ABSTRACT
The circadian clock is synchronized by the day-night cycle to allow plants to anticipate daily environmental changes and to recognize annual changes in day length enabling seasonal flowering. This clock system has been extensively studied in Arabidopsis thaliana and was found to be reset by the dark to light transition at dawn. By contrast, studies on photoperiodic flowering of Pharbitis nil revealed the presence of a clock system reset by the transition from light to dark at dusk to measure the duration of the night. However, a Pharbitis photosynthetic gene was also shown to be insensitive to this dusk transition and to be set by dawn. Thus Pharbitis appeared to have two clock systems, one set by dusk that controls photoperiodic flowering and a second controlling photosynthetic gene expression similar to that of Arabidopsis. Here, we show that circadian mRNA expression of Pharbitis homologs of a series of Arabidopsis clock or clock-controlled genes are insensitive to the dusk transition. These data further define the presence in Pharbitis of a clock system that is analogous to the Arabidopsis system, which co-exists and functions with the dusk-set system dedicated to the control of photoperiodic flowering.
Keywords: photoperiodic flowering, circadian clock, Pharbitis nil, PnLHY, PnTOC1, PnFKF1, PnCDF2, Pn FT
The circadian clock confers endogenous timing information to allow organisms to anticipate daily environmental changes such as light and temperature by triggering behavior and physiology at the appropriate time of the day. The circadian clock is a self-sustaining system that generates an autonomous rhythm with a period length of approximately 24 hours in constant conditions, but this rhythm is also influenced by environmental cues such as changes in light and temperature. In natural conditions the circadian clock therefore synchronizes with day/night cycles.1,2 Synchronization not only allows organisms to accommodate the daily cycles, but also enables them to detect changes in the duration of the day and night that fluctuate regularly throughout the year. Therefore, the circadian clock also represents a seasonal timer that allows organisms to anticipate seasonal climatic changes especially at higher latitudes.3
sRecognition of changes in day length confers seasonal flowering in plants. This mechanism involves a dedicated time-keeping mechanism that integrates information on the light environment to measure duration of the day or night. Its time-keeping activity is derived from the circadian clock. In Arabidopsis, promotion of flowering under long days (LDs) occurs through transcriptional induction of the florigen gene FLOWERING LOCUS T (FT) specifically under these conditions.4,5 The CONSTANS (CO) transcription factor binds to the FT promoter to directly confer its LD-specific induction.6-8 CO represents a photoperiodic-timer gene with its transcript levels being controlled by the circadian clock. CO protein accumulates in response to exposure to light.9,10 Information on light exposure is thereby integrated at CO, leading to its accumulation under LDs and accomplishing measurement of day length.7,8,10–12 Temporal regulation of CO transcription through the clock-controlled blue light photoreceptor FLAVIN-BINDING, KELCH REPEAT, F-BOX1 (FKF1), clock-controlled flowering regulators CYCLING DOF FACTORs (CDFs), and the clock protein GIGANTEA (GI) determines the proper daily accumulation pattern of CO transcript, ensuring that its accumulation coincides with light exposure only under LDs and CO accumulation occurs specifically under these conditions.13-15 These proteins thereby link CO transcription and the circadian clock, which comprises transcriptional negative feedback-loops with particular clock genes such as LATE ELONGATED HYPOCOTYL (LHY) and TIMING OF CAB EXPRESSION 1 (TOC1) in Arabidopsis.16
