ABSTRACT
Infestation of Triticum (wheat) plants by their pest Rhopalosiphum maidis (corn leaf aphid) causes severe vegetative damage. Despite the agro-economic importance of wheat, the metabolic diversity of Triticum turgidum (tetraploid wheat) in response to aphid attack has not been sufficiently addressed. In this study, we compared the metabolic diversity of two tetraploid wheat genotypes, domesticated and wild emmer. The plants were grown in a control growth room and infested with aphids for 96 h. Our untargeted metabolic analysis performed on plants with and without aphids revealed massive differences between the two genotypes. The targeted metabolic analysis highlighted the differences in the biosynthesis of phytohormones. The aphid progeny was lower in the cultivated durum wheat than in the wild emmer wheat. Overall, these observations emphasize the potential of using the natural diversity of wheat species to better understand the metabolic responses to pest damage.
Keywords: Triticum turgidum, wheat domestication, aphid, Rhopalosiphum maidis, insect, phytohormones, defense, metabolism
Introduction
Wheat (Triticum) is one of the world’s most cultivated small grain crops, with 728 million metric tons harvested from more than 220 million hectares annually. This staple crop provides 20% of the world’s calorie and protein consumption.1 However, biotic stresses, such as pathogen infection and herbivore feeding, can reduce yield dramatically. The introduction of insect resistance through plant breeding is a powerful tool to reduce the aphid population. However, breeding for resistance and the deployment of aphid-resistant wheat cultivars are not yet fully established practices. By identifying the metabolites that are responsible for defense against insect infestation, the development of more informed resistance breeding strategies can occur.2 A major approach to improving agriculturally important traits involves screening the natural variation (wild and domesticated genotypes) within the same plant species, which potentially exposes new alleles and markers for crop improvement.3,4 Indeed, the wild emmer wheat gene pool harbors a rich allelic repertoire for improving essential agronomical traits,5 and previous studies used the natural variation between Triticum genotypes to discover the genetic elements related to biotic resistance.6–8
Aphids (Hemiptera: Aphididae) are piercing/sucking insects that cause damage to plants by acquiring phloem nutrients, thus reducing growth, photosynthetic efficiency and yield.9–12 These insects also act as extremely efficient vectors of several plant viruses that cause economically significant diseases in cereal crops and forage grasses.13,14 Although there are approximately 5,000 species of aphids across the globe, only a handful – commonly termed “cereal aphids” – pose a threat to cereal production.15,16 The most common cereal aphid species include the grain aphid (Sitobion avenae Fabricius), the bird cherry-oat aphid (Rhopalosiphum padi L.), the corn leaf aphid (Rhopalosiphum maidis Fitch), the Russian wheat aphid (Diuraphis noxia Kudjumov), the Indian grain aphid (Sitobion miscanthi Takahashi), the rice root aphid (Rhopalosiphum rufiabdominalis Sasaki), the apple grass aphid (Rhopalosiphum insertum Walker), and the greenbug aphid (Schizaphis graminum Rondani).17 Research conducted since the 1970s has led to the identification of wheat cultivars with resistances to varied aphid species. Nevertheless, new biotypes of these pests, which overcome single-gene resistance mechanisms, have emerged.18,19 Moreover, aphid and other pest populations are expanding into new regions due to climate change, which further emphasizes the need for an in-depth investigation of plant defense mechanisms.
In this study, we explore the effect of corn leaf aphid (R. maidis) feeding on two tetraploid wheat genotypes: i) a durum wheat cultivar (Triticum turgidum ssp. durum) named Svevo and ii) a wild emmer genotype, the progenitor of the most economically important wheat varieties (Triticum turgidum ssp. dicoccoides), named Zavitan. Both genotypes have been intensively investigated and sequenced, and they serve as a source for discovering resistance genes and markers.20,21 Upon aphid attack, the tetraploid genotypes demonstrated massive metabolic variation (Figure 1). Evaluating these genotypes’ resistance to aphids by measuring aphid progeny revealed that Svevo is more resistant than Zavitan (Figure 2). We quantified the levels of several selected metabolites by targeted analysis, including phytohormones such as jasmonic acid (JA), auxin (IAA), fatty acids (oleic-, linoleic- and linolenic acids) and the benzoxazinoid degradation product, 6-methoxy-benzoxazolin-2-one (MBOA). Based on this analysis, we propose a cross-talk between defense and phytohormone responses.22,23 Additionally, these metabolites were significantly affected by aphid feeding, and their levels altered in different manners in the two wheat genotypes (Figure 3). Our results indicate that the differences in the global metabolic profiles and levels of resistance to aphids could be due to the biosynthesis of defense metabolites and the involvement of phytohormone regulation and signaling. Moreover, we hypothesize that during the domestication of the durum tetraploid wheat, this genotype gained alleles responsible for aphid resistance. Our study suggests the need to further explore this metabolic diversity in wheat under controlled growth conditions and in the field to improve plant resistance and to elucidate genetic sources for breeders.
