Abstract
Despite significant clinical and basic science advancements, cancer remains a devastating disease that affects people of all ages, races, and backgrounds. The pathogenesis of cancer has recently been described to result from eight biological capabilities or hallmarks and two enabling characteristics. These eight hallmarks are: deregulation of cellular energetics, avoiding immune destruction, enabling replicative immortality, inducing angiogenesis, sustaining proliferative signaling, evading growth suppressors, resisting cell death, and activating invasion and metastasis. The enabling characteristics are: genome instability and mutation and tumor-promoting inflammation. Survivin, the fourth most common transcript found in cancer cells, is a protein that is thought to be involved in the enhanced proliferation, survival, and metastasis and possibly other key hallmarks of cancer cells. Understanding how this gene is turned on and off is vitally important for attempt improving cancer management and therapy. Our work has identified a novel transcriptional regulator of survivin called Yin Yang 1 (YY1), which has been observed to activate some gene promoters and repress others and is gaining increasing interest as a target of cancer therapy. Our work shows for the first time that YY1 represses survivin transcription by physically interacting with the survivin promoter. Furthermore, YY1 appears to contribute to basal survivin transcriptional activity, indicating that disruption of its binding may in part contribute to survivin overexpression after cellular stress events including chemotherapy and radiotherapy.
Keywords: survivin, YY1, metastasis, cancer, transcription factor
I. INTRODUCTION
Cancer is one of the most devastating diseases in the world and one that has touched the lives of nearly every family and individual in the United States. The American Cancer Society and the National Cancer Institute estimated in 2016 that there would be an estimated 15.5 million individuals living in the United States with a personal history of cancer and the projected number of new cases being 1,685,210.1 Cancer is the second most common cause of death, accounting for approximately one in four deaths in the United States. Furthermore, 595,690 individuals are projected to suffer death as a result of cancer in 2016,2 imposing an enormous financial burden on the United States. The National Institutes of Health estimated the overall cost of all cancers in the United States in 2013 at $74.8 billion and approximately $226.8 billion worldwide: $103.8 billion for direct medical costs and $123 billion for indirect costs related to premature death and lost productivity.3
In an attempt to create an organizing principle for rationalizing the complexities of cancer, Hanahan and Weinberg have produced seminal work summarizing the hallmarks of cancer.4 These hallmarks include: (1) evasion of programmed cell death, (2) insensitivity to growth-inhibitory signals, (3) limitless replicative potential, (4) sustained angiogenesis, (5) self-sufficiency in growth signals, and (6) tissue invasion and metastatic spread. They more recently proposed two emerging hallmarks, evasion of immune destruction and deregulation of cellular energetics, and two enabling characteristics, genome instability and mutation and tumor-promoting inflammation.5 Where the eight hallmarks attempt to conceptualize the diseases that are cancer, the enabling characteristics speak to the tumor microenvironment5 and how those supposed normal cells that exist among the diseased ones contribute to pathogenesis.
Altered metabolic and redox states associated with stress such as hypoxia and pH imbalances are often found in the microenvironmental mixture of tumor and nontumor cells. The inhibitor of apoptosis (IAP) proteins have been shown to provide protection of cancer cells under these conditions6 and to be released from tumor cells into this microenvironment, where they can be taken up by both tumor and nontumor cells, resulting in a changing phenotype that has been documented to include enhanced proliferation, invasion, therapeutic resistance, and immune modulation.6–8
II. INHIBITOR OF APOPTOSIS SURVIVIN
Survivin controls diverse cellular functions, including the regulation of mitosis, surveillance checkpoints, suppression of cell death, and the adaptation to unfavorable environments.9–12 Its suppression of cell death activities and the baculovirus IAP repeat (BIR) domain characterize it as a member of the IAP family of proteins.13 However, its lack of a COOH-terminal RING finger domain and the caspase recruitment domain14 make it structurally unique among the mammalian IAPs. The overall multifaceted functionality of survivin is still being intensely scrutinized, although it appears that protein compartmentalization plays an important role. Survivin has been shown to localize in the mitochondria, where it abolishes tumor cell apoptosis and promotes tumorigenesis in immunocompromised animals.15 It therefore may possess a role in apoptosis similar to the pro-apoptotic Bcl-2 family of proteins. Survivin has also been found in the nucleus and cytosol, where it has roles in mitosis regulation and apoptosis inhibition, respectively.16 Survivin has been observed to be expressed in most common human cancers and, although present during embryonic and fetal development, is undetectable in a variety of adult tissues.17 Its aberrant, high protein expression in cancer cells and concomitantly low expression in most normal tissues make survivin an important anticancer target.18
The accumulated data from the characterization of survivin expression in human cancer tissues reveals an overwhelming consistent observation that the expression of survivin is enhanced in various human cancers compared with the adjacent normal tissues. Multiple therapeutic strategies have been investigated successfully, including the molecular antagonists such as antisense oligos, RNA inhibition, dominant-negative mutants, survivin-specific cytolytic T cells, a nonphosphorylatable survivin mutant Thr34→Ala (T34A), and, most recently, binding interface mimetics.19–26 The observation that a pool of survivin is localized extracellularly and is linked to erosive joint disease in a significant fraction of rheumatoid arthritis patients and that an autoimmune response (survivin-targeting antibodies) to survivin correlates with protection from joint disease27,28 provide evidence that anti-survivin therapy may be possible in other pathologies such as cancer. In addition, survivin expression favors cell migration, invasion, and metastasis through the association with upregulated WNT/β-catenin and PI3K/AKT, whereas survivin knockdown or inhibition results in the loss of metastatic properties.29–31 Work in our laboratory is currently defining the role of exosomal survivin in the regulation of the tumor microenvironment.6
III. SURVIVIN TRANSCRIPTION
Although many different therapeutic approaches have been used, few have been aimed at the regulation of survivin transcription. This is due, in large part, to the complexity of mechanisms involved in the epigenetic, transcriptional, and posttranscriptional gene regulations. It will likely require uncovering of all or most aspects of the machinery involved in aberrant gene regulation and personalized medical approaches to then treating each unique cancer.
Survivin transcription is critical in embryogenesis, but is normally turned off in adult life.32 However, survivin can be transcriptionally upregulated in adult life and often results in disease, particularly cancer. Survivin is the fourth-most frequently overexpressed transcript in most human cancers33 and the specificity of the survivin promoter for regulation in cancerous tissue has been demonstrated numerous times. It is currently being investigated as a means of driving the expression of therapeutic genes34–36 because of its high degree of specificity to malignant cells, which could decrease off-target expression of a suicide gene or other forms of gene therapy. Survivin’s robust and specific upregulation in cancer implies that the transcription factors involved in survivin transcription must be present and themselves upregulated in cancerous tissue. Table 1 summarizes the role of several key transcription factors in survivin transcriptional regulation.
TABLE 1.
Summary of key transcriptional regulators of survivin
| Mechanism | Pathway | Key information and current status |
|---|---|---|
| Transcription | STAT3 | Inhibition of STAT3 signaling activation downregulates survivin expression |
| Transcription | NF-κB | Regulates survivin, but mechanism unclear |
| Transcription | p53 | Transcriptionally downregulates survivin |
| Transcription | APC/β-catenin/TCF-4 | APC downregulates survivin by inhibiting β-catenin/TCF-4 |
| Transcription | HIF-1α | Transcriptionally upregulates survivin |
| Transcription | Sp1-DNA | Interference of Sp1 interaction; survivin interaction downregulates survivin |
Modified from Zhang et al., 2006.37
IV. ACTIVATORS OF SURVIVIN TRANSCRIPTION
Survivin transcription is induced, in part, by the presence of cellular stress such as that induced by chemotherapeutic agents, radiotherapy, and aspects of the tumor microenvironment. One such aspect common to the tumor microenvironment of most solid tumors is hypoxia and subsequent induction of neovascularization via vascular endothelial growth factor (VEGF) and hypoxia-inducible factor 1α (HIF-1α) activation. This has led to the investigation of a possible relationship between the hypoxia-responsive gene HIF-1α and survivin. A study from Wei et al.38 found a strong correlation between HIF-1α and survivin expression in immunohistochemically analyzed pancreatic cancer samples. Follow-up studies found that the use of antisense HIF-1α in pancreatic cancer BxPc-3 cells inhibited survivin expression and induced apoptosis.39 Peng et al. found an association between epidermal growth factor (EGF) overexpression and survivin overexpression.40 This EGF-related upregulation was mediated by HIF-1α transcriptional activation of the survivin gene, even under normoxic conditions. Bai et al. more recently identified a strong relationship between survivin overexpression and HIF-1α overexpression in cervical cancer.41 They showed HIF-1α-responsive element-independent upregulation of survivin reporter constructs, specifically in the first 158 bp of the survivin promoter. Indeed, HIF-1α-mediated upregulation of survivin has now been observed in many cancer types, including pancreatic,39 prostate,42 cervical,41 non-small cell lung,43 laryngeal,44 and colorectal45 cancers. Efforts are under way to evaluate the effectiveness of disruption of HIF-1α expression as a means to sensitize cells to therapeutic modalities.