The fact that CO is stabilized by light exposure indicates that Arabidopsis triggers photoperiodic flowering responses by measuring the duration of the day. Transcription of FT also occurs only during the day.13 On the other hand, Pharbitis nil, a member of the Convolvulaceae and widely used as a model short day plants (SDP), measures the duration of the night.17 In Pharbitis, darkness is required for inducing expression of the florigen-related gene FT1 (P. nil FT1 or Pn FT1), the functional ortholog of Arabidopsis FT.17 Moreover, the circadian phase of Pn FT1 expression in Pharbitis is strongly set by the light-to-dark transition at dusk or light-off in day/night cycles, which allows its expression to proceed only when plants are exposed to sufficiently long nights and eventually to accumulate specifically under SDs.17 Such a phase setting manner in Pn FT1 expression indicates presence of a dusk-set photoperiodic time-keeping rhythm and a clock system reset by the light-to-dark transition. However, the clock in Arabidopsis appears to be rather reset by the light-on at dawn without being affected by the light-to-dark transition at dusk. Circadian phases in expression rhythms of the photosynthetic-gene CAB is strongly set by light exposure at dawn without being strongly affected by the timing of lights off.18,19 Therefore, Pharbitis is likely to contain a clock system that controls photoperiodic flowering whose manner of light resetting is inconsistent with that of Arabidopsis.17
On the other hand, like in Arabidopsis the circadian phase in expression of Pharbitis CAB is not strongly affected by light-to-dark transitions, suggesting that other than the clock system reset by light-off, Pharbitis retains another clock system of the Arabidopsis type.17 Here, we strengthen and extend this idea by further illustrating effects of light-to-dark transitions on circadian expression of Pharbitis homologs of a series of Arabidopsis clock and clock-controlled genes. Circadian phases in expression of these genes are set by the dark-to-light transition at dawn without being affected by dusk transitions, further supporting the concept that Pharbitis retains a clock system similar to that in Arabidopsis as well as the system a differently regulated system dedicated to control of photoperiodic flowering.
In Arabidopsis both LHY and TOC1 act in the clock to confer circadian rhythms in expression of a wide range of genes, including that of photosynthetic genes such as CAB.16 We therefore checked the effects of light-to-dark transitions on circadian phases in expression of LHY- and TOC1- related genes in Pharbitis. The Pharbitis TOC1-related gene or PnTOC1 was identified by a BLAST search against the database “Japanese Morning Glory Genome Database” (http://ipomoeanil.nibb.ac.jp/). The LHY-related gene, termed PnLHY, was previously isolated and its circadian expression with a phase similar to that of LHY reported, although its protein function is still unknown.20 Comparison of the deduced amino acid sequence of the protein encoded by PnTOC1 against TOC1 and the Arabidopsis paralogs is illustrated in the Supplementary Figure 1. The highest similarity of PnTOC1 is to TOC1, with the pseudo-receiver and CO, COL and TOC1 (CCT) domains also being conserved in its amino acid sequence.6,8,21–23
We entrained Pharbitis plants in light/dark cycles and transferred them to constant light (LL), and released populations into continuous darkness (DD) at 8 h intervals to check the rhythm of PnLHY and PnTOC1 expression. If the clock system that comprises these genes or controls their transcription is reset by dark-to-light transition at dawn, then expression of these genes should rise at a constant time after the transfer to LL regardless of when DD begins. However, if the clock system is reset by the light-to-dark transition at dusk, expression of these genes should rise at a constant time after transfer to DD regardless of when LL begins. Expression of PnLHY and PnTOC1 exhibited circadian rhythms peaking approximately at circadian time (CT) 0 in the subjective morning and CT 8 in the subjective evening, respectively, consistent with peak times in circadian expression of LHY and TOC1 in Arabidopsis,24,25 (Figure 1B,E, H, K, Supplementary Figure 1A-D). Expression of both of these genes also began to rise constantly from transfer to LL, indicating that circadian phases in expression of PnLHY and PnTOC1 are set by the dark-to-light transition without being affected by light-to-dark transitions (Figure 1B,E, H, K, Supplementary Figure 1A-D). For the control experiment we also checked circadian expression of Pn FT1 and its paralog Pn FT2 in the same RNA samples. Pn FT2 has been reported to be expressed with a circadian pattern similar to Pn FT1,17 although its role in flowering has not been uncovered. Consistent with previous data, expression of these genes consistently began to rise approximately 12–16 h after transfer to DD, clearly demonstrating that light-off sets expression of these rhythms (Figure1A, D, G, J).
Figure 1.
