Abstract
Exposure to low concentrations of antibiotics found in aquatic environments can increase susceptibility to infection in adult fish due to microbiome disruption. However, little is known regarding the effect of antibiotic pollution on fish larvae. Here, we show that exposure to streptomycin, a common antibiotic used in medicine and aquaculture, disrupts the normal composition of zebrafish larvae microbiomes, significantly reducing the microbial diversity found in the fish. Exposure to streptomycin also significantly increased early mortality among fish larvae, causing full mortality within a few days of exposure at 10 μg/mL. Finally, we found that subclinical concentrations of streptomycin also increased the abundance of class 1 integrons, an integrase-dependent genetic system associated to the horizontal transfer of antibiotic resistance genes, in the larvae microbiomes. These results suggest that even low concentrations of streptomycin associated with environmental pollution could impact fish populations and lead to the creation of antibiotic resistance reservoirs.
Keywords: antimicrobial, pollution, 16S rRNA, metagenomic, Danio rerio
Low concentrations of streptomycin found in aquatic environments disrupt the microbiomes of zebrafish larvae, increasing early mortality and favoring the spread of antibiotic resistance markers.
INTRODUCTION
The global use of antibiotics increased steadily over the past decades not only in human medicine but also in other sectors of commercial activities (Klein et al.2018). For example, antibiotic consumption in livestock reached 63,151 tons in 2010 and is predicted to increase by 67% by 2030 (Van Boeckel et al.2015). Antibiotic use is also rising in aquaculture as intensive farming expands as the fastest growing food sector worldwide (Henriksson et al.2017). Consequently, antimicrobials of pharmaceutical origin can be found in terrestrial, freshwater and marine environments (Boy-Roura et al.2018). Antibiotic residues can even be found in atmospheric particulates near urban areas (Ferrey et al.2018), contributing to the burden of antibiotic pollution (Martinez 2009).
In addition to agricultural (Li et al.2018) and aquaculture runoffs (Henriksson et al.2017), antibiotics are released into the environment through discharge of human sewage (Kostich, Batt and Lazorchak 2014) or industrial discharges (Bielen et al.2017). Concentrations of antimicrobials found in the environment vary depending on the source, the antibiotic's persistence and the location affected. For example, antibiotic concentration found in treated sewage effluents can range from a few nanograms per liter for common antibiotics such as streptomycin (Kostich, Batt and Lazorchak 2014) to orders of magnitude greater than therapeutic concentrations in effluents affected by industrial plants (Cardoso, Porcher and Sanchez 2014; Larsson 2014). Even though concentrations of antimicrobials found in the environment usually do not exceed therapeutic dose (Kostich, Batt and Lazorchak 2014; Kulkarni et al.2017), low concentrations of antibiotics can still lead to the evolution of resistant bacteria (Gullberg et al.2011; Sandegren 2014). Indeed, antibiotic-resistant bacteria and genetic elements linked to antibiotic resistance are often detected in effluents near water treatment facilities, hospitals, pharmaceutical plants or more generally near urban areas (Gaze et al.2011; Czekalski, Gascon Diez and Burgmann 2014). Unregulated antibiotic pollution in streams and rivers around the world is now considered to be an important driver of multidrug-resistance evolution (Wellington et al.2013; Bengtsson-Palme, Kristiansson and Larsson 2018). Therefore, it is argued that future action plans to combat antibiotic resistance should consider the fate of antibiotics in the environment (Larsson 2014; Rosi-Marshall and Kelly 2015).
Antibiotic pollution in aquatic environments can also impact on different trophic levels in local ecosystems. For one, antibiotics can alter local microbial populations naturally present in sediments or water tables, disrupting phylogenetic structuration and nutrient cycling (Ding and He 2010; Martínez 2017; Grenni, Ancona and Barra Caracciolo 2018). Antibiotics inhibiting bacterial protein synthesis such as streptomycin can also exhibit toxic effects against unicellular algae, an important driver of aquatic ecosystem productivity (Fu et al.2017). Similarly, low concentrations of antibiotics were shown to impact on the survival and behaviors of micro-invertebrates such as Daphnia magna (Flaherty and Dodson 2005) and macro-fauna such as zebrafish embryos (Bielen et al.2017), via changes in gene transcription and expression (Zhang et al.2016).
Finally, antibiotics can also affect microbial populations associated with animal hosts. Such microbial populations carry different functional roles for the hosts ranging from efficient nutrient metabolism to promoting bone formation (Yan et al.2016; Raymann and Moran 2018). The disruption of a host microbiome, known as dysbiosis, can therefore lead to important health consequences (Lynch and Pedersen 2016). For example, streptomycin, an antibiotic commonly used in human medicine and agriculture, interferes with the intestinal microbiomes, leading to increased susceptibility to enteric infection (Sekirov et al.2008; Schubert, Sinani and Schloss 2015) and modifying lymphocytes expression (Bazett, Bergeron and Haston 2016) in murine model systems. Although the role of microbiomes in fish is still relatively unexplored (Austin and Al-Zahrani 1988; Navarrete et al.2008), recent studies showed that antibiotics administered as prophylactic in aquaculture reduced microbial diversity in fish gut (Navarrete et al.2008), and also increased susceptibility to pathogens (Schmidt et al.2017). Increased susceptibility to pathogens was also observed in adult zebrafish chronically exposed to low concentrations or streptomycin, leading to increased mortality (Zhou et al.2018).
Here, we look at the impacts of low concentrations of streptomycin on the microbiomes of zebrafish larvae. Streptomycin is extensively used in human medicine, agriculture and aquaculture and thus is an important source of antibiotic pollution. Fish larvae are especially susceptible to chemical toxicants (Zhang et al.2015) and are commonly used to screen for products toxicity in the environment (Ali, van Mil and Richardson 2011). Using this experimental model system, we investigated whether exposure to low concentrations of the antibiotic lead to dysbiosis and early mortality in the fish larvae. We also studied whether streptomycin favored the spread of integron 1, an integrase-based genetic element that allows the capture and expression of exogenous genes often associated to antibiotic resistance genes (Gillings 2014).
