Abstract
Mitofusins (Mfn) promote fusion-mediated mitochondrial content exchange and subcellular trafficking. Mutations in Mfn2 cause neurodegenerative Charcot Marie Tooth disease type 2A (CMT2A). Here we show that Mfn2 activity can be determined by Met376 and His380 interactions with Asp725 and Leu727 and controlled by PINK1 kinase-mediated phosphorylation of adjacent Mfn2 Ser378. Small molecule mimics of the peptide-peptide interface of Mfn2 disrupted this interaction, allosterically activating Mfn2 and promoting mitochondrial fusion. These first-in-class mitofusin agonists overcame dominant mitochondrial defects provoked in cultured neurons by CMT2A mutants Mfn2 Arg94Gln and Thr105Met, as evidenced by improved mitochondrial dysmotility, fragmentation, depolarization, and clumping. A mitofusin agonist normalized axonal mitochondrial trafficking within sciatic nerves of Mfn2 Thr105Met mice, promising a therapeutic approach for CMT2A and other untreatable diseases of impaired neuronal mitochondrial dynamism/trafficking.
Main Text
Mitochondria are organelles that generate a rich energy source for cells, and continuously traffic and undergo cellular fusion to exchange genomes and promote mutual repair (1). Mitochondrial fusion and subcellular trafficking are mediated in part by mitofusin (Mfn) 1 and mitofusion (Mfn) 2. Genetic mutations in Mfn2 that suppress mitochondrial fusion and motility cause Charcot Marie Tooth Disease 2A (CMT2A), the most common inheritable axonal neuropathy (2). Because no therapeutics exist that directly enhance mitochondrial fusion or trafficking, this disease is unrelenting and irreversible.
Computational modeling based on the closed structure of bacterial dynamin related protein (BDRP) and the more open structure of optic atrophy-1 suggested that Mfn2 can change conformation based how closely the first and second heptad repeat (HR) domains interact (fig. S1). A closed conformation is fusion-incompetent, whereas an open conformation favoring mitochondrial fusion can be induced by a competing peptide analogous to amino acids 367–384 within the Mfn2 HR1domain (3). We identified amino acids controlling these events, first by truncation analysis to define the smallest fusion-promoting minipeptide (Fig. 1A, 1B; residues 374–384), and then through functional interrogation of this minimal peptide using alanine (Ala) scanning. Substitution of Ala for Met376, Ser378, His380 and Met381 that are highly conserved across vertebrate species (fig. S2, S3), impaired minipeptide-stimulated mitochondrial fusion as measured by increased mitochondrial length/width (aspect ratio) (Fig. 1C). The structural model of human Mfn2 in a closed conformation based on homology with BDRP predicted a helical interaction between HR1 and HR2 domains, with alignment of Met376 and His380 side chains in the HR1 domain to Leu727 and Asp725 in the HR2 domain (fig. S1). This suggested that Met376 and His380 stabilize the Mfn2 HR1-HR2 interaction, potentially explaining their critical function as defined by minipeptide Ala scanning. By contrast, Ser378 was modeled extending from the non-interacting surface of the HR1 α-helix (fig. S1), implying a different mechanism for its involvement in mitochondrial fusion.
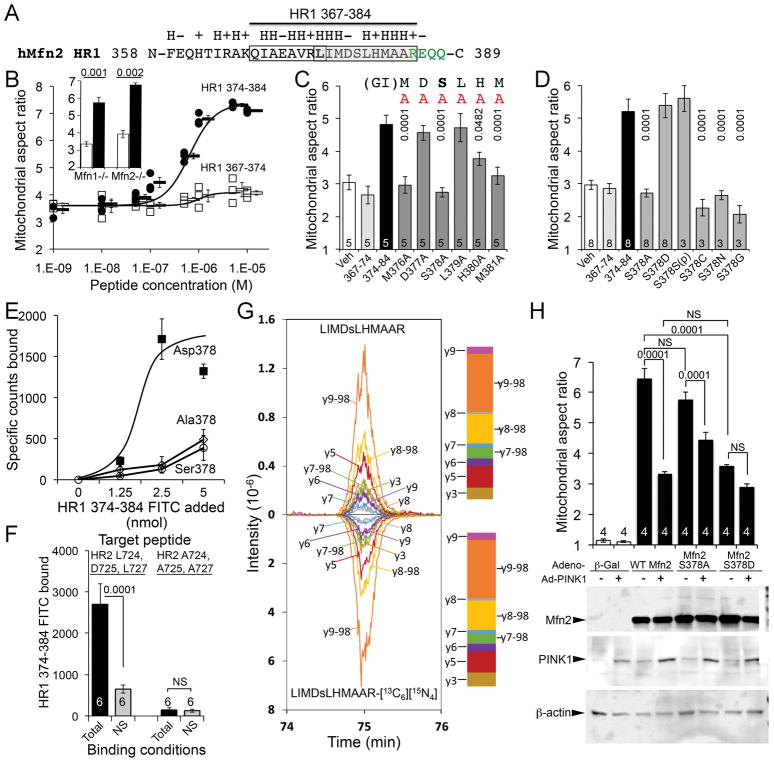
Fig. 1. Mfn2 Ser378 phosphorylation by PINK1 regulates mitochondrial fusion.