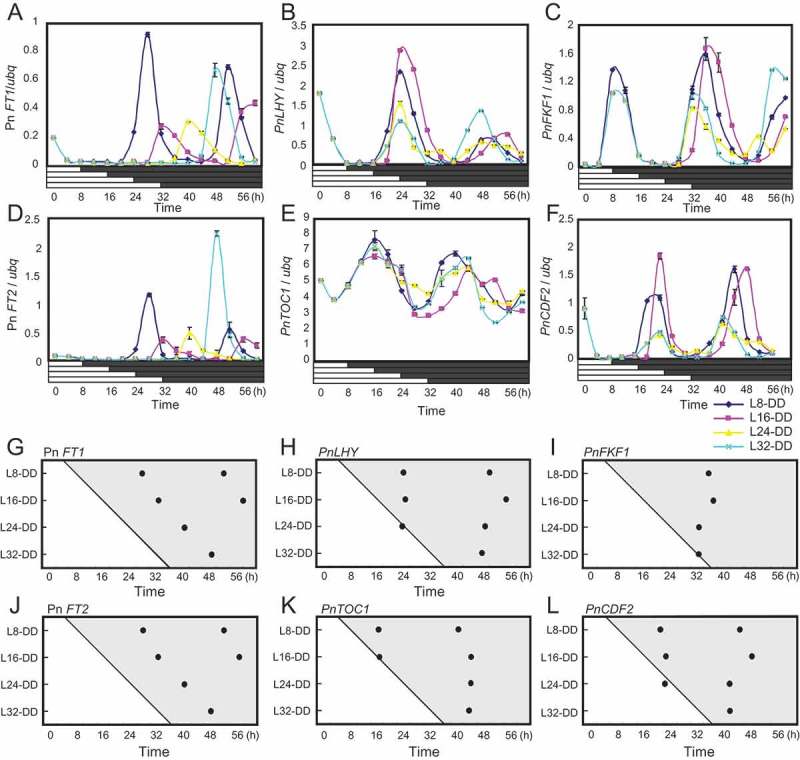
Effects of light-to-dark transitions on circadian expression of Pharbitis clock-controlled genes. Plants were grown under 12h light + 12 h dark for 4 days and were transferred to continuous light at time 0. Plants were then divided into four populations and transferred to continuous dark at 8, 16, 24, and 32 h after the light-on. Cotyledons were harvested from these populations every 4 hours. (A-F) Circadian expression of Pn FT1, PnLHY, PnFKF1, Pn FT2, PnTOC1, and PnCDF2 in DD, respectively. The horizontal axes represent time after dawn from the fifth days. The vertical axes indicate levels of these transcripts relative to ubq. Error bars show SD among two technical replicates in real-time PCR. Two biological replicates were performed (also see supplementary data). (G-L) Diagrams illustrating peak phases in circadian expression of these genes in DD. Peak times in expression of the genes relative to dawn were plotted. The horizontal axes represent time after dawn from the fifth days. The vertical axes represent the time after dawn at which each population was transferred to darkness (e.g., L8-DD spent 8 h in light before transfer to darkness). The results for the four populations of plants shifted to darkness 8, 16, 24, and 32 h after dawn were plotted, and the time that each population was transferred to DD was illustrated in the diagram by the transition from light to shade.
The observed dusk-set circadian phases in Pn FT expression leads to the idea that expression of genes that generate the Pn FT rhythms might also be set by light-to-dark transitions. In Arabidopsis FKF1 and CDFs affect FT expression through transcriptional control of the photoperiodic time-keeping gene CO.13,14 FKF1 and a particular CDF have also been reported to directly affect FT transcription through their association with its promoter.7 We therefore isolated Pharbitis homologs of FKF1 and a CDF by BLAST searches against the database to test effects of light-to-dark transitions on circadian expression of these genes. Comparison of the deduced amino acid sequence of the protein encoded by PnFKF1 with Arabidopsis FKF1 and other related proteins is illustrated in Supplementary Figure 2. The highest similarity of Pn FKF1 is to Arabidopsis FKF1 rather than to other FKF1-related proteins including the Arabidopsis paralogs such as ZEITLUPE (ZTL) and LKP2, which are mainly involved in the control of clock functions in this plant.16,26–28 The deduced amino acid sequence of the protein encoded by PnCDF2 was also most similar to those of Arabidopsis CDFs and related proteins, that also contain the conserved DOF DNA-binding domain in their deduced amino acid sequence14,29 (Supplementary fig. 3).