MATERIALS AND METHODS
Zebrafish husbandry and maintenance
Zebrafish (Danio rerio) were maintained in the laboratories of the Biology Department of Bard College in accordance with standard protocols for zebrafish husbandry (Lawrence and Mason 2012). Experimental populations of zebrafish strain Et20 were raised in a 14-h light:10-h dark cycle in standard recirculating rack water kept at 28.5°C with pH ranging from 7.0 to 7.4. Strain Et20 resulted from multiple outcrossing with wildtype strains in our laboratories and present no phenotypic differences, except for the GFP staining of mantle cells. At 0 days post-fertilization—or dpf, eggs from a single mating were bleached twice in 0.5% hypochlorite solution for 4 min and once in sterile 1 × E3 media (5 mM NaCl, 0.17 mM KCl, 0.33 mM CaCl2, 0.33 mM MgSO4 and 0.5mg/L methylene blue). We then placed 20–30 eggs in sterile petri dishes filled with 60 mL of 1 × E3 media. Every day, 80% of the media was exchanged for fresh media to minimize contamination due to dead cell tissue.
Following larval hatching at five (dfp), we established a control population by randomly picking 24 healthy embryos and allocating each individual in the well of a 24-well plate filled with 2 mL of sterile 1 × E3 media. We then established experimental populations to be exposed to streptomycin by randomly picking and allocating a total of 72 healthy embryos among the cells of three 24-well plates. Each plate was supplemented with either one of the following streptomycin concentrations (Streptomycin sulfate salt, Sigma-Aldrich, St-Louis, MO): 0.1, 1.0, 10.0 μg/mL. Even though streptomycin can degrade in the environment under the effect of light and temperature among other things, all three streptomycin concentrations used in this were previously reported in environmental surveys (Shen et al.2017). Again, approximately 80% of media was exchanged daily for sterile 1 × E3 media with the relevant antibiotic concentration when applicable. Streptomycin solution was prepared and stored at −10°C as per manufacturer protocols (Sigma-Aldrich, St. Louis, MO) before use. Fishes were fed generously once daily with micro-powder food containing rotifers and Paramecium as well as one drop of Roti Feast (Reed Mariculture, CA). We maintained the fishes in experimental conditions for 10 days and measured survival daily by gently poking immobile embryos with a sterile pipette tip to confirm the absence of movement.
At the end of the experiment, five embryos from each treatment were anesthetized on ice for 10 min and sacrificed by overdose of sterile tricaine methane sulfonate (250 mg/l) according to established euthanasia techniques (Matthews and Varga 2012). Only live fishes or fishes that had died within the last 18 h were chosen for downstream application. Fish were removed from tricaine following cessation of opercular movement (∼10 min) and washed three times with nuclease free water to minimize the presence of free-living bacteria. All experimental procedures were approved by the Bard Institutional Animal Care and use Committee (IACUC; most recent approval ID ‘Perron 2018’).
DNA extraction and processing
We extracted and purified microbial DNA from each sacrificed fish larvae using a modified protocol for the DNeasy Blood and Tissue Kit (QIAGEN, Germantown, MD) first described in Hang et al. (2014) and as implemented in Dahan et al. (2018). When necessary, gDNA concentrations were increased using the SpeedVac System (ThermoFisher Scientific, Asheville, NC). Purified gDNA samples were stored in nuclease free H2O at −20°C.
Fish microbiomes were characterized via targeted gene amplification of the 16S rRNA V4 region using Golay-barcoded primers 515F and 806R (Caporaso et al.2012). PCR conditions were previously described (Dahan et al.2018). Following gel-purification, libraries were pooled at equimolar ratios, and sequenced on the MiSeq paired-end Illumina platform adapted for 250-bp paired-end reads (Wright Labs, Juniata College, Huntingdon, PA). All sequence reads are available at the Sequence Read Archive of the NCBI (accession number SRP139123).
Processing of 16S rRNA sequence data
Before processing the 16S rRNA sequence data, phiX control reads were removed by mapping raw sequence reads against an indexed phiX genome provided by Illumina (San Diego, CA). Sequence reads were then processed, aligned and categorized independently using the DADA2 pipeline version 1.6 (Callahan et al.2016) available at (https://github.com/benjjneb/dada2) and implemented in R version 3.2.3 (http://www.r-project.org).
The DADA2 pipeline characterizes microbial communities by identifying unique amplicon sequence variants (ASVs) within the 16S rRNA reads (Callahan, McMurdie and Holmes 2017). In brief, forward and reverse reads are first filtered using the pipeline's recommended parameters. Filtered reads are then de-replicated (i.e. combining identical reads into unique sequences with a consensus quality profile) and de-noised (i.e. inferring and removing errors from samples) using DADA2’s default parameters. After building the ASV table and removing chimeras, taxonomy was assigned using the ‘assignTaxonomy’ function of DADA2 trained against the SILVA ribosomal RNA gene database version 132 (Quast et al.2013; Koo et al.2017). A multiple alignment of all the ASVs and maximum likelihood phylogenetic tree was built using the phangorn package version 2.1.3 (Schliep 2011). Lastly, we removed sequences positively identified as either zebrafish mitochondrial DNA or chloroplast DNA.
