ABSTRACT
Candida albicans surface-attached biofilms such as those formed on intravenous catheters with direct access to the bloodstream often serve as a nidus for continuous release of cells capable of initiating new infectious foci. We previously reported that cells dispersed from a biofilm are yeast cells that originate from the top-most hyphal layers of the biofilm. Compared to their planktonic counterparts, these biofilm dispersal yeast cells displayed enhanced virulence-associated characteristics and drug resistance. However, little is known about their molecular properties. To address that issue, in this study we aimed to define the molecular characteristics of these biofilm dispersal cells. We found that the inducer of dispersal, PES1, genetically interacts with the repressor of filamentation, NRG1, in a manner consistent with the definition of dispersed cells as yeast cells. Further, using a flow biofilm model, we performed comprehensive comparative RNA sequencing on freshly dispersed cells in order to identify unique transcriptomic characteristics. Gene expression analysis demonstrated that dispersed cells largely inherit a biofilm-like mRNA profile. Strikingly, however, dispersed cells seemed transcriptionally reprogrammed to acquire nutrients such as zinc and amino acids and to metabolize alternative carbon sources, while their biofilm-associated parent cells did not induce the same high-affinity transporters or express gluconeogenetic genes, despite exposure to the same nutritional signals. Collectively, the findings from this study characterize cell dispersal as an intrinsic step of biofilm development which generates propagules more adept at colonizing distant host sites. This developmental step anticipates the need for virulence-associated gene expression before the cells experience the associated external signals.
KEYWORDS: Candida albicans, RNA-seq, biofilms, carbon metabolism, dispersal, gene expression, planktonic
IMPORTANCE
Candida albicans surface-attached biofilms serve as a reservoir of cells to perpetuate and expand an infection; cells released from biofilms on catheters have direct access to the bloodstream. Biofilm dispersal yeast cells exhibit enhanced adhesion, invasion, and biofilm formation compared to their planktonic counterparts. Here, we show using transcriptome sequencing (RNA-seq) that dispersed yeast cells are developmentally distinct from the cells in their parent biofilms as well as from planktonic yeast cells. Dispersal cells possess an anticipatory expression pattern that primes them to infect new sites in the host, to survive in nutrient-starved niches, and to invade new sites. These studies identified dispersal cells as a unique proliferative cell type of the biofilm and showed that they could serve as targets for antibiofilm drug development in the future.
INTRODUCTION
Detachment of microorganisms from an established site of proliferation mediates spread of pathogens to new sites or through the bloodstream, resulting in disseminated disease. Frequently, a pathogen’s initial proliferative nidus consists of its presence on a biofilm, on a mucosal or mesothelial surface, or on a foreign body.
The human commensal Candida albicans is the most frequently isolated human fungal opportunistic pathogen. Disseminated candidiasis carries high mortality rates despite appropriate antifungal drug treatment (1, 2). C. albicans is unique in its ability to switch between growth forms in the host, i.e., between budding yeast and filamentous pseudohyphae and hyphae. Morphological switching enhances its ability to adhere and invade and to sustain a community of biofilm cells. Decades of research have elucidated the regulation of hyphal morphogenesis and its association with pathogenesis (3–5). Most recently, candidalysin, a toxin secreted by C. albicans, was shown to be produced only during hyphal growth (6).
Hyphal filaments constitutively produce cells exhibiting lateral compartmentation (lateral yeast cells) on their subapical segments, while apical segments continue to extend as filamentous cells. Lateral yeast cells are formed in vivo, since filamentous cells are typically seen in conjunction with yeast cells in organs of hosts with invasive candidiasis (7, 8). C. albicans biofilms are initiated when yeast cells adhere to a surface and form microcolonies. Over time, the cells differentiate into hyphae that eventually develop into a complex community of basal yeast cells and into layers of hyphal cells encased in a blanket of self-produced polysaccharide matrix (9, 10). Throughout the biofilm growth cycle, hyphae continuously release lateral yeast cells (10). This phenomenon is of great clinical relevance, as cells released from a biofilm formed on an indwelling catheter or an infectious nidus can gain access to the bloodstream, disseminate, and initiate distant foci of infection. Although our current understanding of lateral yeast production and biofilm dispersal is limited, we have previously demonstrated that yeast cells arising from biofilms adhere better to mammalian cells, are more resistant to azole drugs, and display higher virulence in a disseminated mouse model of candidiasis (10). Thus, dispersal of cells from biofilms appears to be a distinct developmental phase of the fungus in which cells are primed for invasion of the host.
To date, Pes1 is the only molecular regulator that has been shown to control production of lateral yeast cells from hyphae and to induce biofilm dispersal (10–12). Pes1 regulates lateral yeast cell production but not hyphal morphology or biofilm architecture and has been shown to be essential for sustained candidiasis (7). In fact, our previous studies demonstrated that curtailing lateral yeast cell release from tissue-invading hyphae by depleting PES1 significantly reduced dissemination and virulence in mice (8). In light of these findings, the current study was designed to elucidate the molecular characteristics of lateral yeast cells released from biofilms.
Here, we used RNA sequencing to delineate biofilm dispersal cell-specific molecular signatures and to identify gene expression patterns that might set the dispersed cells apart from their parent biofilms or their planktonic counterparts. We found that the transcriptional landscape of dispersed yeast largely overlapped that of their parent hyphal biofilms, rather than that of the morphologically similar planktonic yeast cells. Virulence-associated genes were a substantial component of the upregulated set. Despite arising within the same nutritional milieu, dispersed cells and biofilm cells exhibited striking contrasts in their metabolic gene expression profiles which suggested an anticipatory developmental step that takes place during dispersal cell formation, during which cells are prepared for an invasive role before they are released from the biofilm. In a jugular vein-catheterized mouse model, we showed that dispersed yeast cells are the link between biofilm-infected catheters and the foci of infection in a deep organ. In summary, we defined molecular characteristics of biofilm dispersal cells and show that they are in a developmental phase distinct from the biofilm or planktonic state and are specifically equipped for immediate infection in the host.
RESULTS
Transcript profiles of biofilm dispersal cell populations strongly corresponded to those of biofilm cells.
To understand the molecular characteristics of cells dispersed from biofilms, we collected and characterized yeast cells spontaneously released from C. albicans biofilms, using the simple flow biofilm model (13, 14). Age-matched planktonic and biofilm cells were also recovered from biofilms grown for 24 h, and transcript profiles of the 3 cell populations were delineated using transcriptome sequencing (RNA sequencing [RNA-seq]).
To validate our approach, we first compared the expression patterns of biofilm and planktonic cells. Results from the analysis showed a total of 1,524 genes to be significantly altered in expression (using a false-discovery rate [adjusted P value {p-adj}] of <0.05) between the 2 cell types. Our comparison of biofilm cells and free-living cells largely replicated microarray-based studies by others (15, 16), concurrently validating our procedure (Fig. 1; see also Data Set S1 in the supplemental material).
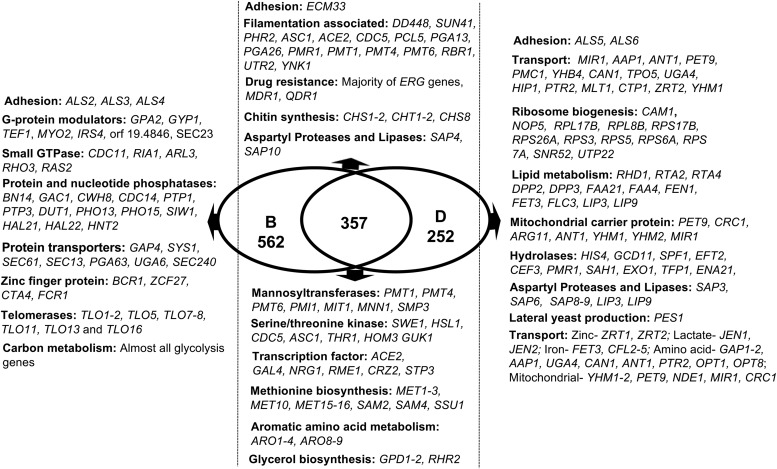
FIG 1 .
Venn diagram of significantly upregulated genes in biofilms and biofilm dispersal cells at 24 h, compared to planktonic cells. Gene expression profiles of C. albicans biofilm (B) and biofilm dispersal cells (D) were each compared individually to age-matched planktonic cells (P). The Venn diagram displays the total number of genes upregulated under each growth condition and those common between biofilms and dispersed cells. The diagram also shows some genes and their functional categories (additional to those mentioned in the body of the manuscript) whose levels are elevated in each group.
Comprehensive differentially expressed data of significantly (P < 0.5) regulated (>1.5-fold) genes. In this Excel sheet, sheet 1 lists genes significantly upregulated in biofilm versus planktonic cells, sheet 2 lists genes significantly downregulated in biofilm versus planktonic cells, sheet 3 lists genes common between our study and other transcriptone studies comparing biofilm expression data to planktonic expression data, sheet 4 lists genes significantly upregulated in dispersed versus planktonic cells, sheet 5 lists genes significantly downregulated in dispersed versus planktonic cells, sheet 6 lists genes showing similar patterns of upregulation in both biofilm and planktonic cells compared to planktonic cells, and sheet 7 lists genes showing similar patterns of downregulation in both biofilm and planktonic cells compared to planktonic cells. Download DATA SET S1, XLSX file, 0.3 MB (310.5KB, xlsx) .
