Abstract
The mechanisms by which organisms adapt to variable environments are a fundamental question in evolutionary biology and are important to protect important species in response to a changing climate. An interesting candidate to study this question is the honey bee Apis cerana, a keystone pollinator with a wide distribution throughout a large variety of climates, that exhibits rapid dispersal. Here, we resequenced the genome of 180 A. cerana individuals from 18 populations throughout China. Using a population genomics approach, we observed considerable genetic variation in A. cerana. Patterns of genetic differentiation indicate high divergence at the subspecies level, and physical barriers rather than distance are the driving force for population divergence. Estimations of divergence time suggested that the main branches diverged between 300 and 500 Ka. Analyses of the population history revealed a substantial influence of the Earth’s climate on the effective population size of A. cerana, as increased population sizes were observed during warmer periods. Further analyses identified candidate genes under natural selection that are potentially related to honey bee cognition, temperature adaptation, and olfactory. Based on our results, A. cerana may have great potential in response to climate change. Our study provides fundamental knowledge of the evolution and adaptation of A. cerana.
Keywords: Apis cerana, honey bee, genetic differentiation, genetic diversity, effective population size, diverse environments, vulnerability, climate change
Introduction
Adaptation to diverse and changing environments is a fundamental question in evolutionary biology. An understanding of the mechanisms by which variable environments influence the genetic diversity of a species will not only illuminate its evolutionary history but also provide information for the development of strategies designed to effectively protect important species in a changing climate (Skelly et al. 2007; Balanyà et al. 2009). In addition, detecting genes under natural selection can help elucidate the underlying mechanisms of adaptation to local environments and facilitate selective breeding.
The eastern honey bee (Apis cerana) is an interesting species to study adaptation to diverse environments, as its natural distribution covers a large variety of environments, ranging from tropical to cold temperate climates and from plains to mountainous areas (Hepburn and Radloff 2011). It also exhibits the ability of fast dispersal (Gloag et al. 2016). However, despite the extensive number of population studies using morphometric characteristics, microsatellites, and mitochondrial DNA segments (Smith et al. 2000; Radloff et al. 2010; Hepburn and Radloff 2011; Tan et al. 2015 and references therein), knowledge of the evolution and adaptation of A. cerana is limited due to the lack of the studies at the genomic level. To date, the most comprehensive studies of A. cerana populations have been conducted by Smith et al. (2000) using mitochondrial DNA and Radloff et al. (2010) using morphometric characteristics, who identified five and six major groups across the range of A. cerana, respectively. Both studies classified the majority of A. cerana in China as one group. However, several studies focusing on different parts of China identified variations in mitochondrial DNA fragments or microsatellite DNA, even in populations from small geographic regions, suggesting large genetic differences among populations in China (Xu et al. 2013; Yin and Ji 2013; Zhao et al. 2014; Tan et al. 2015; Liu et al. 2016; Cao et al. 2017). This disparity may be attributed to the limited information gained from the A. cerana mitochondrial tRNAleu-COII region as most of the noncoding region is missing in many populations (Smith et al. 2000). Population genomics provides powerful toolsets and holds great promise in the study of honey bee populations (Lozier and Zayed 2017). Population studies at the genomic level not only increase the power and resolution of traditional genetic approaches but also identify genetic variation in adaptive and economic traits, as well as yield insights into the genetic architecture. However, population genomics of A. cerana has been hampered by limitations in obtaining adequate samples and the lack of a reference genome until 2015 (Park et al. 2015).
Genomic research of A. cerana is also desirable to fill gaps in knowledge and the need for protection (Teichroew et al. 2017). A. cerana is a keystone species that provides pollination and other valuable ecosystem services that contribute significantly to agricultural production, food security, and nutrition for a growing global population. However, the A. cerana population has experienced a substantial decline in recent decades (Theisen-Jones and Bienefeld 2016). As climate change is one of the major threats (Thomann et al. 2013), studies of aspects of the evolutionary biology of A. cerana are urgently needed to obtain an understanding of how this keystone pollinator may respond to and/or be affected by climate change.
The draft genome of A. cerana (Park et al. 2015) has provided a fundamental resource that enabled the resequencing of genomes and population genomic research. In the present study, we collected samples from 18 areas in China that, although they only constitute a small proportion of the A. cerana range, cover a variety of climates from tropical to temperate areas, with terrains such as plateaus, basins, plains, and islands, and resequenced180 bees with each individual from a different colony. We also downloaded resequenced data obtained from representative subspecies from major lineages of A. mellifera (Harpur et al. 2014; Chen et al. 2016). Using population genomic methods, we identified genetic variations and explored the population structure and differentiation among A. cerana populations. We also investigated the population history to understand climate-driven demography. Finally, we identified genes related to adaptation to local environments in A. cerana. This study provides insights into the evolutionary history and genetic diversity of A. cerana, as well as an example of mechanisms by which a species can adapt to regions with a variety of climates.
Results
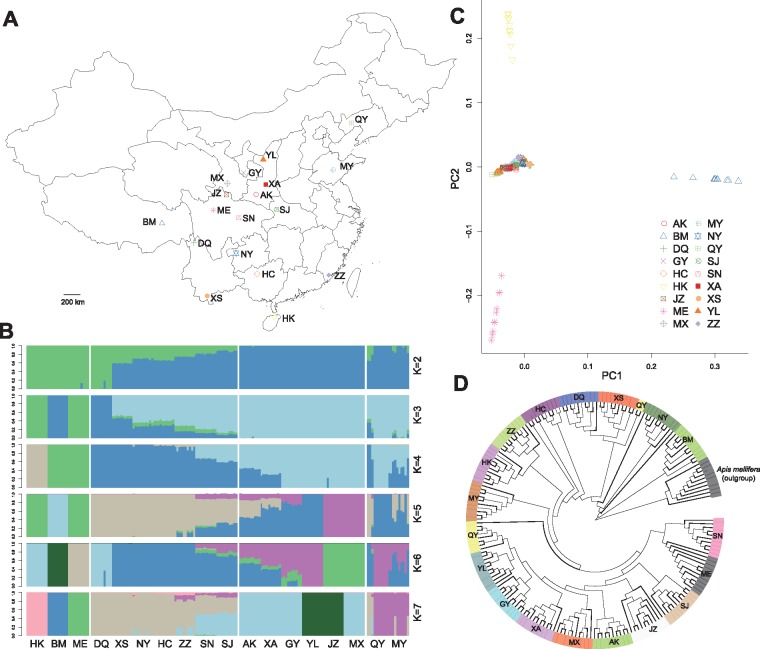
We sampled 180 individuals from 18 populations throughout China (fig. 1A and supplementary table S1, Supplementary Material online). Whole-genome resequencing yielded 361 Gb of sequencing data. Genome alignment resulted in an average depth of 7.8X (supplementary table S2, Supplementary Material online). In total, we identified 2.67 million SNPs after quality control, with 144, 673 in exonic regions and 27, 947 nonsynonymous ones (supplementary table S3, Supplementary Material online).
Fig. 1.
Population structure. (A) Geographic locations of the sampled bees; ten colonies were sampled at each location. (B) Genetic structure of the 180 individuals from 18 populations. Groupings of samples from two to seven ancestral clusters are shown. MY and QY showed signs of impaired local genetic integrity. (C) Scatter plot of principal components 1 versus 2 (PC1 vs. PC2) for the 18 populations. (D) Neighboring-joining phylogenetic tree with Apis mellifera as an outgroup.