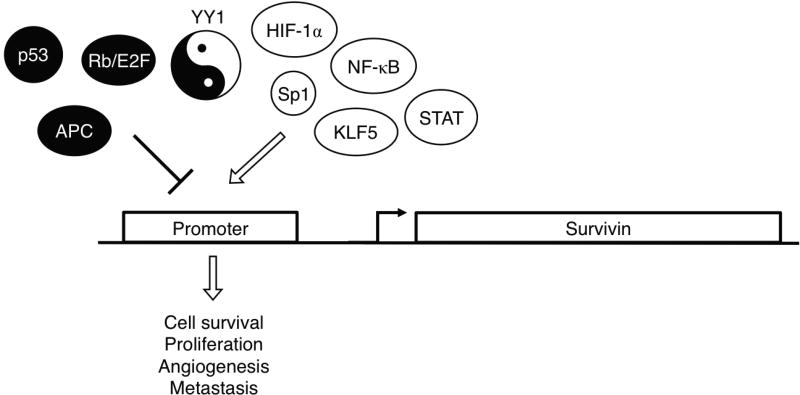
It is now known that basal survivin transcription requires specificity protein 1 (Sp1) or KLF5: Kruppel-like factor 5 (KLF5),46,47 but there are numerous other transcription factors and coactivators that are thought to drive high levels of survivin transcription (Fig. 1). NF-κB is one of these transcription factors. It is a complex of proteins that are involved in inflammation, increased cell proliferation, metastasis, and inhibition of apoptosis. One of the mechanisms by which it contributes to these phenotypes is by transcriptional activation of surviving.48–50 Members of the signal transducers and activators of transcription (STAT) family are also transcription factors capable of binding and activating the survivin promoter.51
FIG. 1.
Many activators and repressors act on survivin’s promoter. Activators: HIF-1α overexpression has been shown to upregulate survivin expression via reporter assays. Sp1 or KLF5 are necessary for basal survivin expression. NF-κB promotes survivin expression at the transcriptional level. Many STAT family proteins are capable of binding the survivin promoter. Repressors: p53 transcriptionally suppresses survivin expression. The Rb/E2F pathway downregulates survivin transcription. Adenomatous polyposis coli (APC) downregulates survivin via β-catenin/transcription factor 4 (TCF-4) inhibition. YY1’s binding site overlaps with that of Sp1 and YY1 can either activate or repress survivin transcription under different circumstances.
V. DOWNREGULATION OF SURVIVIN TRANSCRIPTION
Several key proteins are also able to downregulate survivin transcription. In addition to tumor protein 53’s (p53’s) critical involvement in cell cycle checkpoint regulation, it also prevents the transcription of oncogenes such as survivin. Retinoblastoma (Rb) and E2 transcription factor (E2F) have similar effects on survivin transcription.52 However, these genes are often silenced, mutated, and/or nonfunctional in patients with cancer. Therefore, identification of other transcription factors that may negatively regulate survivin is of importance to cancer therapy. Egr-1, a transcription factor that shares many similarities with Yin Yang-1 (YY1), has previously been noted to be involved in cell cycle, death, and differentiation. Much like YY1, Egr-1 can either act as an activator or repressor depending on the promoter in question and the available coregulators with which it interacts. Egr-1 has a consensus binding site that shares some overlap with the Sp1 transcription factor.53,54 Interestingly, YY1 can also be repressive or activating depending on a number of factors and it also shares some overlap with Sp1-binding sites for some of its targets.55 Our work has shown that the transcription factor YY1 is involved in direct transcriptional repression of surviving.56 Although HIF-1α may be in part responsible for the increased expression of survivin in tumor tissue, YY1 may also be induced under stressful conditions to negatively regulate survivin, suggesting that it is the balance of these transcription factors, and likely others, that may play an important role in the development of cancer and resistance to its treatment.56
VI. NATURAL AGENT TRANSCRIPTION MODULATORS
Natural agents are gaining increasing interest as a means of disrupting oncogene transcription, including survivin. YM155, a small-molecule inhibitor of survivin, has recently been investigated in phase II clinical trials for a variety of cancers, including diffuse large B-cell lymphoma,57 prostate cancer,58 melanoma,59 and non-small cell lung cancer,60 due to its previously observed ability to induce apoptosis and reduce tumor bulk in various in vitro and in vivo models.61 This induction of cell death is thought to be due, at least in part, to its ability to decrease survivin transcription, but the mechanism by which it does this is still under investigation. Nakamura et al.62 recently found a role for interleukin enhancer-binding factor 3 (ILF3/NF110) in this observed inhibition of survivin expression by YM155. They also found that, in luciferase reporter experiments, ILF3-dependent upregulation of reporter activity could be attenuated with YM155, suggesting that ILF3/NF110 is a physiological target of YM155. Current clinical trials show promise for YM155, particularly as a combination therapy to sensitize tumors to existing therapies. Other natural agents also show potential for disruption of survivin transcriptional activity. Specificity proteins Sp1, Sp3, and Sp4 have long been known to be important transcription factors involved in the overexpression of survivin in human cancer. However, little progress has been made in exploiting this relationship for gains in therapeutic approaches to cancer. Recently, curcumin was identified as a natural agent that inhibits the ability of Sp1, Sp3, and Sp4 to activate survivin transcription.63 It appears to do so by generating reactive oxygen species that upregulate repressors of Sp proteins ZBTB10 and ZBTB4 and downregulation of the microRNAs mir-20a, mir-27a, and mir-17-5p, which are regulators of these Sp repressors. Interestingly, curcumin is also showing promise as a sensitizing agent to ionizing radiation in Burkitt’s type lymphoma and non-Hodgkin lymphoma64,65 and alone in its ability to reduce expression of multiple IAPs critical to chemoresistance in pancreatic cancer cell lines.66 The natural agents resveratrol and quercetin in combination (RQ) have also shown a similar downregulation of Sp proteins and their targets, including surviving.67 Interestingly, the investigators cite RQ’s antioxidant capabilities (as opposed to curcumin’s generation of reactive oxygen species) as the potential reason for this observed repression of Sp proteins and their transcriptional targets such as survivin. These data further support the need for continued efforts to develop therapeutic approaches to cancer that include the disruption of survivin transcriptional activation.
VII. MULTIFUNCTIONAL PROTEIN YY1
YY1 is a 65-kDa ubiquitous multifunction transcription factor that is a member of the GLI-Kruppel family of nuclear proteins.68–70 This family of proteins plays roles in development and exerts much of its function through cell cycle regulation. YY1 is a relatively unique transcription factor in that it can act by repressing some genes and activate others by binding to the specific DNA sequence 5′-CGCCATNTT-3′.69,71 This phenomenon was noted first when it was shown that YY1, in the presence of the adenovirus-derived protein called E1A, represses the AAV P5 promoter.72 When E1A is not present, YY1 then activates transcription.73
Reports suggest that YY1 is required for cell survival because complete ablation of YY1 results in lethality.74 Furthermore, array data suggest that YY1 has roles in the cell cycle, cell adhesion, and other markers of disease aggressiveness.75,76 As is true for survivin, YY1 is increasingly found to be involved in cell death regulation via NF-κB. Within the serum amyloid A gene, there is a binding site for NF-κB that was found to overlap with a YY1-binding motif. Lu et al. showed that when YY1 bound to this NF-κB binding site that NF-κB binding and transcriptional activity were lost.36 A similar binding site overlap was observed in a cytomegalovirus promoter.77 This offers some indirect evidence of YY1 involvement in cell death, but more direct evidence is also emerging. Evidence suggests that YY1 transcriptionally represses Fas, which in turns means that YY1 is a significant factor in resistance to Fas-induced apoptosis.78 YY1 also appears to have a direct role in resistance to tumor necrosis factor-related apoptosis inducing ligand (TRAIL). Recent findings show a direct role for YY1 negatively regulating transcription of death receptor 5 (DR5), meaning that YY1 is also a resistance factor for TRAIL-induced apoptosis.79
VIII. YY1 AND HUMAN CANCER
YY1 is gaining increasing interest as a cancer-related transcription factor. The oncogenic role of YY1 has been reviewed numerous times,80–82 yet many questions remain. Consistent with its variable role as a transcription factor depending on a multitude of cellular and molecular conditions, YY1 appears to have a variable role in cancer depending on what type of cancer is being studied. Intriguingly, YY1’s role in some cancers appears to promote longer patient survival, whereas in others, it correlates with poorer outcomes and shorter survival. Table 2 summarizes current findings regarding YY1’s role in various cancer types. A computational analysis of YY1 expression in numerous datasets that looked at a broad array of cancer types indicated a relative increase in YY1 expression compared with its expression in normal tissue. Seligson et al. have shown that YY1 protein levels are higher in metastatic prostate cancer tissue than in primary tumor. However, they also found a correlation with lower YY1 protein levels and survival, suggesting that lower YY1 levels may lend a survival advantage to metastatic cells.83 Further supporting a role for YY1 in prostate cancer formation, Deng et al. found that, in prostate cancer, YY1 interacts with the androgen receptor (AR) to promote the transcription of prostate-specific antigen (PSA).84
TABLE 2.