Data visualization and statistical analyses of 16S rRNA sequence data
Patterns of diversity within the ASV tables were analyzed using a modified version of the pipeline described in Dahan et al. (2018) implemented in R and visualized in ggplot2 (Wickham 2016). A mapping file linking sample names and streptomycin concentration is provided in Supporting Information. Briefly, we identified the core microbiome among control zebrafish using the pipeline described in Dahan et al. (2018). Then, using phyloseq version 1.14.0 (available at https://joey711.github.io/phyloseq/), performed Principle Coordinate Analyses (PCoA) on unweighted UniFrac distance scores (Lozupone and Knight 2005) to identify differences in community composition between samples. We conducted the analyses first by comparing control microbiomes to all microbiomes exposed to different concentrations of streptomycin and then by considering each concentration individually. We then tested the effect of streptomycin as a continuous variable on using a permutational analysis of variance using the adonis function (Oksanen et al.2007) of vegan version 2.3.2. To confirm multivariate homogeneity of variances among and within treatments, we used vegan's package implementation of PERMDISP2 via the betadisper method (Oksanen et al.2007).
We then analyzed community composition using phyloseq and DESeq2 differential abundance comparison (Love, Huber and Anders 2014) adapted for use with microbial count data (McMurdie and Holmes 2014). Core microbiomes were estimated using a custom pipeline (Dahan et al.2018). Finally, we estimated alpha diversity metrics using phyloseq's estimate_richness function. More specifically, we estimated richness S, i.e. observed number of ASVs, Shannon diversity measurements (H΄), as well as Pielou's evenness index (Pielou 1966) from the ASV table subsampled to the lowest sampling depth of 18,477 paired-reads. We then tested whether diversity indices changed with streptomycin concentration using linear modeling and comparing the different statistical models with Akaike's Information Criterion as implemented in R’s stats package.
qPCR of 16S rRNA and int1 genes
To quantify the relative abundance of integron 1, we estimated the abundance of int1 gene, a genetic marker of integron 1 commonly used as an indicator of antibiotic pollution (Gillings et al.2015), in relation to the abundance of 16S rRNA genes found in each sample. We used quantitative real-time PCR conditions as described in Gaze et al. (2011). For each sample, triplicate PCR reactions using the PowerUp SYBR Green Master Mix (Applied Biosystems, Foster City, CA) were cycled using the Bio-Rad CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules, CA). An internal standard curve constructed from a serial dilution of Escherichia coli SK4903 harboring seven 16S rRNA copies and on average six int1 copies was processed with each qPCR run. Strain SK4903 contains plasmid R751, which belong to the IncP1 family, known to be one of the most stably maintained plasmid (Grinter 1984; Adamczyk and Jagura-Burdzy 2003). The relative abundance of int1 genes was normalized to 16S rRNA copies of each sample before being analyzed via linear modeling with int1 relative abundance as the response variable and streptomycin concentrations as the explanatory variable. Statistical analysis and model assumption testing were conducted in R and visualized in ggplot2 (Wickham 2016).
RESULTS
Processing 16S rRNA V4 reads using DADA2
Overall, we sequenced the V4 region of the 16S rRNA genes to characterize the microbiomes of 10 individuals from the control group and five individuals from each of the three streptomycin treatments (i.e. 0.1 μg/mL, 1.0 μg/mL and 10.0 μg/mL) for a total of 25 individuals. We obtained a total of 3487,592 pairs of forward and reverse reads with an average read length of 250 base pairs, totaling ∼1.7 G bases and a median sequencing depth per sample of 151,631 paired-reads. After removing reads mapping as phiX and trimming forward reads at 240 bp and reverse reads at 160 bp, we retained a total of 6078,045 (87.71% of initial) reads. Finally, following de-replication, de-noising and removing predicted chimeric reads, we were left with 2662,658 (76.34% of the initial) paired-reads.
Using the RDP classifier, we identified a total of 344 unique ASVs among the 25 sampled zebrafish larvae. When looking specifically at the larvae from the control group, we found that 12 ASVs were shared amongst every individual (Table 1), suggesting a high instability of the microbiome at early stage of development in zebrafish. As expected from previous work (Roeselers et al.2011; Stephens et al.2016), we found taxa predominantly belonging to the phyla Proteobacteria and Bacteroidetes. Among the most common genera found in the core microbiome of control fishes were genera Flectobacillus sp., 39.81(14.1)%, and Fluviicola sp., 5.88(2.80)%, two Bacteroidetes that were previously associated with zebrafish (Davis et al.2016; Falcinelli et al.2016).
Table 1.
Relative abundance of core phyla and genera in control zebrafish larvae.