(A) Amino acid sequence surrounding fusion-promoting Mfn2 peptide. Side chain characteristics (H, hydrophobic; +, basic; −, acidic) are above. HR1 hinge region amino acids are green. Open box encloses N-terminal 367–374, and grey box encloses C-terminal 378–384 minipeptide. (B) Mitochondrial fusion stimulated by N- and C-terminal minipeptides. Aspect ratio is mitochondrial long axis/short axis. Inset: Fusion in Mfn1- or Mfn2-null MEFs. White elements are HR1 367–374 treated; black elements are HR1 374–384 treated. Data represent mean±SEM of 3 independent experiments; p values by Student’s t-test. (C) Alanine (A) scanning of minipeptide 374–384 fusion activity. Data represent mean±SEM of 3 or 8 independent experiments as indicated. (D) Ser378 substitution analysis of minipeptide 374–384 fusion activity in Mfn2 null MEFs. p values in D and E are vs parent minipeptide 374–384 (ANOVA). (E) Binding of minipeptides with Ser378 substitutions to HR2 target sequence. Data represent mean±SEM of 6 experimental replicates. (F) Binding of Asp378 minipeptide to HR2 target sequence before (left) and after (right) Ala substitution for putative interacting amino acids. Data represent mean±SEM of 6 experimental replicates. p values by Student’s t-test. (G) Ion chromatograms from assigned Mfn2 Ser378 phosphopeptide fragment ions after incubation with PINK1 kinase (top) and stable isotope-labeled synthetic counterpart (bottom); proportional intensities are in adjacent stack plots. The graph shows co-elution of the synthetic phosphopeptide with the PINK1 product, and the identity of the MS2 spectra, which is considered to be positive identification. (H) Mitochondrial fusion promoted by Mfn2 Ser378 mutants with and without PINK1 kinase; immunoblot of protein expression at bottom. Data represent mean±SEM of 4 independent experiments. p values were determined by ANOVA. NS - not significant.
To address whether Ser378 might be phosphorylated, we performed Ser378 substitutions in the minipeptide (to Ala, Cys, Asn or Gly), and found phosphorylation and fusion activity were abrogated. Functionality was restored by substituting with Asp to mimic phosphorylated Ser, or by inserting phospho-Ser [(p)Ser] itself (Fig. 1D, fig. S4). Moreover, in an in vitro binding assay devoid of cellular kinases the Asp378-substituted minipeptide bound to its putative HR2 interacting domain, whereas Ser378 and Ala378 minipeptides did not (Fig. 1E). Elimination of minipeptide binding by replacing HR2 Leu724, Asp725, and Leu727 with Ala confirmed the HR1-HR2 interaction model (Fig. 1F).
Nuclear magnetic resonance spectrometry of the minipeptides showed low conformational stability with a propensity to form helical structures. Ser378 phosphorylation reduced the peptide dynamics most visibly for residues Leu379 to Met381, potentially changing amino acid side chains presented to HR2 (fig. S5; Supplemental dataset 1, Supplemental Movie 1). Indeed, recombinant Mfn2 mutations that replaced Ser378 with Asp (mimicking Mfn2 Ser378 phosphorylation) or substituted Met376 or His380 with Ala (disrupting the putative HR1-HR2 interaction controlled by Ser378 phosphorylation) impaired Mfn2-stimulated mitochondrial fusion (fig. S6). By contrast, replacing Mfn2 Ser378 with Ala (to prevent its phosphorylation), or substitution of Ala for neighboring Val372 that was not critical for HR1-HR2 interactions, did not depress Mfn2-mediated fusion (fig. S6).
Mfn2 can be phosphorylated by mitochondrial PTEN-induced putative kinase 1 (PINK1) (4, 5). Targeted mass spectrometry demonstrated phosphorylation of Mfn2 Ser378 and previously reported Mfn2 Thr111 and Ser442 by PINK1 kinase (Fig. 1G; figs S7 and S8; table S1), but not by software-nominated G-protein receptor kinase 2 (fig. S9). We expressed Mfn2 Ser378 mutants with or without PINK1 in Mfn1/Mfn2-double deficient cells (denoted Mfn1−/−, Mfn2−/−). Fusion-defective mitochondria in these cells were abnormally short at baseline, but forced expression of wild-type Ser378 Mfn2 resulted in elongation from restoration of fusion (Fig. 1H, fig. S10). Co-expression of PINK1 with Mfn2, or mutational substitution of Mfn2 Ser378 to Asp (that mimics PINK1-mediated Ser378 phosphorylation), restrained Mfn2-stimulated elongation (Fig. 1H, fig. S10). By contrast, Mfn2 Ala378 (that cannot be phosphorylated) promoted mitochondrial fusion resistant to PINK1 suppression (Fig. 1H, fig. S10). The effects of Mfn2 Ser378 mutants were recapitulated in in vitro assays of fusion-mediated mitochondrial content exchange (fig. S11).
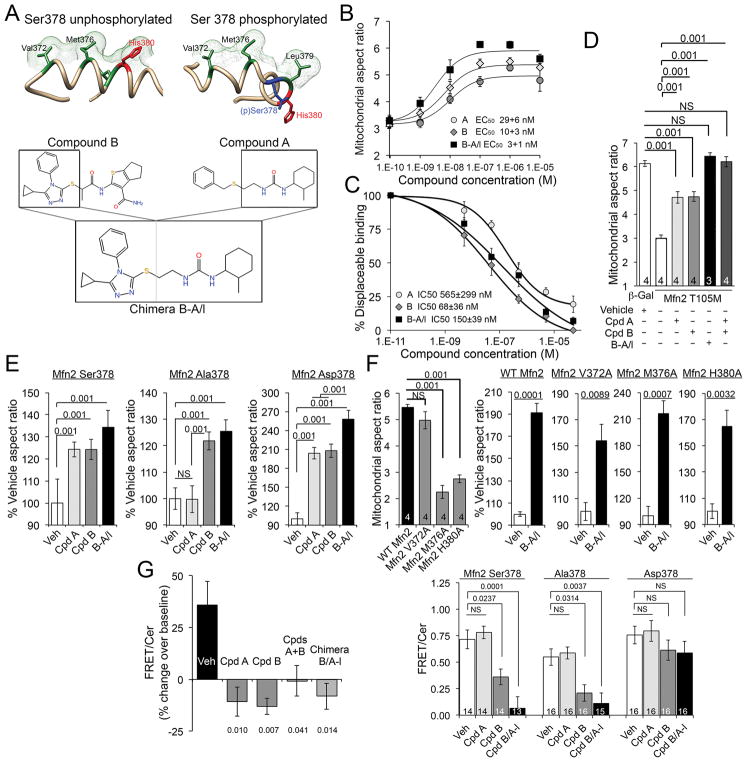
We assessed fusogenic activity of commercially available small molecule candidate pharmacophores (Supplemental dataset 2; Supplemental Materials and Methods), focusing on those having structures that mimicked Ser378 phosphorylated (class A) and unphosphorylated (class B) minipeptide amino acid side chains (fig. S12; Supplemental dataset 3). We reasoned that simultaneous application of class A and B agonists could enhance mitofusin agonism by acting on both Mfn2 Ser378 phosphorylated states. Indeed, lead compounds (Cpd) A and B acted synergistically to promote mitochondrial fusion (fig. S13; compare aspect ratios to fig. S12C). Therefore, we assimilated Cpd A and B functionality into a single molecule by creating Cpd A-B chimeras (Fig. 2A; fig. S14; Supplemental dataset 4). Chimera B-A/long (B-A/l) potently stimulated mitochondrial fusion in Mfn2-deficient cells (Fig. 2B), competed for minipeptide binding at the Mfn2 HR2 interaction site (Fig. 2C), and was as effective as the combination of Cpds A+B in reversing mitochondrial dysmorphology provoked by the fusion-defective CMT2A mutant, Mfn2 T105M (Fig. 2D). Fusogenic effects of Cpd A were specific for the Asp378 mutant of Mfn2 that mimicked Ser378 phosphorylation, whereas Cpd B and chimera B-A/l were non-selective for the phosphomimic Asp378 and non-phosphorylatable Ala378 mutants (Fig. 2E). Because they mimic WT Mfn2 HR1 sequence and interact with HR2, mitofusin agonists evoked fusion to a proportionally similar degree in mitochondria expressing mutants of HR1 that are fusion deficient (Fig. 2F; compare to Fig. 1H and fig. S6). Small molecule mitofusin agonists required endogenous Mfn1 or Mfn2 to promote mitochondrial fusion, exhibited no detectable promiscuous activity for structurally related dynamin, and did not compromise cell viability (fig. S15). Based on FRET analysis of Mfn2 labelled at the N and C termini (Supplemental Materials and Methods), mitofusin agonists promoted an open Mfn2 conformation favoring mitochondrial fusion (3) with a rank order paralleling that for HR2 binding and mitochondrial fusion (Fig. 2G; compare to Fig. 2B and 2C), supporting allosteric activation.
Fig. 2. Small molecule mimetics of Mfn2 HR1 amino acid side chains that interact with HR2 are mitofusin agonists.
(A) (top) Three dimensional representations of hypothetical minipeptide conformations driven by Ser378 phosphorylation, and (bottom) their respective small molecule mimetics. (B) Dose-dependent mitofusin agonism by small molecule agonists. Data represent mean±SEM of 6 independent experiments. (C) Displacement of minipeptide 374–384 from its HR2 binding site by mitofusin agonists. Data represent mean±SEM of 3 independent experiments. (D) Restoration of Mfn2 T105M-impaired mitochondrial fusion in Mfn2−/− MEFs by mitofusin agonists. Data represent mean ± SEM of 3 or 4 independent experiments as indicated. p values were measured by ANOVA. (E) Selectivity of a class A, but not a class B, mitofusin agonist for Ser378-phosphorylated Mfn2.. Data represent mean ± SEM of 4 independent experiments. p values were measured by ANOVA (F) Impaired basal function, but normal proportional agonist responsiveness, of Mfn2 mutations altering HR1-HR2 interacting amino acids. Absolute fusogenicity of these Mfn2 mutants is in fig. S6. Data represent mean±SEM of 4 independent experiments. p values are by ANOVA (left) or Student’s t test. (G) Change in FRET evoked by mitofusin agonists in isolated mitochondria (left) and intact cells (right); decreased FRET reflects conformational opening. Data represent mean±SEM of 3 independent (left) and 14–16 replicate (right) experiments. p values vs vehicle were measured by ANOVA.
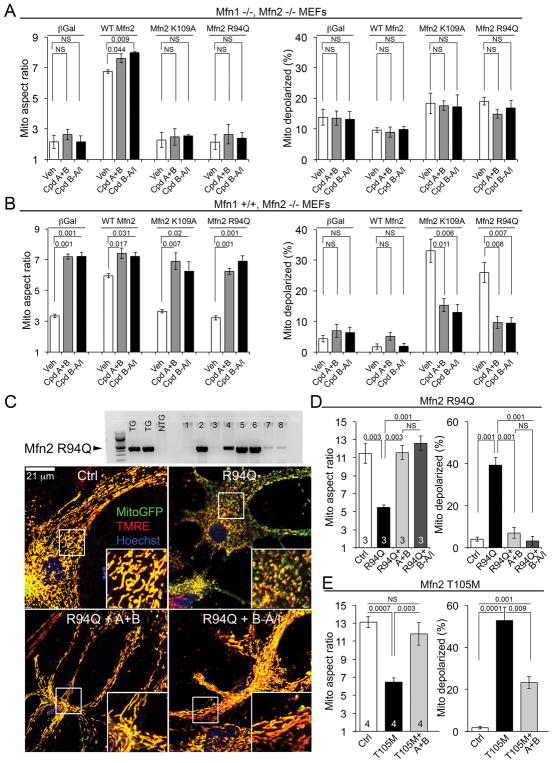
In CMT2A, Mfn2 mutants produce mitochondrial “fragmentation” (defined as decreased aspect ratio) and loss of normal membrane polarization through dominant inhibition of normal mitofusins. Experiments using Mfn1 or Mfn2-deficient murine embryonic fibroblasts (MEFs) showed that in the absence of normal mitofusins, small molecule mitofusin agonists did not improve mitochondria of cells expressing GTPase-crippled Mfn2 R94Q or K109A (Fig. 3A). However, mitofusin agonists corrected mitochondrial dysmorphology and reversed mitochondrial hypopolarization induced by these Mfn2 mutants when Mfn1 was present (Fig. 3B). Mitofusin agonists also reversed mitochondrial fragmentation and hypopolarization in cultured mouse neurons expressing (in addition to endogenous mitofusins) CMT2A mutants Mfn2 R94Q (Fig. 3C, 3D) or Mfn2 T105M (Fig. 3E). Thus, mitofusin agonists do not restore function of CMT2A Mfn2 GTPase domain mutants. Rather, by stabilizing the fusion-permissive open conformation of endogenous normal Mfn1 or Mfn2, mitofusin agonists can overcome dominant suppression of mitochondrial fusion by these disease-causing dysfunctional proteins.
Fig. 3. Mitofusin agonists correct mitochondrial damage induced by nonfunctioning Mfn2 mutants by activating endogenous mitofusins.
(A) Effects of mitofusin agonists in Mfn1−/−, Mfn2−/− cells expressing recombinant WT or mutant Mfn2. Data represent mean ± SEM of 3 independent experiments. p values were measured by ANOVA. (B) Same as (A) in Mfn1+/+, Mfn2−/− cells. Data represent mean ± SEM of 3 independent experiments. p values were measured by ANOVA. (C) (top) Immunoblot showing Mfn2R94Q expression in primary cultured neurons from individual mouse pups. (bottom) Representative confocal images of mitochondrial pathology in cultured neonatal mouse neurons expressing Mfn2 R94Q, and correction by mitofusin agonists. Scale bars are 21 μm; expanded views are from white squares. (D) Group data for mitochondrial aspect ratio and polarization studies in cultured neonatal mouse neurons expressing Mfn2 R94Q, as depicted in (C). Data represent mean ± SEM of 3 independent experiments. p values were measured by ANOVA. (E) Results of mitochondrial aspect ratio and polarization studies in cultured neonatal mouse neurons expressing Mfn2 T105M. Data represent mean ± SEM of 4 independent experiments. p values were measured by ANOVA.
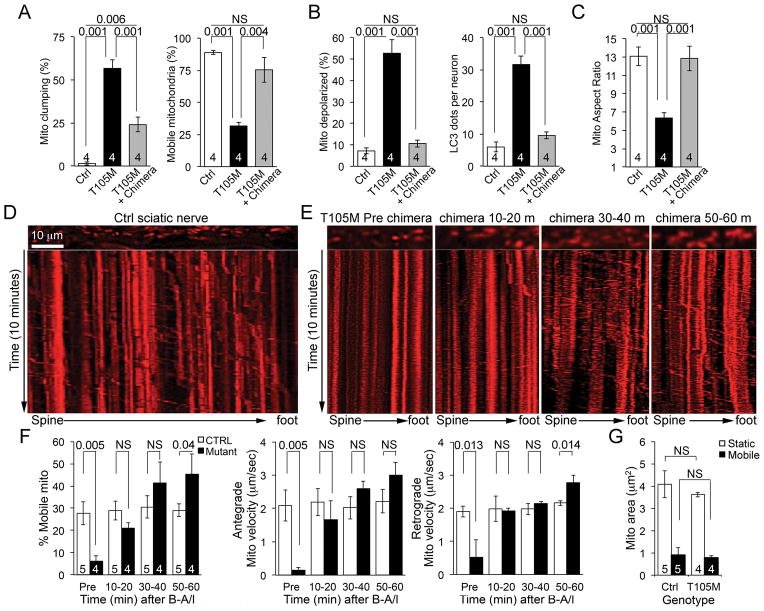
Clinical CMT2A classically affects long nerves innervating the lower and upper limbs (6, 7). It is unclear how a principal defect in mitochondrial fusion would cause length-dependent neuronal disease. Conversely, disruption of axonal mitochondrial trafficking (8) would be predicted to preferentially impact cells requiring mitochondrial transport over the greatest physical distance, such as the sciatic nerves originating in the spine and terminating in the foot. Mfn2 interacts with Miro/Milton to promote mitochondrial motility in neurons (9), so we tested the effects of mitofusin agonism on murine neuronal mitochondrial trafficking. Chimera B-A/l reversed mitochondrial “clumping” (formation of static mitochondrial aggregates) and restored mitochondrial motility in cultured mouse neurons expressing the CMT2A mutant Mfn2 T105M (Fig. 4A; Supplemental movie 2). Mitochondrial hypopolarization and increased autophagy (Fig. 4B, fig. S16) and mitochondrial dysmorphology (Fig. 4C, fig. S16) were concomitantly improved. Thus, a small molecule mitofusin agonist enhanced organelle and cell fitness in CMT2A neurons by promoting mitochondrial fusion and subcellular transport.
Fig. 4. A mitofusin agonist restores axonal mitochondrial trafficking suppressed by CMT2A mutant Mfn2 T105M.
(A–C) Chimera B-A/l effects on mitochondrial mobility (A), function (B), and morphology (C) in cultured CMT2A Mfn2 T105M-expressing mouse neurons. Data represent mean ± SEM of 4 independent experiments. p values were measured by ANOVA. (D) Representative kymograph of mitochondrial trafficking in a Ctrl mouse sciatic nerve. Scale bar is 10μm. (E) Representative serial kymographs of mitochondria in a Mfn2 T105M mouse sciatic nerve before and after chimera B-A/l. (F) Quantitative data for sciatic nerve mitochondrial motility studies. (G) Size of motile and static mitochondria in Ctrl and B-A/l-treated (60 minutes) sciatic nerves. Data in (F) and (G) represent mean ± SEM of 4 or 5 independent experiments. p values were measured by ANOVA.
We evaluated the concept of activating mitofusins to stimulate in vivo axonal mitochondrial trafficking in sciatic nerves of mice expressing the CMT2A mutant Mfn2 T105M. In normal sciatic nerves ~30% of axonal mitochondria exhibited robust bidirectional transport (Fig. 4D; Supplemental movie 3). Mitochondria of Mfn2 T105M sciatic nerves were severely hypomotile (Fig. 4E, Supplemental movie 4), but application of chimera B-A/l to Mfn2 T105M sciatic nerves restored mitochondrial motility to within normal levels (Fig. 4F, Supplemental movie 4). Mobile mitochondria in wild-type and B-A/l-treated Mfn2 T105M axons were smaller (Fig. 4G), supporting in vitro observations discriminating between Mfn2-mediated mitochondrial dysmotility and defective fusion in CMT2A (10).
Here, we found that PINK1 phosphorylation of Mfn2 at Ser378 can alter the positions of Met376 and His380 (in the HR1 domain) that normally interact with HR2 domain amino acids to orchestrate Mfn2 toggling between conformations that modulate mitochondrial fusion. These findings establish a mechanistic basis for clinical observations that Mfn2 Met376 mutations to Ile, Thr and Val could cause CMT2A (7, 11, 12).
Based on in silico pharmacophore modeling complemented by functional interrogation of Mfn2 HR1 domain-derived minipeptides we developed a novel small molecule mitofusin agonist that reversed neuronal mitochondrial dysmorphometry and impaired mobility evoked by 2 CMT2A Mfn2 mutants. CMT2A is the prototypical clinical disorder of defective mitochondrial fusion, but impaired mitochondrial trafficking may play as great a role as mitochondrial fragmentation in CMT2A axonal degeneration (8–10). Individuals with CMT2A express one mutant Mfn2 allele in combination with one normal Mfn2 allele, and harbor two normal Mfn1 alleles (13). It is therefore possible that a therapeutic substrate exists for Mfn agonists to “supercharge” normal mitofusins and overcome dominant inhibition by Mfn2 mutants. Our observation that in vivo mitochondrial dysmotility provoked by a CMT2A mutant can be normalized by mitofusin agonism mechanistically links abnormal mitochondrial trafficking in experimental CMT2A to Mfn2 dysfunction. Mitofusin agonists may also have therapeutic potential for neurological conditions other than CMT2A, such as Alzheimer’s, Parkinsons’ and Huntington’s diseases, wherein mitochondrial dysmotility and fragmentation are contributing factors (14–16).
Supplementary Material
NMR assignments, shift peaks, and constraints for Ser378 phosphorylated and unphosphorylated minipeptide studies.
Fusogenicity screening results, characteristics, and commercial sourcing of 55 candidate mitofusin agonists.
Characteristics and commercial sourcing of 12 (6 each) class A and class B mitofusin agonists.
Characteristics and sourcing of 4 novel newly synthesized chimeric classA/B mitofusin agonists.
Hypothetical HR1 MP374–384 conformation before and after S378 phosphorylation.
In vitro mouse mitochondrial mobility in Ctrl neuron, Mfn2 T105M neuron, and Mfn2 T105M neuron treated with compounds A+B (24 hours).
Mitochondrial mobility in a neuronal axon of a control mouse sciatic nerve.
Mitochondrial mobility in axons of a Mfn2 T105M mouse sciatic nerve before and at serial 15 minute periods after application of chimera B-A/l.