YY1 expression in human cancers and its clinical relevance
| Tumor type | Methods | Clinical relevance of YY1 overexpression | Reference |
|---|---|---|---|
| Prostate cancer | IHC | Positive correlation with metastasis and inverse relationship with poor outcome | 83 |
| Ovarian cancer | Microarray | Positive correlation with long-term survival | 89 |
| Ovarian cancer | Microarray, IHC, RT-PCR | Positive correlation with survival and response to taxanes | 90 |
| Cervical neoplasms | RT-PCR | Positive correlation with disease progression | 85 |
| Osteosarcoma | RT-PCR, IHC, WB | Positive correlation with more malignant phenotype | 91 |
| Myeloid Leukemia | RT-PCR | Positive correlation with t(8;21) | 92 |
| Non-Hodgkin Lymphoma | RT-PCR | Positive correlation with poor outcome | 93 |
| Non-Hodgkin Lymphoma | WB, IHC | Positive correlation with more malignant phenotype and poorer outcome | 94 |
| Follicular Lymphoma | IHC | Positive correlation with length of survival | 95 |
| Pancreatic cancer | PCR, cell migration, metastasis model | Positive correlation with invasion and metastasis suppression | 96 |
Modified from Castellano et al.80
A similar association of YY1 with disease progression has been noted in intraepithelial neoplasms and cervical cancer. YY1 expression in high-grade squamous intraepithelial lesions is higher than in low-grade squamous intraepithelial lesions, a finding also consistent with the observation that high expression correlates with the presence of Human Papilloma Virus infection.85
There are also many reports of a direct role for YY1 in cancer’s aberrant cell cycle. Numerous studies show that YY1 is involved in tumorigenesis via interactions with the tumor suppressor p53. The general mechanism it appears to do this by is interference of p53-dependent transcription of its target genes by competing for binding to the ACAT sequence on the gene promoters.86 In addition, YY1 has been shown to be essential for optimal interaction of MDM-2 and p53, which is required for MDM-2 ubiquitination of p53.87 The importance of this finding cannot be overstated because an estimated 50% of all tumors have p53-inactivating mutations.88
A review by Mees et al. has eloquently summarized how a variety of transcription factors play direct roles in each of these hallmarks of cancer in the hopes of furthering a shift in thinking that embraces targeting of transcription factors in cancer therapy.97 Among the many transcription factors discussed, several are worth noting in greater detail here given their relevance to survivin and YY1. Table 3 summarizes several transcription factors with specific relevance to survivin and YY1.
TABLE 3.
Transcription factors involvement in the hallmarks of cancer and their relationship to survivin and YY1
| Transcription factor/ target |
Hallmark of cancer | Rationale |
|---|---|---|
| NF-κB | Sufficiency in growth signals | Constitutively active in many cancers. Positively regulates survivin transcription. |
| Androgen receptor | Sufficiency in growth signals | YY1 interacts directly with AR and enhances AR interaction with PSA promoter. |
| Myc | Insensitivity to growth-inhibitory signals | YY1 activates c-myc promoter |
| p53 | Evasion of programmed cell death | Survivin is downregulated by p53; YY1 downregulates p53 |
| HIF-1α | Sustained angiogenesis | Positively regulates survivin transcription |
| Sp-1 | Sustained angiogenesis/evasion of programmed cell death | Transcriptional activator of surviving; interruption of Sp-1 binding to survivin promoter induces cell death |
Modified from Mees et al.97
IX. THERAPEUTIC POTENTIAL OF YY1 IN CANCER THERAPY
The role of survivin in therapeutic approaches to cancer remains promising because there are ongoing efforts to target it in new and innovative ways. Although the role of YY1 in cancer has been known for a while now, efforts to exploit it for therapy are in their relative infancy. Just as the role of YY1 in cancer biology is controversial, so too is its value in therapeutic approaches to cancer. In ovarian cancer patients, one group found a positive correlation between YY1 expression and response to taxane therapy. In this study, YY1 knockdown led to a significant reduction in cell proliferation and anchorage-independent growth, as well as increased effectiveness of the drug paclitaxel,90 postulated to be because of positive regulation of genes involved in microtubule stabilizing activity. TRAIL is a promising ligand for inducing cell death in clinical applications because it has been shown to induce anti-tumor activity while sparing nonmalignant tissue.98 TRAIL induces by binding to the death receptors DR4 or DR5 while subsequently activating caspases. Baritaki et al.99 showed that siRNA-mediated knockdown of YY1 resulted in increased DR-5 expression and sensitization to TRAIL-mediated apoptosis. YY1 is capable of binding directly to the DR5 promoter to downregulate its expression.100 Baritaki et al.99 also showed that treatment of prostate cancer cells (PC3) with the nitric oxide donor DETANONOate sensitizes cells to TRAIL-induced cell death by downregulating NF-kB downstream of YY1. The same group has shown that inhibition of the anti-apoptotic factor BCLXL is also involved, but it is unknown whether this is via the regulation of BCLXL by YY1. This represents another line of evidence that the interruption of YY1 activity has the potential for sensitization of tumors to chemotherapy and other treatment modalities. Given the controversy as to YY1’s role in cancer, this will result in significant challenges in understanding how to approach individual types of cancer therapy as it relates to the inhibition of YY1. This will require that targeting of YY1 is highly cancer specific to avoid the dysregulation of YY1 in normal tissue.
X. YY1 AND SURVIVIN: BEYOND TRANSCRIPTIONAL REGULATION
Tumor metastasis is the most common cause of death in cancer patients. There are several steps described by Hanahan and Weinberg that are required for a malignant cell to fully metastasize, making it a remarkably complex process.4 The first step is invasion, which involves the loss of cell adhesion molecules. Without the loss of surface adhesion molecules, it is not possible for a cell to begin migration into neighboring tissue. The second step involves intravasation of the invading malignant cells into the blood or lymphatic system. The third step, which only a small percent of intravasated cells are thought to be able to accomplish, is extravasation through capillaries at a site distant to the primary tumor. Once extravasated, cells must then regain adhesion molecules that allow the cell to establish the ability to survive in the new environment. In carcinomas, the metastatic process is thought to consist of a number of distinct steps. The complex mechanisms required to accomplish each of these steps are the subject of intense research.
The epithelial-to-mesenchymal transition (EMT) is a proposed mechanism by which malignant cells initiate the first step described above: the need for a loss of adhesion molecules so that invasion can take place. EMT is characterized by a loss of E-cadherin and β-catenin and a gain in N-cadherin and vimentin expression (reviewed by Huber et al.101). The transcription factors Twist, Snail, and Slug have been identified as the major regulators of these adhesion molecules.102–104 Research from our laboratory indicates that survivin is involved in the invasion step of metastasis.7
Survivin is found to be overexpressed in primary tumors in addition to distant metastatic cells, but no direct involvement in the mechanism of metastasis has been identified. We have recently found a novel means by which survivin promotes cell invasion. Extracellular surviving promotes invasion of HeLa cells through a collagen matrix and antibody depletion of survivin abrogates this increased cell invasion.7 Studies are ongoing to identify mechanisms behind this observation. Very recently, Yie et al.105 found that patients with survivin-expressing circulating non-small cell lung cancer cells had a higher instance of cancer recurrence and increased follow-up lymph node involvement. Other studies have shown that survivin delineates node-positive from node-negative rectal cancer.106 In small-cell adenocarcinoma of the lung, patients with histological evidence of high survivin expression had more evidence of venous invasion of malignant cells and, overall, patients with high survivin expression had decreased survival.107 In squamous cell carcinomas, survivin expression correlates with high-grade, poorly differentiated tumors and with increased lymph node metastasis.108 A common theme in these studies is the presence of almost entirely correlative data with little or no mechanistic information.