Figure 1.
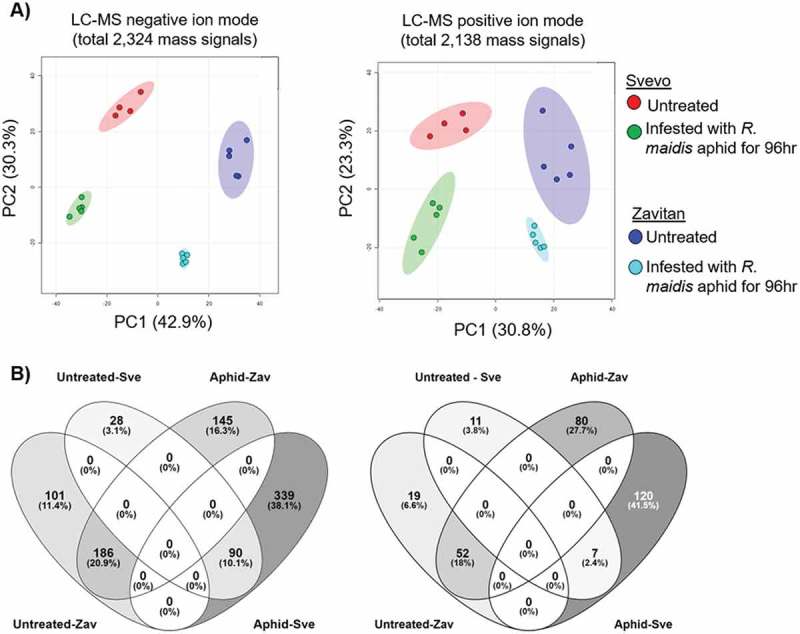
An untargeted metabolic overview of R. maidis feeding on two wheat genotypes. A) PCA plot of negative (2,324 ESI) and positive (2,138 ESI) mass signals, filtered using the Metaboanalyst software. B) Venn diagram illustrating the number of significantly different mass signals comparing the untreated control and aphid infestation between wheat genotypes (P value < 0.05 FDR and fold change > 2 or < 0.5). Untreated-Zav, fold change > 2 relative to Svevo; untreated-Sve, fold change > 2 relative to Zavitan; aphid-Zav, fold change > 2 after treatment with R. maidis relative to Svevo; and aphid-Sve, fold change > 2 after treatment with R. maidis relative to Zavitan.
Figure 2.
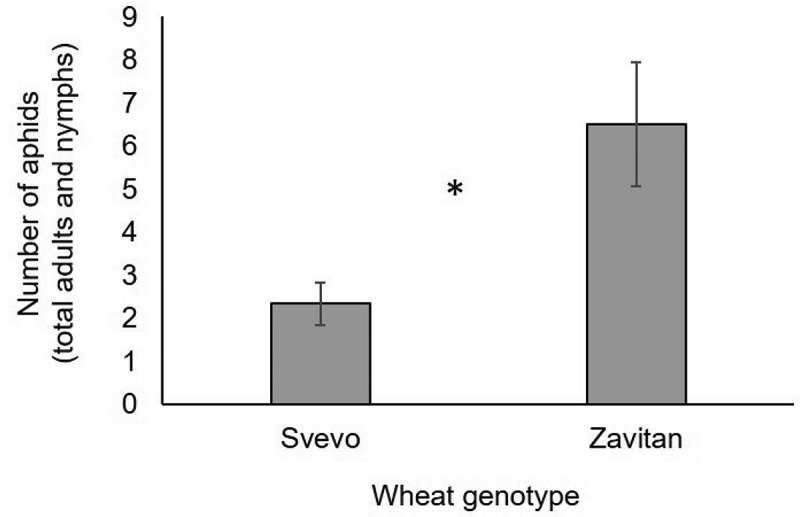
R. maidis progeny production of cultivated Svevo and wild emmer Zavitan wheats after 96 h of infestation. Mean ± SE (n = 6–7). P value < 0.05, Student’s t-test.