Expression of PnFKF1 and PnCDF2 exhibited circadian rhythms peaking approximately at CT8 in the subjective evening and at CT 0 in the subjective morning, respectively (Figure 1C, F, I, L, Supplementary Figure 1E, F, G, H), consistent with circadian expression patterns of FKF1 and CDF2 in Arabidopsis.14,29 Also, expression of both of these genes began to rise constantly from transfer to LL regardless of when plants were shifted to DD (Figure 1C, F, I, L, Supplementary Figure 1E, F, G, H). This indicates that despite dusk-set circadian expression of Pn FT, circadian expression of PnFKF1 and PnCDF2 is not affected by light-to-dark transitions.
The circadian clock in Arabidopsis is reset by red, blue, and far-red light.30 However, the Arabidopsis clock is mainly set by the dark-to-light transition at dawn,18,19 so light qualities that mediate light-off resetting are still unknown in plants. To understand this, we checked the effects of transitions from each of these wavelengths of light to darkness on phase setting in circadian expression of Pn FTs. We entrained plants in LD cycles and the next morning exposed the plants to 8 h white light. We then transferred them to continuous red, far-red, or blue light and released to DD at different times to check expression rhythms of Pn FT1 and Pn FT2. In any light conditions tested, Pn FT1 and Pn FT2 expression constantly began to rise approximately 12–16 h after transfer to DD (Figure 2A-L). These results indicate that any of these light qualities function as determinants for setting the clock to the timing of lights-off. In these experiments reduced peak levels in Pn FT expression in DD were also observed when plants were exposed to prolonged blue or far-red light before DD, although the significance and mechanisms are unclear (Figure 2B, C, E, F, H, I, K, L).
Figure 2.
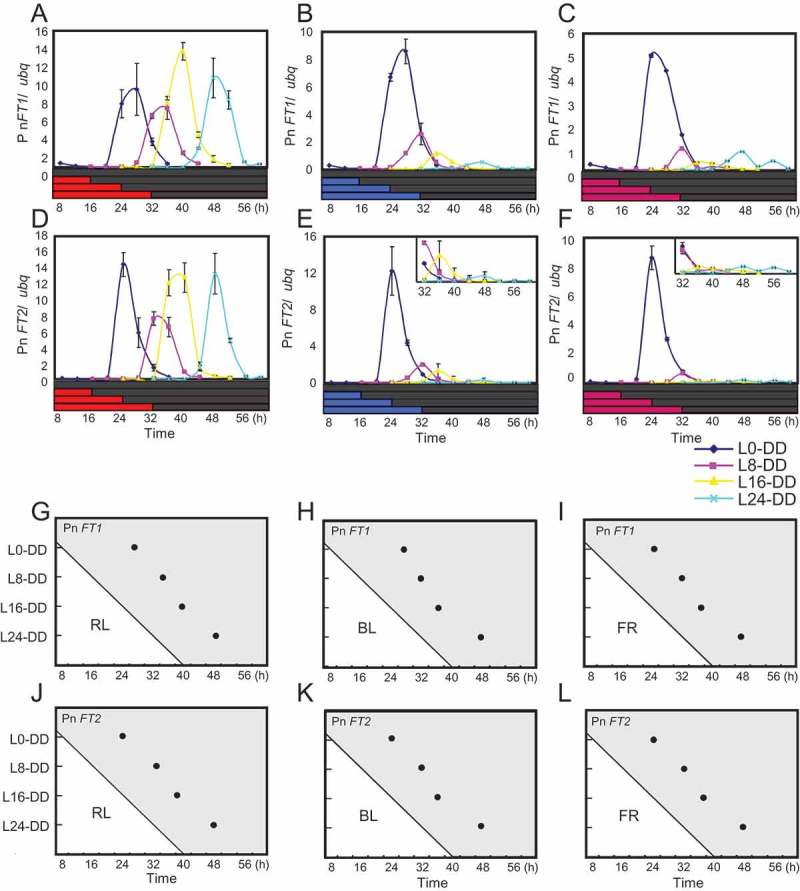
Red, blue and far-red lights function as determinants to reset the clock at lights-off. Plants were grown under 8 h light + 16 h darkness for four days. On the fifth day plants kept in 8 h WL in the morning were further exposed to either red, blue or far-red light of various durations (0, 8, 16, 24 h), and thereafter transferred to DD. Plants were harvested every 4 hours within the period of only 28 hours from transfer to DD. (A-F) Circadian expression of Pn FT1 and Pn FT2 in DD after transfer from red light (A and D), blue light (B and E), or far-red light (C and F). Transcript levels of Pn FT1, Pn FT2, and ubq was analyzed in DD by real-time PCR. The horizontal axes represent hours from dawn on the 5th day. The vertical