Abstract
Paracoccidioides spp. is a thermally dimorphic fungus endemic to Latin America and the etiological agent of paracoccidioidomycosis (PCM), a granulomatous disease acquired through fungal propagule inhalation by its mammalian host. The infection is established after successful mycelia to yeast transition in the host pulmonary alveoli. The challenging environment inside the host exposes the fungus to the need of adaptation in order to circumvent nutritional, thermal, oxidative, immunological and other stresses that can directly affect their survival. Considering that autophagy is a response to abrupt environmental changes and is induced by stress conditions, this study hypothesizes that this process might be crucially involved in the adaptation of Paracoccidioides spp. to the host and, therefore, it is essential for the proper establishment of the disease. By labelling autophagous vesicles with monodansylcadaverine, autophagy was observed as an early event in cells during the normal mycelium to yeast transition, as well as in yeast cells of P. brasiliensis under glucose deprivation, and under either rapamycin or 3-methyladenine (3-MA). Findings in this study demonstrated that autophagy is triggered in P. brasiliensis during the thermal-induced mycelium to yeast transition and by glucose-limited conditions in yeasts, both of which modulated by rapamycin or 3-MA. Certainly, further genetic and in vivo analyses are needed in order to finally address the contribution of autophagy for adaptation. Yet, our data propose that autophagy possibly plays an important role in Paracoccidioides brasiliensis virulence and pathogenicity.
Introduction
Paracoccidioidomycosis (PCM) is a systemic granulomatous disease, geographically confined to Latin America, caused by two species of the genus Paracoccidioides, P. brasiliensis and P. lutzii [1,2]. Subsequently the inhalation of fungus spores by a suitable host, the mycelia (infective form) undergo a thermal-induced differentiation into the yeast parasitic form in the host’s lungs [3]. Once inside the host, the fungus experiences several stresses, such as high temperature, host immune system response and nutrient deprivation [4,5], particularly glucose and amino acids, as suggested by Parente-Rocha et al [6] and Tavares et al [7] in their proteomic and transcriptomic analyses of P. brasiliensis during macrophage interaction in vitro.
Macroautophagy, the prevailing form of autophagy, is one well-known mechanism involved in the response to nutritional deprivation [8]. It aims at counterbalancing the nutrient-scarce environment by reducing the energetic demand of biochemical processes in the cell, as well as providing nutrients to the cell after digesting its own contents inside autophagosomal vesicles [9,10]. Under normal conditions, catabolism and anabolism are two antagonistic mechanisms involved in the maintenance of cells in eukaryotes, both of which controlled by the TOR signalling pathway [11,12]. Macroautophagy (herein named autophagy) represents the cell catabolism of the biological macromolecule triggered when TOR is not active, whereas the activation of the TOR signalling pathway stimulates processes, such as protein synthesis and cell survival, and drives the increase of cell size and mass required to cell growth in an environment that provides enough nutrients to support cells to complete mitosis [11,13].
TOR, a serine/threonine-directed protein kinase found in all eukaryotes, is currently called mTOR, or mechanistic TOR. It constitutes the catalytic subunit of two distinct protein complexes known as mTOR Complex 1 (mTORC1) and mTOR Complex 2 (mTORC2), both of which hold some overlapping functions [14]. They function preponderantly as a sensor indicating the suitability of the environment’s nutritional conditions necessary for the accomplishment of cellular division in response to endocrine stimuli [15]. Specifically, mTORCs control the balance between anabolism and catabolism by stimulating the biosynthesis of macromolecules necessary for cell growth and proliferation, maintaining inactive catabolic pathways such as those leading to autophagy and protein expression of the ubiquitin-proteasome system, thus preventing the activation of pathways which control different types of programmed cell death [16].
Despite the most well-known TOR inhibitor, rapamycin, which also originated the name of this protein kinase (Target-Of-Rapamycin), directly inhibiting mTORC1, it does not suppress mTORC2. While the chronic use of rapamycin may also lead to the inhibition of mTORC2, this effect seems to be more related to the impossibility of assembling new mTORC2 complexes than to the direct inhibition of the formed ones [17–19]. The difference in sensitivity to rapamycin allows differentiating the action of each complex in more defined modules of cellular responses to growth factors. Whereas mTORC2 is primarily involved in the control of cell survival and proliferation, mTORC1 regulates cell metabolism and growth while suppressing autophagy [16]. Thus, it can be said that the effect of rapamycin on eukaryotic cells is mostly associated with mTORC1 and this effect is often related to the stimulation of catabolism and the appearance of autophagic vacuoles by autophagy derepression.
Nevertheless, autophagy is not activated exclusively by nutrient deprivation or in the absence of survival factors. Eukaryotic cells can induce autophagy in several other adverse conditions, such as temperature variation, hypoxia, accumulation of damaged organelles, aggregation of proteins and oxidative stress [20,21]. Essentially, autophagy is a mechanism in which cells adapt to a noxious environment in order to fit a non-efficient cell metabolism to the surrounding demand, particularly when it had changed abruptly. In any case, autophagy allows a rapid change of cell components while it generates nutrients by recycling organelles and other cellular contents to regulate cell adaptation, and then survival, growth, differentiation, and even death [22]. In microorganisms, autophagy clearly has a role in adapting to the environment. While inhibiting autophagy might be deleterious to cells when associated with environmental stresses, stimulating it may accelerate rapidly-dependent cellular processes (turnover) of macromolecules and organelles.
In filamentous fungi, many studies have demonstrated the importance of autophagy for differentiation, adaptation, development and reproduction [23–28]. Studies have shown that autophagy is implicated in the regulation of mitochondrial functions via aerobic respiration of Aspergillus nidulans under carbon deprivation [29], in growth halting, hyphal development and vacuolization of Aspergillus oryzae [30], and in the growth of aerial hyphae, conidia and adequate asexual differentiation of Magnaporthe oryzae [31]. In P. brasiliensis, apoptosis and autophagy-like mechanisms have recently been implicated in a cyclopalladated 7a-mediated cell death [32]. Gontijo et al [33] showed that 21 of 34 autophagy-related genes described in Saccharomyces cerevisiae are present in a basidiomycete, Cryptococcus neoformans, and in Candida albicans and Aspergillus fumigatus, both ascomycetes. Therefore, there is an increasing evidence that autophagy is indeed ubiquitous in the Fungi Kingdom. Considering all the examples found, autophagy allows the cells to adapt to environmental constraints. The recycling of cellular components by autophagy may also occur as part of a fine-tuning to reach the cell physiological homeostasis that is far from being essentially severe or often fully evident.
Several compounds can inhibit autophagy. For instance, 3-methyladenine (3-MA), wortmannin, and LY294002 have been described as compounds capable of suppressing autophagy in its early stages through the inhibition of class III phosphatidylinositol 3-kinases (PI3K) [34–37]. Moreover, chloroquine (CQ) and bafilomycin A1 have also been described as autophagy inhibitor compounds, both of which act on the suppression of lysosomal function, thus, blocking later stages of autophagy [38,39]. Recently, a new compound, MHY1485, has been depicted as an inhibitor of autophagy in mammalian cells by promoting TOR activation and preventing fusion of autophagosomes and lysosomes, though its use has not been characterized in plants and fungi [40].