Figure 3.
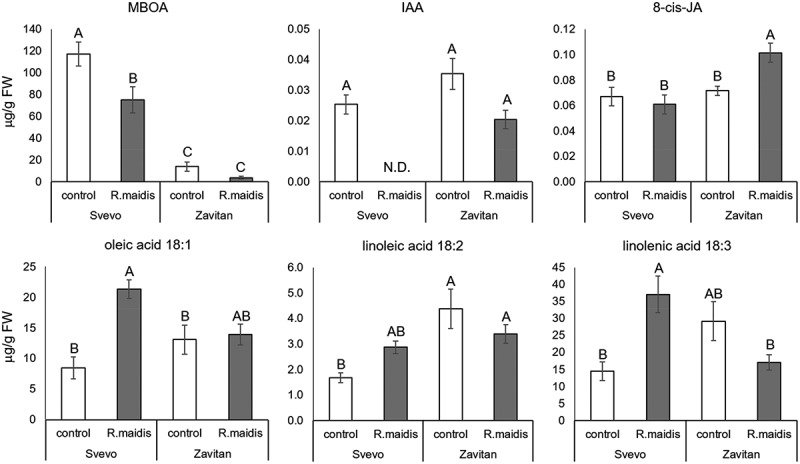
Plant phytohormones and their substrates produced in response to R. maidis feeding on the two wheat genotypes. MBOA, 6-methoxy-benzoxazolin-2-one; IAA, auxin; JA, jasmonic acid. N. D., not detected. Different letters above bars indicate significant differences, P value < 0.05, ANOVA followed by Tukey’s HSD test (n = 5–7).
Results and Discussion
To exploit the natural variation between wheat genotypes, we selected two tetraploid wheat genotypes – Svevo and Zavitan.20 The 10-day-old seedlings were infested with 10 adult corn leaf aphids (R. maidis) using a whole cage bioassay.10,24 After 96 h of aphid infestation, the progeny (nymphs and adults) were counted, and the tip (approximately 10 cm) of the second leaf was harvested for metabolic profiling. First, we performed an untargeted metabolic analysis using a liquid chromatography/time-of-flight/mass spectrometry (LC/TOF/MS) platform. The mass signal levels of both negative and positive ion modes were used to conduct a principal component analysis (PCA; Figure 1A). The results show that all the biological replicates of each genotype were clustered together, which highlights the reproducibility of the experiment. In addition, the PCA results were grouped according to genotype and treatment. This indicates that the metabolic and genetic diversity between the two wheat genotypes is independent of the response to aphid feeding.
We evaluated the distribution of mass signals with significant differences (P value ≤ 0.05, FDR adjusted) and at least two-fold changes between the treated genotypes and their untreated control, using Venn diagrams (Figure 1B). A total of 405 (negative ion mode) and 89 (positive ion mode) mass signals were significantly different between the genotypes of the untreated plants. A higher number of mass signals were modified by the aphid infestation: 760 negative ion mode and 259 positive ion mode in total. Both the PCA clustering patterns and the Venn diagrams reveal the massive metabolic differences between Svevo and Zavitan.
We measured the R. maidis progeny production on Zavitan and Svevo wheat genotypes using whole cage bioassays. The analysis revealed that the cultivated wheat was more resistant to R. maidis than the wild emmer wheat genotype (Figure 2). We also performed a gas chromatography-mass spectrometry (GC-MS) analysis to measure the effect of R. maidis feeding on several molecules such as phytohormones, fatty acids, and a benzoxazinoid degradation compound. Out of the 18 detected metabolites, six were significantly different between genotypes and/or treatment groups (Figure 3). Levels of indole-3-acetic acid (IAA) were reduced below detection levels in the Svevo genotype, and levels of jasmonic acid (JA) increased predominantly in Zavitan. The fatty acids, oleic-, linoleic- and linolenic acids, were mainly increased in the Svevo background, relative to Zavitan, in response to aphid feeding and were not significantly altered in the wild emmer wheat genotype. In addition, accumulation of the benzoxazinoid degradation product 6-methoxy-benzoxazolin-2-one (MBOA) was higher in Svevo than in Zavitan, and was reduced in both genotypes after corn leaf aphid attack. Overall, these data reveal highly significant metabolic differences between the two wheat genotypes at basal levels and also after aphid feeding (Figure 1). They also demonstrate different accumulation patterns for the phytohormones JA, IAA and benzoxazinoid degradation products, in a manner that varies between the two genotypes (Figure 3). Therefore, these metabolic differences may play a role in other cellular functions besides defense and phytohormone regulation.