axes indicate transcript levels of Pn FTs relative to ubq based on obtained data from real-time PCR. Error bars show SD among two biological replicates. (G-L) Diagrams illustrating peak phases in circadian expression of Pn FT1 and Pn FT2 in DD. Peak times in expression of these genes relative to dawn were plotted. The horizontal axes represent time after dawn from the fifth days. The vertical axes represent the duration (h) of red, blue or far-red light that each population received before transfer to darkness (e.g., L8-DD spent 8 h in either one of these lights before transfer to darkness).
In this study we further examined the effects of light-to-dark transitions on phase setting in circadian expression of the Pharbitis homologs of a series of Arabidopsis clock and clock-controlled genes. We found that expression of the Pharbitis homologs of Arabidopsis clock genes LHY and TOC1 is set by the dark-to-light transition at dawn without being affected by light-to-dark transitions, clearly demonstrating the presence in Pharbitis of a clock system that resembles the Arabidopsis clock in terms of the manner of light resetting. Like in Arabidopsis, the circadian rhythm in expression of CAB in Pharbitis is also presumed to be mediated by this circadian system, as in this plant its expression is mainly set by the dark-to-light transition without being strongly affected by light-to-dark transitions.17 Considering that anticipating the light-on at dawn by triggering gene expression during the night is crucial for maximizing photosynthetic activity, and that photosynthesis is vital for plant survival, insensitivity to light-to-dark transitions at dusk might be expected to be a significant and common feature in plant circadian clocks. Especially at higher latitudes where the timing of dusk alters due to fluctuation in day length within the year, this insensitivity has the potential to allow the clock system and its control of photosynthesis to accurately anticipate lights-on at dawn. It is unclear whether the dusk-insensitive Pharbitis clock system comprises genes orthologous to Arabidopsis clock genes. However, since PnLHY and PnTOC1 appeared to be expressed with circadian phases similar to LHY and TOC1, respectively, the clock system could potentially be orthologous to the Arabidopsis system. Related to this, overexpression of the Pharbitis orthologue of Arabidopsis GI causes alterations in various circadian rhythms such as leaf movement in Pharbitis.20 This observation is also consistent with the idea that Pharbitis retains a clock system orthologous to that in Arabidopsis.