| Phylum | Mean % | s.d |
|---|---|---|
| Genus | ||
| Proteobacteria | 48.82 | 12.39 |
| Rhodovarius | 5.75 | 0.55 |
| Bosea | 0.18 | 0.16 |
| Phenylobacterium | 0.37 | 0.28 |
| Shinella | 2.86 | 2.86 |
| Pseudomonas | 0.75 | 0.91 |
| Vibrio | 0.12 | 0.13 |
| Pelomonas | 22.05 | 14.78 |
| Xylophilus | 18.50 | 24.69 |
| Pseudacidovorax | 3.41 | 5.59 |
| Bacteroidetes | 51.18 | 12.39 |
| Fluviicola | 5.88 | 2.80 |
| Chryseobacterium | 5.50 | 5.86 |
| Flectobacillus | 39.81 | 14.10 |
Streptomycin exposure alters microbial community composition
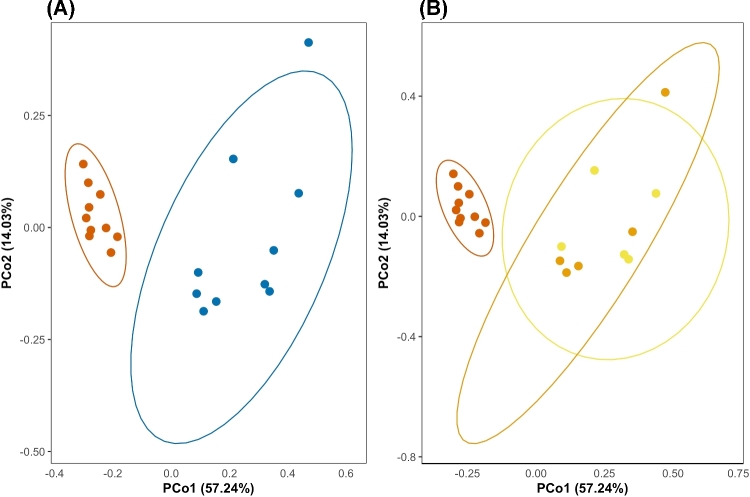
We first tested whether overall streptomycin exposure resulted in changes in the overall microbial community composition of zebrafish larvae. Using PCoA on unweighted UniFrac distances, we found that the two highest-ranked dimensions explained 57.24% and 14.03% of variance respectively, and that microbiomes exposed to any concentration of streptomycin clustered away from control microbiomes (ADONIS; R2 = 0.59; PERM = 999; adj-P = 0.006; Fig. 1A). We found that streptomycin explained less variance when concentration was treated individually (ADONIS; R2 = 0.31; PERM = 999; adj-P = 0.003; Fig. 1B), suggesting that UniFrac scores converged for microbiomes exposed to any concentration of streptomycin when compared to control fishes. We found a small difference in variance homogeneity when comparing controls to streptomycin combined together (F(1,18) = 16.99; adj-P > 0.01), but no differences in sample distances to their treatment centroids when treating streptomycin concentrations individually (F(2,17) = 4.42; adj-P > 0.05).
Figure 1.
Principle coordinates analysis (PCoA) of zebrafish microbiota exposed to different streptomycin concentrations. (A) PCoA of unweighted UniFrac scores shows significant dissimilarity between control microbiomes and microbiomes exposed to streptomycin (ADONIS; R2 = 0.59; PERM = 999; adj-P = 0.006). Control microbiomes are shown in vermilion while microbiomes exposed to streptomycin are in blue. (B) PCoA of unweighted UniFrac scores shows weaker dissimilarity among treatments when streptomycin is treated as a continuous variable (ADONIS; R2 = 0.31; PERM = 999; adj-P = 0.003). Control microbiomes are shown in vermilion while microbiomes exposed to 1.0 and 10 μg/mL streptomycin are shown in orange and yellow, respectively. Ellipses are drawn at 0.95 C.I.
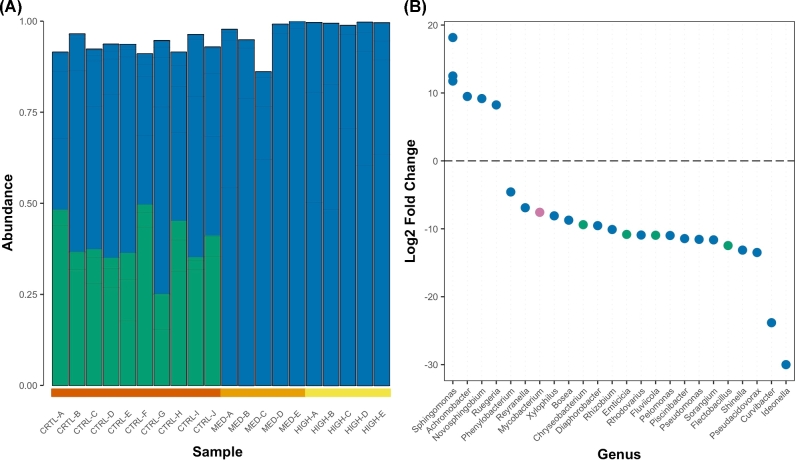
The main change in microbial community was a shift from a more or less equal mixture of Proteobacteria and Bacteroidetes in healthy microbiomes to communities dominated by Proteobacteria when fishes were exposed to streptomycin (Fig. 2A). When looking at changes in ASV specifically, we found that a total of 26 ASVs differed in abundance between control microbiomes and microbiomes exposed to streptomycin (DESeq2; adj-P ≤ 0.01; Fig. 2B). Indeed, six ASVs, classified within four genera, increased in abundance in the presence of streptomycin, while 22 ASVs, including 11 ASVs identified as part of the core microbiome in healthy zebrafish, were significantly less abundant in the presence of streptomycin (Fig. 2B). Three ASVs that increased in abundances were identified as Sphingomonas, a genus commonly isolated from environmental samples using streptomycin as a selective agent (Vanbroekhoven et al.2004). These results taken together suggest that streptomycin exert important selective pressures on the microbiomes of zebrafish larvae, increasing the relative abundance of bacteria resistant or tolerant to the antibiotic.
Figure 2.
Taxa composition in zebrafish microbiomes exposed to different streptomycin concentrations. (A) Relative abundance of Phylum level classification. Each bar within a column represents a unique ASV. Taxa with abundance below 0.1% were removed. (B) Changes in ASVs abundance in the presence of streptomycin (DESeq2; adj-P < 0.01). Genus level classification is provided where available. Genera belonging to Proteobacteria are shown in blue while Bacteroidetes are shown in green and Actinobacteria are shown in lilac.