In summary, the two selected wheat genotypes display massive metabolic differences that are potentially driven by the genetic variation between cultivated and wild wheats. We propose to further utilize these differences in order to understand wheat’s defense mechanisms against corn leaf aphids and other herbivores. We also hope to examine the metabolic and resistance responses under different growth conditions, as well as in field conditions. In addition, the new genome sequence of the wild emmer wheat, coupled with a bi-parental mapping population, will allow us to explore the genes and genetic markers involved in the biosynthesis of these defense metabolites.
Methods
Plant growth conditions. The corn leaf aphid (Rhopalosiphum maidis Fitch) colony was maintained on B73 maize plants as previously described.24 Plants were grown in a Conviron walk-in growth room at 23°C with a 16:8 h light: dark cycle and 180 μmol m-2 s-1 light intensity.
Aphid bioassay. For bioassays measuring aphid progeny reproduction, 10 adult R. maidis aphids were confined on 10-day-old plants with micro-perforated polypropylene bags (15 cm × 61 cm; http://www.pjpmarketplace.com). Adults and nymphs were counted after four days.10,24
Untargeted and targeted metabolite analysis. For analysis of wheat metabolites, approximately 10 cm of leaf material was collected from the second leaf tip. As a control, tissue was caged without aphids and collected for metabolic analysis. Samples were weighed, and all data were normalized relative to the fresh weight. For liquid chromatography/time-of-flight/mass spectrometry (LC/TOF/MS) non-targeted metabolite assays, separation was performed using a Dionex Ultimate 3000 UHPLC system with an Acclaim column (Thermo Scientific), and metabolites were detected using a quadrupole time-time-of flight mass spectrometer (MicrOTOF-Q II; Bruker Daltronics) following the extraction method as previously described.10 Raw mass spectrometry data files were processed using the XCMS25 and CAMERA26 software packages for R. Finally, the positive and negative ionization data sets were transferred to Microsoft Excel. For the targeted gas chromatography-mass spectrometry (GC-MS) analysis, a previously described method27,28 was used for quantification of metabolites in leaf tissue. Samples were solvent-extracted, methylated, collected on a polymeric adsorbent using vapor-phase extraction, and analyzed by GC/isobutane CI-MS using d5-jasmonic acid (C/D/N isotopes Inc., Pointe-Claire, Canada) and U-13C-18:3 (Cambridge Isotope Laboratories, Inc., Tewksbury, MA, USA) as internal standards.28
Statistical analysis. Data for the principal component analysis (PCA) plot were normalized as previously described,29 and data were plotted using MetaboAnalyst 3.0 software.30 Venn diagrams were made using the Venny 2.1.0 drawing tool http://bioinfogp.cnb.csic.es/). Statistical comparisons for insect progeny (Student’s t-test) and metabolite targeted analysis (ANOVA) were made using JMP Pro 12 (SAS; www.jmp.com) and Microsoft Excel for figure representation.
Funding Statement
This work was supported by the Bona Terra Foundation Fund for Promoting Sustainable Agriculture in Drylands to CK and The Ministry of Science and Technology postdoctoral award to RS.
Acknowledgments
We are grateful to Georg Jander for allowing us to conduct this research in his lab by using his equipment and materials. We thank Alon Cna’ani and Zhaniya S. Batyrshina for their comments on this manuscript.
Disclosure of potential conflicts of interest
We declare there are no potential conflicts of interest.
Supplemental material
Supplemental data for this article can be accessed here.