Studies about the mechanisms responsible for triggering autophagy in Paracoccidioides spp, as well as the occurrence of the autophagic process itself, have only been initiated very recently with a single demonstration made by Arruda et al [32] that mechanisms resembling autophagy-like cell death may be behind the cyclopalladated 7a compound killing of P. brasiliensis. This work aims at demonstrating that autophagy is an active process triggered in P. brasiliensis in response to abrupt environmental changes, such as a restrictive temperature or a sudden decrease of glucose availability. Specifically, we present evidence that autophagy may be involved in adaptation processes required by P. brasiliensis to overcome the thermal-induced dimorphism as well as to circumvent nutrient restrictions. Advances in the knowledge of mechanisms controlling autophagy in P. brasiliensis may reveal new promising candidate molecules important for pathogenicity and virulence in this and other fungi.
Materials and methods
Microorganism and growth condition
P. brasiliensis yeasts, isolate 18 (Pb18), were grown in synthetic dextrose medium, SD (0.17% yeast nitrogen base w/o amino acids and ammonium sulphate (Difco), 2% glucose (Difco), 0.5% casamino acids (Difco), 0.5% ammonium sulphate (Synth), pH 4.5). The temperature was kept at 25°C or 36°C, under constant agitation, to grow mycelia or yeast, respectively.
After 5–7 days of growing at a constant temperature of 36°C, yeasts were collected and washed with PBS. The viable cell concentration was adjusted to 1x107 yeasts ml-1, by using a Neubauer chamber and methylene blue was used as a dye. For Pb18 yeasts experiments, 1x106 yeasts.ml-1 were inoculated in different culture media: SD (synthetic medium) as control, SD-G (SD without glucose), SD 0.2% (SD containing 0.2% glucose), SD+R (SD containing 0.2 μg.ml-1 rapamycin), SD+3 (SD containing 10 mM 3-methyladenine), and incubated at 36°C, under constant agitation for 6 days.
For mycelia to yeast transition experiments, mycelia, grown for 10 to 15 days at 25°C, were inoculated in SD medium and SD+R, and later incubated at 36°C for up to 24 hours.
Quantification of the enlarged structures in the hyphal tips of P. brasiliensis during mycelia to yeast transition
Mycelia to yeast transition cells of Pb18 treated in the media SD+R and SD were collected, fixed in paraformaldehyde (4% in PBS, pH 7.2), and photo-documented using a Leica DMLB microscope, coupled to a Leica DFC310 FX camera. The values were described as percentages of enlarged tips over total tips counts. For each experimental condition, at least 200 hyphal tips were counted and distributed between 7 to 10 images taken from different visual fields. According to Campos et al [41], each visible hyphal tip was observed and classified in: hyphae showing enlarged tips with buds (HEB); hyphae with enlarged tips (HE); and hyphae without enlarged tips (H).
Staining of autophagic vesicles with monodansylcadaverine
Monodansylcadaverine (MDC) was used for staining autophagic vesicles [36]. Mycelia or yeasts of Pb18 growing under different conditions were harvested by centrifugation, washed three times with PBS (pH 7.2), and incubated for 15 minutes with 50 μM MDC in the dark at 37°C. After three washes with PBS, the cells were then visualized and photo-documented under fluorescence by using a Leica DMLB microscope, coupled to a Leica DFC310 FX camera. MDC labelling produces green fluorescence (excitation/emission at 488/505 nm) of autophagic vesicles in the cells.
In silico analysis
34 autophagy-related genes (ATG) and the 33 non-ATG genes related to autophagy were obtained from the Saccharomyces Genome Database (SGD) [42] in order to identify which genes related to autophagy described in Saccharomyces cerevisiae would be present in the genomes of P. brasiliensis and P. lutzii.
The obtained sequences were used as a query in ‘tblastn’ search on the online BLAST web interface provided by NCBI [43,44]. All the searches were done using Paracoccidioides (taxid:38946), Aspergillus (taxid:5052), Cryptococcus (taxid:5206) and Candida (taxid:1535326) database. All Paracoccidioides spp. gene sequences were validated by performing a search for each one as a query in the SGD.
Dry weight assay
Pb18 yeasts grown at 36°C in SD-G, SD0.2%, SD, SD+R were recovered after 3 and 6 days had their growth estimated considering their dry weight. Triplicates of 1.5 ml of each culture were collected, and the cells were subsequently washed in 100% ethanol, centrifuged and dried at 60°C. The dry pellet weight was calculated by the subtraction of the weight of the centrifuge vials with the dry cells by the empty ones. The results were plotted in a graph.
Cell viability assay
For six consecutive days, 1.0 ml of each Pb18 yeast culture (SD-G, SD0.2%, SD, SD+R and SD+3) was submitted to serial dilutions (1/2, 1/4 and 1/8). Following, 10 μl of each dilution, as well as the starting culture, were inoculated into Petri dishes containing solid YPD medium (1% Dextrose, 0.5% Yeast Extract, 0.5% peptone and 1.8% bacteriological agar). After 3 and 6 days of incubation at 36°C, the yeast growth was photo documented using a digital camera.
Statistical analysis
Statistical analyses were performed using ANOVA test. The results p<0.05 or p<0.01 were considered statistically significant. Data are either the means or representative results of at least three similar repetitions, each one performed in triplicate.
Results
In the two species of Paracoccidiodes, we used BLAST to identify 21 of the 34 autophagy-related genes that were initially described in S. cerevisiae. This matches the number identified by Gontijo et al [33] for Candida albicans and Aspergillus fumigatus (Table 1).
Table 1. Genes involved in autophagy described in the Saccharomyces Genome Database and their respective homolog one in Paracoccidioides species.
* SGD Systematic Name
Considering the 34 ATG-genes described in S. cerevisiae, there were 9 missing genes among the species of Paracoccidioides, C. albicans and A. fumigatus (ATG10, ATG14, ATG23, ATG31, ATG32, ATG34, ATG36, ATG39, ATG40 and ATG41); 2 shared genes between the species of Paracoccidioides and C. albicans (ATG21, ATG24); 3 shared genes between the species of Paracoccidioides and A. fumigatus (ATG11, ATG17 and ATG22); 1 shared gene between the species of C. albicans and A. fumigatus (ATG27); 1 gene present only in C. albicans (ATG33); 3 genes present only in A. fumigatus (ATG19, ATG29 and ATG38). The other remaining genes were present in the genome of these three microorganisms (ATG1, ATG3, ATG3, ATG4, ATG5, ATG6, ATG7, ATG8, ATG9, ATG12, ATG13, ATG15, ATG16, and ATG18).