Population Structure
We first performed clustering analyses using ADMIXTURE (Alexander et al. 2009) to examine the relationships among the populations (fig. 1B). With a K value of 2, populations from the north (AK, XA, GY, YL, JZ, and MX) formed an ancestral cluster and populations in the west (BM, DQ, and ME) and the Hainan island (HK) formed another cluster, while other populations distributed between these areas showed different degrees of mixed ancestry. As K increases from 3 to 5, HK, BM, and ME showed distinct ancestries from other populations. With K = 6 and K = 7, DQ, JZ, and YL were separated from the other populations. These populations are mainly from island and mountainous areas and are more isolated from the other populations. MY and QY were highly admixed in all cases, with large differences among individuals. Cross-validation errors for different K vales are provided in supplementary table S4, Supplementary Material online. The results of the principal component analysis (PCA) using GCTA (Yang et al. 2011) further supported the patterns. The first and second principal components (PC1 and PC2) separated BM, HK, and ME (fig. 1C), consistent with the ADMIXTURE results at K = 3–5; PC3 further separated JZ, YL, and QY from other populations but was not able to distinguish between YL and QY (supplementary fig. S1, Supplementary Material online); with PC4, DQ was also separated from the other populations, consistent with the ADMIXTURE results at K = 6 and 7. The remaining populations were not separated by PC1 through PC4, further indicating that these populations may be admixed; however, PC3 and PC4 for these populations reflect their distribution along latitude (supplementary figs. S1 and S2, Supplementary Material online).
We performed three population tests of admixture using TreeMix (Pickrell and Pritchard 2012) to further test for signature of admixture among the populations. We calculated the corresponding f3 statistics for all possible combinations of three populations, with a negative f3 statistic indicating admixture (Reich et al. 2009). Although only three populations (AK, HC, and SJ) showed explicit signals of admixture from different source populations (supplementary table S5, Supplementary Material online), the populations from island and mountainous areas (HK, DQ, BM, ME, and JZ) had the highest f3 statistics comparing to the other populations, consistent with the PCA results. Interestingly, the genetic diversity of the island and mountain populations was lower than the other populations (table 1), suggesting that the high genetic diversity of the other populations may be the result of admixture.
Table 1.
Genetic Diversity.
| Pn | Ho | He | F | |
|---|---|---|---|---|
| ZZ | 0.460 | 0.099 | 0.097 | 0.043 |
| MX | 0.409 | 0.096 | 0.093 | 0.074 |
| HC | 0.469 | 0.099 | 0.097 | 0.044 |
| NY | 0.452 | 0.096 | 0.092 | 0.076 |
| SJ | 0.468 | 0.097 | 0.095 | 0.067 |
| HK | 0.300 | 0.090 | 0.083 | 0.138 |
| QY | 0.432 | 0.104 | 0.098 | 0.000 |
| GY | 0.448 | 0.098 | 0.096 | 0.055 |
| JZ | 0.360 | 0.094 | 0.088 | 0.100 |
| ME | 0.326 | 0.084 | 0.079 | 0.196 |
| SN | 0.470 | 0.098 | 0.095 | 0.056 |
| MY | 0.449 | 0.102 | 0.096 | 0.015 |
| AK | 0.479 | 0.101 | 0.098 | 0.030 |
| XA | 0.467 | 0.102 | 0.099 | 0.018 |
| YL | 0.408 | 0.103 | 0.099 | 0.009 |
| BM | 0.272 | 0.090 | 0.080 | 0.131 |
| DQ | 0.362 | 0.093 | 0.090 | 0.104 |
| XS | 0.438 | 0.095 | 0.092 | 0.089 |
Pn, the proportion of loci in a population that have two or more alleles; Ho, observed heterozygosity; He, expected heterozygosity; F, inbreeding coefficient.
Next, we constructed a neighbor-joining tree using A. mellifera as an outgroup. Based on the results of this analysis, the grouping of populations reflected their geographical locations from the north to the south (fig. 1D).
Genetic Differentiation
We calculated pairwise FST between the populations to quantify their genetic differentiation (table 2). Pairwise FST ranged from 0.008 to 0.228, with an average of 0.072, consistent with an overall structured population and heterogeneity in gene flow. FST between the more isolated populations (HK, BM, ME, DQ, and JZ) ranged from 0.099 to 0.228, with an average of 0.162. Compared with A. mellifera, the level of genetic differentiation in A. cerana is higher than among the subspecies of A. mellifera (average FST = 0.10) (Wallberg et al. 2014), but lower than the pairwise FST among A. mellifera lineages (Harpur et al. 2014; Wallberg et al. 2014; Cridland et al. 2017). This result indicates a subspecies level of divergence in these populations.
Table 2.
Pairwise FST Distances between Apis cerana Populations.
| AK | BM | DQ | GY | HC | HK | JZ | ME | MX | MY | NY | QY | SJ | SN | XA | XS | YL | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AK | – | ||||||||||||||||
| BM | 0.12 | – | |||||||||||||||
| DQ | 0.06 | 0.14 | – | ||||||||||||||
| GY | 0.02 | 0.13 | 0.07 | – | |||||||||||||
| HC | 0.02 | 0.10 | 0.05 | 0.03 | – | ||||||||||||
| HK | 0.11 | 0.19 | 0.14 | 0.12 | 0.09 | – | |||||||||||
| JZ | 0.05 | 0.16 | 0.10 | 0.06 | 0.06 | 0.15 | – | ||||||||||
| ME | 0.12 | 0.21 | 0.15 | 0.13 | 0.11 | 0.21 | 0.16 | – | |||||||||
| MX | 0.03 | 0.14 | 0.08 | 0.03 | 0.04 | 0.13 | 0.07 | 0.14 | – | ||||||||
| MY | 0.02 | 0.12 | 0.07 | 0.03 | 0.02 | 0.11 | 0.06 | 0.12 | 0.04 | – | |||||||
| NY | 0.01 | 0.11 | 0.05 | 0.03 | 0.01 | 0.10 | 0.06 | 0.12 | 0.04 | 0.02 | – | ||||||
| QY | 0.03 | 0.13 | 0.08 | 0.04 | 0.04 | 0.13 | 0.08 | 0.13 | 0.05 | 0.03 | 0.03 | – | |||||
| SJ | 0.01 | 0.11 | 0.06 | 0.02 | 0.01 | 0.10 | 0.06 | 0.12 | 0.03 | 0.02 | 0.01 | 0.03 | – | ||||
| SN | 0.01 | 0.11 | 0.05 | 0.02 | 0.01 | 0.10 | 0.06 | 0.11 | 0.03 | 0.02 | 0.01 | 0.03 | 0.01 | – | |||
| XA | 0.01 | 0.12 | 0.07 | 0.02 | 0.02 | 0.11 | 0.06 | 0.12 | 0.04 | 0.02 | 0.02 | 0.03 | 0.01 | 0.02 | – | ||