Copyright © 2018 Uppuluri et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Next, we compared the biofilm dispersal cells to their age-matched planktonic and biofilm sisters. A total of 963 genes showed statistically significant differences between the dispersed and planktonic conditions (P < 0.05). Of the 609 genes upregulated >1.6-fold in dispersed cells, 357 (~59%) were also upregulated in cells from a mature biofilm compared to planktonic cells (Data Set S1). Regulatory patterns of dispersed cells substantially reflected those of their parent biofilms with respect to genes involved in methionine biosynthesis (MET1, MET2, MET3, MET10, MET15, MET16, SAM2, SAM4, and SSU1) (15), aromatic amino acid metabolism (ARO2, ARO3, ARO4, ARO8, and ARO9), ergosterol biosynthesis (a majority of ERG genes), adhesion (ECM33) (17, 18), small-molecule efflux and drug resistance (MDR1 and QDR1) (19, 20), chitin synthesis (CHS1, CHS2, CHS8, CHT1, and CHT2) (21), and glycerol biosynthesis (GPD1, GPD2, and RHR2) (22); almost all ribosome biogenesis genes (15, 23); and many others defined previously as biofilm regulated (24). Expression of the yeast-specific YWP1 gene (25) was 2-fold higher in dispersed cells than in biofilms and was higher in both than in planktonic cells.
Similarly, the majority of the SAP family genes, SAP3, SAP6, SAP8, and SAP9, were found to be upregulated in dispersed cells, while the expression levels of SAP4 and SAP10 were also elevated in biofilm cells. Similarly, LIP3 and LIP9 were upregulated in dispersed cells, with LIP4 and LIP6 expression levels elevated only in biofilm cells. None of the SAP or lipase family genes were expressed in planktonic cells, similarly to previous reports (26).
Dispersed yeast cells expressed genes associated with the hyphal form.
Despite their yeast morphology and the expression of two predominantly yeast-specific genes (YWP1 and RHD3) (27), several genes typically expressed in hyphae rather than in yeast cells, including DDR48, PHR2, ASC1, SUN41, ACE2, CDC5, CHA1, PCL5, PGA13, PGA26, PMR1, PMT1, PMT4, PMT6, RBR1, UTR2, and YNK1, were found to be upregulated in the dispersed cell population. To further characterize the dispersed cells, we developed two separate biofilm assays using two independent C. albicans strains in which the hyphal wall protein HWP1 gene and yeast wall protein YWP1 gene were fused with the red fluorescent protein gene, RFP. These reporter strains showed that ~33% of dispersed yeast cells expressed hypha-specific HWP1, whereas YWP1 was expressed in ~64% of dispersed yeast cells. This discrepancy, seen in a third of dispersed cells, between their growth form and their expression of a cell type-specific cell wall gene raised the issue of whether these cells can be classified as yeast.
PES1, essential in C. albicans yeast, was required for dispersal cell formation in cells induced or repressed for NRG1.
Since the transcriptional profile of dispersed cells most closely resembled that of hyphal biofilm cells, while their morphological appearance corresponded to yeast, we examined dispersed cells by a genetic criterion of cell type: essentiality of the inducer of lateral yeast growth, PES1. PES1, whose homologues are essential in all eukaryotes, is required for C. albicans yeast cell growth, but hyphae tolerate its depletion from repressible promoters (7). Overexpression of PES1 in biofilms induces increased lateral yeast production and dispersal from hyphal layers of the biofilm, while its depletion represses release of dispersal cells (10). Overexpression of NRG1, a negative regulator of filamentation, also results in significantly increased production of dispersed cells (12) from a predominantly hyphal biofilm. PES1 expression was elevated >1.7-fold in dispersed cells compared to biofilm cells. Increased (1.4-fold) NRG1 expression also trended toward significance (P value, 0.03; p-adj, 0.08) in dispersed versus the biofilm cells.
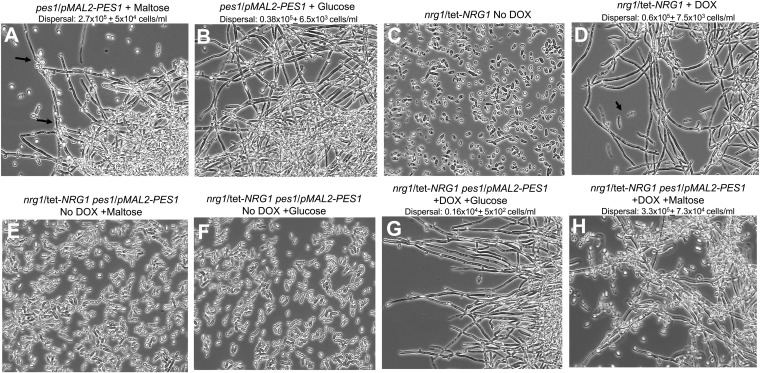
We tested the requirement for PES1 in dispersal cells in which NRG1 expression was exogenously manipulated in a static biofilm model. For this purpose, a pes1/pMAL2-PES1 NRG1/tetO-NRG1 strain was constructed and compared with strains containing similar mutations in PES1 or NRG1 alone. During induction of PES1 in the presence of maltose, the pes1/pMAL2-PES1 strain produced a biofilm with abundant lateral yeast cells (dispersal, ~2.7 × 105 cells/ml) (Fig. 2A), while in glucose (during depletion of PES1), levels of lateral yeast cells were significantly decreased (dispersal, ~0.38 × 105 cells/ml) (Fig. 2B). Upregulation of the hyphal-growth-repressing NRG1 gene abrogated biofilm formation (Fig. 2C), while abundant hyphae and a robust biofilm were produced during NRG1 repression (yeast nitrogen base [YNB] plus doxycycline [DOX]) (Fig. 2D). PES1 up- or downregulation when NRG1 was overexpressed in the absence of doxycycline did not reverse the biofilm defect; cells overexpressing NRG1 remained in the yeast form, rendering them incapable of biofilm formation, regardless of the level of PES1 expression (Fig. 2E and F).
FIG 2 .
Genetic interaction study of PES1 with NRG1 under biofilm conditions. Biofilms were developed overnight in YNB broth in the presence of glucose or maltose as the carbon source and in the presence or absence of DOX. The following strains were used: pes1/pMAL2-PES1, nrg1/tet-NRG1, and nrg1/tet-NRG1 pes1/pMAL2-PES1. Biofilms were visualized by light microscopy. Arrows in panel B and D point to the lateral yeast cells, and the frequency of dispersal is indicated in their respective panels.
In contrast, lateral yeast production was significantly impacted by PES1 induction or repression in NRG1-repressed hyphal biofilms. Downregulation of PES1 abrogated lateral yeast formation and biofilm dispersal (Fig. 2G), while induction of PES1 expression in NRG1-downregulated biofilm cells resulted in prolific lateral yeast production and in a 3-fold increase in biofilm dispersal (Fig. 2H) compared to the NRG1-downregulated biofilm alone (Fig. 2D). Hence, dispersal cell formation is strongly impacted by exogenous up- or downregulation of expression of a gene essential in yeast cells, PES1. These findings indicate that dispersal cells have genetic properties of yeast and that PES1 acts downstream of NRG1 for dispersal cell production.
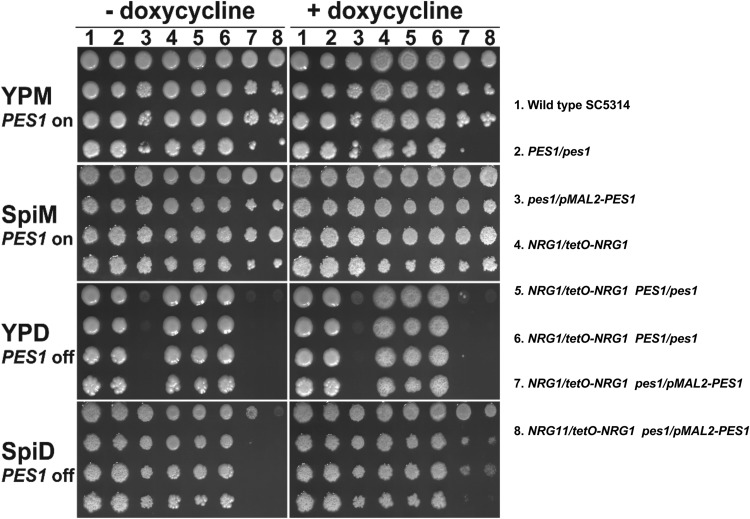
In control experiments, we tested relationships of NRG1 and PES1 for growth on yeast- or hypha-inducing solid media. Consistent with previous findings, a glucose-repressible pes1/pMAL2-PES1 strain failed to grow at 30°C on yeast extract–peptone–2% dextrose (YPD) medium with 2% glucose and yet grew under hypha-inducing conditions, at 37°C on Spider medium containing glucose as the carbon source (Fig. 3, lane 3). NRG1/tetO-NRG1 strains with wild-type (WT) PES1 grew on all media, with the growth comprised primarily of yeast during NRG1 upregulation (− doxycycline), and primarily of hyphal cells during NRG1 repression (+ doxycycline) as previously established by others [28]) (Fig. 3, lanes 4, 5, and 6). When PES1 was depleted in NRG1-overexpressing cells, growth ceased (−doxycycline, lanes 7 and 8), indicating that those cells were PES1 dependent. In contrast, when PES1 was depleted in NRG1-repressed cells, those hyphal cells grew robustly (+ doxycycline, lanes 7 and 8). Hence, during PES1 depletion, growth depended on decreasing genetically determined hyphal repression levels, indirectly supporting the idea that dispersed cells exhibit genetic characteristics of yeast cells.
FIG 3 .