Acknowledgments
We gratefully acknowledge invaluable discussions with Dr. P. Needleman and the specialized assistance of L. Zhang, P. Erdmann-Gilmore, Yiling Mi, and R. Connors.
Funding: Supported by NIH R35HL135736 (GWD) and R01HL128071 (RNK and GWD), a McDonnell Center for Cellular and Molecular Neurobiology Postdoctoral Fellowship (AF), and the Washington University Proteomics Shared Resource supported by NCATS UL1TR000448, NIGMSP41, GM103422 and NCIP30 CA091842. G.W.D. is the Philip and Sima K. Needleman-endowed Professor.
Footnotes
Author contributions: G.W.D., A.M.K, R.R.T., E.G, conceived of or designed the research. G.W.D. wrote the manuscript. J.M.R. and R.R.T. performed phospho-protein mass spectroscopy analyses. A.M.K performed peptide NMR studies. N.B, E.Z., E.G. performed in silico pharmacophore modeling. A.G.R. screened compounds for activity and characterized agonists. A.G.R., A.F., J.M.A., W.C.K. performed mitochondrial studies. A.F. performed cultured neuron and ex vivo sciatic nerve studies. J.W.J. analyzed and purified compounds. R.N.K. and R.H.B. provided essential reagents.
Data and materials availability: All data are available in the manuscript or the supplementary material. There are no MTAs associated with this study.
Competing interests: D.M.R. and G.W.D. are inventors on patent application 15/710,696 submitted by Stanford University that covers the use of peptide regulators of mitochondrial fusion and small molecule peptidomimetcs derived from them. G.W.D. is an inventor on provisional patent applications 62/488,787 and 62/584,515 submitted by Washington University that cover the use of novel small molecule mitofusin agonists to treat chronic neurodegenerative diseases. E.G., R.N.K., N.B., and E.Z. are inventors on patent application 62/573,217 submitted by Albert Einstein College of Medicine that covers compositions of mitofusin agonists and their uses for the treatment of diseases and disorders. D.M.R. is the founder of Mitoconix Bio Ltd., a company focused on improving mitochondrial health as a therapeutic approach for neurodegenerative diseases. None of the research conducted in D.M.R.’s laboratory is supported by or in collaboration with Mitoconix. The other authors declare no competing interests.
References and Notes
- 1.Chan DC. Fusion and fission: interlinked processes critical for mitochondrial health. Annu Rev Genet. 2012;46:265–287. doi: 10.1146/annurev-genet-110410-132529. [DOI] [PubMed] [Google Scholar]
- 2.Liesa M, Palacin M, Zorzano A. Mitochondrial dynamics in mammalian health and disease. Physiol Rev. 2009;89:799–845. doi: 10.1152/physrev.00030.2008. [DOI] [PubMed] [Google Scholar]
- 3.Franco A, et al. Correcting mitochondrial fusion by manipulating mitofusin conformations. Nature. 2016;540:74–79. doi: 10.1038/nature20156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Y, Dorn GW., 2nd PINK1-phosphorylated mitofusin 2 is a Parkin receptor for culling damaged mitochondria. Science. 2013;340:471–475. doi: 10.1126/science.1231031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong G, et al. Parkin-mediated mitophagy directs perinatal cardiac metabolic maturation in mice. Science. 2015;350:aad2459. doi: 10.1126/science.aad2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lawson VH, Graham BV, Flanigan KM. Clinical and electrophysiologic features of CMT2A with mutations in the mitofusin 2 gene. Neurology. 2005;65:197–204. doi: 10.1212/01.wnl.0000168898.76071.70. [DOI] [PubMed] [Google Scholar]
- 7.Verhoeven K, et al. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain. 2006;129:2093–2102. doi: 10.1093/brain/awl126. [DOI] [PubMed] [Google Scholar]
- 8.Baloh RH, Schmidt RE, Pestronk A, Milbrandt J. Altered axonal mitochondrial transport in the pathogenesis of Charcot-Marie-Tooth disease from mitofusin 2 mutations. J Neurosci. 2007;27:422–430. doi: 10.1523/JNEUROSCI.4798-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misko A, Jiang S, Wegorzewska I, Milbrandt J, Baloh RH. Mitofusin 2 is necessary for transport of axonal mitochondria and interacts with the Miro/Milton complex. J Neurosci. 2010;30:4232–4240. doi: 10.1523/JNEUROSCI.6248-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misko AL, Sasaki Y, Tuck E, Milbrandt J, Baloh RH. Mitofusin2 mutations disrupt axonal mitochondrial positioning and promote axon degeneration. J Neurosci. 2012;32:4145–4155. doi: 10.1523/JNEUROSCI.6338-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chung KW, et al. Early onset severe and late-onset mild Charcot-Marie-Tooth disease with mitofusin 2 (MFN2) mutations. Brain. 2006;129:2103–2118. doi: 10.1093/brain/awl174. [DOI] [PubMed] [Google Scholar]