| XS | 0.02 | 0.10 | 0.05 | 0.03 | 0.01 | 0.10 | 0.07 | 0.12 | 0.04 | 0.03 | 0.01 | 0.04 | 0.02 | 0.02 | 0.03 | – | |
| YL | 0.04 | 0.15 | 0.10 | 0.04 | 0.05 | 0.14 | 0.09 | 0.15 | 0.06 | 0.05 | 0.05 | 0.05 | 0.04 | 0.05 | 0.04 | 0.06 | – |
| ZZ | 0.02 | 0.11 | 0.06 | 0.03 | 0.01 | 0.10 | 0.06 | 0.12 | 0.04 | 0.02 | 0.01 | 0.04 | 0.01 | 0.01 | 0.02 | 0.02 | 0.05 |
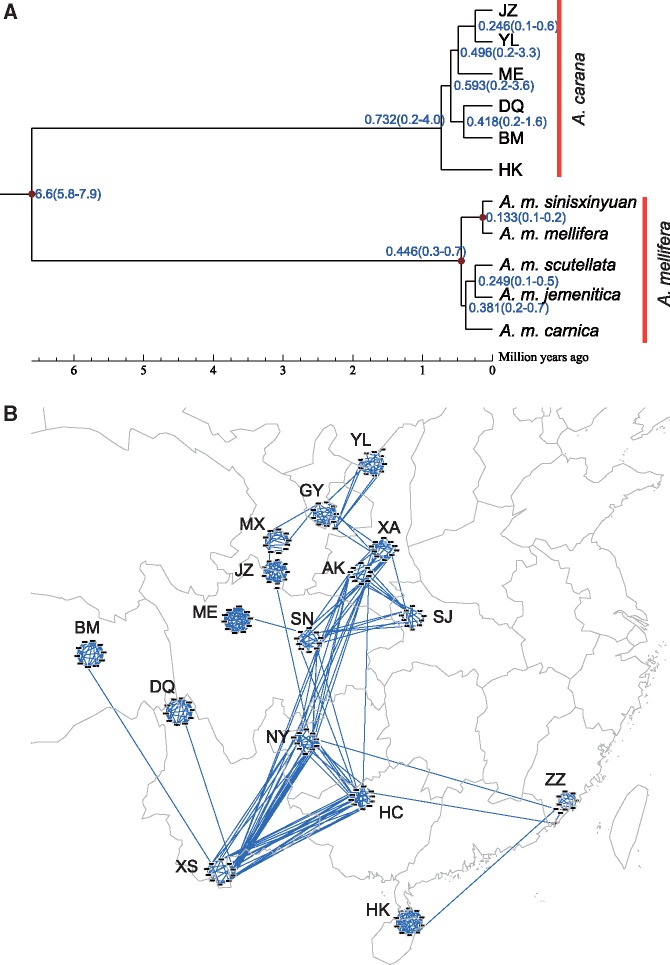
To further test the relationship among the more isolated A. cerana populations, we included representative subspecies from lineages of A. mellifera (Harpur et al. 2014; Chen et al. 2016) and calculated time of divergence using MCMCTree program in the PAML package (Yang 2007). The results suggest an ancient split between the populations (fig. 2A). The divergence time between the branches ranged from 300 to 500 Ka, which is significantly earlier than the time of divergence between A. mellifera subspecies (20–35 Ka) and comparable to divergence time among lineages (150–300 Ka) (Ruttner 1988; Franck et al. 2001; Wallberg et al. 2014). Times of divergence of the A. cerana populations are consistent with a subspecies level of differentiation.
Fig. 2.
Genetic differentiation among populations and individuals. (A) Time of divergence between populations in millions of years. (B) Relationship network among Apis cerana individuals. Individual bees are represented by nodes, while their relationships are represented by edges connecting the nodes. Individuals are placed on the map according to the approximate sampling location for visualization purposes.
To explore the influence of geographic distance on divergence, we performed the Mantel test to test the association between geographic distance matrix and the pairwise FST matrix obtained above, using the R package ade4 (Dray and Dufour 2007). We only identified a weak association between the geographic and genetic distances (P = 0.0837). To reduce the effect of mountains and the Qiong Zhou channel that separate HK from the mainland populations, we removed the island and mountain populations and repeated the Mantel test, but did not observe an association between geographic and genetic distances (P = 0.1521).
In addition to population level differentiation, we also examined the genetic relatedness at the individual level by calculating allele sharing distances among all individuals with PLINK (Purcell et al. 2007), and constructed a network using the NetView pipeline (Neuditschko et al. 2012) (fig. 2B). Individuals are represented by nodes and individual relationships between bees are represented by edges connecting the nodes. The patterns revealed by NetView corroborated the PCA results. Some individuals from distantly located populations are connected (such as individuals in AK and NY), indicating a close relationship among these individuals. In contrast, individuals from geographically close populations are not necessarily connected (MX and JZ individuals, for example), indicating substantial differentiation between these individuals. The NetView results further support the results of the Mantel test showing that the correlation between genetic distances and geographic distances was not significant.
These results of population structure and Mantel tests collectively indicate that strong barriers may have a greater influence on population differentiation in A. cerana than physical distance. For example, the pairwise FST value was high for adjacent populations with barriers such as ME and JZ (277 km, FST = 0.16), and ME and SN (333 km, FST = 0.11), and was low for distant but connected populations, such as between XS in the tropical region (south) and YL in the temperate region (north, 1,875 km, FST = 0.06).
Demographic History
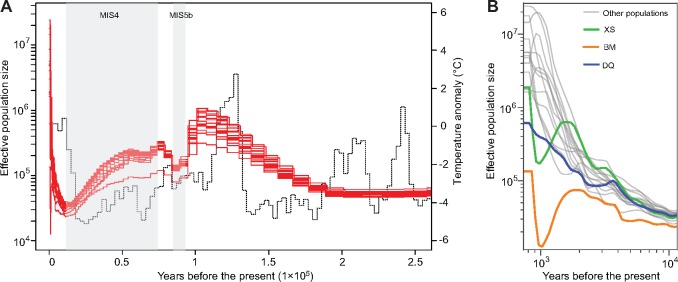
We inferred the demographic history of the A. cerana populations to obtain a better understanding the evolutionary history of this species. We estimated the historical effective population sizes of the populations using SMC++ (Terhorst et al. 2017). Compared with MSMC (Schiffels and Durbin 2014), SMC++ provides more accurate estimates for the recent past, and does not require phasing of the genomic data, thus avoiding the problem of phasing errors for populations that lack a suitable reference panel as is the case in this study (Terhorst et al. 2017). The effective population size history of different A. cerana populations showed a similar pattern (fig. 3A). Surprisingly, changes in effective population size aligned well with changes in the historical global temperature. The effective population size peaked during Marine Isotope Stage 5 (MIS5, ∼80–130 Ka) (Lisiecki and Raymo 2005; Jouzel et al. 2007), the last major interglacial stage in history. During MIS5, the effective population size reached a local minimum during the colder substage MIS5b and recovered during the warmer substage MIS5a. After MIS5, effective population sizes gradually decreased during MIS4, until the beginning of the Holocene (11.7 Ka to the present), when the population started to increase dramatically. The Holocene is an interglacial stage in the current ice age, and the increase in effective population sizes in the warm periods suggested that an elevated global temperature may be advantageous for A. cerana populations. Notably, the effective population sizes of BM and XS experienced temporary decline during ∼1–1.7 Ka, while DQ showed a slower increase compared with the other populations during the same period (fig. 3B). The reason for the decline/slow growth is not clear; however, as BM, DQ, and XS represents the southwest part of China, some local events may have influenced the honey bee population in these areas. After the disturbance, the effective population sizes exhibited a rapid recovery.
Fig. 3.