In addition to the so-called ATG genes, there are other 33 genes (non-ATG genes) known to be involved in the autophagy process in S. cerevisiae; at least 20 of which were present in the species of Paracoccidioides (BET3 / BET5, CUE5, GTR2, GYP6, IRS4 / TAX4, MON1, PTC6, SEC16, SEC17, SEC2, SEC4, SNX3, TRS130, TRS20, TRS23, TRS31, TRS33, UTH1, YPT1, YPT31 / YPT32).
To investigate whether the autophagic process would act on the adaptation of the fungus in regard to the restrictive temperature of 36°C during the dimorphic transition process, we initially sought to observe the emergence of monodansylcadaverine-labelled (MDC-labelled) vesicles typically found in autophagy during the early stages of mycelium to yeast (M-Y) transition.
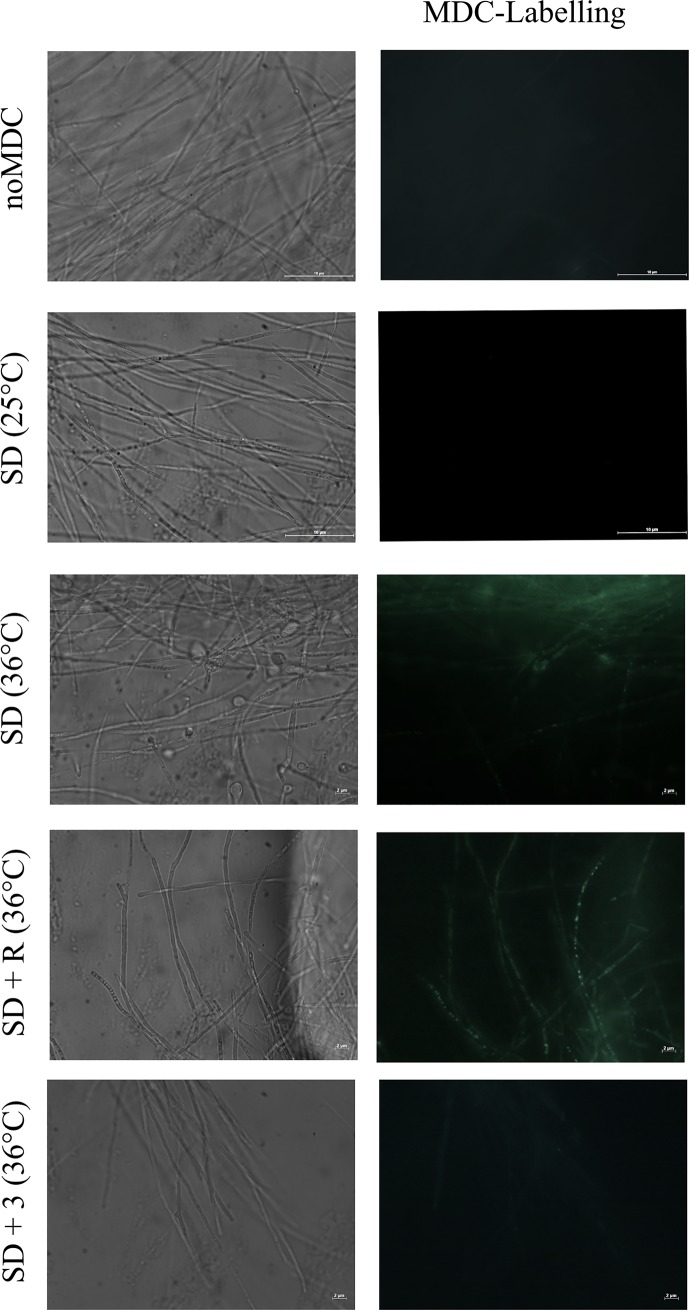
It was observed that Pb18 mycelia grown under standard conditions (Synthetic Dextrose medium—SD) rarely showed MDC-labelled autophagic vesicles (Fig 1, SD-25°C). Nonetheless, an increase in the number of MDC-labelled hyphal structures was observed at 24 hours subsequent to the onset of M-Y differentiation induced by increasing the temperature from 25°C to 36°C (Fig 1, SD-25°C vs. SD-36°C). Although the increase in temperature clearly induced the emergence of autophagic-like vesicles during the regular thermal-induced dimorphism, it was still a mild effect since few cells showed MDC-labelled vesicles.
Fig 1. MDC-labelled vesicles after 24 hours of M-Y transition can be modulated by rapamycin or 3-MA.
Pb18 mycelia were incubated in Synthetic Dextrose medium as control (SD) at 25°C or 36°C or in SD medium containing 0.2 μg.ml-1 rapamycin (SD+R) or 10 mM 3-MA (SD+3) at 36°C. After 24 hours, fungi were incubated with MDC at 50 μM for 15 minutes, washed three times in PBS pH 7.2 and then immediately analyzed under the fluorescence microscope as aforementioned in Materials and Methods. Autophagic vesicles are stained in green. As control was used Pb18 mycelia after 24 hours of mycelium to yeast transition without MDC labelling. Scale bar at 2 μm or 10 μm.
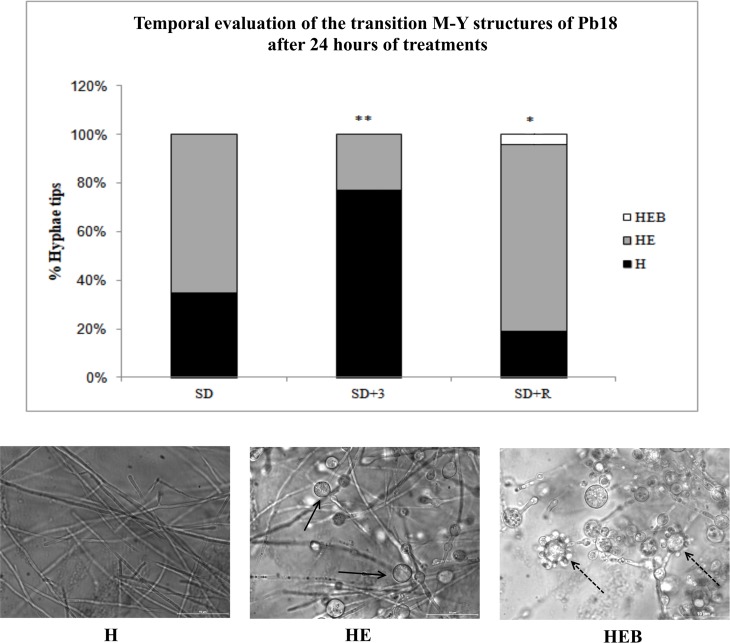
Despite its apparent low frequency, autophagy was required for dimorphism when the 3-methyladenine (3-MA) autophagy inhibitor abolished the emergence of MDC-labelled vesicles (Fig 1, SD + 3) and decreased the incidence of hyphae with enlarged tips (HE) that are typically found during the very early phase of the M-Y transition (Fig 2A). In contrast, rapamycin, a TOR inhibitor and an autophagy-inducing drug, enhanced the emergence of MDC-labelled vesicles during M-Y transition (Fig 1, SD + R), well above the mild induction of autophagic-like vesicles observed (Fig 1, SD 36° C). Interestingly, rapamycin also hastened the emergence of HEB hyphae showing enlarged tips with buds (HEB) after the onset of dimorphism. That is, the typical transition structures, such as the HE (Fig 2B, arrow) and HEB (Fig 2B, dashed arrow), were significantly higher in rapamycin-treated than in non-treated mycelia (SD+R and SD, respectively).