Other than the observed dusk-insensitive clock system that is similar to that of Arabidopsis, Pharbitis also retains a system strongly reset by the dusk transition to generate a dusk-set photoperiodic time-keeping rhythm.17 Based on genetic and molecular studies on light resetting of the circadian clock in Arabidopsis, dusk resetting of the clock in Pharbitis may involve loss of a mechanism that limits the activity of light input to the clock component to dawn, causing its activity to be always on and suspending the activity of the clock at the particular phase during the light period till the dark period begins.18,19 Red, far-red and blue light are likely to be responsible for dusk resetting of the Pharbitis clock, since transitions from any of these wavelengths of light to darkness could set circadian expression of Pn FTs (Figure 2A-L). These light qualities are responsible for entrainment of the Arabidopsis circadian clock as well, but in this plant activities of light input pathways that mediate between these wavelengths of light and the clock components are limited to dawn allowing it to be specifically reset at this time.30
The core clock that mediates photoperiodic flowering in Pharbitis probably shares most of the Arabidopsis clock genes because, as was observed in several Arabidopsis mutants such as early flowering 3 (elf3), 19 the lack of only a single gene function can cause deficiency in the mechanism that temporarily restricts the activity of light input to the clock. Therefore, in Pharbitis an orthologous clock system could remain linked to the same photoperiodic-flowering genes as in Arabidopsis but lack the restriction mechanism against light input, and thereby provide dusk-set rhythms. However, circadian rhythms in expression of PnFKF1 and PnCDF2 were insensitive to light-to-dark transitions in Pharbitis (Figure 1C, F, I, L, Supplementary Figure 1E-H), despite observed clear light-off setting in Pn FT rhythms17,(Figure1A,D, G, J). This inconsistency implies that unlike in Arabidopsis, functions of Pharbitis FKF1 and CDF2 are not coupled with control of FT transcription. Circadian expression of PnCO, a close homolog of CO in Pharbitis whose overexpression can induce flowering in Arabidopsis, has also been reported to be insensitive to light-to-dark transitions,31. The photoperiodic pathway that controls expression of Pn FTs in Pharbitis might therefore involve genes different from FKF1, CDFs and CO in Arabidopsis. Alternatively, there might be other Pharbitis genes whose functions are more closely related to FKF1, CDFs and CO, whose circadian rhythms in expression are affected by light-to-dark transitions.
In this study we further supported the presence of two distinct clock systems in Pharbitis. These presumed clock systems are insensitive to or strongly reset by light-to-dark transitions in daily cycles, carrying the potential to generate distinct circadian rhythms with their phases originating from either the timing of dawn or dusk, respectively. Co-existence of these clock systems could therefore provide significant adaptive advantage to natural day/night cycles especially at higher latitudes. The clock system insensitive to the light-to-dark transition at dusk has the potential to enable accurate anticipation of lights-on at dawn regardless of changes in the timing of dusk within the year. At the same time, the circadian system strongly reset by this transition enables to generate dusk-set circadian rhythms throughout the year without being affected by changes in the timing of dawn. Although, like in Arabidopsis, expression and activities of a number of Pharbitis genes and the physiological processes are presumed to exhibit circadian rhythms, those specifically affected by the clock system reset by the light-to-dark transition at dusk are still poorly understood because genes whose circadian expression was reported to be set by the dusk transition are currently limited to Pn FT1 and Pn FT2. Unmasking genes whose circadian expression is set by lights-off by using genome-wide techniques such as RNAseq and knowledge on the recently uncovered whole genomic sequence in Pharbitis,32 may help us to understand this physiological processes more deeply. Such a study may provide information on the significance of the dusk-set clock in adaptation to natural day/night cycles where the timing of sunrise and sunset annually changes with fluctuating day length.
Material and methods
Plant material and growth condition
Pharbitis nil seed of Ipomoea nil Choisy cv Violet were purchased from Marutane, Japan. Seeds were treated with sulfuric acid for 1 h for, washed with sterilized water, and slowly agitated with water for overnight. Germinated seeds were sown on MS agar media and grown in climate chambers at 22 °C. For experiments with red, blue far-red light LED chambers were used.
RNA isolation, cdna synthesis, and expression analysis
RNA was isolated using RNeasy Plant Mini Kit (Qiagen). cDNA synthesis was performed with SuperScript II reverse transcriptase (Life Technologies) with oligo-dT primer. All protocols were performed according to the manufacture’s instructions. Expression analyses were performed with quantitative real-time PCR using Light Cycler 480 II (Roche) and AriaMx Real-Time PCR System (Agilent Technologies). Each RNA analysis was performed twice with independent plant samples. Primers used for the PCR are Pn FT1 (5ʹ-ACCCTGAGGGAATACCTCCACT-3’ and 5’-GGAAGGAGCAGGGTAATTAATCGG-3ʹ), Pn FT2 (5ʹ-TGAGGGAGTACCTACACTGGTTG-3ʹ and 5ʹ-AGGGTGCGTCATTACGCATT-3’), ubq (5’-GGAGTCGACTCTTCACTTGG- 3’ and 5’-TGGGACATTAGGGGATTCAG-3’), PnLHY (5’-AGGAAGTAGTACGGGCTGTCAA-3’ and 5’-CCTACACGACAAGTTTCGTGATCT-3’), PnFKF1 (5’-TATAGAGATCCCCGGGCTCAAA-3’ and 5’-TAATGGGGTACCGTCCTTCCT-3’), and PnCDF2 (5’-GTGACAGTATGGACACCAAGT-3’ and 5’-ACCAACAGGCAAGTTCCTC-3’).
Funding Statement
This work was supported by a core grant from the Max Planck Society to G.C. and by the Grant-in Aid for Scientific Research on the JSPS to T.M.
Accession numbers of genes isolated and analyzed in this present work
PnPIF4; KJ605441
PnFKF1; KJ605442
PnCDF2; KJ605443
PnTOC1; KM521244
Acknowledgments
We thank Elisabeth Luley for experimental assistances, and the G.C. lab members for many useful discussions.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementry material
Supplemental data for this article can be accessed here.
References
- 1.Boikoglou E, Ma Z, Korff M, Davis AM, Nagy F, Davis SJ.. Environmental memory from a circadian oscillator: the arabidopsis thaliana clock differentially integrates perception of photic vs. thermal entrainment. Genetics. 2011;189:655–664. PMID:21840862. doi: 10.1534/genetics.111.131417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millar AJ. Input signals to the plant circadian clock. J Exp Bot. 2004;55:277–283. PMID:14695902. doi: 10.1093/jxb/erh034. [DOI] [PubMed] [Google Scholar]
- 3.Song YH, Ito S, Imaizumi T. Similarities in the circadian clock and photoperiodism in plants. Curr Opin Plant Biol. 2010;13:594–603. PMID:20620097. doi: 10.1016/j.pbi.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andres F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13:627–639. PMID:22898651. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 5.Song YH, Ito S, Imaizumi T. Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 2013;18:575–583. PMID:23790253. doi: 10.1016/j.tplants.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tiwari SB, Shen Y, Chang HC, Hou Y, Harris A, Sf M, McPartland M, Hymus GJ, Adam L, Marion C, et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010;187:57–66.PMID:20406410. doi: 10.1111/j.1469-8137.2010.03251.x. [DOI] [PubMed] [Google Scholar]
- 7.Song YH, Smith RW, To BJ, Millar AJ, Imaizumi T. FKF1 conveys timing information for CONSTANS stabilization in photoperiodic flowering. Science. 2012;336:1045–1049. PMID:22628657. doi: 10.1126/science.1219644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayama R, Sarid-Krebs L, Richter R, Fernandez V, Jang S, Coupland G. PSEUDO RESPONSE REGULATORs stabilize CONSTANS protein to promote flowering in response to day length. EMBO J. 2017;36:904–918. PMID:28270524. doi: 10.15252/embj.201693907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suarez-Lopez P, Wheatley K, Robson F, Onouchi H, Valverde F, Coupland G. CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature. 2001;410:1116–1120. PMID:11323677. doi: 10.1038/35074138. [DOI] [PubMed] [Google Scholar]
- 10.Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G. Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science. 2004;303:1003–1006. PMID:14963328. doi: 10.1126/science.1091761. [DOI] [PubMed] [Google Scholar]
- 11.Liu LJ, Zhang YC, Li QH, Sang Y, Mao J, Lian HL, Wang L, Yang HQ. COP1-mediated ubiquitination of CONSTANS is implicated in cryptochrome regulation of flowering in Arabidopsis. Plant Cell. 2008;20:292–306. PMID:18296627. doi: 10.1105/tpc.107.057281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jang S, Marchal V, Panigrahi KC, Wenkel S, Soppe W, Deng XW, Valverde F, Coupland G. Arabidopsis COP1 shapes the temporal pattern of CO accumulation conferring a photoperiodic flowering response. EMBO J. 2008;27:1277–1288. PMID:18388858. doi: 10.1038/emboj.2008.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imaizumi T, Tran HG, Swartz TE, Briggs WR, Kay SA. FKF1 is essential for photoperiodic-specific light signalling in Arabidopsis. Nature. 2003;426:302–306. PMID:14628054. doi: 10.1038/nature02090. [DOI] [PubMed] [Google Scholar]