Among the many target genes of YY1 being discovered, some involved in metastasis are now being identified. A report in 2005 hypothesized that the cooperation of YY1 and AP-1 may increase the repression of the galactocerebrosidase (GALC) gene. GALC is an enzyme that is overexpressed on the surface of cancer cells. Suppression of this enzyme leads to an accumulation of galactocerebroside, which results in a decrease in cellular adhesion and inhibition of apoptosis, which in turns leads to increased cell proliferation and migration.109 This observation, although it was largely conjecture, was the first evidence that YY1 may be involved in cancer invasion and metastasis. However, in the search to identify new genes involved in metastasis suppression, Wang et al.110 discovered that HLJ1, a metastasis suppressor, is positively regulated by YY1. High levels of YY1 expression correlated with HLJ1 expression and promoter reporter assays indicated that YY1 was acting directly on the transcription of HLJ1. Subsequent studies found that a synergistic relationship between YY1 and AP1 to a 5 times higher activation of HLJ1 and much more potent in vitro cancer cell invasion.111 Using the osteosarcoma cell line SaOS-2, de Negris et al.76 found that deletion of YY1 to a decrease in cell invasion in vitro and decrease metastasis in vivo. Deletion of YY1 also correlated with a decrease in VEGF and angiogenesis. They also identified a host of genes involved in cell motility, cell cycle, cell adhesion, angiogenesis, and signal transduction that exhibited significant changes when YY1 was deleted.76 One report suggested that YY1 is a regulator of Snail, one of the key transcription factors responsible for regulation of EMT, a key feature of metastasis.112 In all, these data detail the complicated nature of YY1’s involvement in cancer metastasis because it appears that, in some types of cancer, it may serve to inhibit metastasis, whereas in others such as osteosarcoma, it may promote metastasis and aggressiveness of the disease.
XI. FUTURE DIRECTIONS
The work from our laboratory illustrates a role for YY1 in survivin transcriptional repression in the osteosarcoma cell line U2OS.56 However, the role of YY1 in the transcription of survivin in other cancer types has yet to be thoroughly investigated. Our preliminary evidence indicates a similar repressive role for YY1’s observed repression of survivin in the pancreatic cancer cell line PANC-1. This work indicates an important role for survivin expression levels in PANC-1 radioresistance, but it suggested an even larger role for XIAP (Inhibitor of Apoptosis) in the radioresistance of PANC-1 cells.113 Therefore, future studies should broaden the investigation of cancer- specific YY1 regulation of survivin transcription and be expanded to the investigation of transcriptional regulation of IAP’s such as XIAP. In addition, we have shown multiple avenues of evidence for the involvement of YY1 in basal survivin transcription, but future efforts should attempt to identify the role of YY1 in cellular response to stresses in the form of chemotherapeutics, radiotherapy, or natural agent exposure. HIF-1α’s role in survivin transcriptional upregulation is now well established,40,41 but the role that YY1 plays in survivin’s downregulation and basal control must still be more fully and convincingly established. Although YY1 is not clearly established as a stress-response transcriptional factor per se, several studies have indicated that it is involved in unfolded protein response and resulting ER stress114 and may even inhibit the function of p53 in response to genotoxic stress.115
Consistent with the need to better understand YY1’s role in the survivin-mediated cellular stress response, it is also critical for future studies to measure functional outcomes as a consequence of survivin transcriptional modulation. Our preliminary evidence indicates that YY1 overexpression in U2OS tet-off cells may be involved in enhanced cellular proliferation as measured by the Ki-67 assay (unpublished data). However, it is as yet unknown the extent to which YY1 overexpression is specifically involved in this enhanced proliferation or if indeed it is mediated by survivin or one of the other numerous transcriptional targets of YY1. Our reporter data indicate a very robust repression of survivin promoter activity when YY1 is overexpressed, but much more moderate reduction in protein expression.56 A recent study showed a role for YY1 in mammary cell proliferation, migration, clonogenicity, invasion, and tumor formation and identified YY1-mediated p27 degradation as a likely mechanism.116 In a similar fashion, future work should elucidate whether YY1 modulates cellular invasion, migration, proliferation, and other outcomes through its regulation of survivin and other target genes. These studies will be critical in furthering efforts to establish new therapeutic approaches based on survivin targeting.
We have recently described a novel pool of survivin existing in the extracellular space.7 Current studies are exploring ways in which this pool of survivin may contribute to disease in the normal neighboring cells in the tumor microenvironment and antibody depletion of this extracellular pool of survivin may prove to be a valid therapeutic approach for solid tumors. However, at this point, the mechanism for export of survivin is unknown. We are investigating whether this mechanism requires exosomes to export survivin to the extracellular.6 If YY1 is indeed a modulator of survivin transcription, then it stands to reason that YY1 overexpression or knockdown may alter the amount of survivin that is exported to the extracellular space.
XII. SUMMARY AND CONCLUSION
It is becoming increasingly evident that cancer is a constellation of hundreds, if not thousands, of different diseases. This is likely why, despite a multitude of significant advances in our understanding of cancer, current therapies leave much to be desired in terms of patient health and well-being. The future of cancer therapy will likely require personalized approaches to individual disease, but this will require as complete an understanding as is reasonably possible of the underlying factors involved in cancer development and advancement. The advent of molecular biology has given scientists powerful tools with which to understand the mechanisms and architecture involved in cell structure and function and has helped to reveal the true complexity of biological systems. A key feature of this complexity is redundancy, a concept that has plagued therapeutic approaches to cancer. Molecular biology has revealed that virtually no cellular process is without pathway redundancy, including invasion and metastasis, and cancer cells have perhaps even more redundancy than normal cells that better equips them to evade immune response and death. Gene expression, such as that of survivin, is affected by redundant epigenetic, transcriptional, and posttranscriptional regulation factors. To exploit therapy directed against a target such as surviving effectively, it is important to understand the complete picture of how the survivin gene works and how the transcription factors such as YY1, alone or in combination with others, work to facilitate the complexity and redundancy of cancer.
Acknowledgments
The authors thank the Center for Health Disparities and Molecular Medicine for its support and members of the Wall laboratory for careful review of this manuscript.
This work was supported in part by the National Institutes of Health (Grant P20MD006988 to N.R.W.).