Fig 2. M-Y transition showed different rates for the typical transition structures along the first 24 hours.
(A) Graphical summary of the rate of morphological changes in hyphal tips during the first 24 hours of progression through the M-Y transition at 36°C in the absence of modulators (SD) or presence of 0.2 μg ml-1 rapamycin (SD+R) or 10 mM 3-MA (SD+3). (B) H—Hyphae without enlarged tips; HE—Hyphae with enlarged tips; HEB—Hyphae showing enlarged tips with bud. The arrow indicates HE structure, and the dashed arrow indicates HEB structure. Scale bar = 10 μm. * means p>0.05 and ** p>0.01.
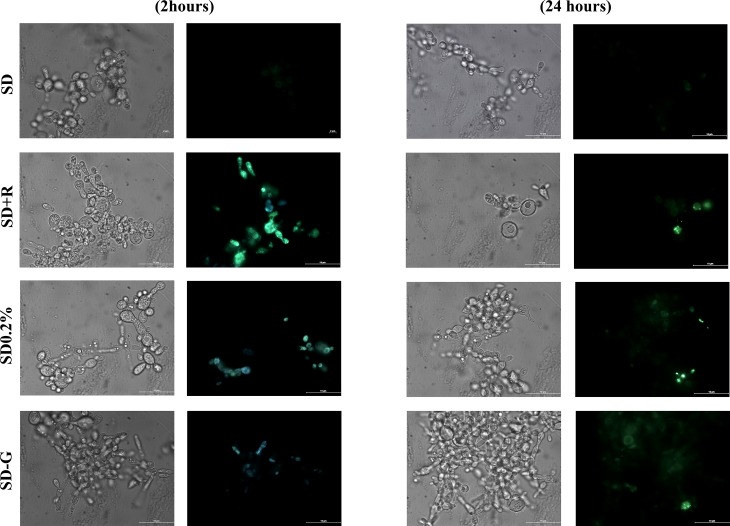
We also aimed at showing that autophagy is a universal adaptation process that P. brasiliensis makes use of under various circumstances, either as a mycelium or as a yeast. Firstly, yeasts can also readily switch on the mechanisms of autophagy induction when in the presence of rapamycin. After 2 hours of TOR inhibition with 0.2 μg ml-1 rapamycin, an expressive increase in the number of MDC-labelled vesicles was observed in the cytoplasm of cells (Fig 3, SD + R). This data unveils that the components (or machinery) necessary to trigger autophagy are likely present in yeast and that they are also kept inactive by the rapamycin-sensitive TOR in a conserved manner in P. brasiliensis, as it is found in other eukaryotes.
Fig 3. MDC-labelled vesicles are induced by rapamycin and glucose deprivation in yeast cells.
Pb18 yeasts growing in SD medium for 4–5 days were washed and incubated (1x106 cell ml-1) in Synthetic Dextrose medium as control (SD), or in SD medium containing 0.2 μg ml-1 rapamycin (SD+R); SD medium containing 0.2% glucose (SD0.2%); SD medium without glucose (SD-G). After 2 and 24 hours at 36°C, cells were incubated with MDC at 50 μM for 15 minutes, washed three times in PBS pH 7.2 and then immediately analyzed by fluorescence microscopy as described in Materials and Methods. Autophagic vesicles are stained in green. Arrow indicates yeasts entirely full of buds. Scale bar at 10 μm.
It is important to highlight that rapamycin-treated yeasts (SD+R) displayed a higher number of buds emerging from the mother cells (Fig 3, arrow), as an indication that rapamycin might favour proliferation and budding.
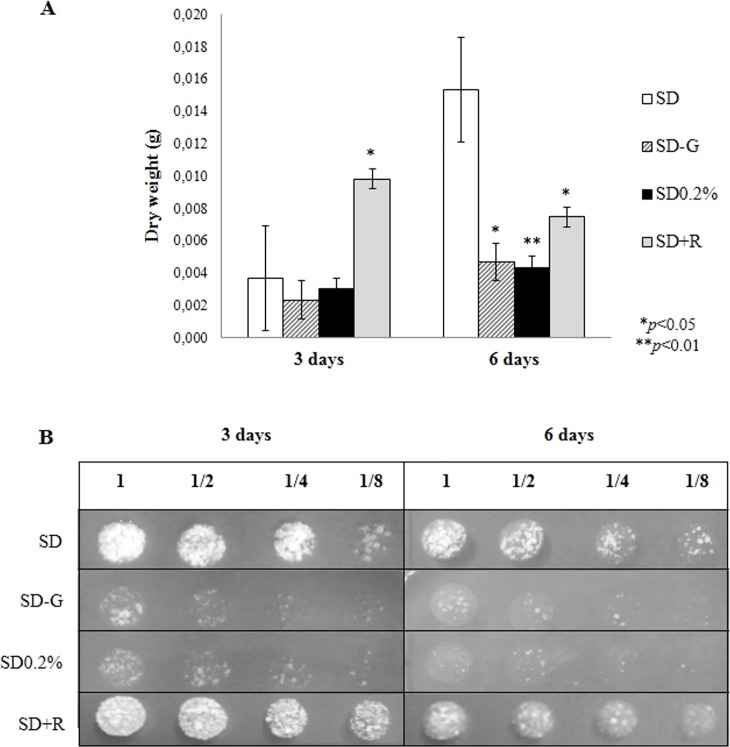
In fact, the appearance of typical MDC-labelled autophagic vesicles observed following the treatment with rapamycin correlates with an increased yeast proliferation up to 3 days being cultured (Fig 4A), but not afterwards. At 6 days, the chronic treatment of yeasts with rapamycin inhibits proliferation, probably leading to the already described paradoxical effect, resulting from the late inhibition of mTORC2 and consequent loss of cellular viability [18,19], as seen in Fig 4B.
Fig 4. Total dry weight and yeast cellular viability of Pb18.
(A) Chart showing total yeast dry weight of yeasts of Pb18 in the different culture treatment. (B) Image of plated Pb18 yeasts after serial dilution (1, 1/2, 1/4 and 1/8), incubated for 3 days, in solid YPD medium. Results were statistically significant in comparison to the control (SD), with p<0.05 * or p<0.01 **.
Cells grown under either low glucose medium (SD 0.2%) or glucose-deprived medium (SD-G) showed an increase in the number of MDC-labelled autophagic structures in their cytoplasm (Fig 3). Although it was only a discrete increase in the incidence of MDC-labelled vesicles, the autophagy triggered in yeasts under glucose limitation can be associated with the adaptation in which yeasts undergo inside macrophage's phagosome, a nutrient-limited environment.