References
- 1.FAOSTAT FAO statistical database: food and agriculture organization of the united nations. 2014; Available from: http://faostat3.fao.org/home/E
- 2.Jaouannet M, Rodriguez PA, Thorpe P, Lenoir CJG, MacLeod R, Escudero-Martinez C, Bos JIB.. Plant immunity in plant–aphid interactions. Front Plant Sci. Internet 2014;5663. doi: 10.3389/fpls.2014.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nevo E, Korol AB, Beiles A, Fahima T. Wild emmer, triticum dicoccoides,wheat progenitor: origin and evolution [internet] evolution of wild emmer and wheat improvement: population genetics, genetic resources, and genome organization of wheat’s progenitor, triticum dicoccoides. In: Nevo E, Korol A.B, Beiles A, Tzion F editors. Berlin, Heidelberg:Springer Berlin Heidelberg; 200211–17. doi: 10.1007/978-3-662-07140-3_2. [DOI] [Google Scholar]
- 4.Tanksley S, McCouch S. Seed banks and molecular maps: unlocking genetic potential from the wild. Science (80-). Internet 1997;2771063–1066. doi: 10.1126/science.277.5329.1063. [DOI] [PubMed] [Google Scholar]
- 5.Chandrasekhar K, Nashef K, Ben-David R. Agronomic and genetic characterization of wild emmer wheat (Triticum turgidum subsp. dicoccoides) introgression lines in a bread wheat genetic background. Genet Resour Crop Evol. Internet 2017;1–10. doi: 10.1007/s10722-016-0481-1. [DOI] [Google Scholar]
- 6.Radchenko E.Resistance of Triticum species to cereal aphids. Czech J Genet Plant Breed. Internet 2011;472009–2012. doi: 10.17221/3257-CJGPB. [DOI] [Google Scholar]
- 7.Migui SM, Lamb RJ. Patterns of resistance to three cereal aphids among wheats in the genus Triticum (Poaceae). Bull Entomol Res. Internet 2003;93323–333. doi: 10.1079/BER2003246. [DOI] [PubMed] [Google Scholar]
- 8.Sela H, Ezrati S, Ben-Yehuda P, Manisterski J, Akhunov E, Dvorak J, Breiman A, Korol A. Linkage disequilibrium and association analysis of stripe rust resistance in wild emmer wheat (Triticum turgidum ssp. dicoccoides) population in Israel. Theor Appl Genet. Internet 2014;1272453–2463. doi: 10.1007/s00122-014-2389-5. [DOI] [PubMed] [Google Scholar]
- 9.Zhou S, Lou Y-R, Tzin V, Jander G. Alteration of plant primary metabolism in response to insect herbivory. Plant Physiol. Internet 2015;169 doi: 10.1104/pp.15.01405%0A%0A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tzin V, Fernandez-Pozo N, Richter A, Schmelz EA, Schoettner M, Schäfer M, Ahern KR, Meihls LN, Huffaker Kaur H A et al. Dynamic Maize Responses to Aphid Feeding Are Revealed by a Time Series of Transcriptomic and Metabolomic Assays. Plant Physiol. Internet 2015;1691727LP–1743. doi: 10.1104/pp.15.01039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bing JW, Novak MG, Obrycki JJ, Guthrie WD. Stylet penetration and feeding sites of Rhopalosiphum maidis (Homoptera: aphididae) on two growth stages of maize. Ann Entomol Soc Am. Internet 1991;84:549. doi: 10.1093/aesa/84.5.549. [DOI] [Google Scholar]
- 12.Rossing WAH, Van De Wiel LAJM. Simulation of damage in winter wheat caused by the grain aphid Sitobion avenae. 1. Quantification of the effects of honeydew on gas exchange of leaves and aphid populations of different size on crop growth. Netherlands J Plant Pathol. Internet 1990;96343–364. doi: 10.1007/BF01998784. [DOI] [Google Scholar]
- 13.El-Muadhidi M, Makkouk K, Kumari S, J M M, Mustafa RS. Survey for legume and cereal viruses in Iraq. Phytopathol Mediterr. Internet 2001;40224–233. . [Google Scholar]
- 14.Hawkes J, Jones R. Incidence and distribution of Barley yellow dwarf virus and Cereal yellow dwarf virus in over-summering grasses in a Mediterranean-type environment. Aust J Agric Res. Internet 2005;56257–270. doi: 10.1071/AR04259. [DOI] [Google Scholar]
