Abstract
Publisher's Note: There is a Blood Commentary on this article in this issue.
TO THE EDITOR:
One uncommon but recurrent alteration identified in ∼1% of Philadelphia chromosome–like acute lymphoblastic leukemia (ALL) is ETV6-NTRK3. ETV6 belongs to the erythroblast transformation–specific family and is an important transcriptional repressor in hematopoiesis.1 It is a common target of alteration in ALL, involved in rearrangements,2 focal deletions, and sequence mutations.3-6 The NTRK3 gene encodes TRKC, a member of the tropomyosin receptor tyrosine kinase (TRK) family. The TRK pathway has been implicated in the pathogenesis of many cancer types, with TRK fusions being the best characterized.7 ETV6-NTRK3 has been identified in a range of malignancies, including secretory breast carcinoma, infantile sarcoma, acute myeloid leukemia, and more recently, pediatric glioma8-11; however, the oncogenic role of ETV6-NTRK3 in ALL has not been investigated. Furthermore, although we and others have shown moderate sensitivity of ETV6-NTRK3 to the ALK inhibitor, crizotinib,12,13 the identification of potent and specific TRK inhibitors is required for clinical efficacy.
We sought to investigate the role of ETV6-NTRK3 in leukemia development using a genetically engineered mouse model, and to assess the efficacy of TRK inhibitors in preclinical models of ETV6-NTRK3 ALL.
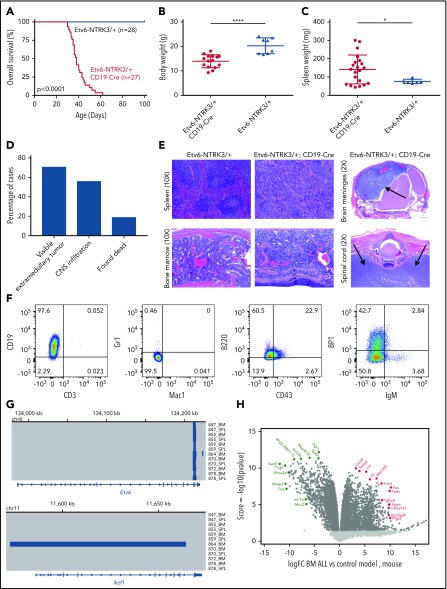
We used a conditional knock-in model of Etv6-NTRK3 with a Rosa-Stop-YFP cassette.14 Expression of Etv6-NTRK3 was achieved using CD19-Cre, which is expressed at the pre–B stage of lymphoid development.15 Etv6-NTRK3/+;CD19-Cre mice developed an aggressive lymphoid leukemia with complete penetrance and a median latency of 38 days (Figure 1A). The average body weight of Etv6-NTRK3/+;CD19-Cre mice was significantly reduced compared with age-matched Etv6-NTRK3/+ controls (13.9 ± 0.7 vs 20.2 ± 1.1 g, P < .0001) (Figure 1B). We also observed increased spleen weight in Etv6-NTRK3/+;CD19-Cre mice (142 ± 17.2 vs 71 ± 1.7 mg, P = .027), but no difference in peripheral blood counts (9.7 ± 1.6 vs 13.4 ± 2.1 × 109/L, P = .3) (Figure 1C; supplemental Figure 1A, available on the Blood Web site). Circulating blasts were confirmed by flow cytometry, and Etv6-NTRK3/+;CD19-Cre mice displayed higher levels of CD19+/B220+ circulating cells compared with Etv6-NTRK3/+ controls (59 ± 3.1 vs 42 ± 1.8%, P = .0008). Expression of Etv6-NTRK3 was confirmed in the bone marrow by reverse transcription polymerase chain reaction (supplemental Figure 1B). Etv6-NTRK3/+;CD19-Cre mice exhibited universal involvement of the bone marrow by leukemic blasts, with variable hematopoietic and nonhematopoietic extramedullary involvement, including cervical and/or inguinal lymphadenopathy and central nervous system (CNS) infiltration evidenced by hindlimb paralysis and doming of the head. This was confirmed by histopathological analysis of bone marrow and spleen, with marked CNS infiltration in the brain meninges and ventral to the thoracic and lumbar vertebra, resulting in compression of the spinal cord (Figure 1D-E). Leukemic blasts gated on YFP+ cells were negative for myeloid (Mac1 and Gr1) and T-cell markers (CD3), with the majority arrested at the pro–B stage (CD43+, B220+, CD19+, BP-1−, and IgM−) (Figure 1F; supplemental Table 3). Gating is provided in supplemental Figure 1. We also identified a subpopulation of cells at the pre–B-cell stage (BP-1+). The leukemia was serially transplantable with a median latency of 25 days. aCGH showed very few additional genetic