Still, even if autophagy played a crucial role in these glucose starving cells (SD0.2% and SD-G), it is possible that autophagy might not be sufficient to sustain cellular proliferation or viability (Fig 4A and 4B), and eventually some other stimulus or cell response may be required to hold yeast cells inside the host.
Discussion
The mycelium to yeast transition in Paracoccidioides spp. is known to be directed by temperature. When the fungus in its mycelial form is subjected to 36°C, it turns into yeast form causing pulmonary and/or disseminated disease [45,46]. Nonetheless, it is important to highlight that not only is temperature variation faced by the fungus during the process of finding a host organism; it also deals with differences in nutritional availability [6,47].
After reaching the pulmonary alveoli, conidia and parts of the fungus mycelium start transitioning into yeast [45]. In addition, it is possible to find yeasts with shoots inside the autophagic vacuoles in the first 18 hours after the initial infection [45]. During this process, the fungus undergoes nutritional stress since it is believed that the mucosal surface of the lungs and the interior of macrophages are poor in nutrients, such as glucose and amino acids [5,48]. Hence, mechanisms involved in cell remodelling, adaptation, and differentiation must be induced in the fungus in order to allow its thermal dimorphic transition [47].
In this work, mycelium of Pb18 grown in nutrient-rich medium and submitted to the dimorphic transition displayed a wide number of autophagic vesicles labelled by monodansylcadaverine after being cultured for 24 hours at 36°C. We hypothesize that stimulation of autophagy may be a requirement for fungal adaptation to the host and for the dimorphic switch from hyphal to yeast growth. To support this assumption, mycelia of Pb18 treated with either rapamycin or 3-MA promoted respectively an increase in the number of MDC-labelled autophagic vesicles or the non-emergence of such vesicles. Moreover, it was shown that the temporal pattern of the emergence of typical structures during M-Y transition, such as the enlargement of the hyphal tips and buds formation, was stimulated by rapamycin and prevented by 3-MA, indicating that dimorphic differentiation was possibly stimulated when autophagy is taking place. Collectively, these data suggest that autophagy may well play a relevant role in the remodelling of the fungus during the thermal dimorphism.
There are no reports on the role of autophagy under any condition in thermally dimorphic fungi. Nevertheless, autophagy appears to play a significant role in filamentous fungi. Not only is it a response mechanism to starvation or other stresses, autophagy is also shown to have an impact on the regulation of fungal growth, morphology, and development, predominantly on the differentiation of cell types [49].
According to Richie et al [50], autophagy in Aspergillus fumigatus is required for conidiation and hyphal foraging as a response to nutrient deficiency and is important to the survival of the organism in its native environment. By studying several autophagy-related (ATG) mutants, Lv et al [51] showed that autophagy plays a critical role in growth, asexual or sexual sporulation and virulence in Fusarium graminearum. In the rice blast fungus Magnaporthe grisea (oryzae) some ATG mutants result in an inability to form the infection structure and loss of the ability of the fungus to infect its plant host [26,27,52–54]. A study carried out by Liu et al [55] showed that deletion of the autophagy-related protein MoAtg14 caused a complete loss of its virulence, besides causing problems in conidiation. Moreover, Nadal and Gold [56] demonstrated that the deletion of ATG1 or ATG8 in the plant-pathogenic fungus Ustilago maydis affected its morphogenesis and resulted in the blockage of the autophagy. Also, a significant reduction of fungal pathogenicity in Ustilago maydis was achieved with the ATG8 mutant [56]. In Beauveria bassiana, the deletion of BbATG1 and BbATG8 affected the levels of conidial protein BbCP15p, reducing conidiation by approximately 90% and 50%, respectively [57]. Additionally, the virulence in these autophagy-deficient mutant fungi was considerably weakened [57].
Similarly, several cell signalling pathways in yeasts, such as those having the kinase TOR, cAMP, Ras/PKA, Sch9 or Snf1 are related to the regulation of some stage of the autophagy [58–62]. Additionally, all these pathways are associated with cellular glucose levels [63], although each of them is controlled by a distinct set of molecules depending on the nutritional and energetic state of the cell [64].
Kim et al [65] showed that Aspergillus nidulans shares some effectors for cellular survival, such as TOR, when subject to rapamycin-induced autophagy and carbon-starvation induced autophagy. Nonetheless, a prolonged cell exposure to carbon deprivation leads to the induction of autophagy by pathways which are not TOR-independent, but PKA-dependent [65].
Considering the results in this work, the autophagy machinery is present also in yeast cells and can be promptly triggered in Pb18 yeasts under different conditions by, for instance, glucose starvation (SD-G), low glucose (SD 0.2%), or rapamycin (SD+R). This latter effect points to the conserved mTORC1 pathway as that involved in the regulation of the fungus autophagy, as it occurs in several eukaryotes [66,67].
Valcourt et al [68] and An et al [69] showed, respectively, that the deprivation of glucose or nitrogen induced morphological changes in yeasts, which became more elongated and presented fewer buds that remained attached to mother cells after cytokinesis. Studies have shown that Saccharomyces cerevisiae yeasts undergo increased levels of autophagy, with cell cycle arrest, in response to the starvation, consequently becoming quiescent [68–70]. Conway et al [71] showed that S. cerevisiae under limited glucose, nitrogen or phosphate, induced a large set of common genes related to growth, cell wall thickening and quiescence.
Regardless of rapamycin inhibition of the nutrient sensor kinase TOR, it should be noted that autophagy in yeasts of P. brasiliensis was noticed at basal level in a Synthetic medium containing 2% glucose, which should, in theory, favour the activation of either parallel or downstream targets or pathways that could bypass the growth constraints induced by the inhibition of TOR.
It is well known that the activation of the pre-initiation complex depends on both its release from the negative regulation promoted by TORC1 and its activation by AMPK [72]. Nevertheless, under the method applied in this study, it could be expected that AMPK, or AMP-dependent kinase, was inactive in SD medium at initial stages of cell culture when glucose levels are still high and, consequently, AMP/ATP ratio should be favourable to cell growth and proliferation. If such was the case, the incomplete activation of the pre-initiation complex–that is to say, the absence of its inhibition and the lack of its stimulation–should not be sufficient to promote autophagy in P. brasiliensis in a nutrient-rich environment.
It is not difficult to realize that this model has been little explored and that this is likely due to experimental limitations for a convenient transformation of yeasts, indicating, therefore, we are still far from understanding the details controlling autophagy mechanisms in this fungus. Although the autophagic process is ubiquitous among eukaryotes, the molecular elements that control them in different organisms have already been quite variable, even within Fungi kingdom. For example, in Cryptococcus neoformans, 21 genes with homology to the autophagy-related (ATG) genes of Saccharomyces cerevisiae [33], which has 34 ATG genes, are known to date [73–76].