- 14.Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. PMID:16002617. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- 15.Sawa M, Nusinow DA, Kay SA, Imaizumi T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science. 2007;318:261–265. PMID:17872410. doi: 10.1126/science.1146994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu PY, Harmer SL. Wheels within wheels: the plant circadian system. Trends Plant Sci. 2013;19:240–249. PMID:24373845. doi: 10.1016/j.tplants.2013.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayama R, Agashe B, Luley E, King R, Coupland G. A circadian rhythm set by dusk determines the expression of FT homologs and the short-day photoperiodic flowering response in Pharbitis. Plant Cell. 2007;19:2988–3000. PMID:17965272. doi: 10.1105/tpc.107.052480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Millar AJ, Kay SA. Integration of circadian and phototransduction pathways in the network controlling CAB gene transcription in Arabidopsis. Proc Natl Acad Sci U S A. 1996;93:15491–15496. PMID:8986839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McWatters HG, Bastow RM, Hall A, Millar AJ. The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature. 2000;408:716–720. PMID:11130072. doi: 10.1038/35047079. [DOI] [PubMed] [Google Scholar]
- 20.Higuchi Y, Sage-Ono K, Sasaki R, Ohtsuki N, Hoshino A, Iida S, Kamada H, Ono M. Constitutive expression of the GIGANTEA ortholog affects circadian rhythms and suppresses one-shot induction of flowering in Pharbitis nil, a typical short-day plant. Plant Cell Physiol. 2011;52:638–650. PMID:21382978. doi: 10.1093/pcp/pcr023. [DOI] [PubMed] [Google Scholar]
- 21.Robson F, Costa MM, Hepworth SR, Vizir I, Pineiro M, Reeves PH, Putterill J, Coupland G. Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 2001;28:619–631. PMID:11851908. [DOI] [PubMed] [Google Scholar]
- 22.Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proc Natl Acad Sci U S A. 2012;109:3167–3172. PMID:22315425. doi: 10.1073/pnas.1200355109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsushika A, Makino S, Kojima M, Mizuno T. Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol. 2000;41:1002–1012. PMID:11100772. [DOI] [PubMed] [Google Scholar]
- 24.Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA. Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science. 2000;289:768–771. PMID:10926537. [DOI] [PubMed] [Google Scholar]
- 25.Schaffer R, Landgraf J, Accerbi M, Simon V, Larson M, Wisman E. Microarray analysis of diurnal and circadian-regulated genes in Arabidopsis. Plant Cell. 2001;13:113–123. PMID:11158533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. PMID:10847686. [DOI] [PubMed] [Google Scholar]
- 27.Mas P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. PMID:14654842. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- 28.Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA. A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell. 2001;13:2659–2670. PMID:11752379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fornara F, Panigrahi KC, Gissot L, Sauerbrunn N, Ruhl M, Jarillo JA, Coupland G. Arabidopsis DOF transcription factors act redundantly to reduce CONSTANS expression and are essential for a photoperiodic flowering response. Dev Cell. 2009;17:75–86. PMID:19619493. doi: 10.1016/j.devcel.2009.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Millar AJ, Straume M, Chory J, Chua NH, Kay SA. The regulation of circadian period by phototransduction pathways in Arabidopsis. Science. 1995;267:1163–1166. PMID:7855596. [DOI] [PubMed] [Google Scholar]
- 31.Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science. 2000;288:1613–1616. PMID:10834834. [DOI] [PubMed] [Google Scholar]
- 32.Hoshino A, Jayakumar V, Nitasaka E, Toyoda A, Noguchi H, Itoh T, Shin-I T, Minakuchi Y, Koda Y, Nagano A, et al. Genome sequence and analysis of the Japanese morning glory Ipomoea nil. Nat Commun. 2017;7:13295.PMID:27824041. doi: 10.1038/ncomms13295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