Genetic interaction of PES1 with NRG1 on solid media. Serial dilutions of strains were spotted onto YP or Spider medium with 2% glucose or maltose as the carbon source, with or without DOX (5 µg/ml). Lane 1, wild-type SC5314; lane 2, PES1/pes1, JKC619; lane 3, pes1/pMAL2-PES1, JKC673; lane 4, NRG1/tetO-NRG1, SSY50-B; lane 5, NRG1/tetO-NRG1 PES1/pes1, JKC869; lane 6, NRG1/tetO-NRG1 PES1/pes1, JKC870; lane 7, NRG1/tetO-NRG1 pes1/pMAL2-PES1, JKC993; lane 8, NRG1/tetO-NRG1 pes1/pMAL2-PES1, JKC996. Plates were photographed after 3 days of incubation at 37°C (Spider) or 30°C (YP).
Genes differentially regulated exclusively in dispersed cells acted in zinc, iron, and amino acid acquisition.
A total of 335 genes (167 upregulated and 168 downregulated) were found to be differentially expressed in dispersed cells compared to both biofilm and planktonic conditions (listed in sheet 2 in Data Set S2). The predominant group of genes upregulated in dispersed cells represented high-affinity transporters for zinc (ZRT1 and ZRT2) (29) (and a regulator of the two zinc transport genes [ZAP1]), lactate (JEN1 and JEN2) (30), and iron (FET3, CFL2, CFL4, and CFL5); for amino acid transport (GAP1, GAP2, AAP1, UGA4, CAN1, ANT1, PTR2, OPT1, and OPT8); and for mitochondrion-associated transport (YHM1, YHM2, PET9, NDE1, MIR1, and CRC1) (Fig. 1).
Data set of genes expressed in dispersed cell populations. In this Excel sheet, sheet 1 shows differential expression of central carbon metabolism genes in both dispersed and planktonic cells versus biofilm cells. Here the dispersed cells show a planktonic-yeast-like expression pattern. Sheet 2 shows a set of genes that are upregulated and downregulated only on dispersed cells. These data were extracted after pairwise comparisons of all the sample conditions to each other, picking largely the genes that were upregulated or downregulated exclusively in dispersed cells. Download DATA SET S2, XLSX file, 0.03 MB (31.3KB, xlsx) .
Copyright © 2018 Uppuluri et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Genes associated with morphogenesis and biofilm growth had an expression pattern in dispersed cells distinct from those seen with biofilm and planktonic cells (CSA1, YNK1, PGA26, GNA1, RBT1, ALS5, ALS6, IFF4, and UTR2). Of the 168 genes downregulated exclusively in the dispersed cells, 60 had unknown biological functions. While the remaining 108 genes were not readily categorized into functional groups, a number either had functions in mRNA binding or coded for proteins with predicted activity in mRNA splicing via the spliceosome, e.g., ZSF1, orf19.285, EXM2, orf19.2261, orf19.7139, and FGR16. Other downregulated dispersed cell-specific genes were predicted to function in retrograde transport from the endosome/endoplasmic reticulum (ER) to the Golgi compartment, including VPS41, VPS17, UFE1, MSO1, HNM3, orf19.2333, and orf19.5114. The significance of downregulation of these genes in dispersed cells remains to be determined.
Telomere-associated (TLO) gene family expression showed stark differences between dispersed and biofilm-associated cells.
A set of genes that exhibited significant differences in expression between biofilm cells, planktonic cells, and dispersed lateral yeast cells was the telomeric open reading frame (TLO) gene family. This gene family, whose products are thought to be components of the transcription-regulating mediator complex (31), is comprised of 13 telomere-adjacent genes (TLO1 to TLO5 [TLO1–5], TLO7–13, and TLO16) (32, 33) and the nontelomeric family member TLO34. In dispersed cells, most TLO genes were downregulated at least 4-fold relative to biofilm cells and 2-fold relative to planktonic cells. Therefore, several TLO genes (TLO1, TLO2, TLO5, TLO7, TLO8, TLO11, TLO13, and TLO16) were most highly expressed in biofilm cells, with TLO2 expressed at a level that was 7-fold higher in biofilms and 2-fold higher in planktonic cells than in dispersed cells.
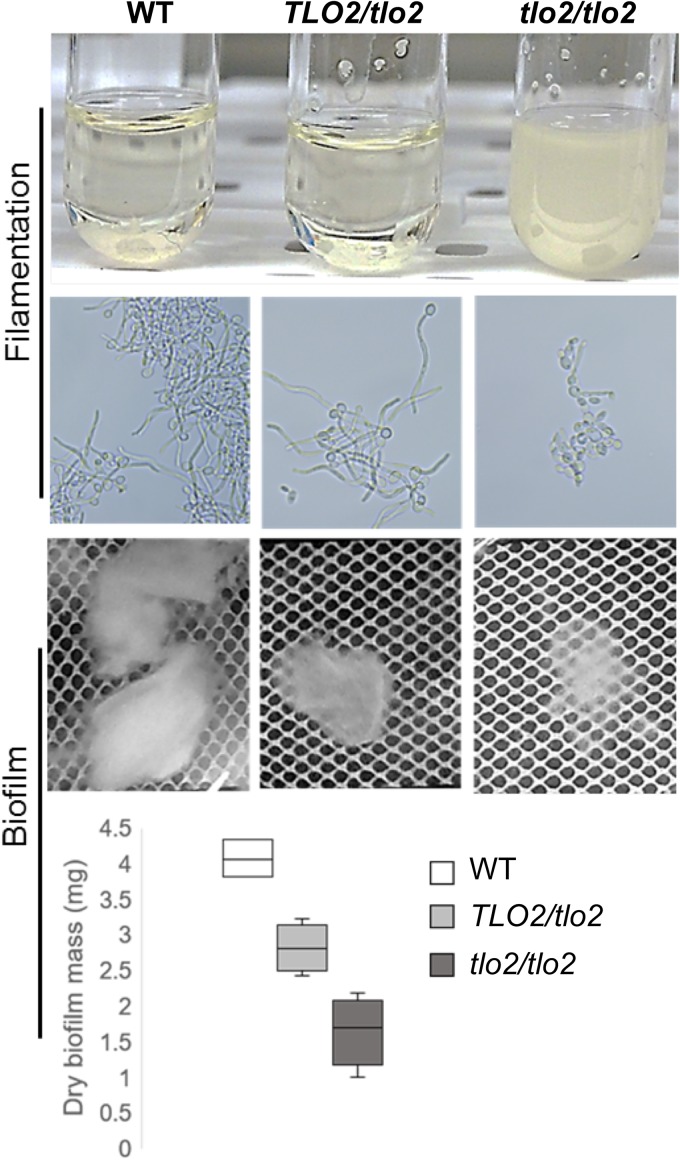
We investigated the morphogenesis, biofilm cell, and dispersed cell phenotypes of a mutant lacking TLO2, since expression of this gene most distinguished biofilm cells from dispersed and planktonic cells. The tlo2−/− strains were found to be defective in filamentation (Fig. 4); cells remained uniformly suspended in liquid Spider medium. This observation was in contrast to the observation of clumps of adherent filaments produced by the wild-type and heterozygous parent strains, which settled to the bottom of the tube. The tlo2−/− strain produced yeast cells almost exclusively (Fig. 4). Under conditions of static biofilm induction, tlo2−/− showed a significant (~60%) decrease in biofilm growth compared to the wild-type strain (Fig. 4), with the TLO2/tlo2− strain having an intermediate phenotype (~31% decrease). Biofilms formed by the null mutant were fragile and tended to disintegrate easily, as a consequence of the deficiency in developing scaffolding filaments. Given their defects in biofilm formation, we were not able to assess the dispersal phenotypes of tlo2−/− mutants. Further experimentation with, e.g., repressible TLO2 alleles, whose expression can be downregulated after a biofilm has formed, will be required to define the distinct roles of Tlo2 in biofilm cells versus dispersed cells. Such experiments could resolve the issue of which transcriptional targets of Tlo2 are differentially regulated in hyphal biofilm cells and their dispersed cell daughters.
FIG 4 .
Filamentation and biofilm-forming capabilities of a C. albicans TLO2 mutant strain. Candida strains (wild type, TLO2 heterozygote, and tlo2/tlo2 homozygote) grown overnight in YPD at 30°C were inoculated into Spider media and incubated at 37°C for 0, 2, 4, and 6 h. Aliquots from each strain were assessed for cell phenotype by microscopy. Biofilms for these strains were also developed under static conditions for 48 h and visualized microscopically, and their dry weight was measured for biomass. The tlo2−/− homozygote showed a drastic reduction in both filamentation and biofilm formation compared to the wild-type strain. The heterozygote displayed normal hyphal development, but the hyphae did not clump together as was seen in the wild-type samples. The tlo2/TLO2 heterozygote also had an intermediate defect on biofilm formation.
Dispersed cells resembled planktonic cells with respect to carbon metabolism.
Under conditions of a continuous flow of fresh medium rich in glucose (1%), biofilm cells expressed much (3-to-9-fold) higher levels of several key genes encoding glycolysis pathway enzymes (GAL4, PGI1, PFK1, PFK2, FBA1, PGK1, ENO1, and CDC19), relative to planktonic cells grown for 24 h. Dispersed cells released from the same biofilms displayed the opposite pattern of expression, where all glycolysis genes were found to be repressed and core genes required for alternative carbon metabolism were highly upregulated (Data Set S2). This pattern of gene expression in dispersed cells resembled that of planktonic cells after 24 h of growth, wherein elements of the gluconeogenesis pathway—PCK1 and FBP1—exhibited 6-fold and 3-fold increases, respectively, relative to biofilm cells. Genes encoding enzymes of the tricarboxylic acid (TCA) pathway presented the greatest differences, as all were upregulated >10-fold in dispersed cells, as well as planktonic cells, versus biofilm cells (Data Set S2, sheet 1). ICL1, which encodes a major enzyme of the glyoxylate pathway, was upregulated 14-fold in dispersed cells and >20-fold in planktonic cells relative to biofilms. Thus, dispersed cells sharply differed from their hyphal mothers. They resembled stationary-phase planktonic cells in their upregulation of alternative carbon metabolism-related genes, despite experiencing a glucose-rich environment.