This significant difference in the number of players acting on controlling or performing autophagy in these distantly related fungi indicates that in order to elucidate even the general players of autophagy, we need to know a variety of response types that represent the general and specific mechanisms of autophagy control in fungi. Although with limitations, P. brasiliensis offers a beautiful model of transition between two morphologies: one induced by a simple temperature change and another accompanied by important biochemical modifications that can be easily followed using cell biology and biochemical techniques. This study was the first to show that autophagy occurring in P. brasiliensis may have a role in the successful establishment of the fungus within its host because it correlates with M-Y dimorphism and is prevented by autophagy inhibitors, thus providing insightful information for future studies aiming at halting paracoccidioidomycosis and other mycoses. Yet, further genetic and in vivo analysis might be carried out to ensure that autophagy, in fact, does play a role in host pathogenesis.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was supported by Fundação de Amparo à Pesquisa no Estado de São Paulo (FAPESP, http://www.fapesp.br/), grant 2012/02138-7. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Martinez R. Epidemiology of paracoccidioidomycosis. Rev Inst Med Trop São Paulo. 2015;57: 11–20. 10.1590/S0036-46652015000700004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teixeira MM, Theodoro RC, Nino-Vega G, Bagagli E, Felipe MSS. Paracoccidioides species Complex: Ecology, Phylogeny, Sexual Reproduction, and Virulence. Heitman J, editor. PLoS Pathog. 2014;10: e1004397 10.1371/journal.ppat.1004397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.San-Blas G, Niño-Vega G, Iturriaga T. Paracoccidioides brasiliensis and paracoccidioidomycosis: molecular approaches to morphogenesis, diagnosis, epidemiology, taxonomy and genetics. Med Mycol. 2002;40: 225–242. [DOI] [PubMed] [Google Scholar]
- 4.Derengowski LS, Tavares AH, Silva S, Procópio LS, Felipe MSS, Silva-Pereira I. Upregulation of glyoxylate cycle genes upon Paracoccidioides brasiliensis internalization by murine macrophages and in vitro nutritional stress condition. Med Mycol. 2008;46: 125–134. 10.1080/13693780701670509 [DOI] [PubMed] [Google Scholar]
- 5.Lima P de S, Casaletti L, Bailão AM, Vasconcelos ATR de, Fernandes G da R, Soares CM de A. Transcriptional and Proteomic Responses to Carbon Starvation in Paracoccidioides. Vinetz JM, editor. PLoS Negl Trop Dis. 2014;8: e2855 10.1371/journal.pntd.0002855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tavares AHFP, Silva SS, Dantas A, Campos ÉG, Andrade RV, Maranhão AQ, et al. Early transcriptional response of Paracoccidioides brasiliensis upon internalization by murine macrophages. Microbes Infect. 2007;9: 583–590. 10.1016/j.micinf.2007.01.024 [DOI] [PubMed] [Google Scholar]
- 7.Parente-Rocha JA, Parente AFA, Baeza LC, Bonfim SMRC, Hernandez O, McEwen JG, et al. Macrophage Interaction with Paracoccidioides brasiliensis Yeast Cells Modulates Fungal Metabolism and Generates a Response to Oxidative Stress. Quinn J, editor. PLOS ONE. 2015;10: e0137619 10.1371/journal.pone.0137619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng Y, He D, Yao Z, Klionsky DJ. The machinery of macroautophagy. Cell Res. 2014;24: 24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stipanuk MH. Macroautophagy and its role in nutrient homeostasis. Nutr Rev. 2009;67: 677–689. 10.1111/j.1753-4887.2009.00252.x [DOI] [PubMed] [Google Scholar]
- 10.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333: 169–174. [DOI] [PubMed] [Google Scholar]
- 11.Noda T. Regulation of Autophagy through TORC1 and mTORC1. Biomolecules. 2017;7 10.3390/biom7030052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonzalez S, Rallis C. The TOR Signaling Pathway in Spatial and Temporal Control of Cell Size and Growth. Front Cell Dev Biol. 2017;5 10.3389/fcell.2017.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang RC, Levine B. Autophagy in cellular growth control. FEBS Lett. 2010;584: 1417–1426. 10.1016/j.febslet.2010.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eltschinger S, Loewith R. TOR Complexes and the Maintenance of Cellular Homeostasis. Trends Cell Biol. 2016;26: 148–159. 10.1016/j.tcb.2015.10.003 [DOI] [PubMed] [Google Scholar]
- 15.Weisman R. 2016. Target of Rapamycin (TOR) Regulates Growth in Response to Nutritional Signals. Microbiol Spectrum 4(5):FUNK-0006-2016. 10.1128/microbiolspec.FUNK-0006-2016 [DOI] [PubMed] [Google Scholar]
- 16.Loewith R, Jacinto E, Wullschleger S, Lorberg A, Crespo JL, Bonenfant D, et al. Two TOR Complexes, Only One of which Is Rapamycin Sensitive, Have Distinct Roles in Cell Growth Control. Mol Cell. 2002;10: 457–468. 10.1016/S1097-2765(02)00636-6 [DOI] [PubMed] [Google Scholar]
- 17.Saxton RA, Sabatini DM. mTOR Signaling in Growth, Metabolism, and Disease. Cell. 2017;169: 361–371. 10.1016/j.cell.2017.03.035 [DOI] [PubMed] [Google Scholar]
- 18.Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, et al. Rapamycin-Induced Insulin Resistance Is Mediated by mTORC2 Loss and Uncoupled from Longevity. Science. 2012;335: 1638–1643. 10.1126/science.1215135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarbassov DD, Ali SM, Sengupta S, Sheen J-H, Hsu PP, Bagley AF, et al. Prolonged Rapamycin Treatment Inhibits mTORC2 Assembly and Akt/PKB. Mol Cell. 2006;22: 159–168. 10.1016/j.molcel.2006.03.029 [DOI] [PubMed] [Google Scholar]
- 20.Abeliovich H, Klionsky D. Sep). Autophagy in yeast: mechanistic insights and physiological function. Microbiol Mol Biol Rev Pp. 2001;65: 463–79. 10.1128/MMBR.65.3.463-479.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kiffin R, Bandyopadhyay U, Cuervo AM. Oxidative stress and autophagy. Antioxid Redox Signal. 2006;8: 152–162. 10.1089/ars.2006.8.152 [DOI] [PubMed] [Google Scholar]
- 22.Yin Z, Pascual C, Klionsky DJ. Autophagy: machinery and regulation. Microb Cell Graz Austria. 2016;3: 588–596. 10.15698/mic2016.12.546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartoszewska M, Kiel J. Jun). The Role of Macroautophagy in Development of Filamentous Fungi. Antioxid Redox Signal Pp. 2011;14: 2271–87. 10.1089/ars.2010.3528 [DOI] [PubMed] [Google Scholar]