ABBREVIATIONS
- AR
androgen receptor
- BIR
baculovirus IAP repeat
- DR5
death receptor 5
- E2F
E2 transcription factor
- EGF
epidermal growth factor
- EMT
epithelial-to-mesenchymal transition
- GALC
galactocerebrosidase
- IAP
inhibitor of apoptosis
- HIF-1α
hypoxia-inducible factor 1α
- ILF3
interleukin enhancer-binding factor 3
- p53
tumor protein 53
- PSA
prostate-specific antigen
- Rb
retinoblastoma
- RQ
resveratrol and quercetin in combination
- Sp1
specificity protein 1
- STAT
signal transducers and activators of transcription
- TRAIL
tumor necrosis factor-related apoptosis inducing ligand
- VEGF
vascular endothelial growth factor
- YY1
Yin Yang 1
Footnotes
Conflict of Interest Statement
The authors declare no competing financial interests.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66(4):271–89. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Tarver T. Cancer Facts and Figures 2012. American Cancer Society (ACS) J of Consumer Health on the Internet. 2012;16(3):366–7. [Google Scholar]
- 4.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 6.Khan S, Jutzy JM, Aspe JR, McGregor DW, Neidigh JW, Wall NR. Survivin is released from cancer cells via exosomes. Apoptosis. 2011;16(1):1–12. doi: 10.1007/s10495-010-0534-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khan S, Aspe JR, Asumen MG, Almaguel F, Odumosu O, Acevedo-Martinez S, De Leon M, Langridge WH, Wall NR. Extracellular, cell-permeable survivin inhibits apoptosis while promoting proliferative and metastatic potential. Br J Cancer. 2009;100(7):1073–86. doi: 10.1038/sj.bjc.6604978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jutzy JM, Khan S, Asuncion-Valenzuela MM, Milford TA, Payne KJ, Wall NR. Tumor-released survivin induces a type-2 t cell response and decreases cytotoxic T cell function, in vitro. Cancer Microenviron. 2013;6(1):57–68. doi: 10.1007/s12307-012-0096-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altieri DC. Survivin, versatile modulation of cell division and apoptosis in cancer. Oncogene. 2003;22(53):8581–9. doi: 10.1038/sj.onc.1207113. [DOI] [PubMed] [Google Scholar]
- 10.Li F, Ambrosini G, Chu EY, Plescia J, Tognin S, Marchisio PC, Altieri DC. Control of apoptosis and mitotic spindle checkpoint by survivin. Nature. 1998;396(6711):580–4. doi: 10.1038/25141. [DOI] [PubMed] [Google Scholar]
- 11.Altieri DC. The case for survivin as a regulator of microtubule dynamics and cell-death decisions. Curr Opin Cell Biol. 2006;18(6):609–15. doi: 10.1016/j.ceb.2006.08.015. [DOI] [PubMed] [Google Scholar]
- 12.Li F. Survivin study: An update of “What is the next wave?”. J Cell Physiol. 2006;208(3):476–86. doi: 10.1002/jcp.20634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3(8):917–21. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 14.Deveraux QL, Reed JC. IAP family proteins—suppressors of apoptosis. Genes Dev. 1999;13(3):239–52. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 15.Dohi T, Beltrami E, Wall NR, Plescia J, Altieri DC. Mitochondrial survivin inhibits apoptosis and promotes tumorigenesis. J Clin Invest. 2004;114(8):1117–27. doi: 10.1172/JCI22222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fortugno P, Wall NR, Giodini A, O’Connor DS, Plescia J, Padgett KM, Tognin S, Marchisio PC, Altieri DC. Survivin exists in immunochemically distinct subcellular pools and is involved in spindle microtubule function. J Cell Sci. 2002;115(Pt 3):575–85. doi: 10.1242/jcs.115.3.575. [DOI] [PubMed] [Google Scholar]
- 17.Adida C, Crotty PL, McGrath J, Berrebi D, Diebold J, Altieri DC. Developmentally regulated expression of the novel cancer anti-apoptosis gene survivin in human and mouse differentiation. Am J Pathol. 1998;152(1):43–9. [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuda S, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006;5(5):1087–98. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- 19.Andersen MH, Pedersen LO, Capeller B, Brocker EB, Becker JC, thor Straten P. Spontaneous cytotoxic T-cell responses against survivin-derived MHC class I-restricted T-cell epitopes in situ as well as ex vivo in cancer patients. Cancer Res. 2001;61(16):5964–8. [PubMed] [Google Scholar]
- 20.Grossman D, Kim PJ, Schechner JS, Altieri DC. Inhibition of melanoma tumor growth in vivo by survivin targeting. Proc Natl Acad Sci U S A. 2001;98(2):635–40. doi: 10.1073/pnas.230450097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mesri M, Wall NR, Li J, Kim RW, Altieri DC. Cancer gene therapy using a survivin mutant adenovirus. J Clin Invest. 2001;108(7):981–90. doi: 10.1172/JCI12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wall NR, O’Connor DS, Plescia J, Pommier Y, Altieri DC. Suppression of survivin phosphorylation on Thr34 by flavopiridol enhances tumor cell apoptosis. Cancer Res. 2003;63(1):230–5. [PubMed] [Google Scholar]
- 23.Plescia J, Salz W, Xia F, Pennati M, Zaffaroni N, Daidone MG, Meli M, Dohi T, Fortugno P, Nefedova Y, Gabrilovich DI, Colombo G, Altieri DC. Rational design of shepherdin, a novel anticancer agent. Cancer Cell. 2005;7(5):457–68. doi: 10.1016/j.ccr.2005.03.035. [DOI] [PubMed] [Google Scholar]
- 24.Olie RA, Simoes-Wust AP, Baumann B, Leech SH, Fabbro D, Stahel RA, Zangemeister-Wittke U. A novel antisense oligonucleotide targeting survivin expression induces apoptosis and sensitizes lung cancer cells to chemotherapy. Cancer Res. 2000;60(11):2805–9. [PubMed] [Google Scholar]
- 25.Kanwar JR, Shen WP, Kanwar RK, Berg RW, Krissansen GW. Effects of survivin antagonists on growth of established tumors and B7-1 immunogene therapy. J Natl Cancer Inst. 2001;93(20):1541–52. doi: 10.1093/jnci/93.20.1541. [DOI] [PubMed] [Google Scholar]
- 26.Andersen MH, Svane IM, Becker JC, Straten PT. The universal character of the tumor-associated antigen survivin. Clin Cancer Res. 2007;13(20):5991–4. doi: 10.1158/1078-0432.CCR-07-0686. [DOI] [PubMed] [Google Scholar]
- 27.Mera S, Magnusson M, Tarkowski A, Bokarewa M. Extracellular survivin up-regulates adhesion molecules on the surface of leukocytes changing their reactivity pattern. J Leukoc Biol. 2008;83(1):149–55. doi: 10.1189/jlb.0507287. [DOI] [PubMed] [Google Scholar]
- 28.Bokarewa M, Lindblad S, Bokarew D, Tarkowski A. Balance between survivin, a key member of the apoptosis inhibitor family, and its specific antibodies determines erosivity in rheumatoid arthritis. Arthritis Res Ther. 2005;7(2):R349–58. doi: 10.1186/ar1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siddharth S, Das S, Nayak A, Kundu CN. SURVIVIN as a marker for quiescent-breast cancer stem cells-An intermediate, adherent, pre-requisite phase of breast cancer metastasis. Clin Exp Meta. 2016;33(7):661–75. doi: 10.1007/s10585-016-9809-7. [DOI] [PubMed] [Google Scholar]
- 30.Zhang W, Liu Y, Li YF, Yue Y, Yang X, Peng L. Targeting of survivin pathways by YM155 inhibits cell death and invasion in oral squamous cell carcinoma cells. Cell Physiol Biochem. 2016;38(6):2426–37. doi: 10.1159/000445594. [DOI] [PubMed] [Google Scholar]
- 31.Hseu YC, Thiyagarajan V, Tsou HT, Lin KY, Chen HJ, Lin CM, Liao JW, Yang HL. In vitro and in vivo anti-tumor activity of CoQ0 against melanoma cells: inhibition of metastasis and induction of cell-cycle arrest and apoptosis through modulation of Wnt/beta-catenin signaling pathways. Oncotarget. 2016;7(16):22409–26. doi: 10.18632/oncotarget.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaux DL, Korsmeyer SJ. Cell death in development. Cell. 1999;96(2):245–54. doi: 10.1016/s0092-8674(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 33.Velculescu VE, Madden SL, Zhang L, Lash AE, Yu J, Rago C, Lal A, Wang CJ, Beaudry GA, Ciriello KM, Cook BP, Dufault MR, Ferguson AT, Gao Y, He TC, Hermeking H, Hiraldo SK, Hwang PM, Lopez MA, Luderer HF, Mathews B, Petroziello JM, Polyak K, Zawel L, Zhang W, Zhang X, Zhou W, Haluska FG, Jen J, Sukumar S, Landes GM, Riggins GJ, Vogelstein B, Kinzler KW. Analysis of human transcriptomes. Nat Genet. 1999;23(4):387–8. doi: 10.1038/70487. [DOI] [PubMed] [Google Scholar]