Effective population size of the Apis cerana populations. (A) Effective population sizes. Each red line represents the effective population size of an A. cerana population. The dotted line indicates the historical global climate. Shaded areas represent Marine Isotope Stage 4 (MIS4, left panel) and MIS5b (right panel). (B) Higher magnification view of the effective population sizes during the Holocene. XS, BM, and DQ populations showed decreased/slower growth during ∼1–1.7 Ka.
We also estimated historical gene flow by calculating relative the cross coalescence rate (CCR) between all pairs of geographically adjacent populations using MSMC (Schiffels and Durbin 2014). The nonisolated populations showed pervasive gene flow in history, while the island and mountain populations showed no or reduced gene flow (supplementary fig. S3, Supplementary Material online). The peaks of historical gene flow appeared between 0 and 5 Ka, corresponding to the period of rapid population growth (fig. 3B).
Candidate Genomic Regions under Natural Selection
A steep spatial gradient of allele frequencies in continuous populations may indicate regions under selection and has been used to identify loci that were not detectable using other methods (Yang et al. 2012). This method is suitable for analyses of populations that cover large geographic areas, as in our samples. We established a subset of six populations (YL, GY, AK, SN, NY, and XS) that were distributed along the latitude and did not have large physical barriers, and obtained SNPs with extreme frequency gradients using SPA software (supplementary fig. S4, Supplementary Material online) (Yang et al. 2012). The enrichment of candidate genes associated with the SNPs in Gene Ontology (GO) categories and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways was tested, and 179 significantly enriched GO terms (supplementary table S6, Supplementary Material online) and 7 significantly enriched KEGG pathways (table 3 and supplementary fig. S5, Supplementary Material online) were identified.
Table 3.
Enriched KEGG Pathways of Genes with Steep Gradients of Allele Frequencies along Latitude.
| Term | # Input | # Background | P Value | Corrected P Valuea |
|---|---|---|---|---|
| Hippo signaling pathway—fly | 19 | 57 | 4.23E-06 | 3.85E-4 |
| Phototransduction—fly | 12 | 28 | 3.64E-05 | 1.66E-3 |
| Neuroactive ligand–receptor interaction | 14 | 44 | 1.18E-4 | 3.59E-3 |
| Phosphatidylinositol signaling system | 13 | 41 | 2.13E-4 | 4.83E-3 |
| ECM-receptor interaction | 6 | 11 | 1.36E-3 | 2.16E-2 |
| Wnt signaling pathway | 18 | 88 | 1.42E-3 | 2.16E-2 |
| Inositol phosphate metabolism | 10 | 36 | 2.59E-3 | 3.37E-2 |
FDR correction method: Benjamini and Hochberg.
The top enriched KEGG pathway is the Hippo signaling pathway, with 19 out of 57 genes showing signals of natural selection. Gradients of allele frequencies of genes in this pathway may reflect adaptation along temperature gradients (supplementary fig. S6, Supplementary Material online). In A. mellifera, the Hippo signaling pathway has been suggested to be important for the adaptation to cold climate (Chen et al. 2016). The involvement of the Hippo signaling pathway in the cold adaptation of both A. mellifera and A. cerana indicates common mechanisms of cold tolerance in honey bees, and suggests convergent evolution between the two species. However, the roles of the candidate genes require further validation. For the enriched GO terms, we further used the REVIGO tool (Supek et al. 2011) to summarize the nonredundant terms (supplementary tables S7–S9, Supplementary Material online). The top enriched category in biological process is “response to stimulus,” as well as other terms such as “response to chemical” and “defense response,” suggesting the importance of the processing of external signals in honey bee’s adaptation to different environments. The enriched KEGG pathways of phototransduction and neuroactive ligand–receptor interaction further indicates that these interactions may be related to the high capacity of the A. cerana perception and cognitive systems. A. cerana has been shown to perform significantly better in learning and memory than A. mellifera in terms of color and grating patterns (Qin et al. 2012). Selected genes in the pathways include Nmdar1, which plays a key role in learning and memory (Xia et al. 2005), inaC, which encodes the eye protein kinase C and is required for inactivation of photoreceptor cells and light adaptation (Hardie et al. 1993), and ninaC, which is required to maintain the stability of inaC (Venkatachalam et al. 2010). Further categorization of the related variants may yield insights into mechanisms underlying honey bee perception and cognition.
We further identified selective sweeps in the genome using 16 populations (excluding MY and QY) by calculating Tajima’s D and FST using VCFtools (Danecek et al. 2011), and the composite likelihood ratio (CLR) using SweepFinder2 (DeGiorgio et al. 2016) along sliding windows; we identified 13.9 M, 40.3 M, and 13.6 M regions under selective sweep, respectively. Genes overlapping the sweep regions were selected, among which 1591 genes were identified by at least two methods and were considered as genes under selective sweep. We performed a gene set enrichment test of KEGG pathways and GO categories, and identified significantly enriched categories (supplementary table S10 and fig. S7, Supplementary Material online). Among the top pathways, a series of signaling pathways were identified, including the Hippo signaling pathway which was also identified in the SPA analysis, the FoxO signaling pathway which is involved in temperature adaptation (Chen et al. 2016), the TGF-beta signaling pathway, the Wnt signaling pathway, the Hedgehog signaling pathway, the Jak-STAT signaling pathway, and the Notch signaling pathway that were shown to be involved in ovary activity (Duncan et al. 2016; Chen et al. 2017). The enrichment of selected genes in different pathways suggests the importance of reaction to external cellular signals in the adaptation of A. cerana.
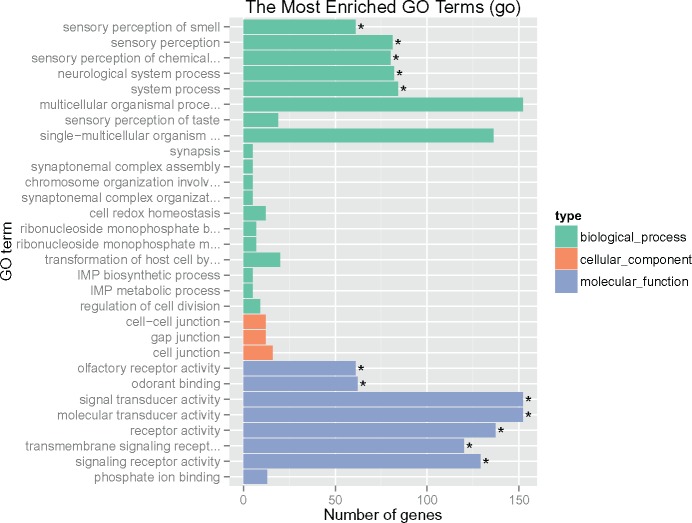
We further identified genes under balancing selection using the top genes with positive Tajima’s D values. Enrichment tests revealed that GO categories related to olfactory and sensory processing were highly enriched (fig. 4), including “olfactory receptor activity,” “odorant binding,” “sensory perception of smell,” and “sensory perception of chemical stimulus.” As balancing selection maintains the diversity of alleles, diversity in olfactory genes may be important for honey bee survival. The olfactory system is involved in communication, recognition of parasites, and foraging, and a high diversity of olfactory genes may be beneficial for a colony to cope with various internal and external chemical signals, but further investigations are required.
Fig. 4.
Enriched GO terms in genes under balancing selection.