alterations in the mouse tumors, with a focal deletion of Ikzf1 and Btg1 in 1 case each (Figure 1G; supplemental Table 4). RNA-sequencing analysis showed upregulation of Jun, Fos, and Fosb, components of the AP1 complex, which has also been observed in mammary tumors driven by Etv6-NTRK3 (Figure 1H; supplemental Table 5).14 ETV6-NTRK3 is transforming in multiple cell lineages, including NIH3T3 mouse fibroblast cells.16 Activation of Etv6-NTRK3 in the mammary glands of mature female mice induces mammary tumors by 4 months of age with complete penetrance.14 We have generated the first genetically engineered mouse model of ETV6-NTRK3 ALL using CD19-Cre to activate fusion gene expression in pre–B cells, recapitulating human B-ALL.
Figure 1.
Etv6-NTRK3/+;CD19-Cre mice develop rapid pre–B acute lymphoblastic leukemia. (A) Kaplan-Meier curve of Etv6-NTRK3/+;CD19-Cre mice (n = 27) compared with Etv6-NTRK3/+ controls (n = 28). (B) Body weight and (C) splenic weight for Etv6-NTRK3/+;CD19-Cre, and Etv6-NTRK3/+ mice. Error bars represent mean ± standard deviation. *P < .05, ****P < .0001 by Student t test. (D) Breakdown of disease signs in Etv6-NTRK3/+;CD19-Cre mice. (E) Tissues of Etv6-NTRK3/+;CD19-Cre mice showing infiltration of leukemic blasts into spleen and bone marrow with disruption of normal architecture by diffuse infiltration of leukemic cells (original magnification ×10; hematoxylin and eosin stain). Sagittal section of the brain showing masslike infiltration of blasts into the meninges, displacing the cortex (arrows). Transverse section of the spinal cord showing infiltration of leukemic blasts ventral to the vertebra, resulting in spinal cord compression (arrows). (F) Representative analysis of bone marrow of diseased animals gated on YFP+ cells by flow cytometry. (G) Microarray-based comparative genomic hybridization (aCGH) on 12 bone marrow or spleen samples from Etv6-NTRK3/+;CD19-Cre mice. A focal deletion at the 3′ end of Etv6 was identified in all samples, resulting from recombination at this locus. One case harbored a focal deletion of Ikzf1. (H) Transcriptome sequencing displaying up- and downregulated genes compared with normal pre–B cells.
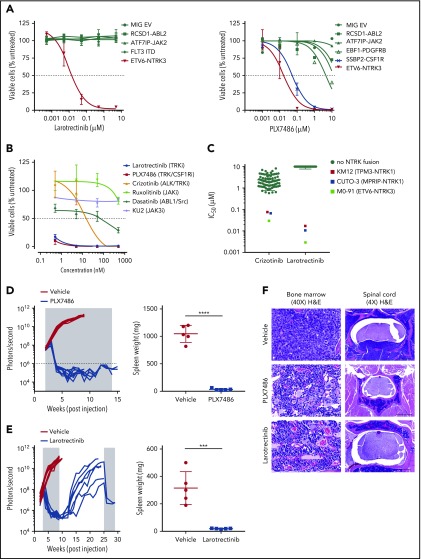
Moderate efficacy of the ALK inhibitor crizotinib against ETV6-NTRK3 has been demonstrated in cell line models.12,13 We characterized the activity of TRK inhibitors, larotrectinib (TRK-specific) and PLX7486 (TRK, CSF1R, AURK) (supplemental Figure 2),17 in Ba/F3 cell-based models representing a spectrum of Philadelphia chromosome–like ALL alterations. Drug sensitivity was determined at 48 hours using the CellTiter-Blue Cell Viability Assay (Promega). Fifty percent inhibitory concentration (IC50) values were determined using nonlinear regression (GraphPad Prism v6.0). Compared with crizotinib (IC50 value 205 nM),18 larotrectinib and PLX7486 were 10 times more potent against Ba/F3-ETV6-NTRK3 cells (IC50 values 17.0 and 17.5 nM, respectively). Larotrectinib at concentrations up to 10 μM had no effect on the viability of cells driven by other kinase alterations (ABL2, JAK2, FLT3). As expected, Ba/F3 cells expressing SSBP2-CSF1R were also sensitive to PLX7486 (IC50 value 47.6 nM) (Figure 2A). Etv6-NTRK3/+;CD19-Cre cells isolated from bone marrow and cultured ex vivo also showed exquisite sensitivity to both larotrectinib and PLX7486 (Figure 2B). Furthermore, larotrectinib was selective for TRKA and C fusions19-21 compared with crizotinib in a cytotoxicity screening of 79 human cancer cell lines (IC50 values 5 to 20 nM) (Figure 2C).
Figure 2.
In vitro and in vivo sensitivity of ETV6-NTRK3 to TRK inhibition. (A) Ba/F3 cells and (B) bone marrow cells harvested from Etv6-NTRK3/+;CD19-Cre mice were incubated in increasing concentrations of drug. Cell viability was measured after 48 hours using CellTiter-Blue viability assay. (C) Seventy-nine human cell lines were incubated in crizotinib or larotrectinib, and cell viability was measured after 72 hours using a cell count microscopy cytotoxicity assay. NTRK fusion cell lines: M0-91 (ETV6-NTRK3, acute myeloid leukemia), CUTO-3 (MPRIP-NTRK1, lung cancer), and KM12 (TPM3-NTRK1, colorectal carcinoma). (D-E) NSG mice (n = 10 per group) were injected with primary leukemic cells. Upon engraftment (108 photons per second), mice were treated with vehicle, PLX7486 (provided as chow), or larotrectinib (200 mg/kg/d once daily gavage). Treatment length is indicated by the shaded area. Five mice from each group were sacrificed when vehicle-treated mice became moribund to evaluate leukemic infiltration. The remaining 5 mice treated with PLX7486 or larotrectinib were assessed for disease growth. Two mice from the larotrectinib-treated group with active disease were retreated at week 25 for 4 weeks. Splenic weights were recorded at the time of euthanasia. Error bars represent mean ± standard deviation. ***P < .001; ****P < .0001 by Student t test. (F) Representative slides of bone marrow showing restoration of normal architecture, hematopoiesis, and vasculature with PLX7486 or larotrectinib treatment (original magnification ×40; hematoxylin and eosin stain). Scale bar, 50 μm. Transverse section of the spinal cord showing infiltration of leukemic blasts in vehicle-treated mice, with clearance of leukemic blasts and restoration of normal architecture and vasculature with PLX7486 or larotrectinib treatment. Scale bar, 500 μm.
The in vivo efficacy of TRK inhibition was assessed using a YFP-luciferase marked PDX model of ETV6-NTRK3 ALL (case ID PASBSK)12 established in NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice. Engraftment was monitored weekly using a Xenogen IVIS-200 system and Living Image software (Caliper Life Sciences).22 Mice were randomized to receive vehicle or TRK-inhibitor when leukemia burden reached 108 photons per second. The mean percentage of human CD45+/CD19+ cells in the peripheral blood, bone marrow, and spleen and splenic weight of vehicle- and inhibitor-treated animals were compared at the indicated time points. Additional details are provided in the supplemental appendix. The administration of PLX7486 as a monotherapy for 12 weeks reduced disease burden to undetectable levels as assessed by bioluminescent imaging, and reduced splenic weight (1046 vs 30.6 mg, P < .0001) (Figure 2D). To confirm the eradication of tumors was a TRK-specific effect, we performed a similar experiment with the more selective inhibitor, larotrectinib (supplemental Figure 3). Treatment with larotrectinib as a monotherapy for 6 weeks also rapidly reduced leukemic burden and splenic weight compared with vehicle-treated mice (316 vs 20 mg, P < .001) (Figure 2E). Leukemic infiltration was reduced to undetectable levels by flow cytometry in the bone marrow (75.8 vs 0% human CD45+/CD19+ cells, P < .0001), spleen (49.5 vs 0%, P = .0001), and peripheral blood (27.7 vs 0%, P = .0018) (supplemental Figure 4). Histopathological analysis showed elimination of leukemic blasts in the bone marrow and CNS with PLX7486 and larotrectinib (Figure 2F). Bone marrow cells harvested 2, 8, and 24 hours after a single in vivo dose of larotrectinib or PLX7486 showed reduced phosphorylation of pERK1/2, pSTAT3, pSTAT5, with the most marked effects observed at 24 hours (supplemental Figure 5).
Several TRK-targeting compounds are currently in clinical development, including entrectinib and larotrectinib.17 In an integrated analysis of 3 studies of larotrectinib, which received US Food and Drug Administration Breakthrough Designation in 2016, the overall response rate in 55 patients was 75%. Seventeen unique tumor types were treated, and efficacy was observed regardless of patient age, tumor type, TRK fusion, and upstream fusion partner. Responses are long lasting, with the median duration of response not yet reached, and 55% of all responding patients remaining progression free at 1 year.23 We describe the first genetically engineered mouse model of ETV6-NTRK3 ALL and report remarkable efficacy of TRK inhibition, with complete suppression of leukemic cell proliferation when administered as a monotherapy. These findings warrant screening for ETV6-NTRK3 in newly diagnosed ALL and testing the clinical efficacy of TRK inhibition.
Supplementary Material
The online version of this article contains a data supplement.
Acknowledgments
The authors thank the staff of the Flow Cytometry and Cell Sorting Core Facility, the Centre for In Vivo Imaging and Therapeutics, and the Hartwell Centre for Bioinformatics and Biotechnology at St. Jude Children’s Research Hospital. They also thank Olga Bridges for technical assistance. The Etv6-NTRK3 conditional knock-in mice were provided by Stuart Orkin from Children’s Hospital Boston. The MSCV-ETV6-NTRK3-ires-Cherry construct was provided by Suzanne Baker at St. Jude Children’s Research Hospital. PLX7486 was provided by Plexxikon.
This work was supported in part by the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital, a National Institutes of Health, National Cancer Institute Comprehensive Cancer Center grant CA012765 and Outstanding Investigator grant R35CA197695 (C.G.M.), an American Society of Hematology Scholar Award (K.G.R.), a St. Baldrick’s Foundation Scholar Award, a Robert J. Arceci Innovation Award, and the Henry Schueler 41 & 9 Foundation (C.G.M.), a Leukemia and Lymphoma Society Specialized Center of Research grant and a St. Baldrick’s Foundation Consortium Award (C.G.M. and S.P.H.), and Loxo Oncology. S.P.H. is the Jeffrey E. Perelman Distinguished Chair in Pediatrics at The Children’s Hospital of Philadelphia.
Authorship
Contribution: K.G.R. designed, directed, and performed research, analyzed data, and wrote the manuscript; L.J.J., J.M., and D.F. analyzed data; S.S. and Y.Z. designed and performed research; A.S., K.E., and B.B.T. designed and directed research and provided key reagents; S.P.H. contributed to research design and provided key reagents; C.G.M. contributed to research design, oversaw studies, and edited the manuscript; and all authors critically reviewed the manuscript prior to submission.
Conflict-of-interest disclosure: C.G.M. has received consulting fees and honoraria from Incyte and Amgen and received funding from Loxo Oncology. S.P.H. has received honoraria from Jazz Pharmaceuticals and Erytech and consulting fees from Novartis. S.S., K.E., and B.B.T. own stocks in Loxo Oncology. The remaining authors declare no competing financial interests.
Correspondence: Charles G. Mullighan, Department of Pathology, 262 Danny Thomas Place, St Jude Children’s Research Hospital, Memphis, TN 38105; e-mail: charles.mullighan@stjude.org.
REFERENCES
- 1.Wang LC, Swat W, Fujiwara Y, et al. The TEL/ETV6 gene is required specifically for hematopoiesis in the bone marrow. Genes Dev. 1998;12(15):2392-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36(8):945-961. [DOI] [PubMed] [Google Scholar]
- 3.Mullighan CG, Goorha S, Radtke I, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446(7137):758-764. [DOI] [PubMed] [Google Scholar]
- 4.Moriyama T, Metzger ML, Wu G, et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: a systematic genetic study. Lancet Oncol. 2015;16(16):1659-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Topka S, Vijai J, Walsh MF, et al. Germline ETV6 Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia. PLoS Genet. 2015;11(6):e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang MY, Churpek JE, Keel SB, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47(2):180-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5(1):25-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eguchi M, Eguchi-Ishimae M, Tojo A, et al. Fusion of ETV6 to neurotrophin-3 receptor TRKC in acute myeloid leukemia with t(12;15)(p13;q25). Blood. 1999;93(4):1355-1363. [PubMed] [Google Scholar]
- 9.Knezevich SR, McFadden DE, Tao W, Lim JF, Sorensen PH. A novel ETV6-NTRK3 gene fusion in congenital fibrosarcoma. Nat Genet. 1998;18(2):184-187. [DOI] [PubMed] [Google Scholar]
- 10.Tognon C, Knezevich SR, Huntsman D, et al. Expression of the ETV6-NTRK3 gene fusion as a primary event in human secretory breast carcinoma. Cancer Cell. 2002;2(5):367-376. [DOI] [PubMed] [Google Scholar]
- 11.Wu G, Diaz AK, Paugh BS, et al. The genomic landscape of diffuse intrinsic pontine glioma and pediatric non-brainstem high-grade glioma. Nat Genet. 2014;46(5):444-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts KG, Li Y, Payne-Turner D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371(11):1005-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taipale M, Krykbaeva I, Whitesell L, et al. Chaperones as thermodynamic sensors of drug-target interactions reveal kinase inhibitor specificities in living cells. Nat Biotechnol. 2013;31(7):630-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Tognon CE, Godinho FJ, et al. ETV6-NTRK3 fusion oncogene initiates breast cancer from committed mammary progenitors via activation of AP1 complex. Cancer Cell. 2007;12(6):542-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rickert RC, Roes J, Rajewsky K. B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res. 1997;25(6):1317-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lannon CL, Sorensen PH. ETV6-NTRK3: a chimeric protein tyrosine kinase with transformation activity in multiple cell lineages. Semin Cancer Biol. 2005;15(3):215-223. [DOI] [PubMed] [Google Scholar]
- 17.Khotskaya YB, Holla VR, Farago AF, Mills Shaw KR, Meric-Bernstam F, Hong DS. Targeting TRK family proteins in cancer. Pharmacol Ther. 2017;173:58-66. [DOI] [PubMed] [Google Scholar]
- 18.Roberts KG, Yang YL, Payne-Turner D, et al. Oncogenic role and therapeutic targeting of ABL-class and JAK-STAT activating kinase alterations in Ph-like ALL. Blood Adv. 2017;1(20):1657-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu TL, Popova L, Reeves C, et al. Phosphoproteomic analysis identifies the M0-91 cell line as a cellular model for the study of TEL-TRKC fusion-associated leukemia. Leukemia. 2007;21(3):563-566. [DOI] [PubMed] [Google Scholar]
- 20.Vaishnavi A, Capelletti M, Le AT, et al. Oncogenic and drug-sensitive NTRK1 rearrangements in lung cancer. Nat Med. 2013;19(11):1469-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardini E, Bosotti R, Borgia AL, et al. The TPM3-NTRK1 rearrangement is a recurring event in colorectal carcinoma and is associated with tumor sensitivity to TRKA kinase inhibition. Mol Oncol. 2014;8(8):1495-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boulos N, Mulder HL, Calabrese CR, et al. Chemotherapeutic agents circumvent emergence of dasatinib-resistant BCR-ABL kinase mutations in a precise mouse model of Philadelphia chromosome-positive acute lymphoblastic leukemia. Blood. 2011;117(13):3585-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med. 2018;378(8):731-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.