Streptomycin exposure alters alpha diversity in fish microbiome
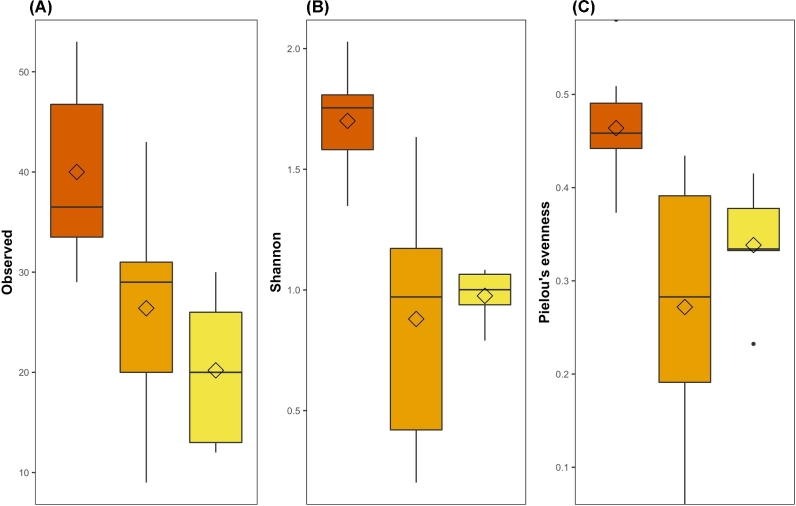
In addition to impact on community composition, we also tested whether streptomycin affected the overall taxonomic diversity present in zebrafish larvae. Overall, we found that taxonomic diversity was lower in microbiomes exposed to streptomycin (Table 2; Fig. 3). Interestingly, while the total number of observed ASVs decreased linearly as streptomycin concentration increased (F(1,18) = 7.58; adj-P = 0.03; Fig. 3A), we found that taxonomic diversity indices that accounts for ASV evenness, even though still lower in treated microbiomes, were slightly larger at 10 μg/mL than at 1 μg/mL. The quadratic relationship showed by the Shannon index (F(1,18) = 7.58; adj-P = 0.03; Fig. 3B) and Pielou's metric (F(1,17) = 9.97; adj-P = 0.01; Fig. 3C) as streptomycin concentration increases suggests that even a small concentration of the antibiotic is sufficient to disrupt the microbiome diversity, allowing relatively few taxa to dominate the microbiomes.
Table 2.
Mean (s.d) of alpha diversity metrics.
| Treatment | Observed | Shannon | Pielou |
|---|---|---|---|
| Control | 40.0 (8.49) | 1.70 (0.22) | 0.46 (0.058) |
| 1 μg/mL | 26.4 (12.72) | 0.88 (0.58) | 0.27 (0.15) |
| 10 μg/mL | 20.2 (7.89) | 0.98 (0.12) | 0.34 (0.068) |
Figure 3.
Alpha diversity metrics in zebrafish microbiomes exposed to different streptomycin concentrations. (A) Observed ASVs (F(1,18) = 7.58; adj-P = 0.03). (B) Shannon diversity (F(1,18) = 7.58; adj-P = 0.03). (C) Pielou's evenness (F(1,18) = 7.58; adj-P = 0.03). All metrics are plotted against streptomycin concentration with controls microbiomes shown in vermilion and microbiomes exposed to 1.0 and 10 μg/mL in orange and yellow, respectively. Diamonds represent population mean while lines represent median and hinges indicate first and third quartiles.
Streptomycin increases int1 abundance in fish microbiome
To investigate the possible link between streptomycin exposure and the spread of integrons in microbial communities, we also measured the abundance of int1 relative to the number of 16S rRNA copy numbers in each microbiome. We found that exposure to streptomycin had a significant effect on the relative abundance of int1 in larvae microbiomes (F(3,20) = 4.33; P < 0.016; Fig. 4). The effect of streptomycin was especially strong at 0.1 and 1.0 μg/mL where we found on average one int1 gene copy number for every four 16s rRNA copy number, a relative abundance 19 times higher than in control populations. Even though int1 relative abundance was on average 13 times higher in fish exposed to 10 μg/mL than in control fish, this difference was not significant due to the large variance in int1 relative abundance in high streptomycin concentration (P = 0.21). Using a dissimilarity matrix comparing PCoA to int1 abundance, we found that int1 weakly correlate with UniFrac clustering between the different samples (ADONIS; R = 0.12 P = 0.068; perm = 999; Fig. S1, Supporting Information), indicating that increases in int1 relative copy number was likely associated with one or few ASVs rather than whole community shifts.
Figure 4.
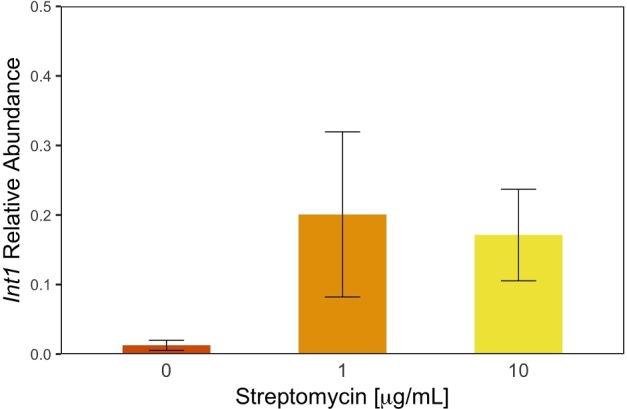
Int1 relative abundance in zebrafish microbiomes exposed to different concentrations of streptomycin. Relative abundance of int1 is calculated as the proportion of int1 copy numbers per 16S rRNA copies and was higher in microbiomes exposed to streptomycin (F(3,20) = 4.33; P < 0.016). Error bars are standard error of the mean and controls microbiomes are shown in vermilion while microbiomes exposed to 1.0 and 10 μg/mL streptomycin are shown in orange and yellow, respectively. For consistency, 0.1 μg/mL treatment was removed from figure.