- 12.Casasnovas C, et al. Phenotypic spectrum of MFN2 mutations in the Spanish population. J Med Genet. 2010;47:249–256. doi: 10.1136/jmg.2009.072488. [DOI] [PubMed] [Google Scholar]
- 13.Bombelli F, et al. Charcot-Marie-Tooth disease type 2A: from typical to rare phenotypic and genotypic features. JAMA Neurol. 2014;71:1036–1042. doi: 10.1001/jamaneurol.2014.629. [DOI] [PubMed] [Google Scholar]
- 14.Hirai K, et al. Mitochondrial abnormalities in Alzheimer’s disease. J Neurosci. 2001;21:3017–3023. doi: 10.1523/JNEUROSCI.21-09-03017.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Winklhofer KF, Haass C. Mitochondrial dysfunction in Parkinson’s disease. Biochim Biophys Acta. 2010;1802:29–44. doi: 10.1016/j.bbadis.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, et al. Mitochondrial loss, dysfunction and altered dynamics in Huntington’s disease. Hum Mol Genet. 2010;19:3919–3935. doi: 10.1093/hmg/ddq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, et al. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Low HH, Low J. A bacterial dynamin-like protein. Nature. 2006;444:766–769. doi: 10.1038/nature05312. [DOI] [PubMed] [Google Scholar]
- 20.Qi Y, et al. Structures of human mitofusin 1 provide insight into mitochondrial tethering. J Cell Biol. 2016;215:621–629. doi: 10.1083/jcb.201609019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan L, et al. Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission. J Mol Cell Biol. 2011;3:378–381. doi: 10.1093/jmcb/mjr032. [DOI] [PubMed] [Google Scholar]
- 22.Ford MG, Jenni S, Nunnari J. The crystal structure of dynamin. Nature. 2011;477:561–566. doi: 10.1038/nature10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fröhlich C, et al. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goujon C, Greenbury RA, Papaioannou S, Doyle T, Malim MH. A triple-arginine motif in the amino-terminal domain and oligomerization are required for HIV-1 inhibition by human MX2. J Virol. 2015;89:4676–4680. doi: 10.1128/JVI.00169-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 26.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fiesel FC, Hudec R, Springer W. Non-radioactive in vitro PINK1 kinase assays using ubiquitin or Parkin as substrate. Bio Protoc. 2016;6 doi: 10.21769/BioProtoc.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobieski C, Jiang X, Crawford DC, Mennerick S. Loss of local astrocyte support disrupts action potential propagation and glutamate release synchrony from unmyelinated hippocampal axon terminals in vitro. J Neurosci. 2015;35:11105–11117. doi: 10.1523/JNEUROSCI.1289-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bannerman P, Burns T, Xu J, Miers L, Pleasure D. Mice hemizygous for a pathogenic Mitofusin-2 allele exhibit hindlimb/foot gait deficits and phenotypic perturbations in nerve and muscle. PLoS One. 2016;11:e0167573. doi: 10.1371/journal.pone.0167573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang X, Arber S, Willian C, Li L, Tanabe Y, Jessell TM, Birchmeier C, Burden SJ. Patterning of muscle acetylcholine receptor gene expression in the absence of motor innervation. Neuron. 2001;30:399–410. doi: 10.1016/s0896-6273(01)00287-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
NMR assignments, shift peaks, and constraints for Ser378 phosphorylated and unphosphorylated minipeptide studies.
Fusogenicity screening results, characteristics, and commercial sourcing of 55 candidate mitofusin agonists.
Characteristics and commercial sourcing of 12 (6 each) class A and class B mitofusin agonists.
Characteristics and sourcing of 4 novel newly synthesized chimeric classA/B mitofusin agonists.
Hypothetical HR1 MP374–384 conformation before and after S378 phosphorylation.
In vitro mouse mitochondrial mobility in Ctrl neuron, Mfn2 T105M neuron, and Mfn2 T105M neuron treated with compounds A+B (24 hours).
Mitochondrial mobility in a neuronal axon of a control mouse sciatic nerve.
Mitochondrial mobility in axons of a Mfn2 T105M mouse sciatic nerve before and at serial 15 minute periods after application of chimera B-A/l.