- 34.Bao R, Connolly DC, Murphy M, Green J, Weinstein JK, Pisarcik DA, Hamilton TC. Activation of cancer-specific gene expression by the survivin promoter. J Natl Cancer Inst. 2002;94(7):522–8. doi: 10.1093/jnci/94.7.522. [DOI] [PubMed] [Google Scholar]
- 35.Zhu ZB, Makhija SK, Lu B, Wang M, Kaliberova L, Liu B, Rivera AA, Nettelbeck DM, Mahasreshti PJ, Leath CA, Barker S, Yamaoto M, Li F, Alvarez RD, Curiel DT. Transcriptional targeting of tumors with a novel tumor-specific survivin promoter. Cancer Gene Ther. 2004;11(4):256–62. doi: 10.1038/sj.cgt.7700679. [DOI] [PubMed] [Google Scholar]
- 36.Lu B, Makhija SK, Nettelbeck DM, Rivera AA, Wang M, Komarova S, Zhou F, Yamamoto M, Haisma HJ, Alvarez RD, Curiel DT, Zhu ZB. Evaluation of tumor-specific promoter activities in melanoma. Gene Ther. 2005;12(4):330–8. doi: 10.1038/sj.gt.3302385. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M, Yang J, Li F. Transcriptional and post-transcriptional controls of survivin in cancer cells: novel approaches for cancer treatment. J Exp Clin Cancer Res. 2006;25(3):391–402. [PMC free article] [PubMed] [Google Scholar]
- 38.Wei H, Wang C, Chen L. Proliferating cell nuclear antigen, survivin, and CD34 expressions in pancreatic cancer and their correlation with hypoxia-inducible factor 1alpha. Pancreas. 2006;32(2):159–63. doi: 10.1097/01.mpa.0000202961.71600.9b. [DOI] [PubMed] [Google Scholar]
- 39.Chang Q, Qin R, Huang T, Gao J, Feng Y. Effect of antisense hypoxia-inducible factor 1alpha on progression, metastasis, and chemosensitivity of pancreatic cancer. Pancreas. 2006;32(3):297–305. doi: 10.1097/00006676-200604000-00010. [DOI] [PubMed] [Google Scholar]
- 40.Peng XH, Karna P, Cao Z, Jiang BH, Zhou M, Yang L. Cross-talk between epidermal growth factor receptor and hypoxia-inducible factor-1alpha signal pathways increases resistance to apoptosis by up-regulating survivin gene expression. J Biol Chem. 2006;281(36):25903–14. doi: 10.1074/jbc.M603414200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bai H, Ge S, Lu J, Qian G, Xu R. Hypoxia inducible factor-1alpha-mediated activation of survivin in cervical cancer cells. J Obstet Gynaecol Res. 2013;39(2):555–63. doi: 10.1111/j.1447-0756.2012.01995.x. [DOI] [PubMed] [Google Scholar]
- 42.Yun YJ, Li SH, Cho YS, Park JW, Chun YS. Survivin mediates prostate cell protection by HIF-1alpha against zinc toxicity. Prostate. 2010;70(11):1179–88. doi: 10.1002/pros.21152. [DOI] [PubMed] [Google Scholar]
- 43.Chen YQ, Zhao CL, Li W. Effect of hypoxia-inducible factor-1alpha on transcription of survivin in non-small cell lung cancer. J Exp Clin Cancer Res. 2009;28:29. doi: 10.1186/1756-9966-28-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li DW, Zhou L, Jin B, Xie J, Dong P. Expression and significance of hypoxia-inducible factor-1alpha and survivin in laryngeal carcinoma tissue and cells. Otolaryngol Head Neck Surg. 2013;148(1):75–81. doi: 10.1177/0194599812464759. [DOI] [PubMed] [Google Scholar]
- 45.Fan LF, Dong WG, Jiang CQ, Qian Q, Yu QF. Role of Hypoxia-inducible factor-1 alpha and Survivin in colorectal carcinoma progression. Int J Colorectal Dis. 2008;23(11):1057–64. doi: 10.1007/s00384-008-0511-3. [DOI] [PubMed] [Google Scholar]
- 46.Li F, Altieri DC. Transcriptional analysis of human survivin gene expression. Biochem J. 1999;344(Pt 2):305–11. doi: 10.1042/0264-6021:3440305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu N, Gu L, Findley HW, Chen C, Dong JT, Yang L, Zhou M. KLF5 interacts with p53 in regulating survivin expression in acute lymphoblastic leukemia. J Biol Chem. 2006;281(21):14711–8. doi: 10.1074/jbc.M513810200. [DOI] [PubMed] [Google Scholar]
- 48.Angileri FF, Alfredo MA, Domenico C, Salvatore L, Rosalia C, Tomasello CC, Germano A, Vita G, Tomasello F. Nuclear factor-kappaB activation and differential expression of survivin and Bcl-2 in human grade 2–4 astrocytomas. Cancer. 2008;112(10):2258–66. doi: 10.1002/cncr.23407. [DOI] [PubMed] [Google Scholar]
- 49.Zucchini C, Rocchi A, Manara MC, De Sanctis P, Capanni C, Bianchini M, Carinci P, Scotlandi K, Valvassori L. Apoptotic genes as potential markers of metastatic phenotype in human osteosarcoma cell lines. Int J Oncol. 2008;32(1):17–31. [PubMed] [Google Scholar]
- 50.Kawakami H, Tomita M, Matsuda T, Ohta T, Tanaka Y, Fujii M, Hatano M, Tokuhisa T, Mori N. Transcriptional activation of survivin through the NF-kappaB pathway by human T-cell leukemia virus type I tax. Int J Cancer. 2005;115(6):967–74. doi: 10.1002/ijc.20954. [DOI] [PubMed] [Google Scholar]
- 51.Gritsko T, Williams A, Turkson J, Kaneko S, Bowman T, Huang M, Nam S, Eweis I, Diaz N, Sullivan D, Yoder S, Enkemann S, Eschrich S, Lee JH, Beam CA, Cheng J, Minton S, Muro-Cacho CA, Jove R. Persistent activation of stat3 signaling induces survivin gene expression and confers resistance to apoptosis in human breast cancer cells. Clin Cancer Res. 2006;12(1):11–9. doi: 10.1158/1078-0432.CCR-04-1752. [DOI] [PubMed] [Google Scholar]
- 52.Raj D, Liu T, Samadashwily G, Li F, Grossman D. Survivin repression by p53, Rb and E2F2 in normal human melanocytes. Carcinogenesis. 2008;29(1):194–201. doi: 10.1093/carcin/bgm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ackerman SL, Minden AG, Williams GT, Bobonis C, Yeung CY. Functional significance of an overlapping consensus binding motif for Sp1 and Zif268 in the murine adenosine deaminase gene promoter. Proc Natl Acad Sci U S A. 1991;88(17):7523–7. doi: 10.1073/pnas.88.17.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Skerka C, Decker EL, Zipfel PF. A regulatory element in the human interleukin 2 gene promoter is a binding site for the zinc finger proteins Sp1 and EGR-1. J Biol Chem. 1995;270(38):22500–6. doi: 10.1074/jbc.270.38.22500. [DOI] [PubMed] [Google Scholar]
- 55.Dong X-P, Pfister H. Overlapping YY1- and aberrant SP1-binding sites proximal to the early promoter of human papillomavirus type 16. J Gen Virol. 1999;80(8):2097–101. doi: 10.1099/0022-1317-80-8-2097. [DOI] [PubMed] [Google Scholar]
- 56.Galloway NR, Diaz Osterman CJ, Reiber K, Jutzy JM, Li F, Sui G, Soto U, Wall NR. Yin Yang 1 regulates the transcriptional repression of Survivin. Biochem Biophys Res Commun. 2014;445(1):208–13. doi: 10.1016/j.bbrc.2014.01.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheson BD, Bartlett NL, Vose JM, Lopez-Hernandez A, Seiz AL, Keating AT, Shamsili S, Papadopoulos KP. A phase II study of the survivin suppressant YM155 in patients with refractory diffuse large B-cell lymphoma. Cancer. 2012;118(12):3128–34. doi: 10.1002/cncr.26510. [DOI] [PubMed] [Google Scholar]
- 58.Tolcher AW, Quinn DI, Ferrari A, Ahmann F, Giaccone G, Drake T, Keating A, de Bono JS. A phase II study of YM155, a novel small-molecule suppressor of survivin, in castration-resistant taxane-pretreated prostate cancer. Ann Oncol. 2012;23(4):968–73. doi: 10.1093/annonc/mdr353. [DOI] [PubMed] [Google Scholar]
- 59.Lewis KD, Samlowski W, Ward J, Catlett J, Cranmer L, Kirkwood J, Lawson D, Whitman E, Gonzalez R. A multicenter phase II evaluation of the small molecule survivin suppressor YM155 in patients with unresectable stage III or IV melanoma. Investig New Drugs. 2011;29(1):161–6. doi: 10.1007/s10637-009-9333-6. [DOI] [PubMed] [Google Scholar]
- 60.Giaccone G, Zatloukal P, Roubec J, Floor K, Musil J, Kuta M, van Klaveren RJ, Chaudhary S, Gunther A, Shamsili S. Multicenter phase II trial of YM155, a small-molecule suppressor of survivin, in patients with advanced, refractory, non-small-cell lung cancer. J Clin Oncol. 2009;27(27):4481–6. doi: 10.1200/JCO.2008.21.1862. [DOI] [PubMed] [Google Scholar]