Exposure to streptomycin increases early mortality in zebrafish larvae
Finally, we investigated whether streptomycin affected larval survival by monitoring the growth of 24 independent larvae exposed to 0, 0.1, 1.0 and 10 mg/mL for a period of 10 days. We found that early mortality was strongly affected by the presence of streptomycin (χ2(3, N = 240) = 161; P < 0.001). While we observed a total of five deaths in control fishes, we observed mortality in nine larvae out of 24 fish exposed to 10 μg/mL of streptomycin by the fourth day of treatment and complete mortality by day seven. Fish exposed to 0.1 and 1.0 μg/mL also showed increased mortality compared to the control population with a total of 11 (45.8%) and 20 (83.3%) mortalities respectively by the end of the experiment. Whether increased mortality resulted from dysbiosis or the possible toxicological effect of streptomycin (see Owens et al. (2009)) remains to be determined. Because streptomycin is commonly used in standard zebrafish husbandry protocols to minimize contamination without apparent effect on survival (Nusslein-Volhard and Dahm 2002; Melancon et al.2017), however, we believe that the decrease in survival rate observed in this study is, at least partly, explained by the consequences of dysbiosis.
DISCUSSION
The possible effects of antibiotic pollution found in rivers and streams on the microbial populations associated with animal hosts are mostly unknown. Here, we show that low concentrations of streptomycin associated with antibiotic pollution can result in important changes in the microbial communities associated with zebrafish larvae, an important model system. In addition, we found that low concentrations of streptomycin increased the relative abundance of integron 1 in the host microbiomes and increased the onset of early mortality of larvae. These results suggest that even low concentrations of antibiotics found in polluted aquatic environments could have important consequences on animal populations.
Compared to mammals, fishes show a wider variability in microbiome composition, differing greatly within a species depending on diet and environmental conditions (Bolnick et al.2014; Smith et al.2015; Schmidt et al.2017). In line with previous findings, we found that the larval zebrafish microbiomes are highly heterogeneous and likely influenced by their environment. For example, many of the genera identified as part of the core microbiomes in healthy zebrafish larvae were previously associated with aquatic environments rather than with animal hosts. When considering ASVs that were present in at least 8 out of 10 larvae, we found additional core taxa such as Fusobacterium sp. that are associated with zebrafish larvae and other teleost fishes (Roeselers et al.2011).
Given the lack of information on fish microbiomes, it is hard to predict the possible effect of antibiotics on fish microbiomes. The few studies done on the topic found that fathead minnows recovered quickly from changes in gut microbiomes following exposure to triclosan (Narrowe et al.2015), while other studies showed that exposure to different antibiotics caused early mortality in adult black molly, Poecilia sphenops, without significant changes in microbiome composition (Schmidt et al.2017) and in adult zebrafish (Zhou et al.2018). Here, despite high heterogeneity at the genus level, we found that streptomycin had a predictable effect on the overall community composition of zebrafish larvae (Fig. 2). Namely, we found that even the lowest concentrations of streptomycin caused a reduction in overall diversity and caused a shift from a mixture of Proteobacteria and Bacteroidetes in healthy fish to communities dominated by Proteobacteria in fish exposed to the antibiotic. Even though zebrafish microbiomes normally develop towards communities dominated by Proteobacteria, this shift is not expected until much later in the life cycle of the animal (Stephens et al.2016).
The most abundant Proteobacteria in our study was identified as Sphingomonas, a genus not normally associated with healthy microbiomes in either larvae or adult zebrafish (Stephens et al.2016). In fact, the Sphingomonas genus contains species known to be pathogenic in humans and to harbor resistance against streptomycin (Vanbroekhoven et al.2004). Similarly, we observed an increase of integron 1 genetic elements in microbiomes treated with streptomycin, an important indicator of antibiotic resistance in the environment (Gillings et al.2015). We found that the presence of integrons did not correlate specifically with any ASV in our study, suggesting that integrons either spread horizontally across multiple ASVs or that we did not have the statistical power to detect such correlation. Either way, our results thus demonstrate that even low concentrations of antibiotics could contribute to the creation of environmental reservoirs of pathogenic bacteria, pathobionts and antibiotic resistance in animal hosts (Perron, Quessy and Bell 2008).
Finally, mortality following antibiotic exposure was previously observed in fishes. In two cases, mortality was due to the host's increased susceptibility to pathogens (Schmidt et al.2017; Zhou et al.2018). While it is likely that the increase in early mortality observed in this study is due at least in part to dysbiosis, the effect could be due to streptomycin toxicity (Klis et al.2014). While a previous studies showed only limited acute toxicity of streptomycin on Daphnia (Wollenberger, Halling-Sørensen and Kusk 2000) and no acute effects on mammals and birds (Seyler and Extension 1994), zebrafish embryos at 3 dpf absorb more drug than at any other developmental periods (Zhang et al.2015) and could therefore be more sensitive to the effect of streptomycin.
The extent to which low concentrations of antibiotics affect fishes and other aquatic organisms remains to be fully investigated. For example, further study is required to understand whether exposure to low concentrations of antibiotics conveys similar effects in adult. Still, our results demonstrate that antibiotic pollution in aquatic environments can not only contribute to the burden of antibiotic resistance but also can have important consequences on the health of exposed organisms. The significant increase in early mortality observed in this study could have a dramatic effect on endangered species with precariously small population size.
Supplementary Material
Acknowledgements
We thank Brooke Jude and Michael Tibbetts for guidance with experimental design and Maureen O’Callaghan-Scholl for assistance with zebrafish husbandry.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSLE online.
FUNDING
The work conducted in this study was supported by the Biology Department and the Bard Summer Research Institute of Bard College.
Conflicts of interest. None declare.