- 15.Parry HR.Cereal aphid movement: general principles and simulation modelling. Mov Ecol. Internet 2013;114. doi: 10.1186/2051-3933-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raccah B, Gal-On A, Eastop VF. The role of flying aphid vectors in the transmission of cucumber mosaic virus and potato virus Y to peppers in Israel. Ann Appl Biol. Internet 1985;106451–460. doi: 10.1111/j.1744-7348.1985.tb03135.x. [DOI] [Google Scholar]
- 17.Blackman R, Eastop V. Aphids on the world’s crops: an identification and information guide. London:John Wiley & Sons; 2000. [Google Scholar]
- 18.Puterka GJ, Nicholson SJ, Brown MJ, Cooper WR, Peairs FB, Randolph TL. Characterization of eight Russian wheat aphid (Hemiptera: aphididae) biotypes using two-category resistant — susceptible plant responses. J Econ Entomol. Internet 2014;1071274–1283.doi: 10.1603/EC13408. [DOI] [PubMed] [Google Scholar]
- 19.Shufran KA, Wilde GE. Clonal diversity in overwintering populations of Schizaphis graminum (Homoptera: aphididae). Bull Entomol Res. Internet 1994;84105–114. doi: 10.1017/S0007485300032272. [DOI] [Google Scholar]
- 20.Avni R, Nave M, Eilam T, Sela H, Alekperov C, Peleg Z, Dvorak J, Korol A, Distelfeld A. Ultra-dense genetic map of durum wheat x wild emmer wheat developed using the 90K iSelect SNP genotyping assay. Mol Breed. Internet 2014;341549–1562. doi: 10.1007/s11032-014-0176-2. [DOI] [Google Scholar]
- 21.Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science (80-). Internet 2017;35793LP–97. doi: 10.1126/science.aan0032. [DOI] [PubMed] [Google Scholar]
- 22.Villagrasa M, Guillamón M, Labandeira A, Taberner A, Eljarrat E, Barceló D. Benzoxazinoid allelochemicals in wheat: distribution among foliage, roots, and seeds. J Agric Food Chem. Internet 2006;541009–1015. doi: 10.1021/jf050898h. [DOI] [PubMed] [Google Scholar]
- 23.Tzin V, Hojo Y, Strickler SR, Bartsch LJ, Archer CM, Ahern KR, Zhou S, Christensen SA, Galis I, Mueller LA et al. Rapid defense responses in maize leaves induced by Spodoptera exigua caterpillar feeding. J Exp Bot. Internet 2017;684709–4723.Available from http://academic.oup.com/jxb/article/doi/10.1093/jxb/erx274/4091015/Rapid-defense-responses-in-maize-leaves-induced-by. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Lipka AE, Gershenzon J, Buckler ES, Erb M et al. Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell. Internet 2013;251–16. doi: 10.1105/tpc.113.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith CA, Want EJ, O’Maille G, Abagyan R, Siuzdak G. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem. Internet 2006;78779–787.doi: 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- 26.Kuhl C, Tautenhahn R, Bottcher C, Larson T, Neumann S. CAMERA: an integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal Chem. Internet 2012;84283–289.doi: 10.1021/ac202450g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schmelz EA, Engelberth J, Tumlinson JH, Block A, Alborn HT. The use of vapor phase extraction in metabolic profiling of phytohormones and other metabolites. Plant J. Internet 2004;39790–808. doi: 10.1111/j.1365-313X.2004.02168.x. [DOI] [PubMed] [Google Scholar]
- 28.Tzin V, Lindsay PL, Christensen SA, Meihls LN, Blue LB, Jander G. Genetic mapping shows intraspecific variation and transgressive segregation for caterpillar-induced aphid resistance in maize. Mol Ecol. Internet 2015;5739–50. doi: 10.1111/mec.13418. [DOI] [PubMed] [Google Scholar]
- 29.Tzin V, Malitsky S, Aharoni A, Galili G. Expression of a bacterial bi-functional chorismate mutase/prephenate dehydratase modulates primary and secondary metabolism associated with aromatic amino acids in Arabidopsis. Plant J. Internet 2009;60156–167. doi: 10.1111/j.1365-313X.2009.03945.x. [DOI] [PubMed] [Google Scholar]
- 30.Xia J, Psychogios N, Young N, Wishart DS. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. Internet 2009;37W652–60. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