Discussion
Variable climates have profound effects on the genetic diversity of a species. Analyses at the genomic level not only provide detailed information about the structure, dynamics, and adaptation to a variety of climates but also facilitate protection and selective breeding. The first study of the population genomics of A. cerana showed that populations in China harbor considerable genetic diversity, with a divergence at subspecies level or higher between HK, BM, DQ, ME, and possibly YL and JZ populations. Landscape factors such as mountains and channels, rather than physical distances, appear to play an important role in population differentiation. Analyses of the population history suggest that the historical global temperature had a substantial influence on the effective population size, with a warmer climate facilitating population growth. Genes related to adaptation to local environments were identified.
Previously, understanding of the differentiation of A. cerana was quite limited. The most recent comprehensive study was based on morphometric characters (Radloff et al. 2010), which identified six main morphoclusters, and showed that most of China belongs to morphocluster I. Other studies have categorized several ecotypes of A. cerana in China but conflicting results were reported (Gong 2000; Yang 2001). Currently, the most generally accepted hypothesis is that nine A. cerana ecotypes are present in China, including Hainan, Yungui Plateau, Tibet, Aba, Changbai mountains, south Yunnan, north China, south China, and central China (The National Animal Genetic Resources Committee 2011). However, these studies mainly relied on the results obtained using no more than 12 morphological characters that were traditionally developed for A. mellifera, and therefore may be missing some crucially different characters when applied to A. cerana (Meixner et al. 2013). Researchers have attempted to use the mitochondrial tRNAleu-COII region, which was also developed for characterization of A. mellifera populations (De La Rúa et al. 2000; Smith et al. 2000; Tan et al. 2006, 2007, 2015; Zhao et al. 2014); however, these studies only provided limited information about A. cerana due to the short fragment of A. cerana tRNAleu-COII. In contrast, our genome resequencing approach provided 2.67 million polymorphic sites distributed throughout the genome, with which fine scale population structures can be obtained. This study may serve as a useful reference in future studies.
Analyses of population structures and connectivity showed that populations analyzed in this study belonged to one of two categories: the more physically isolated populations located in islands or mountainous areas and the connected populations. The more isolated populations, namely, HK, BM, DQ, ME, and JZ, exhibited high differentiation from the other populations and lower genetic diversity. Conversely, the less isolated populations showed less differentiation, higher gene flow and higher genetic diversity. MY and QY showed inconsistent individual genetic compositions, and these differences may be attributed to human activities that introduced nonlocal populations (Zhang 2012), which potentially impaired the local genetic integrity.
The results of the Mantel tests are consistent with the results of small-scale studies using microsatellite markers (Yin et al. 2008; Ji et al. 2009). Our results identified a relationship among physical barriers, genetic differentiation, gene flow, and local genetic diversity. Barriers rather than physical distances appeared to exert a substantial effect on population differentiation, based on both the population structure and the results of Mantel tests. This observation may be the result of the rapid migration of honey bees that promotes gene flow among populations in the absence of physical barriers. The potential of honey bees for rapid migration was best illustrated by the invasive A. cerana in Australia (Gloag et al. 2016) and the Africanized Honey Bees in the American continents, which spread from Brazil to the southwestern United States within a few decades (Winston 1992; Schneider et al. 2004), averaging a speed of >250 km/year. The rapid migration of honey bees in a landscape without strong physical barriers may promote gene flow and reduces genetic differentiation. In addition, population expansion/growth may facilitate the gene flow between populations as illustrated by the high relative CCR during the period of rapid population growth (fig. 3B).
The high genetic diversity observed in the connected populations is another possible result of high gene flow. In A. mellifera, human mediated gene flow has resulted in a higher genetic diversity in managed honey bee colonies (Harpur et al. 2012; Harpur, Minaei, et al. 2013). Genetic variability is important for a population to survive in a changing climate. For honey bees, high genetic diversity at the colony level can increase the fitness of colonies, as colonies that display higher diversity exhibit enhanced homeostasis (Jones et al. 2004; Oldroyd and Fewell 2007), are more productive (Oldroyd et al. 1992; Mattila and Seeley 2007) and are more resistant to diseases (Tarpy 2003). In addition, complementary sex determination of honey bees confers a high genetic load in populations that lack genetic diversity at the sex-determining locus (Harpur, Sobhani, et al. 2013). Consequently, factors that reduce gene flow, such as habitat fragmentation, may result in a reduced local genetic variability and colony fitness. In China, due to human activities such as urbanization, expansion of agricultural lands, and the invasion of A. mellifera, the distribution of the A. cerana population has been reduced by over 75% (Yang 2005). A. cerana populations has not only disappeared in many areas but the genetic variability in the remaining areas may also be at risk due to serious habitat fragmentation. Conservation efforts should consider population connectivity, and corridors of habitats may be important for the effective conservation of genetic diversity. However, high diversity due to human mediated gene flow is not necessarily beneficial because it can result in a loss of genetic integrity and adaptive alleles to the local environment, and thus the usage of endemic populations for apiculture should be promoted (De la Rúa et al. 2013; Harpur, Minaei, et al. 2013). Human mediated gene flow can also facilitate the spread of diseases. An outbreak of Sacbrood virus disease originated from QY in the year after the introduction of A. cerana from southern provinces into QY (Zhang 2012). The introduction of nonlocal populations must be prohibited to protect local A. cerana populations.
According to our data, differentiation among the more isolated populations (ecotypes) is occurred at the subspecies level, based on both the FST and divergence time analyses. FST for the more isolated A. cerana populations averaged 0.162, ranging from 0.099 to 0.228. In comparison, FST between A. mellifera subspecies from the same lineage ranged from 0.05 to 0.15, with an average of 0.10 (Wallberg et al. 2014). FST between different lineages of A. mellifera range from 0.20 to 0.56 as estimated by Wallberg et al. (2014), 0.324 to 0.540 as estimated by Harpur et al. (2014), and 0.134 to 0.423 as estimated by Cridland et al. (2017). Overall, FST for the more isolated populations in our study indicate differentiation at the subspecies level, corresponding to the differentiation level of more distantly related subspecies in A. mellifera. The divergence time also suggest early differentiation among the populations between 300 and 500 Ka (fig. 2A), which is comparable to the time of divergence between different A. mellifera lineages. Therefore, differentiation among the more isolated A. cerana populations in our study occurred at the subspecies level.
In general, a species can survive climate changes in three ways: migration, plasticity, and adaptive evolution (Williams et al. 2008). A. cerana shows the potential to utilize all three strategies. Firstly, as discussed earlier, honey bees showed rapid dispersal across habitats to track a suitable climate space. However, researchers have not clearly determined how habitat fragmentation affects this ability. Based on our results, populations in islands and mountainous areas have low/no gene flow, indicating that migration may not be an option for these populations. Secondly, the eusocial honey bees may have high plasticity because they can maintain inner nest homeostasis to counteract change in the external environments (Seeley 1985; Schmickl and Crailsheim 2004). Thirdly, A. cerana may have a high intrinsic capacity to adapt to future climates through high genetic diversity. Finally, according to the population history, warmer climates may be beneficial for A. cerana, as the effective population size historically increased during warmer periods, and showed a rapid increase in the recent millennia. Historical population sizes also indicates the resilience of A. cerana, as rapid recovery of populations was observed after the disturbance between 1 and 1.7 Ka in southwestern China. In summary, as a species with a wide range in size, A. cerana shows good potential to cope with climate change as a whole, but some populations may face greater challenges than others. Moreover, global warming is only one of the many stresses that honey bees are currently facing, and bees are also vulnerable to stresses such as pesticides, pollutants, pathogens, parasites, and limited floral resources (Goulson et al. 2015; Klein et al. 2017).