REFERENCES
- Adamczyk M, Jagura-Burdzy G. Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim Pol 2003;50:425–53. [PubMed] [Google Scholar]
- Ali S, van Mil HGJ, Richardson MK. Large-scale assessment of the zebrafish embryo as a possible predictive model in toxicity testing. PLoS One 2011;6:e21076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin B, Al-Zahrani AMJ. The effect of antimicrobial compounds on the gastrointestinal microflora of rainbow trout, Salmo gairdneri Richardson. J Fish Biol 1988;33:1–14. [Google Scholar]
- Bazett M, Bergeron M-E, Haston CK. Streptomycin treatment alters the intestinal microbiome, pulmonary T cell profile and airway hyperresponsiveness in a cystic fibrosis mouse model. Sci Rep 2016;6:19189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Kristiansson E, Larsson DGJ. Environmental factors influencing the development and spread of antibiotic resistance. FEMS Microbiol Rev 2018;42:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielen A, Simatovic A, Kosic-Vuksic J et al. Negative environmental impacts of antibiotic-contaminated effluents from pharmaceutical industries. Water Res 2017;126:79–87. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Snowberg LK, Hirsch PE et al. Individuals' diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch). Ecol Lett 2014;17:979–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy-Roura M, Mas-Pla J, Petrovic M et al. Towards the understanding of antibiotic occurrence and transport in groundwater: Findings from the Baix Fluvià alluvial aquifer (NE Catalonia, Spain). Sci Total Environ 2018;612:1387–406. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Holmes SP. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J 2017;11:2639–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 2012;6:1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso O, Porcher J-M, Sanchez W. Factory-discharged pharmaceuticals could be a relevant source of aquatic environment contamination: review of evidence and need for knowledge. Chemosphere 2014;115:20–30. [DOI] [PubMed] [Google Scholar]
- Czekalski N, Gascon Diez E, Burgmann H. Wastewater as a point source of antibiotic-resistance genes in the sediment of a freshwater lake. ISME J 2014;8:1381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan D, Jude BA, Lamendella G, Keesing F, Perron GG. Exposure to arsenic alters the microbiome of larval zebrafish. Frontiers in Microbiol 2018;9:1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis DJ, Bryda EC, Gillespie CH et al. 16S rRNA amplicon sequencing dataset for conventionalized and conventionally raised zebrafish larvae. Data Brief 2016;8:938–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding C, He J. Effect of antibiotics in the environment on microbial populations. Appl Microbiol Biotechnol 2010;87:925–41. [DOI] [PubMed] [Google Scholar]
- Falcinelli S, Rodiles A, Unniappan S et al. Probiotic treatment reduces appetite and glucose level in the zebrafish model. Sci Rep 2016;6:18061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrey ML, Coreen Hamilton M, Backe WJ et al. Pharmaceuticals and other anthropogenic chemicals in atmospheric particulates and precipitation. Sci Total Environ 2018;612:1488–97. [DOI] [PubMed] [Google Scholar]
- Flaherty CM, Dodson SI. Effects of pharmaceuticals on Daphnia survival, growth, and reproduction. Chemosphere 2005;61:200–7. [DOI] [PubMed] [Google Scholar]
- Fu L, Huang T, Wang S et al. Toxicity of 13 different antibiotics towards freshwater green algae Pseudokirchneriella subcapitata and their modes of action. Chemosphere 2017;168:217–22. [DOI] [PubMed] [Google Scholar]
- Gaze WH, Zhang L, Abdouslam NA et al. Impacts of anthropogenic activity on the ecology of class 1 integrons and integron-associated genes in the environment. ISME J 2011;5:1253–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings MR, Gaze WH, Pruden A et al. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J 2015;9:1269–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillings MR. Integrons: past, present, and future. Microbiol Mol Biol Rev 2014;78:257–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenni P, Ancona V, Barra Caracciolo A. Ecological effects of antibiotics on natural ecosystems: A review. Pharmacol ResAnal Approach 2018;136:25–39. [Google Scholar]
- Grinter NJ. Replication control of IncP plasmids. Plasmid 1984;11:74–81. [DOI] [PubMed] [Google Scholar]
- Gullberg E, Cao S, Berg OG et al. Selection of resistant bacteria at very low antibiotic concentrations. PLoS Pathog 2011;7:e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hang J, Desai V, Zavaljevski N et al. 16S rRNA gene pyrosequencing of reference and clinical samples and investigation of the temperature stability of microbiome profiles. Microbiome 2014;2:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson PJG, Rico A, Troell M et al. Unpacking factors influencing antimicrobial use in global aquaculture and their implication for management: a review from a systems perspective. Sustain Sci 2017. DOI: 10.1007/s11625-017-0511-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein EY, Van Boeckel TP, Martinez EM et al. Global increase and geographic convergence in antibiotic consumption between 2000 and 2015. Proc Natl Acad Sci USA 2018;5:201717295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis S, Stienstra Y, Phillips RO et al. Long term streptomycin toxicity in the treatment of Buruli Ulcer: follow-up of participants in the BURULICO drug trial. PLoS Negl Trop Dis 2014;8:e2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H, Hakim JA, Morrow CD et al. Comparison of two bioinformatics tools used to characterize the microbial diversity and predictive functional attributes of microbial mats from Lake Obersee, Antarctica. J Microbiol Methods 2017;140:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostich MS, Batt AL, Lazorchak JM. Concentrations of prioritized pharmaceuticals in effluents from 50 large wastewater treatment plants in the US and implications for risk estimation. Environ Pollut 2014;184:354–9. [DOI] [PubMed] [Google Scholar]
- Kulkarni P, Olson ND, Raspanti GA et al. Antibiotic concentrations decrease during wastewater treatment but persist at low levels in reclaimed water. Int J Environ Res Public Health 2017;14:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson DGJ. Pollution from drug manufacturing: review and perspectives. Philos Trans R Soc Lond B Biol Sci 2014;369:20130571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence C, Mason T. Zebrafish housing systems: a review of basic operating principles and considerations for design and functionality. ILAR J 2012;53:179–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu C, Chen Y et al. Antibiotic residues in liquid manure from swine feedlot and their effects on nearby groundwater in regions of North China. Environ Sci Pollut Res Int 2018;8:251. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 2005;71:8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SV, Pedersen O. The Human intestinal microbiome in health and disease. N Engl J Med 2016;375:2369–79. [DOI] [PubMed] [Google Scholar]
- Martinez JL. Environmental pollution by antibiotics and by antibiotic resistance determinants. Environ Pollut 2009;157:2893–902. [DOI] [PubMed] [Google Scholar]
- Martínez JL. Effect of antibiotics on bacterial populations: a multi-hierachical selection process. F1000Res 2017;6:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews M, Varga Z. Anesthesia and Euthanasia in Zebrafish. ILAR journal/National Research Council. Institute of Laboratory Animal Resources 2012;53:192–204. [DOI] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 2014;10:e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melancon E, Gomez De La Torre Canny S, Sichel S et al. Best practices for germ-free derivation and gnotobiotic zebrafish husbandry. Methods in Cell Biol 2017;138:61–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narrowe AB, Albuthi-Lantz M, Smith EP et al. Perturbation and restoration of the fathead minnow gut microbiome after low-level triclosan exposure. Microbiome 2015;3:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarrete P, Mardones P, Opazo R et al. Oxytetracycline treatment reduces bacterial diversity of intestinal microbiota of Atlantic salmon. J Aquat Anim Health 2008;20:177–83. [DOI] [PubMed] [Google Scholar]
- Nusslein-Volhard C, Dahm R. Zebrafish. Oxford University Press, Oxford, UK, 2002. [Google Scholar]
- Owens KN, Coffin AB, Hong LS et al. Response of mechanosensory hair cells of the zebrafish lateral line to aminoglycosides reveals distinct cell death pathways. Hear Res 2009;253:32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen Jari. Multivariate Analysis of Ecological Communities in R: Vegan Tutorial. 2007http://cc. oulu. fi/∼jarioksa/opetus/metodi/vegantutor.pdf.1. [Google Scholar]
- Perron GG, Quessy S, Bell G. A reservoir of drug-resistant pathogenic bacteria in asymptomatic hosts. PLoS One 2008;3:e3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pielou EC. The measurement of diversity in different types of biological collections. J Theor Biol 1966;13:131–144. [Google Scholar]
- Quast C, Pruesse E, Yilmaz P et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymann K, Moran NA. The role of the gut microbiome in health and disease of adult honey bee workers. Curr Opin Insect Sci 2018;26:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ et al. Evidence for a core gut microbiota in the zebrafish. ISME J 2011;5:1595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi-Marshall EJ, Kelly JJ. Antibiotic stewardship should consider environmental fate of antibiotics. Environ Sci Technol 2015;49:5257–8. [DOI] [PubMed] [Google Scholar]
- Sandegren L. Selection of antibiotic resistance at very low antibiotic concentrations. Ups J Med Sci 2014;119:103–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep KP. phangorn: phylogenetic analysis in R. Bioinformatics 2011;27:592–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt V, Gomez-Chiarri M, Roy C et al. Subtle microbiome manipulation using probiotics reduces antibiotic-associated mortality in fish. mSystems 2017;2:e00133–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert AM, Sinani H, Schloss PD. Antibiotic-induced alterations of the murine gut microbiota and subsequent effects on colonization resistance against Clostridium difficile. mBio 2015;6:e00974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Tam NM, Jogova M et al. Antibiotic-induced perturbations of the intestinal microbiota alter host susceptibility to enteric infection. Infect Immun 2008;76:4726–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyler LA, Extension CC. EXTOXNET: Extension Toxicology Network: Pesticide Information Notebook. University of California - Davis, Davis, CA, 1994. [Google Scholar]
- Shen Y, Zhao W, Zhang C et al. Degradation of streptomycin in aquatic environment: kinetics, pathway, and antibacterial activity analysis. Environ Sci Pollut Res 2017;24:14337–45. [DOI] [PubMed] [Google Scholar]
- Smith CCR, Snowberg LK, Gregory Caporaso J et al. Dietary input of microbes and host genetic variation shape among-population differences in stickleback gut microbiota. ISME J 2015;9:2515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens WZ, Burns AR, Stagaman K et al. The composition of the zebrafish intestinal microbial community varies across development. ISME J 2016;10:644–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Boeckel TP, Brower C, Gilbert M et al. Global trends in antimicrobial use in food animals. Proc Natl Acad Sci USA 2015;112:5649–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanbroekhoven K, Ryngaert A, Bastiaens L et al. Streptomycin as a selective agent to facilitate recovery and isolation of introduced and indigenous Sphingomonas from environmental samples. Environ Microbiol 2004;6:1123–36. [DOI] [PubMed] [Google Scholar]
- Wellington EM, Boxall AB, Cross P et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis 2013;13:155–65. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York, 2016. [Google Scholar]
- Wollenberger L, Halling-Sørensen B, Kusk KO. Acute and chronic toxicity of veterinary antibiotics to Daphnia magna. Chemosphere 2000;40:723–30. [DOI] [PubMed] [Google Scholar]
- Yan J, Herzog JW, Tsang K et al. Gut microbiota induce IGF-1 and promote bone formation and growth. Proc Natl Acad Sci USA 2016;113:E7554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Qin W, Zhang J-P et al. Antibiotic toxicity and absorption in zebrafish using liquid chromatography-tandem mass spectrometry. PLoS One 2015;10:e0124805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang X, Yin X et al. Toxicity assessment of combined fluoroquinolone and tetracycline exposure in zebrafish (Danio rerio). Environ Toxicol 2016;31:736–50. [DOI] [PubMed] [Google Scholar]
- Zhou L, Limbu SM, Shen M et al. Environmental concentrations of antibiotics impair zebrafish gut health. Environ Pollut 2018;235:245–54. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.