- 24.Deng YZ, Ramos-Pamplona M, Naqvi NI. Autophagy-assisted glycogen catabolism regulates asexual differentiation in Magnaporthe oryzae. Autophagy. 2009;5: 33–43. 10.4161/auto.5.1.7175 [DOI] [PubMed] [Google Scholar]
- 25.Kohda TA, Tanaka K, Konomi M, Sato M, Osumi M, Yamamoto M. Fission yeast autophagy induced by nitrogen starvation generates a nitrogen source that drives adaptation processes. Genes Cells Devoted Mol Cell Mech. 2007;12: 155–170. 10.1111/j.1365-2443.2007.01041.x [DOI] [PubMed] [Google Scholar]
- 26.Liu X-H, Yang J, He R-L, Lu J-P, Zhang C-L, Lu S-L, et al. An autophagy gene, TrATG5, affects conidiospore differentiation in Trichoderma reesei. Res Microbiol. 2011;162: 756–763. 10.1016/j.resmic.2011.06.011 [DOI] [PubMed] [Google Scholar]
- 27.Liu X-H, Lu J-P, Lin F-C. Autophagy During Conidiation, Conidial Germination and Turgor Generation in Magnaporthe grisea. Autophagy. 2007;3: 472–473. 10.4161/auto.4339 [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Berges MS, Rispail N, Prados-Rosales RC, Di Pietro A. A Nitrogen Response Pathway Regulates Virulence Functions in Fusarium oxysporum via the Protein Kinase TOR and the bZIP Protein MeaB. PLANT CELL ONLINE. 2010;22: 2459–2475. 10.1105/tpc.110.075937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krohn NG, Brown NA, Colabardini AC, Reis T, Savoldi M, Dinamarco TM, et al. The Aspergillus nidulans ATM Kinase Regulates Mitochondrial Function, Glucose Uptake and the Carbon Starvation Response. G3 GenesGenomesGenetics. 2014;4: 49–62. 10.1534/g3.113.008607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollack JK, Li ZJ, Marten MR. Fungal mycelia show lag time before re-growth on endogenous carbon. Biotechnol Bioeng. 2008;100: 458–465. 10.1002/bit.21779 [DOI] [PubMed] [Google Scholar]
- 31.Deng YZ, Naqvi NI. A vacuolar glucoamylase, Sga1, participates in glycogen autophagy for proper asexual differentiation in Magnaporthe oryzae. Autophagy. 2010;6: 455–461. 10.4161/auto.6.4.11736 [DOI] [PubMed] [Google Scholar]
- 32.Arruda DC, Matsuo AL, Silva LS, Real F, Leitão NP, Pires JHS, et al. Cyclopalladated Compound 7a Induces Apoptosis- and Autophagy-Like Mechanisms in Paracoccidioides and Is a Candidate for Paracoccidioidomycosis Treatment. Antimicrob Agents Chemother. 2015;59: 7214–7223. 10.1128/AAC.00512-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gontijo F de A, de Melo AT, Pascon RC, Fernandes L, Paes HC, Alspaugh JA, et al. The role of Aspartyl aminopeptidase (Ape4) in Cryptococcus neoformans virulence and authophagy. Nielsen K, editor. PLOS ONE. 2017;12: e0177461 10.1371/journal.pone.0177461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blommaart EF, Krause U, Schellens JP, Vreeling-Sindelárová H, Meijer AJ. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur J Biochem. 1997;243: 240–246. [DOI] [PubMed] [Google Scholar]
- 35.Pasquier B. Autophagy inhibitors. Cell Mol Life Sci. 2016;73: 985–1001. 10.1007/s00018-015-2104-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seglen PO, Gordon PB. 3-Methyladenine: specific inhibitor of autophagic/lysosomal protein degradation in isolated rat hepatocytes. Proc Natl Acad Sci U S A. 1982;79: 1889–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang C, Hu Q, Shen H-M. Pharmacological inhibitors of autophagy as novel cancer therapeutic agents. Pharmacol Res. 2016;105: 164–175. 10.1016/j.phrs.2016.01.028 [DOI] [PubMed] [Google Scholar]
- 38.Wang X, Proud CG. mTORC1 signaling: what we still don’t know. J Mol Cell Biol. 2011;3: 206–220. 10.1093/jmcb/mjq038 [DOI] [PubMed] [Google Scholar]
- 39.Yamamoto A, Tagawa Y, Yoshimori T, Moriyama Y, Masaki R, Tashiro Y. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct Funct. 1998;23: 33–42. [DOI] [PubMed] [Google Scholar]
- 40.Choi YJ, Park YJ, Park JY, Jeong HO, Kim DH, Ha YM, et al. Inhibitory Effect of mTOR Activator MHY1485 on Autophagy: Suppression of Lysosomal Fusion. Tajmir-Riahi H-A, editor. PLoS ONE. 2012;7: e43418 10.1371/journal.pone.0043418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campos CBL, Di Benedette JPT, Morais FV, Ovalle R, Nobrega MP. Evidence for the Role of Calcineurin in Morphogenesis and Calcium Homeostasis during Mycelium-to-Yeast Dimorphism of Paracoccidioides brasiliensis. Eukaryot Cell. 2008;7: 1856–1864. 10.1128/EC.00110-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stanford University, Stanford. Saccharomyces Genome Database. In: Saccharomyces Genome Database (SGD) [Internet]. Available: https://www.yeastgenome.org/
- 43.NCBI. Basic Local Alignment Search Tool (BLAST). In: Basic Local Alignment Search Tool [Internet]. Available: https://blast.ncbi.nlm.nih.gov/Blast.cgi
- 44.Madden T. The BLAST Sequence Analysis Tool The NCBI Handbook [Internet]. Chapter 16: Bethesda (MD): National Center for Biotechnology Information; 2002. Available: http://www.ncbi.nlm.nih.gov/books/NBK21097/ [Google Scholar]
- 45.McEwen JG, Bedoya V, Patiño MM, Salazar ME, Restrepo A. Experimental murine paracoccidiodomycosis induced by the inhalation of conidia. J Med Vet Mycol Bi-Mon Publ Int Soc Hum Anim Mycol. 1987;25: 165–175. [DOI] [PubMed] [Google Scholar]
- 46.Queiroz-Telles F, Escuissato DL. Pulmonary paracoccidioidomycosis. Semin Respir Crit Care Med. 2011;32: 764–774. 10.1055/s-0031-1295724 [DOI] [PubMed] [Google Scholar]
- 47.Tavares AH, Fernandes L, Bocca AL, Silva-Pereira I, Felipe MS. Transcriptomic reprogramming of genus Paracoccidioides in dimorphism and host niches. Fungal Genet Biol. 2015;81: 98–109. 10.1016/j.fgb.2014.01.008 [DOI] [PubMed] [Google Scholar]
- 48.Lorenz MC, Fink GR. The glyoxylate cycle is required for fungal virulence. Nature. 2001;412: 83–86. 10.1038/35083594 [DOI] [PubMed] [Google Scholar]
- 49.Pollack J, Harris S, Marten M. Autophagy in filamentous fungi. Fungal Genet Biol. 2009;46: 1–8. 10.1016/j.fgb.2008.10.010 [DOI] [PubMed] [Google Scholar]