- 61.Nakahara T, Kita A, Yamanaka K, Mori M, Amino N, Takeuchi M, Tominaga F, Hatakeyama S, Kinoyama I, Matsuhisa A, Kudoh M, Sasamata M. YM155, a novel small-molecule survivin suppressant, induces regression of established human hormone-refractory prostate tumor xenografts. Cancer Res. 2007;67(17):8014–21. doi: 10.1158/0008-5472.CAN-07-1343. [DOI] [PubMed] [Google Scholar]
- 62.Nakamura N, Yamauchi T, Hiramoto M, Yui M, Naito M, Takeuchi M, Yamanaka K, Kita A, Nakahara T, Kinoyama I, Matsuhisa A, Kaneko N, Koutoku H, Sasamata M, Yokota H, Kawabata S, Furuichi K. Interleukin enhancer-binding factor 3/NF110 is a target of YM155, a suppressant of survivin. Mol Cell Proteomics. 2012;11(7):M111. doi: 10.1074/mcp.M111.013243. 013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gandhy SU, Kim K, Larsen L, Rosengren RJ, Safe S. Curcumin and synthetic analogs induce reactive oxygen species and decreases specificity protein (Sp) transcription factors by targeting microRNAs. BMC Cancer. 2012;12:564. doi: 10.1186/1471-2407-12-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiao Q, Jiang Y, Li G. Inhibition of the PI3K/AKT-NF-kappaB pathway with curcumin enhanced radiation-induced apoptosis in human Burkitt’s lymphoma. J Pharmacol Sci. 2013;121(4):247–56. doi: 10.1254/jphs.12149fp. [DOI] [PubMed] [Google Scholar]
- 65.Qiao Q, Jiang Y, Li G. Curcumin enhances the response of non-Hodgkin’s lymphoma cells to ionizing radiation through further induction of cell cycle arrest at the G2/M phase and inhibition of mTOR phosphorylation. Oncol Rep. 2013;29(1):380–6. doi: 10.3892/or.2012.2091. [DOI] [PubMed] [Google Scholar]
- 66.Diaz Osterman CJ, Gonda A, Stiff T, Sigaran U, Valenzuela MM, Ferguson Bennit HR, Moyron RB, Khan S, Wall NR. Curcumin induces pancreatic adenocarcinoma cell death via reduction of the inhibitors of apoptosis. Pancreas. 2016;45(1):101–9. doi: 10.1097/MPA.0000000000000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Del Follo-Martinez A, Banerjee N, Li X, Safe S, Mertens-Talcott S. Resveratrol and quercetin in combination have anticancer activity in colon cancer cells and repress oncogenic microRNA-27a. Nutr Cancer. 2013;65(3):494–504. doi: 10.1080/01635581.2012.725194. [DOI] [PubMed] [Google Scholar]
- 68.Ficzycz A, Ovsenek N. The Yin Yang 1 transcription factor associates with ribonucleoprotein (mRNP) complexes in the cytoplasm of Xenopus oocytes. J Biol Chem. 2002;277(10):8382–7. doi: 10.1074/jbc.M110304200. [DOI] [PubMed] [Google Scholar]
- 69.Galvin KM, Shi Y. Multiple mechanisms of transcriptional repression by YY1. Mol Cell Biol. 1997;17(7):3723–32. doi: 10.1128/mcb.17.7.3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Y, Seto E, Chang LS, Shenk T. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67(2):377–88. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 71.Yao YL, Yang WM, Seto E. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol. 2001;21(17):5979–91. doi: 10.1128/MCB.21.17.5979-5991.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chang LS, Shi Y, Shenk T. Adeno-associated virus P5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J Virol. 1989;63(8):3479–88. doi: 10.1128/jvi.63.8.3479-3488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim Biophys Acta. 1997;1332(2):F49–66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 74.Donohoe ME, Zhang X, McGinnis L, Biggers J, Li E, Shi Y. Targeted disruption of mouse Yin Yang 1 transcription factor results in peri-implantation lethality. Mol Cell Biol. 1999;19(10):7237–44. doi: 10.1128/mcb.19.10.7237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Affar EB, Gay F, Shi Y, Liu H, Huarte M, Wu S, Collins T, Li E, Shi Y. Essential dosage-dependent functions of the transcription factor Yin Yang 1 in late embryonic development and cell cycle progression. Mol Cell Biol. 2006;26(9):3565–81. doi: 10.1128/MCB.26.9.3565-3581.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Nigris F, Rossiello R, Schiano C, Arra C, Williams-Ignarro S, Barbieri A, Lanza A, Balestrieri A, Giuliano MT, Ignarro LJ, Napoli C. Deletion of Yin Yang 1 protein in osteosarcoma cells on cell invasion and CXCR4/angiogenesis and metastasis. Cancer Res. 2008;68(6):1797–808. doi: 10.1158/0008-5472.CAN-07-5582. [DOI] [PubMed] [Google Scholar]
- 77.Chan YJ, Chiou CJ, Huang Q, Hayward GS. Synergistic interactions between overlapping binding sites for the serum response factor and ELK-1 proteins mediate both basal enhancement and phorbol ester responsiveness of primate cytomegalovirus major immediate-early promoters in monocyte and T-lymphocyte cell types. J Virol. 1996;70(12):8590–605. doi: 10.1128/jvi.70.12.8590-8605.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garban HJ, Bonavida B. Nitric oxide inhibits the transcription repressor Yin-Yang 1 binding activity at the silencer region of the Fas promoter: a pivotal role for nitric oxide in the up-regulation of Fas gene expression in human tumor cells. J Immunol. 2001;167(1):75–81. doi: 10.4049/jimmunol.167.1.75. [DOI] [PubMed] [Google Scholar]
- 79.Vega MI, Jazirehi AR, Huerta-Yepez S, Bonavida B. Rituximab-induced inhibition of YY1 and Bcl-xL expression in Ramos non-Hodgkin’s lymphoma cell line via inhibition of NF-kappa B activity: role of YY1 and Bcl-xL in Fas resistance and chemoresistance, respectively. J Immunol. 2005;175(4):2174–83. doi: 10.4049/jimmunol.175.4.2174. [DOI] [PubMed] [Google Scholar]
- 80.Castellano G, Torrisi E, Ligresti G, Malaponte G, Militello L, Russo AE, McCubrey JA, Canevari S, Libra M. The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle. 2009;8(9):1367–72. doi: 10.4161/cc.8.9.8314. [DOI] [PubMed] [Google Scholar]
- 81.Deng Z, Cao P, Wan MM, Sui G. Yin Yang 1: a multifaceted protein beyond a transcription factor. Transcription. 2010;1(2):81–4. doi: 10.4161/trns.1.2.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2005;25(8):1125–42. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- 83.Seligson D, Horvath S, Huerta-Yepez S, Hanna S, Garban H, Roberts A, Shi T, Liu X, Chia D, Goodglick L, Bonavida B. Expression of transcription factor Yin Yang 1 in prostate cancer. Int J Oncol. 2005;27(1):131–41. [PubMed] [Google Scholar]
- 84.Deng Z, Wan M, Cao P, Rao A, Cramer SD, Sui G. Yin Yang 1 regulates the transcriptional activity of androgen receptor. Oncogene. 2009;28(42):3746–57. doi: 10.1038/onc.2009.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Baritaki S, Sifakis S, Huerta-Yepez S, Neonakis IK, Soufla G, Bonavida B, Spandidos DA. Overexpression of VEGF and TGF-beta1 mRNA in Pap smears correlates with progression of cervical intraepithelial neoplasia to cancer: implication of YY1 in cervical tumorigenesis and HPV infection. Int J Oncol. 2007;31(1):69–79. [PubMed] [Google Scholar]
- 86.Yakovleva T, Kolesnikova L, Vukojevic V, Gileva I, Tan-No K, Austen M, Luscher B, Ekstrom TJ, Terenius L, Bakalkin G. YY1 binding to a subset of p53 DNA-target sites regulates p53-dependent transcription. Biochem Biophys Res Commun. 2004;318(2):615–24. doi: 10.1016/j.bbrc.2004.04.065. [DOI] [PubMed] [Google Scholar]
- 87.Sui G, Affar el B, Shi Y, Brignone C, Wall NR, Yin P, Donohoe M, Luke MP, Calvo D, Grossman SR, Shi Y. Yin Yang 1 is a negative regulator of p53. Cell. 2004;117(7):859–72. doi: 10.1016/j.cell.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 88.Lane DP. p53 and human cancers. Br Med Bull. 1994;50(3):582–99. doi: 10.1093/oxfordjournals.bmb.a072911. [DOI] [PubMed] [Google Scholar]
- 89.Berchuck A, Iversen ES, Lancaster JM, Pittman J, Luo J, Lee P, Murphy S, Dressman HK, Febbo PG, West M, Nevins JR, Marks JR. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin Cancer Res. 2005;11(10):3686–96. doi: 10.1158/1078-0432.CCR-04-2398. [DOI] [PubMed] [Google Scholar]