Expression patterns shifted in lateral yeast cells prior to their release from the biofilm.
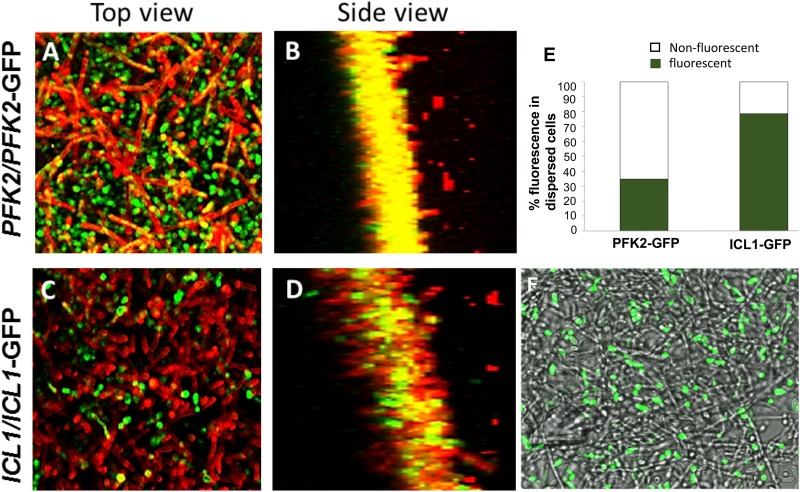
To visualize the disparity of carbon metabolism gene expression between biofilm cells and dispersed cells at the protein level, we used strains expressing green fluorescent protein gene (GFP)-tagged PFK2 (a glycolysis gene) or ICL1 (a glyoxylate cycle gene). Biofilms were developed under the same growth conditions as those of cells analyzed by RNA-seq. Following a 24-h growth period, biofilms were stained using Alexa 594-conjugated concanavalin A (ConA; a lectin that binds to mannans of the fungal cell wall) and visualized by confocal microscopy. A top-down view of PFK2-GFP biofilms displayed a high intensity of green fluorescence (with yellow streaks in hyphae due to overlapped red ConA signal), suggesting that biofilm cells were undergoing glycolysis. Merging signals of the side view of the biofilm showed overlapping signals, indicating a potential for glycolytic activity in hyphae stained by ConA (Fig. 5A and B; see also slide 1 of Fig. S1 for alternative images). In contrast, only a small proportion of cells in the ICL1-GFP biofilms displayed green fluorescence (Fig. 5C and D), suggesting that the glyoxylate cycle was largely inactive in the biofilms.
FIG 5 .
Carbon source utilization signature of biofilm and biofilm dispersal cells. C. albicans cells harboring a GFP-tagged glyoxylate pathway gene, ICL1 (ICL1/ICL1-GFP), and a GFP-tagged glycolytic gene (PFK2/PFK2-GFP) were allowed to form biofilms individually, for 48 h. The biofilm cells were stained with ConA, which stains the cell walls of the fungal cell red, and z-stacks were collected at two wavelengths, 488 nm (GFP) and 594 nm (ConA). (A to D) Top and side-scatter images reveal that the topmost hyphal layers of the biofilm expressed Pfk2 (A and B, respectively) rather than Icl1 (C and D, respectively). (E) In contrast, the cells dispersed from the biofilms expressed Icl1 at a significantly higher frequency than Pfk2. (F) Those results can be visualized by overlaying a bright-field image and GFP fluorescence image of the biofilm, with the result showing brightly fluorescent yeast cells on top of a mat of nonfluorescent ICL1-GFP biofilms.
Figures related to Fig. 2 in the manuscript. Slide 1 shows that the topmost layers of the flow biofilm expressed green fluorescent Pfk2. Slide 2 shows that the cells attached to the silicone substrate (innermost portion of the flow biofilm) expressed green fluorescent Icl1. Download FIG S1, TIF file, 1 MB (1MB, tif) .
Copyright © 2018 Uppuluri et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Interestingly, dispersed cells expressed ICL1 at a significantly higher rate than PFK2 (Fig. 5E). The dispersed cells from the PFK2-GFP biofilms displayed an overall dim fluorescence, and only about 35% of cells showed a clear fluorescent signal. In contrast, ~78% of the dispersed cells from the ICL1-GFP biofilms showed bright green fluorescence. A merged bright-field–GFP fluorescence image of the ICL1-GFP biofilm surface clearly demonstrated that GFP fluorescence was evident only in lateral yeast cells, i.e., those poised for release from the hyphal surface, and not in the hyphal mat (Fig. 5F).
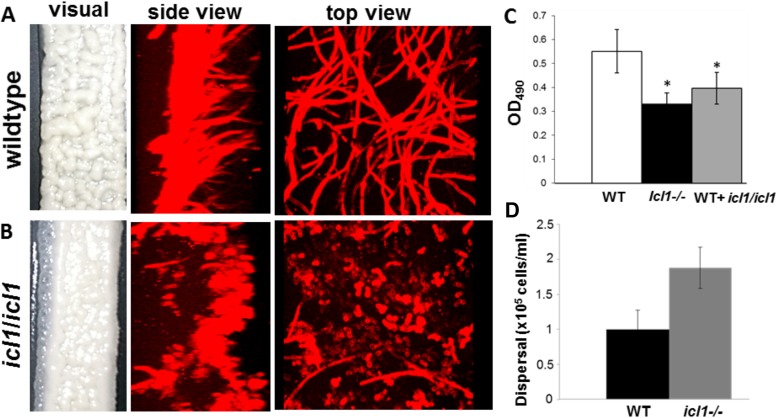
We wanted to examine whether Icl1 contributes to dispersal. However, a C. albicans icl1−/− mutant appeared to form a significantly less robust biofilm by macroscopic inspection. Confocal microscopy confirmed that ici1−/− cells had a defect in filamentation under biofilm conditions (Fig. 6A and B); however, no defects were noted during planktonic growth. Yeast cell dispersal from icl1−/− biofilms was increased 2-fold over that from the wild-type biofilms, indicating that induction of dispersal, but not biofilm formation, proceeds independently of this protein (Fig. 6D). Although ICL1 contributes to the metabolic activity of biofilm growth, its detection was perhaps below the limit of detection under the experimental conditions used for RNA-seq. Given this paradox, we detached the biofilm in its entirety (by using forceps), inverted it, and imaged its underside as well as the thin basal layer of cells still attached to the silicone material. Images revealed ICL1-GFP signal in several patches within the innermost layers of the biofilm, indicating expression of Icl1 in this environment (Fig. S1, slide 2). All the more striking, ICL1 was expressed in lateral yeast cells at the surface of the biofilm, where ambient nutrients are abundant. Taken together, these results suggest that a biofilm harbors subpopulations of cells which have their own unique gene expression pattern. This was shown by ICL1 expression within the deeper layers of the biofilm, where the cells experience substantial nutrient limitation. This finding implies that lateral yeast cells undergo metabolic reprogramming before release from the biofilm.
FIG 6 .
Impact of ICL1 loss on biofilm formation and dispersal. Biofilms of C. albicans wild-type and icl1/icl1 strains were allowed to form for 48 h. (A to C) Macroscopic and microscopic observations (biofilms stained with ConA and imaged by confocal laser scanning microscopy [CLSM]) revealed that the biofilms formed by the mutant (B) were less robust than those formed by the wild-type strain (A), which was confirmed by XTT metabolic assay (C). (D) Nonetheless, the level of dispersal from icl1 mutant biofilms was higher than that from the wild-type biofilms.
Control of biofilm dispersal inhibited C. albicans dissemination in jugular vein-catheterized mice.
To investigate whether modulation of dispersal from biofilms in vivo controls biofilm-mediated invasive disease, catheters were inoculated intraluminally with a pes1/tetO-PES1 strain in which the expression level of the major regulator of lateral yeast production, PES1, was controlled by addition or removal of DOX in the environment (7).
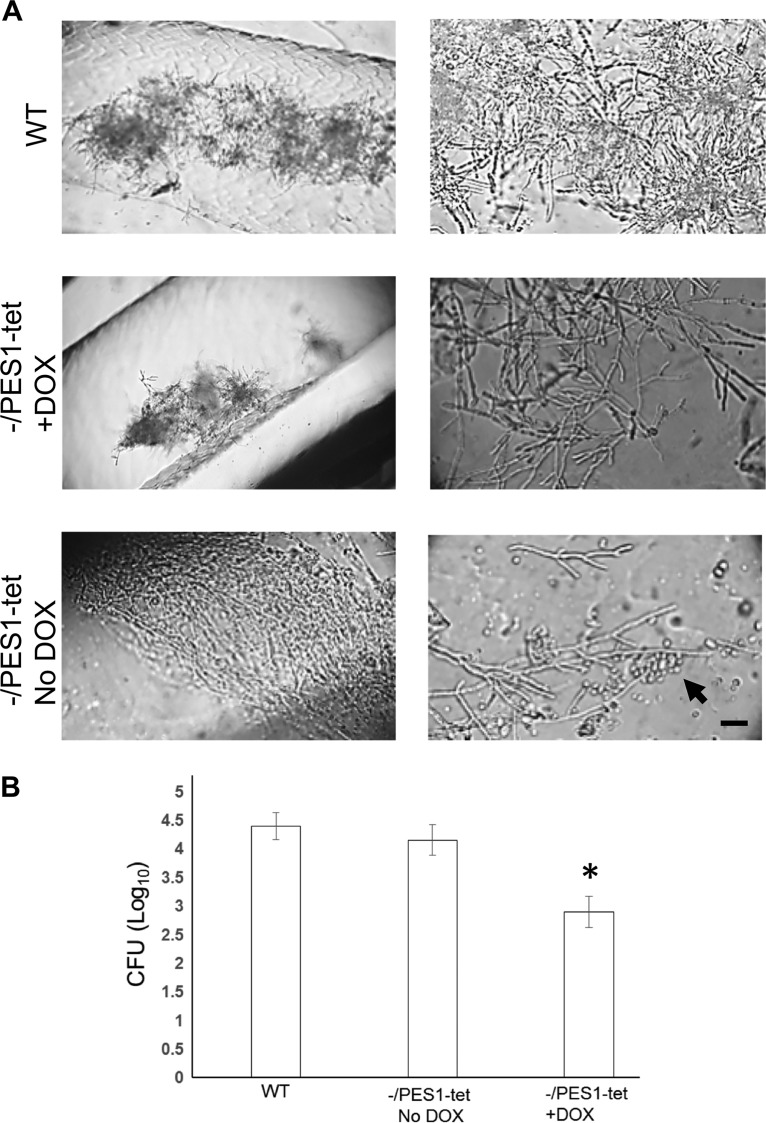
After 3 days, catheters were recovered and the distal 2 cm of the catheter was cut into pieces to measure the level of metabolic activity by using the XTT [2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide salt] assay. Regardless of the presence or absence of DOX in the catheter, the pes1/tetO-PES1 biofilms and the WT biofilms displayed the same XTT readout, indicating equivalent results with respect to biofilm formation on catheters (data not shown). Microscopy of the biofilms revealed that all catheters harbored biofilms containing hyphal filaments; catheters in which PES1 was overexpressed (in the absence of DOX) produced a high number of lateral yeast cells from the hyphal filaments (Fig. 7A). The levels of dissemination and kidney colonization were comparable between these catheters and wild-type-biofilm-containing catheters. In contrast, biofilms in catheters treated with DOX to repress PES1 lacked lateral yeast cells, and the kidneys of these mice showed a 1.5 log decrease in C. albicans dissemination and kidney colonization (Fig. 7).
FIG 7 .
Biofilm formation and dispersal in jugular vein-catheterized mice. (A) Germinating C. albicans tetracycline-regulatable PES1 strain pes1/PES1-tet grown in YNB medium with and without doxycycline was instilled in the lumen of the catheters (full catheter volume) at a concentration of 5 × 106 cells/ml. Biofilms were allowed to develop for 3 days, after which the catheters were harvested, cut laterally, and examined under a phase-contrast microscope. While all 3 strains developed biofilms in the catheters in the presence or absence of DOX, the Pes1-tet hyphae displayed increased lateral yeast production in the absence of DOX (arrow). Bar, 20 µm. (B) The extent of biofilm-mediated dissemination in mice harboring the catheters containing the two strains was also determined by measuring CFU levels in the kidney. In the presence of DOX, dissemination and kidney colonization were reduced significantly (15-fold). *, P < 0.01 (compared to the mice with catheters infected with the wild-type strain or with Pes1-tet without DOX).
DISCUSSION
Biofilm formation is the predominant mode of growth for most microorganisms in natural and clinical environments (34). For many pathogenic organisms, biofilm dispersal plays a critical role in the transmission of cells between hosts and in the propagation and spread of infection within a single host (35). For example, Streptococcus mutans can detach from dental biofilms in a mother’s mouth and be transmitted to an infant by direct or indirect contact (36). Examples of intrahost spread encompass hospital-acquired pneumonia caused by bacteria detached from biofilms in a patient’s endotracheal tube (37); an extreme case of hematogenous spread of C. albicans into the eye during urinary sepsis arising from an infected ureteric stent (38); and dissemination of C. albicans from biofilms on intravenous catheters, as we modeled in this study. Hence, understanding the characteristics of dispersed cells is important to understand how C. albicans adapts once released from the biofilm.
We have previously reported that dispersed cells exhibit a number of characteristics associated with pathogenesis compared to planktonically grown yeast cells (10), indicating measurable differences between the molecular signatures of these two different populations of yeast cells. To address that issue, in this study we performed global comparative transcriptional profiling on dispersed cells. The findings from the RNA sequencing-based analysis of gene expression patterns defined characteristics that largely overlapped those of their biofilm mothers and contrasted with those of planktonic yeast. However, these cells exhibited striking differences from the biofilm cells with which they shared similar growth conditions with respect to the genes required for attachment and invasion of a host. In addition, RNA sequencing data replicated our previous studies comparing biofilm cells and planktonic cells (15, 16). Collectively, our findings provide evidence for the presence of a developmental program that is activated during the cell dispersal process, as detailed below.
Given gene expression patterns that were obviously different from those of planktonic yeast, we wished to define the cell type of dispersal cells beyond their morphological appearance. To do this, we examined the genetic relationship between PES1 and NRG1 in these cells, because PES1 is known to be essential in C. albicans yeast. We previously reported that expression levels of PES1 and NRG1 regulate the rate of C. albicans biofilm dispersal cell production (10, 12). We have now found that in cells locked in the yeast form during NRG1 overexpression, PES1 depletion became synthetically lethal. In contrast, hyphal induction through extracellular signals or through NRG1 repression permitted biofilm growth during PES1 depletion. PES1 overexpression suppressed the filament-locked phenotype of NRG1-depleted biofilms to generate a profusion of lateral yeast cells, suggesting that PES1 acts downstream of NRG1 in regulating this developmental step. Further experiments will be needed to understand the molecular basis of these observations.
We note that the yeast-specific YWP1 gene was lowest in expression in the planktonic yeast cells. In order to maintain the nutritional conditions of biofilm cells and planktonic cells to be as similar as possible, planktonic cells were grown in YNB medium at 37°C, which may produce a mixed population of yeast and pseudohyphal cells that as a result do not express YWP1 robustly, thereby resulting in some atypical expression patterns for yeast.
Differential gene expression studies in our flow biofilms compared to age-matched planktonic cells revealed similarities in more than 180 core genes known to have roles in biofilm development (15, 16, 39) (see Data Set S1 in the supplemental material). Compared to planktonic cells, several genes related to adhesion and filamentation were upregulated in dispersed cells to an extent comparable to that seen in biofilm cells. This provided a molecular basis for our previous observation that dispersed cells are virulent entities (9, 10). Biofilm cells secrete products of a number of SAP genes at high levels as biofilm-specific proteases (candidapepsins) (26), and several of these genes (SAP3, SAP6, SAP8, and SAP9) were also found in the present study to be highly expressed in biofilm dispersal cells. SAP genes in C. albicans have several virulence functions, including host tissue adhesion, invasion and damage, or destruction of cells and molecules of the host immune system to resist antimicrobial attack (40). Thus, with the continued expression of SAP genes, the dispersal cells remain “armed” to invade the host.
In variable environments, microbes may enhance their fitness by predicting and preparing for a coming change (41). As an example, the enteric bacterium Escherichia coli elicits a transcriptional response for hypoxia when shifted from growth at 30°C to 37°C (42), as the increased temperature may indicate its arrival in the gut, where oxygen tends to be scarce. Similarly, C. albicans responds to higher pH by expressing genes involved in iron and zinc uptake via the alkaline-induced transcription factor Rim101 (43). Since the two metals are less soluble at high pH, the fungus predicts metal starvation based on its current growth environment. We posit, therefore, that dispersed C. albicans cells similarly express genes required for adhesion and tissue invasion in an anticipatory manner, based not on external cues but on a developmental program. Genes controlling zinc (ZAP1, ZRT1, and ZRT2), iron, and amino acid acquisition were found to be exclusively upregulated in the dispersed cells, though they were exposed to the same ample abundance of nutrients as their biofilm-associated mothers.
Zinc-dependent signaling has been shown previously to affect the ratio of hyphae to yeast in a biofilm; reduced expression of ZAP1—a regulator of zinc transport genes ZRT1 and ZRT2—in a biofilm leads to yeast accumulation (44). Perhaps lower ZAP1 expression levels in biofilm cells trigger yeast accumulation and dispersal. Upregulation of ZRT1 and ZRT2 in dispersed cells might follow ZAP1 expression elevation in this population.
Strikingly, dispersed cells strongly induced expression of genes encoding elements of the gluconeogenesis and glyoxylate pathways, required during carbon source starvation, while in the same rich medium, their biofilm mothers expressed a metabolic gene profile reflective of glycolysis. We conjecture that this discordant relationship between nutrient availability and expression of genes required during starvation such as those encoding transporters and glyoxylate cycle components reflects an anticipatory developmental step during production of cells primed to infect new sites in the host with low availability of glucose and other crucial nutrients. In summary, it is tempting to speculate that increased expression of adhesin genes, hypha-specific genes, and secretory aspartyl protease genes as well as of gluconeogenesis-associated and glyoxylate cycle-associated genes in biofilm dispersal cells or planktonic cells suggests that in biofilm-released cells, gene products required during or after tissue invasion are induced before the actual invasion process is initiated.
Unlike the yeast-to-hypha transition, which requires a specific external trigger (such as heat, serum, specific nutrients, alkaline pH, hypoxia, etc.), the hypha-to-lateral-yeast developmental process occurs constitutively. In nutritionally rich or poor media (7, 45, 46), in host tissues (8), or in biofilms formed in catheters (10), C. albicans hyphae consistently produce lateral yeast cells. Hence, this process appears to reflect an intrinsic developmental program. TLO genes, which are broad transcriptional regulators, are differentially upregulated in biofilm cells. Whether the mechanism underlying differential gene expression in a biofilm mother cell and its dispersed daughter consists of selective mRNA transport between the cytoplasm of these cells or, e.g., of reprogramming of gene expression after mitosis of the dispersed cell remains to be determined in future work.
In conclusion, our findings confirm the importance of PES1 for C. albicans yeast growth and its significant impact on lateral yeast production and biofilm dispersal. Regulation of PES1 could have important implications in clinical settings, where biofilms formed on catheters have direct access to the bloodstream. This was seen in our in vivo studies using the jugular vein catheter mouse model where overexpression of PES1 in biofilms growing in the lumen of the catheters resulted in enhanced dispersal and disseminated infection, while repression of PES1 resulted in abrogation of biofilm dispersal and a 15-fold decrease in Candida infection of distal organs. These findings may lay the foundation for discovery of novel inhibitors that interfere with the regulatory signals controlling dispersal from biofilms. A better understanding of the molecular mechanisms of biofilm dispersal might pave the way for control of biofilm-mediated disseminated diseases in humans.
MATERIALS AND METHODS
Strains and culture conditions.
Stock cultures of all strains were stored in 15% glycerol at −80°C. Strains were routinely grown under yeast conditions (media at 30°C) in YPG (1% yeast extract, 2% Bacto peptone, 2% glucose) or under filament-inducing conditions using RPMI medium (Sigma, St. Louis, MO) with MOPS (morpholinepropanesulfonic acid) buffer or Spider medium. The strains used in this study, their origin, and their construction are briefly listed in Table 1.
TABLE 1 .
List of strains used in this study
| Strain | Genotype/construction | Reference or source |
|---|---|---|
| SC5314 | Wild type | 54 |
| CLM3-2 | ura3::λ imm434/ura3::λ imm434, pPFK2-GFP | 55 |
| CJB-3 | ura3::λ imm434/ura3::λ imm434, pICL1-GFP | 55 |
| RM1000 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG | 56 |
| CLM18-1 | ura3::λ imm434/ura3::λ imm434, his1::hisG/his1::hisG, icl1::HIS1/icl1::URA3 | 55 |
| JKC619 | pes1::FLP-NAT1/PES1 (parent SC5314) | 7 |
| JKC673 | pes1::FRT/FLP-NAT1-pMAL2-PES1 (parent JKC619) | 7 |
| SSY50B |
NRG1/tetO-NRG1-URA3 ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/eno1::ENO1-tetR-ScHAP4AD-3× HA-ADE2 |
57 |
| JKC869 |
pes1::FLP-NAT1/PES1 NRG1/tetO-NRG1-URA3 ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/ eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2; transformant 3; SSY50B was transformed with pJK890 (deletion plasmid to replace PES1 with FLP-NAT1 as previously described [7]); parent strain, SSY50-B |
This work |
| JKC870 |
pes1::FLP-NAT1/PES1 NRG1/tetO-NRG1-URA3 ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/ eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2; transformant 4; SSY50B was transformed with pJK890 (deletion plasmid to replace PES1 with FLP-NAT1 as previously described [7]); parent strain, SSY50-B |
This work |
| JKC993 |
pes1::FRT/FLP-NAT1-pMAL2-PES1 NRG1/tetO-NRG1-URA3 ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/ eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2; FLP-NAT1 was excised from JKC869 by inducing flippase, and the resulting strain was transformed with pJK896 (promoter replacement plasmid to replace PES1 promoter with MAL2 promoter as previously described [7]); parent strain, JKC869 |
This work |
| JKC996 |
pes1::FRT/FLP-NAT1-pMAL2-PES1 NRG1/tetO-NRG1-URA3 ade2::hisG/ade2::hisG ura3::imm434/ura3::imm434 ENO1/ eno1::ENO1-tetR-ScHAP4AD-3×HA-ADE2; FLP-NAT1 was excised from JKC869 by inducing flippase, and the resulting strain was transformed with pJK896 (promoter replacement plasmid to replace PES1 promoter with MAL2 promoter as previously described [7]); parent strain, JKC870 |
This work |
| TLOβ2/tloβ2 | Strain 12822 (haploid strain from Yue Wang, Singapore) was transformed with the PCR product of pGEM-URA3 (56); transformants were selected for on SDC-ura and confirmed by PCR for pGFP-NAT1 (58); ransformants were selected for on YPAD plus neurseothricin and confirmed; PCR with TLO2 primers confirmed that no wild-type copy remained |
This work |
Cell collection and RNA extraction.
Biofilms were developed in a simple flow biofilm model as described previously (13, 14). Biofilms were allowed to grow for 24 h in YNB–1% glucose media. The biofilms were collected from the SE strip (Cardiovascular Instruments Corp., Wakefield, MA) and flash frozen in liquid nitrogen followed by short-term storage at −80°C. Prior to harvesting, cells released from the biofilm in the flowthrough media were collected at the completion of the 24-h growth period. These cells were microscopically confirmed to be yeast cells. As biofilms were previously shown to disperse at the rate of ~5 × 105 cells/ml in YNB medium (10), to obtain enough cells for RNA extraction, we collected the flowthrough several times from replicate biofilms (3 ml at a time in tubes placed on ice). The cells were pelleted by cold ultracentrifugation, and the total time from collection of the dispersed cells to freezing of the pellet on dry ice at −80°C was 8 min. For some experiments, dispersed cells were enumerated by hemocytometer and by colony counts on solid medium (YPD-agar plates). Planktonic cells growing for 24 h in flasks containing YNB–1% glucose at 37°C were centrifuged, and the pellets were stored at −80°C. RNA was extracted from all frozen pellets using a RiboPure yeast kit (22, 44).
Static biofilm formation.
Biofilms were grown under static conditions in microtiter plates and visualized by bright-field or confocal scanning laser microscopy, as previously described (47), with minor modifications. Briefly, 1 ml of C. albicans cells (1 × 106 cells/ml) was added to the wells of a 24-well microtiter plate and incubated overnight in YNB with or without DOX and in glucose or maltose as the carbon source, and then the biofilms formed were measured by an XTT assay as previously described (47). For enumeration of dispersed cells, static biofilm supernatants (and one wash) were collected and cells counted with a hemocytometer. Biofilms were also grown on catheter material and quantified by dry weight measurements, as previously published (39, 47, 48). Statistical significance data (P values) were calculated with a Student’s t test.
Comparative transcriptomic analysis by RNA sequencing.
RNA was extracted from a total of six samples: two replicates each of the planktonic cells, the biofilm cells, and the biofilm dispersal cells. RNA sequencing and bioinformatic analysis were performed at the Genome Sequencing Facility of Greehey Children’s Cancer Research Institute at the University of Texas Health Science Center at San Antonio, Briefly, 5 µg total RNA was used for mRNA isolation with Dynabeads OligodT (Invitrogen, Carlsbad, CA), and then about 30 to 50 ng isolated mRNA was used for mRNA-seq library preparation by following the sample preparation guide provided with a BIOO Scientific NEXTflex Directional RNA-seq kit (dUTP-based). The first step in the workflow involves purifying the poly(A)-containing mRNA molecules using Dynabeads poly(T) oligonucleotide-attached magnetic beads. Following purification, the mRNA is fragmented into small pieces using divalent cations and elevated temperature (95°C for 10 min). The cleaved RNA fragments are copied into first-strand cDNA using reverse transcriptase and random primers. This is followed by second-strand cDNA synthesis using DNA polymerase I and RNase H. These cDNA fragments then go through an end repair process, the addition of a single “A” base, and then ligation of the adapters. The products are then purified and enriched by PCR to create the final RNA-seq library. Directionality is retained by adding dUTP during the second-strand synthesis step and by subsequent cleavage of the uridine-containing strand using uracil-DNA glycosylase. After the RNA-seq libraries were subjected to the quantification process and pooled for cBot amplification, subsequent 100-bp-paired-end sequencing was run with an Illumina HiSeq 2000 platform (San Diego, CA). After the sequencing run, demultiplexing with CASAVA was employed to generate the fastq file for each sample. All sequencing reads were filtered, trimmed, and aligned with a C. albicans reference genome using TopHat2 (49) default settings, and the Bam files from alignment were processed using HTSeq-count (50) to obtain the counts per gene in all samples. A statistical analysis of differential gene expression was performed using the DESeq package from Bioconductor (51), and a gene was considered significantly altered if the false-discovery rate for differential expression was ≤0.05. The processes associated with differentially expressed genes were identified using Gene Ontology Slim Mapper (52).
Epistasis study on solid media.
Cells were grown on yeast extract–peptone–2% maltose (YPM) plates containing 5 µg/ml doxycycline (DOX) for 48 h at 30°C. Following two washes with 0.9% NaCl, 5-fold cell dilutions (beginning at an optical density at 600 nm [OD600] of 0.5) were spotted with a calibrated replicator (V&P Scientific, San Diego, CA) onto YPM, yeast extract–2% dextrose (YPD), or Spider medium without mannitol with 2% maltose (SpiM) or with 2% dextrose (SpiD). Plates were incubated for 3 days at 30°C (YP) or 37°C (Spi). Plates contained DOX (5 µg/ml) or vehicle.