- 50.Richie DL, Fuller KK, Fortwendel J, Miley MD, McCarthy JW, Feldmesser M, et al. Unexpected link between metal ion deficiency and autophagy in Aspergillus fumigatus. Eukaryot Cell. 2007;6: 2437–2447. 10.1128/EC.00224-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lv W, Wang C, Yang N, Que Y, Talbot NJ, Wang Z. Genome-wide functional analysis reveals that autophagy is necessary for growth, sporulation, deoxynivalenol production and virulence in Fusarium graminearum. Sci Rep. 2017;7: 11062 10.1038/s41598-017-11640-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong B, Liu X-H, Lu J-P, Zhang F-S, Gao H-M, Wang H-K, et al. MgAtg9 trafficking in Magnaporthe oryzae. Autophagy. 2009;5: 946–953. 10.4161/auto.5.7.9161 [DOI] [PubMed] [Google Scholar]
- 53.Lu J-P, Liu X-H, Feng X-X, Min H, Lin F-C. An autophagy gene, MgATG5, is required for cell differentiation and pathogenesis in Magnaporthe oryzae. Curr Genet. 2009;55: 461–473. 10.1007/s00294-009-0259-5 [DOI] [PubMed] [Google Scholar]
- 54.Veneault-Fourrey C, Barooah M, Egan M, Wakley G, Talbot NJ. Autophagic fungal cell death is necessary for infection by the rice blast fungus. Science. 2006;312: 580–583. 10.1126/science.1124550 [DOI] [PubMed] [Google Scholar]
- 55.Liu X-H, Zhao Y-H, Zhu X-M, Zeng X-Q, Huang L-Y, Dong B, et al. Autophagy-related protein MoAtg14 is involved in differentiation, development and pathogenicity in the rice blast fungus Magnaporthe oryzae. Sci Rep. 2017;7: 40018 10.1038/srep40018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nadal M, Gold SE. The autophagy genes ATG8 and ATG1 affect morphogenesis and pathogenicity in Ustilago maydis. Mol Plant Pathol. 2010;11: 463–478. 10.1111/j.1364-3703.2010.00620.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ying S-H, Liu J, Chu X-L, Xie X-Q, Feng M-G. The autophagy-related genes BbATG1 and BbATG8 have different functions in differentiation, stress resistance and virulence of mycopathogen Beauveria bassiana. Sci Rep. 2016;6 10.1038/srep26376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bermejo C, Haerizadeh F, Sadoine MSC, Chermak D, Frommer WB. Differential regulation of glucose transport activity in yeast by specific cAMP signatures. Biochem J. 2013;452: 489–497. 10.1042/BJ20121736 [DOI] [PubMed] [Google Scholar]
- 59.Budovskaya YV, Stephan JS, Reggiori F, Klionsky DJ, Herman PK. The Ras/cAMP-dependent Protein Kinase Signaling Pathway Regulates an Early Step of the Autophagy Process in Saccharomyces cerevisiae. J Biol Chem. 2004;279: 20663–20671. 10.1074/jbc.M400272200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem. 1998;273: 3963–3966. [DOI] [PubMed] [Google Scholar]
- 61.Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol Cell Biol. 2001;21: 5742–5752. 10.1128/MCB.21.17.5742-5752.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yorimitsu T, Zaman S, Broach JR, Klionsky DJ. Protein Kinase A and Sch9 Cooperatively Regulate Induction of Autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18: 4180–4189. 10.1091/mbc.E07-05-0485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broach JR. Nutritional Control of Growth and Development in Yeast. Genetics. 2012;192: 73–105. 10.1534/genetics.111.135731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taylor J Robert, Chen P-H, Chou C-C, Patel J, Jin SV. KCS1 deletion in Saccharomyces cerevisiae leads to a defect in translocation of autophagic proteins and reduces autophagosome formation. Autophagy. 2012;8: 1300–1311. 10.4161/auto.20681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim Y, Islam N, Moss BJ, Nandakumar MP, Marten MR. Autophagy induced by rapamycin and carbon-starvation have distinct proteome profiles in Aspergillus nidulans. Biotechnol Bioeng. 2011;108: 2705–2715. 10.1002/bit.23223 [DOI] [PubMed] [Google Scholar]
- 66.Cutler NS, Heitman J, Cardenas ME. TOR kinase homologs function in a signal transduction pathway that is conserved from yeast to mammals. Mol Cell Endocrinol. 1999;155: 135–142. [DOI] [PubMed] [Google Scholar]
- 67.Díaz-Troya S, Pérez-Pérez ME, Florencio FJ, Crespo JL. The role of TOR in autophagy regulation from yeast to plants and mammals. Autophagy. 2008;4: 851–865. [DOI] [PubMed] [Google Scholar]
- 68.Valcourt JR, Lemons JMS, Haley EM, Kojima M, Demuren OO, Coller HA. Staying alive: metabolic adaptations to quiescence. Cell Cycle Georget Tex. 2012;11: 1680–1696. 10.4161/cc.19879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.An Z, Tassa A, Thomas C, Zhong R, Xiao G, Fotedar R. Oct 1). Autophagy is required for G1/G0 quiescence in response to nitrogen starvation in Saccharomyces cerevisiae. Autophagy Pp. 2014;10: 1702–1711. 10.4161/auto.32122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, Werner-Washburne M. “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol Mol Biol Rev MMBR. 2004;68: 187–206. 10.1128/MMBR.68.2.187-206.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Conway MK, Grunwald D, Heideman W. Glucose, Nitrogen, and Phosphate Repletion in Saccharomyces cerevisiae: Common Transcriptional Responses to Different Nutrient Signals. G3amp58 GenesGenomesGenetics. 2012;2: 1003–1017. 10.1534/g3.112.002808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wirth M, Joachim J, Tooze SA. Autophagosome formation—the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23: 301–309. 10.1016/j.semcancer.2013.05.007 [DOI] [PubMed] [Google Scholar]
- 73.Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl Aminopeptidase Is Imported from the Cytoplasm to the Vacuole by Selective Autophagy in Saccharomyces cerevisiae. J Biol Chem. 2011;286: 13704–13713. 10.1074/jbc.M110.173906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451: 1069–1075. 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shintani M, Sangawa A, Yamao N, Miyake T, Kamoshida S. Immunohistochemical analysis of cell death pathways in gastrointestinal adenocarcinoma. Biomed Res. 2011;32: 379–386. 10.2220/biomedres.32.379 [DOI] [PubMed] [Google Scholar]
- 76.Suzuki K, Morimoto M, Kondo C, Ohsumi Y. Selective Autophagy Regulates Insertional Mutagenesis by the Ty1 Retrotransposon in Saccharomyces cerevisiae. Dev Cell. 2011;21: 358–365. 10.1016/j.devcel.2011.06.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper.