Abstract
In this study, using NextGen sequencing of the collective 16S rRNA genes obtained from two sets of samples collected from Lake Obersee, Antarctica, we compared and contrasted two bioinformatics tools, PICRUSt and Tax4Fun. We then developed an R script to assess the taxonomic and predictive functional profiles of the microbial communities within the samples. Taxa such as Pseudoxanthomonas, Planctomycetaceae, Cyanobacteria Subsection III, Nitrosomonadaceae, Leptothrix, and Rhodobacter were exclusively identified by Tax4Fun that uses SILVA database; whereas PICRUSt that uses Greengenes database uniquely identified Pirellulaceae, Gemmatimonadetes A1–B1, Pseudanabaena, Salinibacterium and Sinobacteraceae. Predictive functional profiling of the microbial communities using Tax4Fun and PICRUSt separately revealed common metabolic capabilities, while also showing specific functional IDs not shared between the two approaches. Combining these functional predictions using a customized R script revealed a more inclusive metabolic profile, such as hydrolases, oxidoreductases, transferases; enzymes involved in carbohydrate and amino acid metabolisms; and membrane transport proteins known for nutrient uptake from the surrounding environment. Our results present the first molecular-phylogenetic characterization and predictive functional profiles of the microbial mat communities in Lake Obersee, while demonstrating the efficacy of combining both the taxonomic assignment information and functional IDs using the R script created in this study for a more streamlined evaluation of predictive functional profiles of microbial communities.
Keywords: PICRUSt, Tax4Fun, SILVA, Greengenes, R Code, NextGen sequencing
1. Introduction
Significant efforts using both culture-dependent and culture-independent methods have been used to understand taxonomic diversity as well as functional profiles of microbial communities in a wide range of environments, including extreme ecosystems (Fuhrman, 2009; Huse et al., 2008). However, cultivation-based approaches are limited by their inability to culture most microorganisms using routine techniques, with estimates suggesting that fewer than 1% of the taxa from microbial communities in a given ecosystem are amenable to being cultured (Amann et al., 1995; Pace, 1997).
Our knowledge of the “unseen majority” has significantly advanced with the rapid progress of cultivation-independent NextGen sequencing (NGS) technology and the concurrent development of bioinformatics software (Horner et al., 2010; van Dijk et al., 2014). Traditionally, NGS technology has targeted bacterial 16S rRNA genes (V1–V9 segments) within metacommunity DNA revealing the taxonomic diversity of microbial communities in ecosystems of interest at the highest possible coverage (Chakravorty et al., 2007; Huse et al., 2008; Martinez-Porchas et al., 2017; Sanschagrin and Yergeau, 2014; Shah et al., 2011). Powerful bioinformatics tools such as PICRUSt, Tax4Fun, and Piphillin have been recently introduced that utilize 16S rRNA gene-based taxonomic information to predict the functional attributes of microbial assemblages (Aßhauer et al., 2015; Iwai et al., 2016; Langille et al., 2013). These tools are designed to derive functional gene content of microorganisms based on the known genome information of bacteria closest to their taxonomic lineage. Bioinformatics packages offer an assessment of functional metagenomics by assigning gene IDs from gene data repositories (e.g., Kanehisa and Goto, 2000; Kanehisa et al., 2014) to the Operational Taxonomic Units (OTUs) identified through a microbiome analysis. Importantly, to match gene content to observed taxa, OTUs defined in the microbiome analysis require a representative sequence and ID from 16S rRNA gene repositories such as Grenegenes (DeSantis et al., 2006) or SILVA (Quast et al., 2012). The application of NextGen sequencing technology has been particularly advantageous for the study of microbial community composition in extreme environments where microorganisms have adapted to unique physicochemical conditions that are difficult to recreate in the laboratory (Ramganesh et al., 2014). In this study, using both Greengenes and SILVA taxonomic databases used by the PICRUSt and Tax4Fun software packages, respectively, we present the first data describing microbial community composition of the microbial mats found beneath the thick, perennial ice of Lake Obersee Antarctica. We compared PICRUSt and Tax4Fun, which use 16S rRNA gene-based taxonomic information and the Kyoto Encyclopedia of Genes and Genomes (KEGG) database to predict functional attributes of microbial communities in Lake Obsersee. In conjunction with the comparative analyses by these software packages, we developed a customized R script that allowed us to streamline the process of retrieving as well as revealing a comprehensive outlook of the KEGG functional categories from the KEGG database.
2. Material and methods
2.1. Study site
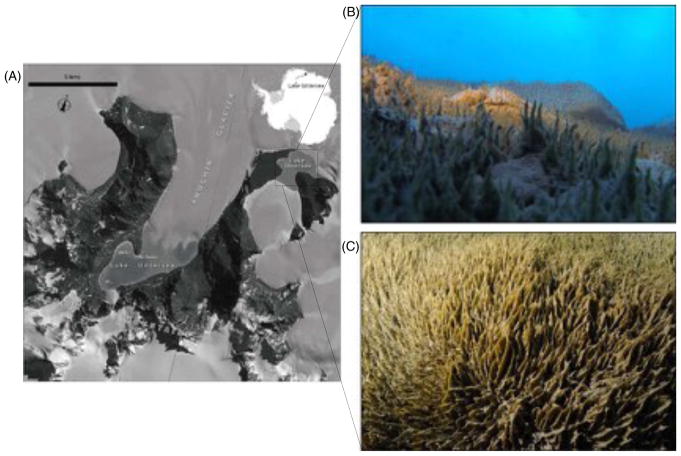
Perennially ice-covered Lake Obersee is located at 71°17'S 13°39'E in the Grüber Mountains of Central Queen Maud Land, East Antarctica (Loopmann et al., 1988) (Fig. 1). The lake is located 180 km from the coastal ice shelf at an elevation of 756 m above sea level. Lake Obersee has a surface area of approximately 3.4 km2 and a maximum depth of 83 m. The water column temperature ranges between 0.2 °C just below the ice-cover to a maximum of 0.6 °C. Although the hydrochemistry, bathymetry and chlorophyll data of Lake Obersee have been documented (Martin, 1988, Andersen unpublished data), there is no information about the composition and function of the microbial community in the benthic mats which were first observed during this study.
Fig. 1.
Geographical location and underwater photographs of the benthic microbial mats of Lake Obersee, East Antarctica. (A) Satellite image map of Lake Obersee (71.17° S 13.39° E) in the NE corner of Untersee Oasis, Antarctica. (Satellite imagery copyright DigitalGlobe, Inc. Provided by NGA Commercial Imagery Program); (B) Oblique view of the microbial mats in Lake Obersee, depth 30 m; (C) Microbial mats in Lake Obersee, depth 15 m.
2.2. Sample collection, DNA extraction, and sequence file analyses
In November–December 2012, benthic microbial mats at a depth of 15 m were imaged and sampled by scientific divers from a single dive hole employing sampling and diving techniques developed for the studies of Antarctic lakes (Andersen, 2007). Two core samples of the laminated microbial mats in Lake Obersee (herein referred to as OB12 and OB13) were collected by gently inserting a 50 mm diameter sterile polycarbonate tube into the mats and then sealing both ends with rubber stoppers before returning them to the surface. After collection, the set of intact cores were kept frozen (−20 °C) and returned to the University of Alabama at Birmingham (UAB) for DNA extraction and bioinformatics analyses.
Three subsamples (~1 cm2 each) from different laminae within each core were subjected to DNA purification using the MoBio PowerSoil DNA Isolation Kit (MoBio Laboratories Inc., CA; www.mobio.com; cat # 12888-100). DNA concentrations of each purified DNA sample were determined using the Eppendorf BioPhotometer Plus (Hamburg, Germany). An equal amount of high-quality DNA from each sample was pooled into a single sample (~1 μg) (Koo et al., 2016; White et al., 2016). A single pooled DNA sample from each mat was submitted to the Microbiome Resource Core Facility at UAB for 16S rRNA gene (V4) targeted metagenomics using the Illumina Miseq platform (paired-end, 2 by 250 bp).
The raw fastq sequence files were uploaded into Quantitative Insight into Microbial Ecology (QIIME, ver. 1.8.0) (Caporaso et al., 2010). The fastq-formatted sequences were quality-checked, and ambiguous sequences were filtered (> 25 quality score, 80% coverage, and chimera sequences) by using FastQC (Andrews, 2010) and FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), respectively. After pre-processing, paired-end reads were merged by using USEARCH (Edgar, 2010) based on overlapping regions. The quality of sequences was checked again using FastQC to ensure sufficient quality for downstream analysis, and grouped into OTUs using UCLUST (Edgar, 2010) at a 97% sequence similarity. After representative sequences were selected for each OTU, these sequences were used for further analyses using two different approaches: 1) Silva (Quast et al., 2012; Yilmaz et al., 2013) with Tax4Fun (Aßhauer et al., 2015); and 2) Greengenes (DeSantis et al., 2006) with PICRUSt (Langille et al., 2013).
2.3. Comparison of taxonomic information using Silva and Greengenes
In order to conduct the Silva-Tax4Fun approach, representative sequences were assigned to reference sequences in the SILVA database (release 123). However, to conduct the Greengenes-PICRUSt approach, the same representative sequences were used for taxonomic assignments based on Ribosomal Database Project (RDP) classifier (Wang et al., 2007), trained using the Greengenes (v 13.5) 16S rRNA database. The OTU table was then generated from each approach to visualize taxonomic information (up to genus level) and summarized in a stacked column bar graph.
2.4. Comparison of predictive functions using Tax4Fun and PICRUSt
We obtained and compared the predictive functional attributes of microbial communities in the OB12 and OB13 mat samples using the Silva-Tax4Fun and the Greengenes-PICRUSt approaches. For the Silva-Tax4Fun approach, the SILVA-labeled OTU table was used by Tax4Fun (Aßhauer et al., 2015), which is an open-source R (http://www.R-project.org/) package, to investigate predictive functional attributes of microbial communities in the mat. For this approach, Tax4Fun converted the SILVA-labeled OTUs into prokaryotic KEGG organisms, and then normalized these predictions by the 16S rRNA copy number (obtained from the NCBI genome annotations) (Aßhauer et al., 2015; Kaiser et al., 2016). The predictive functions of the microbial communities were determined by linearly combining the normalized taxonomic abundances into the precomputed association matrix of KEGG Ortholog reference profiles to Silva defined microorganisms constructed by Tax4Fun.
For the Greengenes-PICRUSt approach, the Greengenes-labeled OTU table was uploaded into PICRUSt (Langille et al., 2013). Similar to Tax4Fun, PICRUSt utilized the Greengenes-labeled OTU table to predict metabolic functions from the KEGG database. However, PICRUSt performed these assignments by referencing a pre-calculated file that contains Greengenes IDs and their associated KEGG functional categories, defined through ancestral state reconstruction (Langille et al., 2013). In accordance with the suggested practices of the PICRUSt manual, the OTU table was first normalized by the known or predicted 16S copy number abundance (Langille et al., 2013), and the resultant OTU table was used to derive predictive metagenomes by using the “predict_metagenomes.py” command. In addition, accuracy of the PI-CRUSt predictions was estimated using the Nearest Sequenced Taxon Index (NSTI) values of the two mat samples used in this study. The NSTI value validates the taxonomy-based PICRUSt-predicted KEGG functional categories of the microbial communities (Langille et al., 2013; Staley et al., 2014; Lopes et al., 2016; Kim et al., 2017). Since the output of the functional profiles generated by PICRUSt was not compatible with Tax4Fun, we have written an R script for the comparative analyses of the taxonomy-based metabolic functional predictions of the microbial communities in the two mat samples (Supplementary Materials, S1.1, S1.2, and S2).
3. Results
3.1. Total sequence reads, quality trimming, and OTU information
A total of 306,339 raw sequences reads were generated from the OB12 and OB13 mat samples (Table 1). After merging the forward and reverse raw sequences, quality-based trimming and filtering produced 248,430 sequences, which were used for further bioinformatics analyses. Overall, with OTUs clustered at a 97% sequence similarity, a higher number of OTUs were generated in both samples using the Silva database (release 123) as compared to the Greengenes database (v.13.5).
Table 1.
Raw and trimmed sequence reads following NextGen sequencing of the V4 region of the 16S rRNA gene. The number of OTUs based on both Greengenes (v. 13.5) and Silva (release 123) databases and calculated Shannon- and Simpson-diversity indices of the microbial mat samples (OB12 and OB13) in Lake Obersee are listed.
| OB12 | OB13 | Total | |
|---|---|---|---|
| Number of raw sequences | 129,660 | 176,679 | 306,339 |
| Number of sequences after trimming and filtering processes | 106,450 | 141,980 | 248,430 |
| Number of OTUs based on Greengenes (v 13.5) database | 1307 | 1336 | 2643 |
| Number of OTUs based on Silva (release 123) database | 1839 | 1742 | 3581 |
| Shannon diversity | 5.213 | 5.394 | – |
| Simpson diversity | 0.903 | 0.944 | – |
3.2. Comparison of microbial diversity assigned by using Silva and Greengenes databases
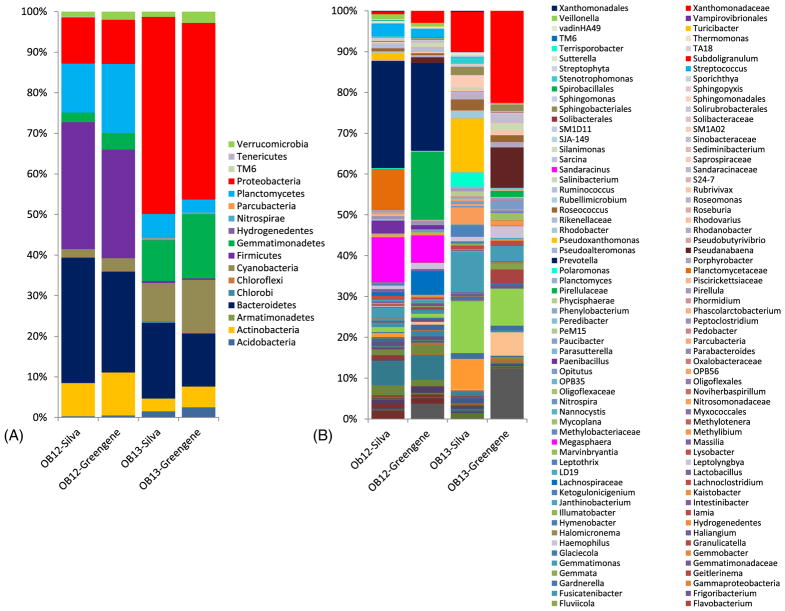
Using both databases, the relative abundances of microbial taxa in the OB12 and OB13 mat samples were determined to the most resolvable level (up to family or genus) (Fig. 2). A total of 12 phyla and 68 genera were found in both Silva and Greengenes databases. As compared to the Greengenes database, five additional phyla (Armatimonadetes, Hydrogenedentes, Parcubacteria, TM6, Tenericutes) were detected by the Silva database. These phyla had a combined relative abundance totaling < 0.2% in each sample.
Fig. 2.
Stacked column bar graph revealing the distribution and abundances of bacterial communities in Lake Obersee mat samples (OB12 and OB13) analyzed using both the Silva and Greengenes databases. The bar graphs show the bacterial distribution up to (A) Phylum level and (B) Genus level. OB12-Silva: OB12 microbial mat sample analyzed by using Silva; OB12-Greengene: OB12 microbial mat sample analyzed by using Greengenes; OB13-Silva: OB13 microbial mat sample analyzed by using Silva; OB13-Greengene: OB13 microbial mat sample analyzed by using Greengenes.
The microbial profiles revealed a relatively similar taxa distribution at the phylum level in the OB12 mat, based on both the Silva and Greengenes databases (Fig. 2a). Firmicutes and Bacteroidetes were highly abundant, followed by Planctomycetes, Proteobacteria, Actinobacteria, Gemmatimonadetes, and Cyanobacteria. However, both databases yielded significantly different microbial profiles in the OB13 sample at the phylum level (Fig. 2a). Proteobacteria was present at higher abundances in both databases. Using the Silva database, Bacteroidetes was found to be the second-most abundant taxon, followed by Gemmatimonadetes, Cyanobacteria, Planctomycetes, Actinobacteria, Acidobacteria, Verrucomicrobia, and Firmicutes. In contrast, by using the Grenegenes database, Gemmatimonadetes was found to be the second most abundant taxon, followed by Bacteroidetes, Cyanobacteria, Actinobacteria, Planctomycetes, Verrucomicrobia, and Acidobacteria.
At the most resolvable level (either family or genus), a total 146 taxa were identified by the Silva database and 103 taxa were identified by the Greengenes database (Fig. 2b). Although there was congruency between the two taxonomic assignment strategies, an additional 80 taxa were exclusively identified by Silva, but not by Greenegenes, whereas 32 taxa were identified by Greengenes and not by Silva. While Prevotella, Megasphaera, Bifidobacterium, Blautia, Streptococcus, Bacteroides, Leptolyngbya, Paenibacillus, and Acidaminococcus were found using both databases in the OB12 mat sample, the Silva database revealed Planctomycetaceae, Gemmatimonas, and Pseudoxanthomonas at relatively high abundances, and conversely for Greengenes, taxa identified as Pirellulaceae, Lachnospiraceae, A1–B1, and Xanthomonadaceae were detected as more abundant (Fig. 2b). In the OB13 mat sample, Cytophagaceae, Gemmatimonas, Xanthomonadaceae Roseococcus, Sphingobacteriales, Leptolyngbya were detected by both databases. Only the Silva database revealed Pseudoxanthomonas, Cyanobacteria; SubsectionIII, Nitrosomonadaceae, Polaromonas, SM1A02, and Leptothrix, while Greengenes showed A1–B1, Pseudanabaena, Comamonadaceae, Ellin6067, and Opitutus to be represented (Fig. 2b). A detailed list of the distribution of taxonomic groups up to genus level for the Silva and Greengenes databases in OB12 and OB13 samples is elaborated in Supplementary Material S3.
3.3. Comparison of predicted functional attributes
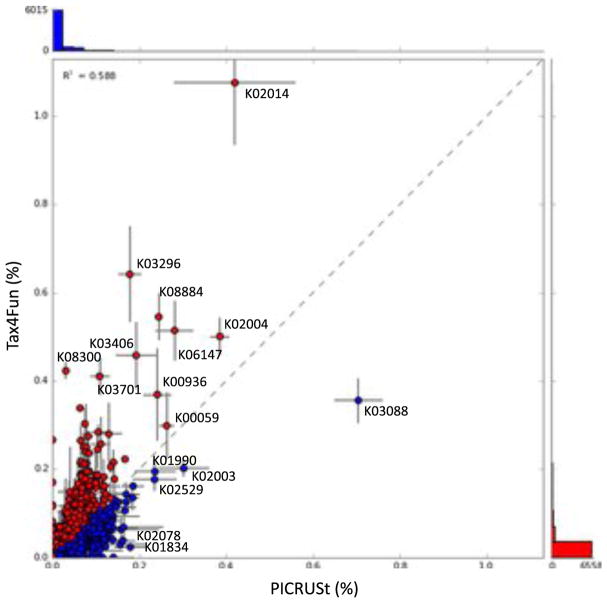
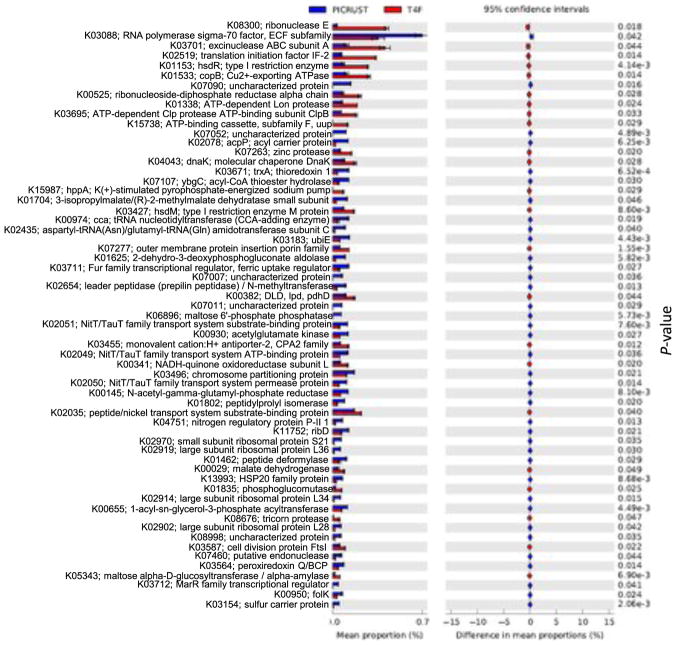
Comparison of the predicted metabolic functions of OB12 and OB13 showed highly abundant KEGG categories between the Tax4Fun and PICRUSt (Figs. 3 and 4). The NSTI value was estimated for each mat sample (0.10 in OB12 and 0.13 in OB13) and the mean value was calculated to be 0.12 ± 0.025 s.d. In general, RNA polymerase sigma-70 factor (K03088), putative ABC transport system ATP-binding protein (K02003), Acyl carrier protein (K02078), LacI family transcriptional regulator (K02529), ABC-2 type transport system ATP-binding protein (K01990), acyl-CoA thioester hydrolase (K07107), 3-isopropylmalate/ (R)-2-methylmalate dehydratase small subunit (K01704), tRNA nucleotidyltransferase (K00974), and aspartyl-tRNA(Asn)/glutamyl-tRNA(Gln) amidotransferase subunit C (K02435) were highly abundant in mat samples analyzed by the PICRUSt method (Figs. 3 and 4). In contrast, mat samples analyzed by Tax4Fun showed a higher abundance of pathways related to excinuclease ABC subunit A (K03701), ribonuclease E (K08300), translation initiation factor IF-2 (K02519), type I restriction enzyme, hsdR (K01153), Cu2+-exporting ATPase (K01533), ribonucleoside-diphosphate reductase alpha chain (K00525), ATP-dependent Lon protease (K01338), ATP-dependent Clp protease ATP-binding subunit ClpB (K03695), and ATP-binding cassette (K15738) (Figs. 3 and 4). Of the total number of KEGG categories assigned to OB12 and OB13, 297 KEGG categories were not identified using Tax4Fun, whereas 476 KEGG categories were missed by PICRUSt (Supplementary Material S4). Detailed information of each KEGG category identified using both PICRUSt and Tax4Fun are listed in Supplementary Material S5.
Fig. 3.
Scatter plot comparison using STAMP of the KEGG functional categories identified by Tax4Fun and PICRUSt. The dotted line shows equal distribution of KEGG functional categories between the two analyses. Circles above this dotted line represent KEGG functional categories generated by Tax4Fun, whereas the circles below generated by PICRUSt. Circles distributed closer to the dotted line represent a similar relative abundance of KEGG functional categories. Labeled circles indicate the greatest proportional differences of KEGG functional categories between Tax4Fun and PICRUSt. The KEGG functional profiles detected by both analyses represented in this figure are elaborated in the Supplementary Material (S5).
Fig. 4.
A comparison of the KEGG functional categories between PICRUSt and Tax4Fun are represented in an extended error plot. Total mean proportions in the KEGG categories are represented by the bar graph (left column); the upper bar graph (blue) represents the PICRUSt results, whereas the lower bar graph (red) resulted from the Tax4Fun analysis. The colored circles corresponding to the right column (blue and red) represent 95% confidence intervals calculated by Welch's t-test (Bluman, 2007). KEGG functional categories were filtered by p-value (0.05) and effect size (0.04). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
16S rRNA gene-based sequencing technology is widely used to elucidate microbial community composition in various ecosystems including extreme environments. Knowledge of the predictive functional capabilities of these microbial communities through comparisons with gene data repositories could therefore be highly beneficial (Aßhauer et al., 2015). Recently, a comparative analysis of Piphillin software with PICRUSt and Tax4Fun showed that although the Piphillin performed better in predicting gene composition and disease associated with specific gene orthologs in human clinical samples, it underperformed on environmental samples including microbial mats from hypersaline environments (Iwai et al., 2016). Thus, we used PICRUSt and Tax4Fun along with the R script to generate a comprehensive taxonomic listing, as well as predictive functional gene compositions in Lake Obersee mat samples. Results of the two taxa assignment strategies showed differences in microbial compositions, revealing five additional phyla along with 80 genera using the Silva database. In contrast, the use of the Greengenes database resulted in no additional phyla and 32 genera (Fig. 2 and Supplementary Material S3). Subsequent assignment of functional categories through KEGG also showed that PICRUSt assigned 297 extra KEGG IDs, whereas Tax4Fun recorded 476 additional KEGG IDs. To account for this variation, we wrote a customized R script to consolidate all KEGG IDs, generated by the two software packages (Supplementary Materials S1 and S2). With this script we were able to retrieve a comprehensive list of KEGG IDs as opposed to using each of these software packages alone (Supplementary Material S4 and S5).
The two core samples (OB12 and OB13) from Lake Obersee mats were dominated by Cytophagaceae, Prevotella, Gemmatimonas, Megasphaera, Xanthomonadaceae, Bifidobacterium, Polaromonas, Roseococcus, Streptococcus, Bacteroides, and Sphingobacteriales based on both the Silva and Greengenes databases. These groups of heterotrophic organisms are commonly identified in Antarctic soils, ice cores, and freshwater samples (Huang et al., 2014; Niederberger et al., 2015; Rampelotto, 2016; Segawa et al., 2010), In particular, members of Cytophaga of the phylum Bacteroidetes are chemoorganotrophic and known to enzymatically lyse Cyanobacteria and other Gram-positive bacteria (Madigan et al., 2008; Marshall, 1989; Seckbach and Oren, 2010). Therefore, these groups likely play a significant role in nutrient recycling in Lake Obersee. Whether or not this could be a reason for the low relative abundances of Cyanobacteria species in the mats, requires additional investigation. In addition to mutually different bacterial profiles, each database (SILVA and Greengenes) provided unique taxa information. The Silva database detected a relatively high abundance of 1) filamentous-shaped, sulfur-oxidizing bacteria (Leptothrix, Rhodobacter), which could be involved in the formation of the core structure of the mats (Drewniak et al., 2016); and 2) anammox bacteria (Planctomycetaceae), which could be co-occurring with the dominating members within Cytophaga and Gammaproteobacteria (i.e. Pseudoxanthomonas found in our study) by utilizing ammonium produced by the both groups (Wöbken, 2007) (Supplementary Material S3). In contrast, the Greengenes database identified relatively high abundances of 1) ammonia-oxidizing bacteria (Pirellulaceae), which could generate nitrites by oxidizing ammonium (Lawler et al., 2016); 2) benthic Cyanobacteria (Pseudanabaena), which is generally dominant in benthic microbial mats of polar freshwater ecosystems (Jungblut et al., 2010), and 3) family A1–B1 from phylum Gemmatimonadetes, of which little physiological information is available, and have rarely been found in the Antarctic continent (Foong et al., 2010).
The divergent taxonomic profiles generated by the Silva (release 123) and Greengenes (v13.5) databases may be due to the update frequency of the microbial taxa-respective databases. The current reference genomes supported by the Silva database was released in 2015, which includes 1,756,783 bacterial, archaeal, and eukaryotic sequences. However, the Greengenes database was released in 2013, which contains 1,262,986 archaeal and bacterial sequences. Certain genomes that were observed in our study, such as Hydrogenedentes and Parcubacteria, were only found in the Silva database, and did not have reference genomes in the Greengenes database. Thus, by coupling the taxonomic information from these two databases using the R script, we were able to obtain a more inclusive microbial profile than using either Silva or Greengenes alone.
The predictive functional profiles of microbial communities determined by combining the Tax4Fun and PICRUSt outputs using the R script revealed a relatively higher abundance of enzymes such as serine/threonine protein kinase, acyl-carrier protein reductase (Bechet et al., 2009; Toomey and Wakil, 1966); components necessary for carbohydrate and amino acid metabolisms; and importantly, the Uup protein belonging to the subfamily of ATP-binding cassette of the ABC transporter system (Davidson et al., 2008; Wilkins et al., 2013). Serine/ threonine kinases have been found in various bacteria and appear to be involved in the regulation of cellular functions, including cell development processes (Bechet et al., 2009; Bakal and Davies, 2000). In addition, the acyl-carrier protein reductase was firstly reported in E. coli, and is shown to be involved in the bacterial fatty acid synthesis system (Toomey and Wakil, 1966; Cukier et al., 2013). Moreover, ABC transporters play crucial roles in bacteria in nutritionally poor environments, as these systems function to move organic and inorganic molecules across the cell membrane to regulate several physiological processes (Davidson et al., 2008; Wilkins et al., 2013). ABC transporters mobilize a variety of substrates across the cell membrane, from smaller to larger molecules such as amino acids, nucleotides, metal clusters, lipid molecules, and oligonucleotides (Gerday and Glansdorff, 2009; Horikoshi et al., 2010). Hence, its presence may confer an advantage to the microbial communities in the oligotrophic environment of Lake Obersee. Additionally, the high abundance of heterotrophic communities correlates with the relatively dominant carbohydrate and amino acid metabolism pathways (Vincent and Laybourn-Parry, 2008).
The R-script used in this study was able to identify functional categories that were missed by either PICRUSt or Tax4Fun (Supplementary Material S4). The results showed that Tax4Fun was able to detect additional categories of polyketide biosynthesis proteins, transcription factors, membrane transport, and energy metabolism (nitrate/nitrite, and a sulfonate transport systems, and a methane metabolism), which were not identified by PICRUSt. These functional categories are related to energy metabolisms, and are known to play key roles in the biogeochemical cycles, adaptation, and survival of bacteria in the extreme Antarctic ultraoligotrophic environment (Laybourn-Parry and Pearce, 2016; Vincent and Laybourn-Parry, 2008).
The mean NSTI value of our mat samples using the PICRUSt showed 0.12 ± 0.025 s.d., which is better than previously reported studies on various environmental samples such as: soil samples from cold deserts of the Antarctic McMurdo Dry Valleys and hot deserts of the Southwestern United States (mid-range NSTI = 0.17 ± 0.02 s.d.), rhizosphere microbial communities (mean NSTI = 0.23 ± 0.02 s.d.), hypersaline mat microbiome (mean NSTI = 0.23 ± 0.07 s.d.), and surface soil samples from the Austre Lovénbreen glacier in High Arctic (mean NSTI = 0.18 ± 0.03 s.d.) (Langille et al., 2013; Staley et al., 2014; Lopes et al., 2016; Kim et al., 2017). Thus, the mean NSTI values in our study suggest that the predicted metabolic functions of the microbial communities in Lake Obersee mat samples are close to the known microbial reference genome databases, implying higher accuracy of the predictions. To the best of our knowledge, like PICRUSt, Tax4Fun does not provide a means to calculate the NSTI values for the taxonomic-based predicted metabolic functions of the organisms to the known microbial reference genome database. Therefore we were unable to compare the NSTI values from the PICRUSt analysis with the Tax4Fun-predicted metabolic functions of the microbial communities.
Overall, our study revealed that the functional profile predicted by Tax4Fun using the Silva database produced 14.95% higher KEGG functional IDs as compared to the PICRUSt method which uses the Greengenes database. However, it is important to note that PICRUSt identified certain KEGG functional categories that were not identified through Tax4Fun. Therefore, the use of both bioinformatics software packages was necessary for a more comprehensive outlook of the metabolic functions of mat communities in Lake Obersee. Moreover, the proposed R script allowed us to streamline the comparative aspects of the analysis of the KEGG functional categories generated by the two software packages to consolidate the functional genes.
Supplementary Material
Acknowledgments
Funding
Primary support for this research was provided by the Tawani Foundation of Chicago (6803099), the Trottier Family Foundation, NASA's Exobiology and Astrobiology Programs (NNX08AO19G), and the Arctic and Antarctic Research Institute/Russian Antarctic Expedition.
The following are acknowledged for their support of the Microbiome Resource at the University of Alabama at Birmingham: Comprehensive Cancer Center (P30AR050948), Center for AIDS Research (5P30AI027767), Center for Clinical and Translational Science (UL1TR001417), University Wide Institutional Core and Heflin Center for Genomic Sciences.
We thank UAB Heflin Center for Genomic Sciences for the next-generation sequencing, UAB Cheaha HPC and HTC grid for NGS data analyses, and Matthew Pace, Mathew Thompson and T.D. Todd of CAS IT for computer support. Antarctic logistics support was provided by the Antarctic Logistics Centre International (ALCI), Cape Town, South Africa.
Footnotes
Accession codes
All NextGen raw sequence data files are being processed by the NCBI SRA for public access for an accession number.
Conflict of interests
The authors declare that the research conducted in this study had no commercial or financial ties with industry or other organizations, thus does not have any conflicts of interest.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.mimet.2017.06.017.
References
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59(1):143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen DT. Antarctic inland waters: scientific diving in the perennially ice-covered lakes of the McMurdo Dry Valleys and Bunger Hills. In: Lang MA, Sayer MDJ, editors. Proceedings of the International Polar Diving Workshop. Smithsonian Institution; Washington, DC. 2007. pp. 163–170. [Google Scholar]
- Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. [Google Scholar]
- Aßhauer KP, Wemheuer B, Daniel R, Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31(17):2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakal CJ, Davies JE. No longer an exclusive club: eukaryotic signalling domains in bacteria. Trends Cell Biol. 2000;10(1):32–38. doi: 10.1016/s0962-8924(99)01681-5. [DOI] [PubMed] [Google Scholar]
- Bechet E, Guiral S, Torres S, Mijakovic I, Cozzone AJ, Grangeasse C. Tyrosine-kinases in bacteria: from a matter of controversy to the status of key regulatory enzymes. Amino Acids. 2009;37(3):499–507. doi: 10.1007/s00726-009-0237-8. [DOI] [PubMed] [Google Scholar]
- Bluman AG. Elementary Statistics: A Step by Step Approach. 6. McGraw Hill Higher Education; New York, New York: 2007. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravorty S, Helb D, Burday M, Connell N, Alland D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. 2007;69(2):330–339. doi: 10.1016/j.mimet.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukier CD, Hope AG, Elamin AA, Moynie L, Schnell R, Schach S, et al. Discovery of an allosteric inhibitor binding site in 3-Oxo-acyl-ACP reductase from Pseudomonas aeruginosa. ACS Chem Biol. 2013;8(11):2518–2527. doi: 10.1021/cb4005063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AL, Dassa E, Orelle C, Chen J. Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev. 2008;72(2):317–364. doi: 10.1128/MMBR.00031-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, … Andersen GL. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Auger H, Jaszczyszyn Y, Thermes C. Ten years of next-generation sequencing technology. Trends Genet. 2014;30(9):418–426. doi: 10.1016/j.tig.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Drewniak L, Krawczyk PS, Mielnicki S, Adamska D, Sobczak A, Lipinski L, Burec-Drewniak W, Sklodowska A. Physiological and metagenomic analyses of microbial mats involved in self-purification of mine waters contaminated with heavy metals. Front Microbiol. 2016;7:1252. doi: 10.3389/fmicb.2016.01252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Foong CP, Ling CMWV, González M. Metagenomic analyses of the dominant bacterial community in the Fildes Peninsula, King George Island (South Shetland Islands) Polar Sci. 2010;4(2):263–273. [Google Scholar]
- Fuhrman JA. Microbial community structure and its functional implications. Nature. 2009;459(7244):193–199. doi: 10.1038/nature08058. [DOI] [PubMed] [Google Scholar]
- Gerday C, Glansdorff N. Extremophiles. Eolss Publishers; 2009. [Google Scholar]
- Horikoshi K, Antranikian G, Bull AT, Robb FT, Stetter KO. Extremophiles Handbook. Springer Science & Business Media; 2010. [Google Scholar]
- Horner DS, Pavesi G, Castrignanò T, De Meo PDO, Liuni S, Sammeth M, Picardi E, Pesole G. Bioinformatics approaches for genomics and post genomics applications of next-generation sequencing. Brief Bioinform. 2010;11(2):181–197. doi: 10.1093/bib/bbp046. [DOI] [PubMed] [Google Scholar]
- Huang JP, Swain AK, Andersen DT, Bej AK. Bacterial diversity within five unexplored freshwater lakes interconnected by surface channels in East Antarctic Dronning Maud Land (Schirmacher Oasis) using amplicon pyrosequencing. Polar Biol. 2014;37(3):359–366. [Google Scholar]
- Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet. 2008;4(11):e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai S, Weinmaier T, Schmidt BL, Albertson DG, Poloso NJ, Dabbagh K, DeSantis TZ. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One. 2016;11(11):e0166104. doi: 10.1371/journal.pone.0166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungblut AD, Lovejoy C, Vincent WF. Global distribution of cyanobacterial ecotypes in the cold biosphere. ISME J. 2010;4(2):191–202. doi: 10.1038/ismej.2009.113. [DOI] [PubMed] [Google Scholar]
- Kaiser K, Wemheuer B, Korolkow V, Wemheuer F, Nacke H, Schöning I, Schrumpf M, Daniel R. Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci Rep. 2016:6. doi: 10.1038/srep33696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42(D1):D199–D205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Jung JY, Laffly D, Kwon HY, Lee YK. Shifts in bacterial community structure during succession in a glacier foreland of the High Arctic. FEMS Microbiol Ecol. 2017;93(1):fiw213. doi: 10.1093/femsec/fiw213. [DOI] [PubMed] [Google Scholar]
- Koo H, Hakim JA, Fisher PR, Grueneberg A, Andersen DT, Bej AK. Distribution of cold adaptation proteins in microbial mats in Lake Joyce, Antarctica: analysis of metagenomic data by using two bioinformatics tools. J Microbiol Methods. 2016;120:23–28. doi: 10.1016/j.mimet.2015.11.008. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Thurber RLV, Knight R, Beiko RG. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31(9):814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawler SN, Kellogg CA, France SC, Clostio RW, Brooke SD, Ross SW. Coral-associated bacterial diversity is conserved across two deep-sea Anthothela species. Front Microbiol. 2016:7. doi: 10.3389/fmicb.2016.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn-Parry J, Pearce D. Heterotrophic bacteria in Antarctic lacustrine and glacial environments. Polar Biol. 2016;39(12):2207–2225. [Google Scholar]
- Loopmann A, Kaup E, Klokov V, Simonov I, Haendel D. The bathymetry of some lakes of the Antarctic oases Schirmacher and Untersee. In: Martin J, editor. Limnological Studies in Queen Maud Land (East Antarctica) Eesti NSV Teaduste Akadeemia; Tallinn: 1988. pp. 6–14. [Google Scholar]
- Lopes L, De Silva MDCP, Andreote FD. Bacterial abilities and adaptation toward the rhizosphere colonization. Front Microbiol. 2016;7:1341. doi: 10.3389/fmicb.2016.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM, Dunlap PV, Clark DP. Brock Biology of mi-croorganisms 12th edn. Int Microbiol. 2008;11:65–73. [Google Scholar]
- Marshall K. Cyanobacterial-Heterotrophic Bacterial Interaction 1989 [Google Scholar]
- Martin J. Limnological Studies in Queen Maud Land (East Antarctica) Valgus 1988 [Google Scholar]
- Martinez-Porchas M, Villalpando-Canchola E, Suarez LEO, Vargas-Albores F. How conserved are the conserved 16S-rRNA regions? PeerJ. 2017;5:e3036. doi: 10.7717/peerj.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederberger TD, Sohm JA, Gunderson TE, Parker AE, Tirindelli J, Capone DG, Carpenter EJ, Cary SC. Microbial community composition of transiently wetted Antarctic Dry Valley soils. Front Microbiol. 2015;6:9. doi: 10.3389/fmicb.2015.00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace NR. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2012:gks1219. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramganesh S, Maredza A, Tekere M. Microbial exploration in extreme conditions: metagenomic analysis and future perspectives. In: Benedetti C, editor. Metagenomics Methods, Applications, and Perspectives. Nova Science Publishers; 2014. pp. 157–181. [Google Scholar]
- Rampelotto PH. Biotechnology of Extremophiles. Springer; 2016. [Google Scholar]
- Sanschagrin S, Yergeau E. Next-generation sequencing of 16S ribosomal RNA gene amplicons. J Vis Exp. 2014;90:e51709. doi: 10.3791/51709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckbach J, Oren A. Microbial Mats: Modern and Ancient Microorganisms in Stratified Systems. Vol. 14. Springer Science & Business Media; 2010. [Google Scholar]
- Segawa T, Ushida K, Narita H, Kanda H, Kohshima S. Bacterial communities in two Antarctic ice cores analyzed by 16S rRNA gene sequencing analysis. Polar Sci. 2010;4(2):215–227. [Google Scholar]
- Shah N, Tang H, Doak TG, Ye Y. Comparing Bacterial Communities Inferred from 16S rRNA Gene Sequencing and Shotgun Metagenomics. Paper Presented at the Pacific Symposium on Biocomputing; 2011. [DOI] [PubMed] [Google Scholar]
- Staley C, Gould TJ, Wang P, Phillips J, Cotner JB, Sadowsky MJ. Core functional traits of bacterial communities in the Upper Mississippi River show limited variation in response to land cover. Front Microbiol. 2014;5:414. doi: 10.3389/fmicb.2014.00414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomey RE, Wakil SJ. Studies on the mechanism of fatty acid synthesis XV. Preparation and general properties of β-ketoacyl acyl carrier protein reductase from Escherichia coli. Biochim Biophys Acta, Lipids Lipid Metab. 1966;116(2):189–197. doi: 10.1016/0005-2760(66)90001-4. [DOI] [PubMed] [Google Scholar]
- Vincent WF, Laybourn-Parry J. Polar Lakes and Rivers: Limnology of Arctic and Antarctic Aquatic Ecosystems. Oxford University Press; 2008. [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, III, Chan AM, Gavelis GS, Leander BS, Brady AL, Slater GF, Lim DS, Suttle CA. Metagenomic analysis suggests modern freshwater micro-bialites harbor a distinct core microbial community. Front Microbiol. 2016;6:1531. doi: 10.3389/fmicb.2015.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins D, Yau S, Williams TJ, Allen MA, Brown MV, DeMaere MZ, Lauro FM, Cavicchioli R. Key microbial drivers in Antarctic aquatic environments. FEMS Microbiol Rev. 2013;37(3):303–335. doi: 10.1111/1574-6976.12007. [DOI] [PubMed] [Google Scholar]
- Wöbken D. Diversity and Ecology of Marine Planctomycetes. Staats-und Universitätsbibliothek Bremen; 2007. [Google Scholar]
- Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glöckner FO. The SILVA and “all-species living tree project (LTP)” taxonomic frameworks. Nucleic Acids Res. 2013:gkt1209. doi: 10.1093/nar/gkt1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.