Staphylococcus aureus employs a complex regulatory network to coordinate the expression of various virulence genes to achieve successful infections. How virulence genes are coordinately regulated is still poorly understood. We have been studying capsule regulation as a model system to explore regulatory networking in S. aureus. In this study, we found that XdrA and CodY have broad binding sites that overlap extensively in the capsule promoter region. Our results also suggest that XdrA assists CodY in the repression of capsule. As capsule gene regulation by DNA-binding regulators has not been fully investigated, the results presented here fill an important knowledge gap, thereby further advancing our understanding of the global virulence regulatory network in S. aureus.
KEYWORDS: CodY, Staphylococcus aureus, XdrA, capsule, virulence regulation
ABSTRACT
Capsule is one of many virulence factors produced by Staphylococcus aureus, and its expression is highly regulated. Here, we report the repression of capsule by direct interaction of XdrA and CodY with the capsule promoter region. We found, by footprinting analyses, that XdrA repressed capsule by binding to a broad region that extended from upstream of the −35 region of the promoter to the coding region of capA, the first gene of the 16-gene cap operon. Footprinting analyses also revealed that CodY bound to a large region that overlapped extensively with that of XdrA. We found that repression of the cap genes in the xdrA mutant could be achieved by the overexpression of codY but not vice versa, suggesting codY is epistatic to xdrA. However, we found XdrA had no effect on CodY expression. These results suggest that XdrA plays a secondary role in capsule regulation by promoting CodY repression of the cap genes. Oxacillin slightly induced xdrA expression and reduced cap promoter activity, but the effect of oxacillin on capsule was not mediated through XdrA.
IMPORTANCE Staphylococcus aureus employs a complex regulatory network to coordinate the expression of various virulence genes to achieve successful infections. How virulence genes are coordinately regulated is still poorly understood. We have been studying capsule regulation as a model system to explore regulatory networking in S. aureus. In this study, we found that XdrA and CodY have broad binding sites that overlap extensively in the capsule promoter region. Our results also suggest that XdrA assists CodY in the repression of capsule. As capsule gene regulation by DNA-binding regulators has not been fully investigated, the results presented here fill an important knowledge gap, thereby further advancing our understanding of the global virulence regulatory network in S. aureus.
INTRODUCTION
Staphylococcus aureus is an important human and animal pathogen that can cause various serious infections. The organism is capable of producing a plethora of virulence factors. A successful staphylococcal infection depends on the coordinate regulation of these virulence factors by a complex network of a similarly large number of regulators (1–3). Capsular polysaccharides are surface virulence factors that endow the bacteria the ability to resist phagocytosis by the host's immune system. Type 5 and type 8 are the predominant capsule serotypes in clinical isolates (4, 5). The genetic loci of the two serotypes are allelic. Sixteen cap genes involved in capsule biosynthesis are arranged within a long operon in which the central four genes (capHIJK) are type specific (6). The cap operon is mainly controlled at the primary promoter upstream of the first gene, which is common to the two serotypes, indicating that the two serotypes are regulated similarly (7). A 10-bp inverted repeat just upstream of the promoter is essential for cap gene expression (8). As the cap promoter (Pcap) has been characterized in detail and capsule plays an important role in pathogenesis, we have been using capsule as a model virulence factor to further understand virulence regulation in S. aureus by unraveling the regulatory pathways and mechanisms of regulation. Despite the efforts, the regulatory pathways affecting capsule production are still not completely understood. In particular, the regulators that directly interact with the Pcap have not been well studied.
Recently, we identified several regulators that directly interact with the Pcap region and defined the binding site of one of the regulators, RbsR, by DNase I footprinting analysis (9). In the present study, to further understand cap regulation, we chose to study XdrA and CodY, two negative regulators of capsule that interact directly with the Pcap region. XdrA, which belongs to the Xre (xenobiotic response element) family of helix-turn-helix (Xre-HTH) DNA-binding proteins, was previously characterized by its activation of the spa gene encoding staphylococcal protein A (10, 11). Most recently, XdrA was found to affect biofilm formation (12). We found that XdrA repressed capsule expression by binding to Pcap with a broad binding site that extended from upstream of the cap promoter into the coding region of capA, the first gene of the cap operon. CodY is a global regulator found in most low-GC Gram-positive bacteria (13, 14). In S. aureus, CodY has been shown to be a key regulator linking metabolism and virulence (15–22). CodY represses capsule expression and was previously shown to bind the Pcap region, but the binding site has not been determined (15, 16). Here, we report that CodY also binds to a large region in the Pcap region that overlaps extensively with that of XdrA. Further analyses of the XdrA-CodY interaction suggest that CodY plays a more important role than XdrA in repressing capsule expression.