We have identified genes that may related to the adaptation to temperature or other environmental factors. These genes can be potential candidates for further exploration of the underlying mechanisms of the important traits and will be useful for conservation of honey bees in the presence of various challenges.
Conclusions
Our study takes advantage of the widely distributed A. cerana species and provides insights into the differentiation, adaptation, and history using population genomic approaches. A. cerana exhibits high genetic diversity, and physical barriers rather than distance are the driving factor of divergence in this highly migratory species. Our analyses highlight the role of historical global climates in the population dynamics of A. cerana, with warm climates favoring population growth. We identified adaptive genes along environmental gradients and other stresses. Our results provide insights into the evolution of A. cerana to diverse environments and advance our understanding of its vulnerability to climate change.
Materials and Methods
Sampling
A. cerana samples were collected in 18 locations in China (fig. 1A and supplementary table S1, Supplementary Material online). Bees from ten colonies were collected from each location, and one worker bee was randomly chosen from each colony for further sequencing.
Sequencing, Read Mapping, and Quality Control
For each sample, we prepared a paired-end library with high quality DNA and sequenced the DNA on the Illumina HiSeq 2500 sequencing platform following the standard procedures. We also download sequences for A. m. sinisxinyuan as an outgroup (Chen et al. 2016) and sequences for A. m. mellifera, A. m. jementica, A. m. carnica, and A. m. scutellata (Harpur et al. 2014) to calculation the divergence time. We used a custom Perl script to filter out low quality reads pairs, including reads with >50% of low quality bases (Q ≤ 5) in either of the paired reads, and reads containing >10% Ns. Clean reads were then mapped to the reference genome of A. cerana (Park et al. 2015) using the BWA-MEM aligner (Li and Durbin 2009) with the option “-t 4 -k 32 –M.” The resulting bam files were sorted using SAMtools (Li et al. 2009) and duplicated reads were removed.
For variant calling, we first used SAMtools to collect summary information from BAM files and compute the likelihood of possible genotypes, and called variants using the call function in Bcftools (Li et al. 2009). The reference genome was subdivided into segments and analyzed in parallel. Raw SNPs were then filtered based on quality scores and SNPs with a quality score <20 removed. Furthermore, a SNP was discarded if the total coverage of the SNP was less than one-third or greater than five times the overall coverage. If two SNPs were <5 bp apart, both SNPs were removed.
Genetic Diversity and Differentiation
The inbreeding coefficient (F), expected heterozygosity (He), and observed heterozygosity (Ho) were calculated using PLINK (Purcell et al. 2007). The minor allele frequency (MAF) and the proportion of polymorphic SNPs (Pn) were calculated using a custom Perl script. Pairwise FST were calculated using SNPRelate package (Zheng et al. 2012). We then performed Mantel tests with FST matrixes and distance matrixes using the ade4 package (Dray and Dufour 2007).
Population Structure
We used ADMIXTURE (Alexander et al. 2009) to investigate the population structure, with coancestry clusters ranging from two to seven. The principal component analysis was performed using GCTA software (Yang et al. 2011). Furthermore, we calculated the genome-wide allele sharing distance using PLINK (Purcell et al. 2007), and constructed a network using the procedure described in the NetView pipeline (Neuditschko et al. 2012). We also calculated f3 statistics for each possible combination of three populations using the threepop program in the TreeMix package (Pickrell and Pritchard 2012). A neighbor-joining phylogenetic tree was constructed using TreeBest software (http://treesoft.sourceforge.net/treebest.shtml), with A. mellifera as an outgroup. We included QY and MY populations in the analyses of population structure; however, due to the presence of questionable samples, we excluded QY and MY from analyses of population history and genes under selective sweep.
Population History
To estimate divergence time between populations, we first obtained single copy genes in A. cerana and A. mellifera genome based on the result of blastp (E-value cutoff of 1 × 10−7) and treefam clustering (Li et al. 2006), and obtained 5, 854 single copy genes. Next, we constructed a supergene for each population using putative sequences of coding regions for single copy genes with SNPs, and calculated the divergence time using the MCMCtree package in PAML 4.5 software (Yang 2007) with the HKY85 model and two calibrations: 1) a divergence time of 6 to 8 Ma between A. mellifera and A. cerana (Arias and Sheppard 1996), and 2) a divergence time of 0.03 to 0.165 Ma between A. m. sinisxinyuan and A. m. mellifera (Chen et al. 2016).
We used SMC++ (Terhorst et al. 2017) to estimate the historical effective population size with a mutation rate set to 5.3 × 10−9 (Wallberg et al. 2014) and a polarization error of 0.5 as the identity of the ancestral allele is not known. To estimate historical gene flow between populations, we first performed haplotype phasing using Shapeit2 package (https://mathgen.stats.ox.ac.uk/genetics_software/shapeit/shapeit.html; last accessed February 20, 2017) and calculated relative cross coalescence rate using MSMC based on eight haplotypes (four haplotypes for each population) (Schiffels and Durbin 2014) for each pair of populations.
Genes under Selective Sweep
We used SPA software (Yang et al. 2012) to identify SNPs with extreme frequency gradients. A subset of data including YL, GY, AK, SN, NY, and XS was obtained from the original VCF file. Loci were first pruned for linkage disequilibrium using PLINK (–indep 50 5 1.1) (Purcell et al. 2007) and then filtered to only retain biallelic loci as required by SPA. The SPA analysis was performed under an unsupervised scenario.
F ST and Tajima’s D statistics for sliding windows were calculated using VCFtools (Danecek et al. 2011), with a window size of 10, 000. The top 10% of windows from the distribution of FST and the negative end of Tajima’s D values were selected as regions under selective sweep. CLRs were calculated using SweepFinder2 (DeGiorgio et al. 2016) with a window size of 200 bp, and the top 10% of windows were selected as sweep region. Genes overlapping the sweep regions were identified. Additionally, the top 10% of windows from the positive end of Tajima’s D values were identified, and genes overlapping the windows were categorized as genes under balancing selection. To perform enrichment analysis, we first assigned genes to GO terms and KEGG pathways based on their orthologues in the fruit fly (Drosophila melanogaster). The enrichment of selected genes in KEGG pathways was performed using the KOBAS system (Mao et al. 2005). Multiple comparisons were corrected using the FDR method. We used the REVIGO tool (Supek et al. 2011) to summarize nonredundant GO terms, with an allowed similarity of 0.4 based on the D. melanogaster database.
Supplementary Material
Acknowledgments
This work was supported by The Agricultural Science and Technology Innovation Program (CAAS-ASTIP-2018-IAR); the earmarked fund for Modern Agro-industry Technology Research System (CARDS-45-KXJ1); and the Fundamental Research Funds for Central Non-profit Scientific Institution. We thank Baohua Xu, Xinfang Song, Shaoyu He, Wenzheng Zhao, Baolong Wang, Zhaluo, Baoguo Gao, Biao Wang, Lianguo Ye, and Chunying Yuan for their help in collecting the samples. All sequencing data generated during the current study are deposited in the Sequence Read Archive (PRJNA418874).