Jugular vein catheter mouse model of infection.
A C. albicans mouse catheter biofilm model was used for in vivo experiments as previously described (53) with minor modifications. These in vivo experiments were approved by the Los Angeles Biomedical Research Institute, Harbor-UCLA IACUC. Briefly, we used catheterized 8-week-old C57BL/6 male mice, purchased from laboratories of Charles River, Inc. (Wilmington, MA), where the surgery was performed. The surgery involves insertion of a Silastic catheter into the jugular vein of the mice. Patency is tested, and the catheter is filled with heparin lock solution and sealed with a plug. Following receipt of the jugular vein-catheterized mice, the catheters were instilled with 25 µl of a C. albicans inoculum of 5 × 106 cells/ml (the entire catheter volume) using a 23-gauge blunt-ended needle after removal of the plug and the lock solution (the plug was put back in place after inoculation).
The wild-type and pes1/PES1-tet strains were allowed to germinate for 90 min in the absence of DOX in YNB medium. Mice were divided into the following 3 groups with 5 mice in each group: (i) mice with catheters infected with the wild-type strain in YNB, (ii) mice with catheters infected with the pes1/PES1-tet strain in YNB, and (iii) mice with catheters infected with the pes1/PES1-tet strain in YNB containing 25 µM DOX. Cells were allowed to adhere in the catheter lumen for 3 days, after which the mice were euthanized and catheters aseptically removed. The catheters were cut laterally and imaged under a phase-contrast microscope to visualize the morphology of the cells growing within the catheters of the individual groups. The distal 2 cm of the catheters was cut first laterally and then into small pieces and introduced into a 96-well plate containing XTT solution for 90 min, to measure the metabolic activity of cells in the biofilm. Additionally, kidneys were harvested, weighed, homogenized, and plated on YPD for CFU enumeration. Differences in organ fungal burden between the 3 groups are presented as log CFU per gram tissue, and results from a two-tailed t test with a P value of <0.05 were considered significant.
Footnotes
Citation Uppuluri P, Acosta Zaldívar M, Anderson MZ, Dunn MJ, Berman J, Lopez Ribot JL, Köhler JR. 2018. Candida albicans dispersed cells are developmentally distinct from biofilm and planktonic cells. mBio 9:e01338-18. https://doi.org/10.1128/mBio.01338-18.
REFERENCES
- 1.Gudlaugsson O, Gillespie S, Lee K, Vande Berg J, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. 2003. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis 37:1172–1177. doi: 10.1086/378745. [DOI] [PubMed] [Google Scholar]
- 2.Morgan J. 2005. Global trends in candidemia: review of reports from 1995–2005. Curr Infect Dis Rep 7:429–439. doi: 10.1007/s11908-005-0044-7. [DOI] [PubMed] [Google Scholar]
- 3.Brown AJ, Gow NA. 1999. Regulatory networks controlling Candida albicans morphogenesis. Trends Microbiol 7:333–338. doi: 10.1016/S0966-842X(99)01556-5. [DOI] [PubMed] [Google Scholar]
- 4.Gow NA, van de Veerdonk FL, Brown AJ, Netea MG. 2011. Candida albicans morphogenesis and host defence: discriminating invasion from colonization. Nat Rev Microbiol 10:112–122. doi: 10.1038/nrmicro2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noble SM, Gianetti BA, Witchley JN. 2017. Candida albicans cell-type switching and functional plasticity in the mammalian host. Nat Rev Microbiol 15:96–108. doi: 10.1038/nrmicro.2016.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, Murciano C, Blagojevic M, Thavaraj S, Förster TM, Hebecker B, Kasper L, Vizcay G, Iancu SI, Kichik N, Häder A, Kurzai O, Luo T, Krüger T, Kniemeyer O, Cota E, Bader O, Wheeler RT, Gutsmann T, Hube B, Naglik JR. 2016. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature 532:64–68. doi: 10.1038/nature17625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shen J, Cowen LE, Griffin AM, Chan L, Köhler JR. 2008. The Candida albicans pescadillo homolog is required for normal hypha-to-yeast morphogenesis and yeast proliferation. Proc Natl Acad Sci U S A 105:20918–20923. doi: 10.1073/pnas.0809147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uppuluri P, Chaturvedi AK, Jani N, Pukkila-Worley R, Monteagudo C, Mylonakis E, Köhler JR, Lopez Ribot JL. 2012. Physiologic expression of the Candida albicans pescadillo homolog is required for virulence in a murine model of hematogenously disseminated candidiasis. Eukaryot Cell 11:1552–1556. doi: 10.1128/EC.00171-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nett J, Andes D. 2006. Candida albicans biofilm development, modeling a host-pathogen interaction. Curr Opin Microbiol 9:340–345. doi: 10.1016/j.mib.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Köhler JR, Kadosh D, Lopez-Ribot JL. 2010. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog 6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nobile CJ, Fox EP, Hartooni N, Mitchell KF, Hnisz D, Andes DR, Kuchler K, Johnson AD. 2014. A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. mBio 5:e01201-14. doi: 10.1128/mBio.01201-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Uppuluri P, Pierce CG, Thomas DP, Bubeck SS, Saville SP, Lopez-Ribot JL. 2010. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot Cell 9:1531–1537. doi: 10.1128/EC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uppuluri P, Chaturvedi AK, Lopez-Ribot JL. 2009. Design of a simple model of Candida albicans biofilms formed under conditions of flow: development, architecture, and drug resistance. Mycopathologia 168:101–109. doi: 10.1007/s11046-009-9205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uppuluri P, Lopez-Ribot JL. 2010. An easy and economical in vitro method for the formation of Candida albicans biofilms under continuous conditions of flow. Virulence 1:483–487. doi: 10.4161/viru.1.6.13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.García-Sánchez S, Aubert S, Iraqui I, Janbon G, Ghigo JM, d’Enfert C. 2004. Candida albicans biofilms: a developmental state associated with specific and stable gene expression patterns. Eukaryot Cell 3:536–545. doi: 10.1128/EC.3.2.536-545.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nett JE, Lepak AJ, Marchillo K, Andes DR. 2009. Time course global gene expression analysis of an in vivo Candida biofilm. J Infect Dis 200:307–313. doi: 10.1086/599838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nobile CJ, Schneider HA, Nett JE, Sheppard DC, Filler SG, Andes DR, Mitchell AP. 2008. Complementary adhesin function in C. albicans biofilm formation. Curr Biol 18:1017–1024. doi: 10.1016/j.cub.2008.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rouabhia M, Semlali A, Chandra J, Mukherjee P, Chmielewski W, Ghannoum MA. 2012. Disruption of the ECM33 gene in Candida albicans prevents biofilm formation, engineered human oral mucosa tissue damage and gingival cell necrosis/apoptosis. Mediators Inflamm 2012:398207. doi: 10.1155/2012/398207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mukherjee PK, Chandra J, Kuhn DM, Ghannoum MA. 2003. Mechanism of fluconazole resistance in Candida albicans biofilms: phase-specific role of efflux pumps and membrane sterols. Infect Immun 71:4333–4340. doi: 10.1128/IAI.71.8.4333-4340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nett JE, Sanchez H, Cain MT, Ross KM, Andes DR. 2011. Interface of Candida albicans biofilm matrix-associated drug resistance and cell wall integrity regulation. Eukaryot Cell 10:1660–1669. doi: 10.1128/EC.05126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cabral V, Znaidi S, Walker LA, Martin-Yken H, Dague E, Legrand M, Lee K, Chauvel M, Firon A, Rossignol T, Richard ML, Munro CA, Bachellier-Bassi S, d’Enfert C. 2014. Targeted changes of the cell wall proteome influence Candida albicans ability to form single- and multi-strain biofilms. PLoS Pathog 10:e1004542. doi: 10.1371/journal.ppat.1004542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desai JV, Bruno VM, Ganguly S, Stamper RJ, Mitchell KF, Solis N, Hill EM, Xu W, Filler SG, Andes DR, Fanning S, Lanni F, Mitchell AP. 2013. Regulatory role of glycerol in Candida albicans biofilm formation. mBio 4:e00637-12. doi: 10.1128/mBio.00637-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murillo LA, Newport G, Lan CY, Habelitz S, Dungan J, Agabian NM. 2005. Genome-wide transcription profiling of the early phase of biofilm formation by Candida albicans. Eukaryot Cell 4:1562–1573. doi: 10.1128/EC.4.9.1562-1573.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nobile CJ, Johnson AD. 2015. Candida albicans biofilms and human disease. Annu Rev Microbiol 69:71–92. doi: 10.1146/annurev-micro-091014-104330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Granger BL, Flenniken ML, Davis DA, Mitchell AP, Cutler JE. 2005. Yeast wall protein 1 of Candida albicans. Microbiology 151:1631–1644. doi: 10.1099/mic.0.27663-0. [DOI] [PubMed] [Google Scholar]
- 26.Winter MB, Salcedo EC, Lohse MB, Hartooni N, Gulati M, Sanchez H, Takagi J, Hube B, Andes DR, Johnson AD, Craik CS, Nobile CJ. 2016. Global identification of biofilm-specific proteolysis in Candida albicans. mBio 7:e01514-16. doi: 10.1128/mBio.01514-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Boer AD, de Groot PW, Weindl G, Schaller M, Riedel D, Diez-Orejas R, Klis FM, de Koster CG, Dekker HL, Gross U, Bader O, Weig M. 2010. The Candida albicans cell wall protein Rhd3/Pga29 is abundant in the yeast form and contributes to virulence. Yeast 27:611–624. doi: 10.1002/yea.1790. [DOI] [PubMed] [Google Scholar]
- 28.Giacometti R, Kronberg F, Biondi RM, Passeron S. 2011. Candida albicans Tpk1p and Tpk2p isoforms differentially regulate pseudohyphal development, biofilm structure, cell aggregation and adhesins expression. Yeast 28:293–308. doi: 10.1002/yea.1839. [DOI] [PubMed] [Google Scholar]
- 29.Nobile CJ, Nett JE, Hernday AD, Homann OR, Deneault JS, Nantel A, Andes DR, Johnson AD, Mitchell AP. 2009. Biofilm matrix regulation by Candida albicans Zap1. PLoS Biol 7:e1000133. doi: 10.1371/journal.pbio.1000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vieira N, Casal M, Johansson B, MacCallum DM, Brown AJ, Paiva S. 2010. Functional specialization and differential regulation of short-chain carboxylic acid transporters in the pathogen Candida albicans. Mol Microbiol 75:1337–1354. doi: 10.1111/j.1365-2958.2009.07003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haran J, Boyle H, Hokamp K, Yeomans T, Liu Z, Church M, Fleming AB, Anderson MZ, Berman J, Myers LC, Sullivan DJ, Moran GP. 2014. Telomeric ORFs (TLOs) in Candida spp. encode mediator subunits that regulate distinct virulence traits. PLoS Genet 10:e1004658. doi: 10.1371/journal.pgen.1004658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson MZ, Baller JA, Dulmage K, Wigen L, Berman J. 2012. The three clades of the telomere-associated TLO gene family of Candida albicans have different splicing, localization, and expression features. Eukaryot Cell 11:1268–1275. doi: 10.1128/EC.00230-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van het Hoog M, Rast TJ, Martchenko M, Grindle S, Dignard D, Hogues H, Cuomo C, Berriman M, Scherer S, Magee BB, Whiteway M, Chibana H, Nantel A, Magee PT. 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol 8:R52. doi: 10.1186/gb-2007-8-4-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caldwell DE, Atuku E, Wilkie DC, Wivcharuk KP, Karthikeyan S, Korber DR, Schmid DF, Wolfaardt GM. 1997. Germ theory vs. community theory in understanding and controlling the proliferation of biofilms. Adv Dent Res 11:4–13. doi: 10.1177/08959374970110011501. [DOI] [PubMed] [Google Scholar]
- 35.Kaplan JB. 2010. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res 89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berkowitz RJ, Jones P. 1985. Mouth-to-mouth transmission of the bacterium Streptococcus mutans between mother and child. Arch Oral Biol 30:377–379. doi: 10.1016/0003-9969(85)90014-7. [DOI] [PubMed] [Google Scholar]
- 37.Adair CG, Gorman SP, Feron BM, Byers LM, Jones DS, Goldsmith CE, Moore JE, Kerr JR, Curran MD, Hogg G, Webb CH, McCarthy GJ, Milligan KR. 1999. Implications of endotracheal tube biofilm for ventilator-associated pneumonia. Intensive Care Med 25:1072–1076. doi: 10.1007/s001340051014. [DOI] [PubMed] [Google Scholar]
- 38.Hassan A, Poon W, Baker M, Linton C, Mühlschlegel FA. 2012. Confirmed Candida albicans endogenous fungal endophthalmitis in a patient with chronic candidiasis. Med Mycol Case Rep 1:42–44. doi: 10.1016/j.mmcr.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nobile CJ, Fox EP, Nett JE, Sorrells TR, Mitrovich QM, Hernday AD, Tuch BB, Andes DR, Johnson AD. 2012. A recently evolved transcriptional network controls biofilm development in Candida albicans. Cell 148:126–138. doi: 10.1016/j.cell.2011.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hube B, Naglik J. 2001. Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 147:1997–2005. doi: 10.1099/00221287-147-8-1997. [DOI] [PubMed] [Google Scholar]
- 41.Brunke S, Hube B. 2014. Adaptive prediction as a strategy in microbial infections. PLoS Pathog 10:e1004356. doi: 10.1371/journal.ppat.1004356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tagkopoulos I, Liu YC, Tavazoie S. 2008. Predictive behavior within microbial genetic networks. Science 320:1313–1317. doi: 10.1126/science.1154456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bensen ES, Martin SJ, Li M, Berman J, Davis DA. 2004. Transcriptional profiling in Candida albicans reveals new adaptive responses to extracellular pH and functions for Rim101p. Mol Microbiol 54:1335–1351. doi: 10.1111/j.1365-2958.2004.04350.x. [DOI] [PubMed] [Google Scholar]
- 44.Ganguly S, Bishop AC, Xu W, Ghosh S, Nickerson KW, Lanni F, Patton-Vogt J, Mitchell AP. 2011. Zap1 control of cell-cell signaling in Candida albicans biofilms. Eukaryot Cell 10:1448–1454. doi: 10.1128/EC.05196-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bahn YS, Staab J, Sundstrom P. 2003. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol Microbiol 50:391–409. doi: 10.1046/j.1365-2958.2003.03692.x. [DOI] [PubMed] [Google Scholar]
- 46.Bahn YS, Sundstrom P. 2001. CAP1, an adenylate cyclase-associated protein gene, regulates bud-hypha transitions, filamentous growth, and cyclic AMP levels and is required for virulence of Candida albicans. J Bacteriol 183:3211–3223. doi: 10.1128/JB.183.10.3211-3223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pierce CG, Uppuluri P, Tristan AR, Wormley FL Jr, Mowat E, Ramage G, Lopez-Ribot JL. 2008. A simple and reproducible 96-well plate-based method for the formation of fungal biofilms and its application to antifungal susceptibility testing. Nat Protoc 3:1494–1500. doi: 10.1038/nport.2008.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uppuluri P, Busscher HJ, Chakladar J, van der Mei HC, Chaffin WL. 2017. Transcriptional profiling of C. albicans in a two species biofilm with Rothia dentocariosa. Front Cell Infect Microbiol 7:311. doi: 10.3389/fcimb.2017.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Anders S, Pyl PT, Huber W. 2015. Seq—a python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Inglis DO, Arnaud MB, Binkley J, Shah P, Skrzypek MS, Wymore F, Binkley G, Miyasato SR, Simison M, Sherlock G. 2012. The Candida genome database incorporates multiple Candida species: multispecies search and analysis tools with curated gene and protein information for Candida albicans and Candida glabrata. Nucleic Acids Res 40:D667–D674. doi: 10.1093/nar/gkr945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lazzell AL, Chaturvedi AK, Pierce CG, Prasad D, Uppuluri P, Lopez-Ribot JL. 2009. Treatment and prevention of Candida albicans biofilms with caspofungin in a novel central venous catheter murine model of candidiasis. J Antimicrob Chemother 64:567–570. doi: 10.1093/jac/dkp242. [DOI] [PubMed] [Google Scholar]
- 54.Fonzi WA, Irwin MY. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barelle CJ, Priest CL, Maccallum DM, Gow NA, Odds FC, Brown AJ. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol 8:961–971. doi: 10.1111/j.1462-5822.2005.00676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilson RB, Davis D, Mitchell AP. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J Bacteriol 181:1868–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saville SP, Lazzell AL, Monteagudo C, Lopez-Ribot JL. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot Cell 2:1053–1060. doi: 10.1128/EC.2.5.1053-1060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gerami-Nejad M, Forche A, McClellan M, Berman J. 2012. Analysis of protein function in clinical C. albicans isolates. Yeast 29:303–309. doi: 10.1002/yea.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comprehensive differentially expressed data of significantly (P < 0.5) regulated (>1.5-fold) genes. In this Excel sheet, sheet 1 lists genes significantly upregulated in biofilm versus planktonic cells, sheet 2 lists genes significantly downregulated in biofilm versus planktonic cells, sheet 3 lists genes common between our study and other transcriptone studies comparing biofilm expression data to planktonic expression data, sheet 4 lists genes significantly upregulated in dispersed versus planktonic cells, sheet 5 lists genes significantly downregulated in dispersed versus planktonic cells, sheet 6 lists genes showing similar patterns of upregulation in both biofilm and planktonic cells compared to planktonic cells, and sheet 7 lists genes showing similar patterns of downregulation in both biofilm and planktonic cells compared to planktonic cells. Download DATA SET S1, XLSX file, 0.3 MB (310.5KB, xlsx) .
Copyright © 2018 Uppuluri et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data set of genes expressed in dispersed cell populations. In this Excel sheet, sheet 1 shows differential expression of central carbon metabolism genes in both dispersed and planktonic cells versus biofilm cells. Here the dispersed cells show a planktonic-yeast-like expression pattern. Sheet 2 shows a set of genes that are upregulated and downregulated only on dispersed cells. These data were extracted after pairwise comparisons of all the sample conditions to each other, picking largely the genes that were upregulated or downregulated exclusively in dispersed cells. Download DATA SET S2, XLSX file, 0.03 MB (31.3KB, xlsx) .
Copyright © 2018 Uppuluri et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Figures related to Fig. 2 in the manuscript. Slide 1 shows that the topmost layers of the flow biofilm expressed green fluorescent Pfk2. Slide 2 shows that the cells attached to the silicone substrate (innermost portion of the flow biofilm) expressed green fluorescent Icl1. Download FIG S1, TIF file, 1 MB (1MB, tif) .
Copyright © 2018 Uppuluri et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.