- 90.Matsumura N, Huang Z, Baba T, Lee PS, Barnett JC, Mori S, Chang JT, Kuo WL, Gusberg AH, Whitaker RS, Gray JW, Fujii S, Berchuck A, Murphy SK. Yin yang 1 modulates taxane response in epithelial ovarian cancer. Mol Cancer Res. 2009;7(2):210–20. doi: 10.1158/1541-7786.MCR-08-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.de Nigris F, Botti C, de Chiara A, Rossiello R, Apice G, Fazioli F, Fiorito C, Sica V, Napoli C. Expression of transcription factor Yin Yang 1 in human osteosarcomas. Eur J Cancer. 2006;42(15):2420–4. doi: 10.1016/j.ejca.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 92.Grubach L, Juhl-Christensen C, Rethmeier A, Olesen LH, Aggerholm A, Hokland P, Ostergaard M. Gene expression profiling of Polycomb, Hox and Meis genes in patients with acute myeloid leukaemia. Eur J Haematol. 2008;81(2):112–22. doi: 10.1111/j.1600-0609.2008.01083.x. [DOI] [PubMed] [Google Scholar]
- 93.Sakhinia E, Glennie C, Hoyland JA, Menasce LP, Brady G, Miller C, Radford JA, Byers RJ. Clinical quantitation of diagnostic and predictive gene expression levels in follicular and diffuse large B-cell lymphoma by RT-PCR gene expression profiling. Blood. 2007;109(9):3922–8. doi: 10.1182/blood-2006-09-046391. [DOI] [PubMed] [Google Scholar]
- 94.Bonavida B, Huerta-Yepez S, Baritaki S, Vega M, Liu H, Chen H, Berenson J. Overexpression of Yin Yang 1 in the pathogenesis of human hematopoietic malignancies. Crit Rev Oncog. 2011;16(3–4):261–7. doi: 10.1615/critrevoncog.v16.i3-4.90. [DOI] [PubMed] [Google Scholar]
- 95.Naidoo K, Clay V, Hoyland JA, Swindell R, Linton K, Illidge T, Radford JA, Byers RJ. YY1 expression predicts favourable outcome in follicular lymphoma. J Clin Pathol. 2011;64(2):125–9. doi: 10.1136/jcp.2010.078188. [DOI] [PubMed] [Google Scholar]
- 96.Zhang JJ, Zhu Y, Xie KL, Peng YP, Tao JQ, Tang J, Li Z, Xu ZK, Dai CC, Qian ZY, Jiang KR, Wu JL, Gao WT, Du Q, Miao Y. Yin Yang-1 suppresses invasion and metastasis of pancreatic ductal adenocarcinoma by downregulating MMP10 in a MUC4/ErbB2/p38/MEF2C-dependent mechanism. Mol Cancer. 2014;13:130. doi: 10.1186/1476-4598-13-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mees C, Nemunaitis J, Senzer N. Transcription factors: their potential as targets for an individualized therapeutic approach to cancer. Cancer Gene Ther. 2009;16(2):103–12. doi: 10.1038/cgt.2008.73. [DOI] [PubMed] [Google Scholar]
- 98.Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA, Blackie C, Chang L, McMurtrey AE, Hevert A, DeForge L, Koumenis IL, Lewis D, Harris L, Bussiere J, Koeppen H, Shahrokh Z, Schwall RW. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest. 1999;104(2):155–62. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Baritaki S, Huerta-Yepez S, Sakai T, Spandidos DA, Bonavida B. Chemotherapeutic drugs sensitize cancer cells to TRAIL-mediated apoptosis: up-regulation of DR5 and inhibition of Yin Yang 1. Mol Cancer Ther. 2007;6(4):1387–99. doi: 10.1158/1535-7163.MCT-06-0521. [DOI] [PubMed] [Google Scholar]
- 100.Huerta-Yepez S, Vega M, Escoto-Chavez SE, Murdock B, Sakai T, Baritaki S, Bonavida B. Nitric oxide sensitizes tumor cells to TRAIL-induced apoptosis via inhibition of the DR5 transcription repressor Yin Yang 1. Nitric Oxide. 2009;20(1):39–52. doi: 10.1016/j.niox.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 101.Huber MA, Kraut N, Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr Opin Cell Biol. 2005;17(5):548–58. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 102.Kurrey NK, K A, Bapat SA. Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol Oncol. 2005;97(1):155–65. doi: 10.1016/j.ygyno.2004.12.043. [DOI] [PubMed] [Google Scholar]
- 103.Medici D, Hay ED, Olsen BR. Snail and Slug promote epithelial- mesenchymal transition through beta-catenin-T-cell factor-4-dependent expression of transforming growth factor-beta3. Mol Biol Cell. 2008;19(11):4875–87. doi: 10.1091/mbc.E08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117(7):927–39. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 105.Yie SM, Lou B, Ye SR, He X, Cao M, Xie K, Ye NY, Lin R, Wu SM, Xiao HB, Gao E. Clinical significance of detecting survivin-expressing circulating cancer cells in patients with non-small cell lung cancer. Lung Cancer. 2009;63:284–90. doi: 10.1016/j.lungcan.2008.05.024. [DOI] [PubMed] [Google Scholar]
- 106.Yang D, Schneider S, Azuma M, Iqbal S, El-Khoueiry A, Groshen S, Agafitei D, Danenberg KD, Danenberg PV, Ladner RD, Lenz HJ. Gene expression levels of epidermal growth factor receptor, survivin, and vascular endothelial growth factor as molecular markers of lymph node involvement in patients with locally advanced rectal cancer. Clin Colorectal Cancer. 2006;6(4):305–11. doi: 10.3816/CCC.2006.n.049. [DOI] [PubMed] [Google Scholar]
- 107.Ikehara M, Oshita F, Kameda Y, Ito H, Ohgane N, Suzuki R, Saito H, Yamada K, Noda K, Mitsuda A. Expression of survivin correlated with vessel invasion is a marker of poor prognosis in small adenocarcinoma of the lung. Oncol Rep. 2002;9(4):835–8. [PubMed] [Google Scholar]
- 108.Lo Muzio L, Staibano S, Pannone G, Mignogna MD, Mariggio A, Salvatore G, Chieffi P, Tramontano D, De Rosa G, Altieri DC. Expression of the apoptosis inhibitor survivin in aggressive squamous cell carcinoma. Exp Mol Pathol. 2001;70(3):249–54. doi: 10.1006/exmp.2001.2367. [DOI] [PubMed] [Google Scholar]
- 109.Ulf Henning Beier TG. Implications of galactocerebrosidase and galactosylcerebroside metabolism in cancer cells. Int J Cancer. 2005;115(1):6–10. doi: 10.1002/ijc.20851. [DOI] [PubMed] [Google Scholar]
- 110.Wang CC, Tsai MF, Hong TM, Chang GC, Chen CY, Yang WM, Chen JJ, Yang PC. The transcriptional factor YY1 upregulates the novel invasion suppressor HLJ1 expression and inhibits cancer cell invasion. Oncogene. 2005;24(25):4081–93. doi: 10.1038/sj.onc.1208573. [DOI] [PubMed] [Google Scholar]
- 111.Wang CC, Tsai MF, Dai TH, Hong TM, Chan WK, Chen JJ, Yang PC. Synergistic activation of the tumor suppressor, HLJ1, by the transcription factors YY1 and activator protein 1. Cancer Res. 2007;67(10):4816–26. doi: 10.1158/0008-5472.CAN-07-0504. [DOI] [PubMed] [Google Scholar]
- 112.Palmer MB, Majumder P, Cooper JC, Yoon H, Wade PA, Boss JM. Yin yang 1 regulates the expression of snail through a distal enhancer. Mol Cancer Res. 2009;7(2):221–9. doi: 10.1158/1541-7786.MCR-08-0229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Galloway NR, Aspe JR, Sellers C, Wall NR. Enhanced antitumor effect of combined gemcitabine and proton radiation in the treatment of pancreatic cancer. Pancreas. 2009;38(7):782–90. doi: 10.1097/MPA.0b013e3181a85999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baumeister P, Luo S, Skarnes WC, Sui G, Seto E, Shi Y, Lee AS. Endoplasmic reticulum stress induction of the Grp78/BiP promoter: activating mechanisms mediated by YY1 and its interactive chromatin modifiers. Mol Cell Biol. 2005;25(11):4529–40. doi: 10.1128/MCB.25.11.4529-4540.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gronroos E, Terentiev AA, Punga T, Ericsson J. YY1 inhibits the activation of the p53 tumor suppressor in response to genotoxic stress. Proc Natl Acad Sci U S A. 2004;101(33):12165–70. doi: 10.1073/pnas.0402283101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wan M, Huang W, Kute TE, Miller LD, Zhang Q, Hatcher H, Wang J, Stovall DB, Russell GB, Cao PD, Deng Z, Wang W, Zhang Q, Lei M, Torti SV, Akman SA, Sui G. Yin Yang 1 plays an essential role in breast cancer and negatively regulates p27. Am J Pathol. 2012;180(5):2120–33. doi: 10.1016/j.ajpath.2012.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]