References
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 199:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias MC, Sheppard WS.. 1996. Molecular phylogenetics of honey bee subspecies (Apis mellifera L.) inferred from mitochondrial DNA sequence. Mol Phylogenet Evol. 53:557–566. [DOI] [PubMed] [Google Scholar]
- Balanyà J, Huey RB, Gilchrist GW, Serra L.. 2009. The chromosomal polymorphism of Drosophila subobscura: a microevolutionary weapon to monitor global change. Heredity 1035:364–367. [DOI] [PubMed] [Google Scholar]
- Cao L, Gu P, Lin Y.. 2017. Genetic diversity of Apis cerana cerana based on mitochondrial DNA in Lishui, Zhejiang, China. J Zhejiang Univ Agric Life Sci. 43:425–430. [Google Scholar]
- Chen C, Liu Z, Pan Q, Chen X, Wang H, Guo H, Liu S, Lu H, Tian S, Li R, et al. 2016. Genomic analyses reveal demographic history and temperate adaptation of the newly discovered honey bee subspecies Apis mellifera sinisxinyuan n. ssp. Mol Biol Evol. 335:1337–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ma C, Chen C, Lu Q, Shi W, Liu Z, Wang H, Guo H.. 2017. Integration of lncRNA–miRNA–mRNA reveals novel insights into oviposition regulation in honey bees. PeerJ 5:e3881.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cridland JM, Tsutsui ND, Ramírez SR.. 2017. The complex demographic history and evolutionary origin of the western honey bee, Apis mellifera. Genome Biol Evol. 92:457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 2715:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Rúa P, Jaffé R, Muñoz I, Serrano J, Moritz RFA, Kraus FB.. 2013. Conserving genetic diversity in the honeybee: comments on Harpur et al. (2012). Mol Ecol. 2212:3208–3210. [DOI] [PubMed] [Google Scholar]
- De La Rúa P, Simon UE, Tilde AC, Moritz RFA, Fuchs S.. 2000. MtDNA variation in Apis cerana populations from the Philippines. Heredity 841:124–130. [DOI] [PubMed] [Google Scholar]
- DeGiorgio M, Huber CD, Hubisz MJ, Hellmann I, Nielsen R.. 2016. SweepFinder 2: increased sensitivity, robustness and flexibility. Bioinformatics 3212:1895–1897. [DOI] [PubMed] [Google Scholar]
- Dray S, Dufour A-B.. 2007. The ade4 Package: implementing the duality diagram for ecologists. J Stat Softw. 224:1–20. [Google Scholar]
- Duncan EJ, Hyink O, Dearden PK.. 2016. Notch signalling mediates reproductive constraint in the adult worker honeybee. Nat Commun. 7:12427.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franck P, Garnery L, Loiseau A, Oldroyd BP, Hepburn HR, Solignac M, Cornuet J-M.. 2001. Genetic diversity of the honeybee in Africa: microsatellite and mitochondrial data. Heredity 86(Pt 4):420–430. [DOI] [PubMed] [Google Scholar]
- Gloag R, Ding G, Christie JR, Buchmann G, Beekman M, Oldroyd BP.. 2016. An invasive social insect overcomes genetic load at the sex locus. Nat Ecol Evol. 11:11.. [DOI] [PubMed] [Google Scholar]
- Gong Y. 2000. Taxonomy and evolution of honeybees (in Chinese). Fuzhou: Fujian Science and Technology Press. [Google Scholar]
- Goulson D, Nicholls E, Botias C, Rotheray EL.. 2015. Bee declines driven by combined stress from parasites, pesticides, and lack of flowers. Science 3476229:1255957.. [DOI] [PubMed] [Google Scholar]
- Hardie RC, Peretz A, Suss-Toby E, Rom-Glas A, Bishop SA, Selinger Z, Minke B.. 1993. Protein kinase C is required for light adaptation in Drosophila photoreceptors. Nature 3636430:634–637. [DOI] [PubMed] [Google Scholar]
- Harpur BA, Kent CF, Molodtsova D, Lebon JMD, Alqarni AS, Owayss AA, Zayed A.. 2014. Population genomics of the honey bee reveals strong signatures of positive selection on worker traits. Proc Natl Acad Sci U S A. 1117:2614–2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harpur BA, Minaei S, Kent CF, Zayed A.. 2012. Management increases genetic diversity of honey bees via admixture: genetic diversity in the honeybee. Mol Ecol. 2118:4414–4421. [DOI] [PubMed] [Google Scholar]
- Harpur BA, Minaei S, Kent CF, Zayed A.. 2013. Admixture increases diversity in managed honey bees: reply to De la Rúa et al. (2013). Mol Ecol. 2212:3211–3215. [DOI] [PubMed] [Google Scholar]
- Harpur BA, Sobhani M, Zayed A.. 2013. A review of the consequences of complementary sex determination and diploid male production on mating failures in the Hymenoptera. Entomol Exp Appl. 1461:156–164. [Google Scholar]
- Hepburn HR, Radloff SE.. 2011. Honeybees of Asia. Dordrecht, London, New York: Springer-Verlag Berlin Heidelberg. [Google Scholar]
- Ji T, Yin L, Liu M, Chen G.. 2009. Genetic diversity and genetic differentiation of six geographic populations of Apis cerana cerana (Hymenoptera: Apidae) in East China. Acta Entomol Sin. 52:413–419. [Google Scholar]
- Jones JC, Myerscough MR, Graham S, Oldroyd BP.. 2004. Honey bee nest thermoregulation: diversity promotes stability. Science 3055682:402–404. [DOI] [PubMed] [Google Scholar]
- Jouzel J, Masson-Delmotte V, Cattani O, Dreyfus G, Falourd S, Hoffmann G, Minster B, Nouet J, Barnola JM, Chappellaz J, et al. 2007. Orbital and millennial Antarctic climate variability over the past 800, 000 years. Science 3175839:793–796. [DOI] [PubMed] [Google Scholar]
- Klein S, Cabirol A, Devaud J-M, Barron AB, Lihoreau M.. 2017. Why bees are so vulnerable to environmental stressors. Trends Ecol Evol. 324:268–278. [DOI] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Hériché J-K, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, et al. 2006. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 3490001:D572–D580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2514:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R.. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 2516:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisiecki LE, Raymo ME.. 2005. A Pliocene‐Pleistocene stack of 57 globally distributed benthic δ18O records. Paleoceanography 201:PA1003. [Google Scholar]
- Liu F, Shi T, Huang S, Yu L, Bi S.. 2016. Genetic structure of Mount Huang honey bee (Apis cerana) populations: evidence from microsatellite polymorphism. Hereditas 153:8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozier JD, Zayed A.. 2017. Bee conservation in the age of genomics. Conserv Genet. 183:713–729. [Google Scholar]
- Mao X, Cai T, Olyarchuk JG, Wei L.. 2005. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2119:3787–3793. [DOI] [PubMed] [Google Scholar]
- Mattila HR, Seeley TD.. 2007. Genetic diversity in honey bee colonies enhances productivity and fitness. Science 3175836:362–364. [DOI] [PubMed] [Google Scholar]
- Meixner MD, Pinto MA, Bouga M, Kryger P, Ivanova E, Fuchs S.. 2013. Standard methods for characterising subspecies and ecotypes of Apis mellifera. J Apic Res. 524:1–28. [Google Scholar]
- Neuditschko M, Khatkar MS, Raadsma HW.. 2012. NetView: a high-definition network-visualization approach to detect fine-scale population structures from genome-wide patterns of variation. PLoS One 710:e48375.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldroyd BP, Fewell JH.. 2007. Genetic diversity promotes homeostasis in insect colonies. Trends Ecol Evol. 228:408–413. [DOI] [PubMed] [Google Scholar]
- Oldroyd BP, Rinderer TE, Harbo JR, Buco SM.. 1992. Effects of intracolonial genetic diversity on honey bee (Hymenoptera: Apidae) colony performance. Ann Entomol Soc Am. 853:335–343. [Google Scholar]
- Park D, Jung JW, Choi B-S, Jayakodi M, Lee J, Lim J, Yu Y, Choi Y-S, Lee M-L, Park Y, et al. 2015. Uncovering the novel characteristics of Asian honey bee, Apis cerana, by whole genome sequencing. BMC Genomics 16:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK.. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 811:e1002967.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 813:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, He X, Tian L-Q, Zhang SW, Zeng Z-J.. 2012. Comparison of learning and memory of Apis cerana and Apis mellifera. J Comp Physiol A 19810:777–786. [DOI] [PubMed] [Google Scholar]
- Radloff SE, Hepburn C, Hepburn HR, Fuchs S, Hadisoesilo S, Tan K, Engel MS, Kuznetsov V.. 2010. Population structure and classification of Apis cerana. Apidologie 416:589–601. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L.. 2009. Reconstructing Indian population history. Nature 4617263:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttner F. 1988. Biogeography and taxonomy of honeybees. Berlin: Springer-Verlag. [Google Scholar]
- Schiffels S, Durbin R.. 2014. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 46:919–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmickl T, Crailsheim K.. 2004. Inner nest homeostasis in a changing environment with special emphasis on honey bee brood nursing and pollen supply. Apidologie 35:249–263. [Google Scholar]
- Schneider SS, DeGrandi-Hoffman GR, Smith D.. 2004. The African honey bee: factors contributing to a successful biological invasion. Annu Rev Entomol. 49:351–376. [DOI] [PubMed] [Google Scholar]
- Seeley TD. 1985. Honeybee ecology: a study of adaptation in social life. Princeton ( NJ: ): Princeton University Press [Google Scholar]
- Skelly DK, Joseph LN, Possingham HP, Freidenburg LK, Farrugia TJ, Kinnison MT, Hendry AP.. 2007. Evolutionary responses to climate change. Conserv Biol. 21:1353–1355. [DOI] [PubMed] [Google Scholar]
- Smith DR, Villafuerte L, Otis G, Palmer MR.. 2000. Biogeography of Apis cerana F. and A. nigrocincta Smith: insights from mtDNA studies. Apidologie 31:265–279. [Google Scholar]
- Supek F, Bošnjak M, Škunca N, Šmuc T.. 2011. REVIGO Summarizes and visualizes long lists of gene ontology terms. PLoS One 6:e21800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, Meixner MD, Fuchs S, Zhang X, He S, Kandemir I, Sheppard WS, Koeniger N.. 2006. Geographic distribution of the eastern honeybee, Apis cerana (Hymenoptera: Apidae), across ecological zones in China: morphological and molecular analyses. Syst Biodivers. 4:473–482. [Google Scholar]
- Tan K, Qu Y, Wang Z, Liu Z, Engel MS.. 2015. Haplotype diversity and genetic similarity among populations of the Eastern honey bee from Himalaya-Southwest China and Nepal (Hymenoptera: Apidae). Apidologie 47:197–205. [Google Scholar]
- Tan K, Warrit N, Smith DR.. 2007. Mitochondrial DNA diversity of Chinese Apis cerana. Apidologie 38:238–246. [Google Scholar]
- Tarpy DR. 2003. Genetic diversity within honeybee colonies prevents severe infections and promotes colony growth. Proc R Soc Lond B Biol Sci. 270:99–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teichroew JL, Xu J, Ahrends A, Huang ZY, Tan K, Xie Z.. 2017. Is China’s unparalleled and understudied bee diversity at risk? Biol Conserv. 201:19–28. [Google Scholar]
- Terhorst J, Kamm JA, Song YS.. 2017. Robust and scalable inference of population history from hundreds of unphased whole genomes. Nat Genet. 49:303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The National Animal Genetic Resources Committee. 2011. Animal genetic resources in China bees. Beijing: China Agriculture Press. [Google Scholar]
- Theisen-Jones H, Bienefeld K.. 2016. The Asian honey bee (Apis cerana) is significantly in decline. Bee World 93:90–97. [Google Scholar]
- Thomann M, Imbert E, Devaux C, Cheptou P-O.. 2013. Flowering plants under global pollinator decline. Trends Plant Sci. 18:353–359. [DOI] [PubMed] [Google Scholar]
- Venkatachalam K, Wasserman D, Wang X, Li R, Mills E, Elsaesser R, Li H-S, Montell C.. 2010. Dependence on a retinophilin/myosin complex for stability of PKC and INAD and termination of phototransduction. J Neurosci. 30:11337–11345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallberg A, Han F, Wellhagen G, Dahle B, Kawata M, Haddad N, Simões ZLP, Allsopp MH, Kandemir I, De la Rúa P, et al. 2014. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat Genet. 46:1081–1088. [DOI] [PubMed] [Google Scholar]
- Williams SE, Shoo LP, Isaac JL, Hoffmann AA, Langham G.. 2008. Towards an integrated framework for assessing the vulnerability of species to climate change. PLoS Biol. 6:e325.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winston ML. 1992. The Biology and management of Africanized Honey Bees. Annu Rev Entomol. 37:173–193. [Google Scholar]
- Xia S, Miyashita T, Fu T-F, Lin W-Y, Wu C-L, Pyzocha L, Lin I-R, Saitoe M, Tully T, Chiang A-S.. 2005. NMDA Receptors mediate olfactory learning and memory in Drosophila. Curr Biol. 15:603–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Zhu X, Zhou S, Wu X, Zhou B.. 2013. Genetic differentiation between Apis cerana cerana populations from Damen Island and adjacent mainland in China. Acta Ecol Sin. 33:122–126. [Google Scholar]
- Yang G. 2001. Chinese honeybee (in Chinese). Beijing: China Agricultural Science and Technology Press. [Google Scholar]
- Yang G. 2005. Harm of introducing the western honeybee Apis mellifera L. to the Chinese honeybee Apis cerana F. and its ecological impact (In Chinese). Acta Entomol Sin. 48:401–406. [Google Scholar]
- Yang J, Lee SH, Goddard ME, Visscher PM.. 2011. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 88:76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W-Y, Novembre J, Eskin E, Halperin E.. 2012. A model-based approach for analysis of spatial structure in genetic data. Nat Genet. 44:725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Yin L, Ji T.. 2013. Genetic diversity of the honeybee Apis cerana in Yunnan, China, based on mitochondrial DNA. Genet Mol Res. 12:2002–2009. [DOI] [PubMed] [Google Scholar]
- Yin L, Ji T, Liu M, Bao W, Chen G.. 2008. Genetic diversity and relationship between genetic distance and geographical distance of 6 Apis cerana cerana populations in China. Res J Anim Sci. 2:183–187. [Google Scholar]
- Zhang D. 2012. Survey and analyses of Sacbrood virus disease of the Chinese honey bee in Liaoning Province (In Chinese). Apic China 63:19–20. [Google Scholar]
- Zhao W, Tan K, Zhou D, Wang M, Cheng C, Yu Z, Miao Y, He S.. 2014. Phylogeographic analysis of Apis cerana populations on Hainan Island and southern mainland China, based on mitochondrial DNA sequences. Apidologie 45:21–33. [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS.. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.