med Chem 32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wirth T, Falush D, Lan R, Colles F, Mensa P, Wieler LH, Karch H, Reeves PR, Maiden MC, Ochman H, Achtman M. 2006. Sex and virulence in Escherichia coli: an evolutionary perspective. Mol Microbiol 60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kieser T. 1984. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid 12:19–36. doi: 10.1016/0147-619X(84)90063-5. [DOI] [PubMed] [Google Scholar]
- 36.Jaidane N, Bonnin RA, Mansour W, Girlich D, Creton E, Colleton G, Chaouch C, Boujaafar N, Bouallegue O, Naas T. 2018. Genomic insights into colistin-resistant Klebsiella pneumoniae from a Tunisian teaching hospital. Antimicrob Agents Chemother 62:e01601-17. doi: 10.1128/AAC.01601-17. [DOI] [PMC free article] [PubMed] [Google Scholar]