RESULTS
XdrA is a repressor of capsule.
To understand how cap genes are transcriptionally regulated by direct DNA-binding regulators, we previously identified XdrA as one of the putative regulators by direct in-gel proteomic analysis of proteins bound to a Pcap fragment (9). A transposon insertion in the xdrA gene (CYL12837 strain) resulted in increased capsule production, suggesting that XdrA is a capsule repressor (9). To further confirm that XdrA represses capsule, we performed complementation experiments. The xdrA gene was cloned under the control of the Pxyl-tetO promoter (pML4155). As shown in Fig. 1, the increase in capsule production in the xdrA::bursa mutant (CYL12837) was complemented by pML4155 to a level comparable to that of the wild-type strain CYL11481, even without the inducer anhydrotetracycline (ATc). This is not surprising, as the Pxyl-tetO promoter has been shown to be leaky (23). In the presence of the inducer ATc, capsule production was repressed further to a level lower than that of the wild type, likely due to the overproduction of XdrA. As the insert fragment in pML4155 contained the xdrA gene only, these results indicate that XdrA is a repressor of capsule.
FIG 1.
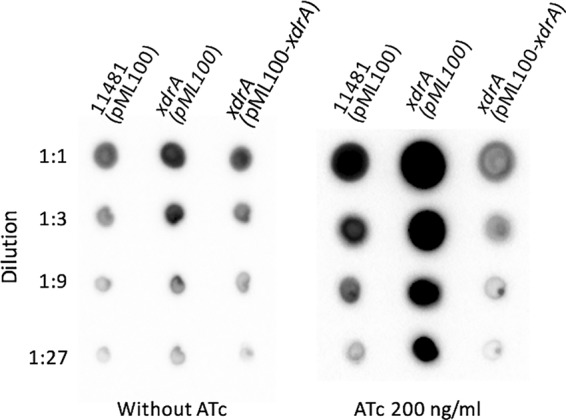
Complementation of xdrA::bursa mutant with pML4155 (pML100-xdrA) by capsule assay. Capsules were isolated from cultures grown in TSB-0G in the presence of 10 μg/ml chloramphenicol either for 4 h without ATc (left) or for 2 h and then induced with 200 ng/ml of ATc for 2 h (right). Isolated capsules were serially diluted, spotted on membranes, and blotted with an anticapsule antibody. The difference in the intensities of the two panels is primarily due to the fact that these results were obtained from different experiments with unequal exposures to the developing reagents.
XdrA represses capsule by direct promoter binding.
XdrA was identified by its ability to bind the Pcap region (9), and it contains a helix-turn-helix DNA-binding motif near its N-terminal end (10, 11). Thus, it is most likely that it affects capsule expression by direct DNA binding. To confirm this, we performed electrophoretic mobility shift assays (EMSAs) using a purified His6-XdrA fusion protein from Escherichia coli and a 156-bp Pcap fragment (Fig. 2A, fragment A) as a probe for binding to XdrA. As shown in Fig. 2B, fragment A was readily upshifted by His6-XdrA, which was effectively competed away with nonlabeled fragment A (cold DNA) but not with a fragment containing the icaR promoter (Pica) or herring sperm DNA (gDNA). Interestingly, the shifted band was competed away by the 75-bp fragment C but only slightly affected by the 91-bp fragment B. These results indicate that XdrA binds to the Pcap fragment specifically and that the binding site is likely located in the region upstream of the ATG start codon of the capA gene to the −35 region of the promoter. To further determine the approximate XdrA-binding region of the Pcap, we used the larger 312-bp fragment D as a probe and a nonlabeled shorter fragments as shown in Fig. 2A for competition. The results (Fig. 2C) showed that the D fragment was readily shifted up and was competed effectively by the cold fragment D as well as the cold shorter fragments, although fragments F and G were not as effective. However, the DNA fragments further downstream (fragment H and a DNA fragment within the downstream capD gene) showed much less effective competition. These EMSA results indicate that XdrA binds to a broad region in the cap promoter area and to sequences well into the coding sequence of the capA gene (approximately 100 bp of the capA coding region). The results in Fig. 2C also suggest that fragments B and H and capD may contain XdrA binding sites, as partial competition was observed. It should be noted here that we also carried out a control EMSA by using a similarly purified fraction of cell extracts of E. coli containing the expression vector. As shown in Fig. S1 in the supplemental material, no shifted band was observed when the control E. coli fraction was used, suggesting that the shifted bands by the His6-XdrA protein are not due to contamination of E. coli proteins during purification.
FIG 2.
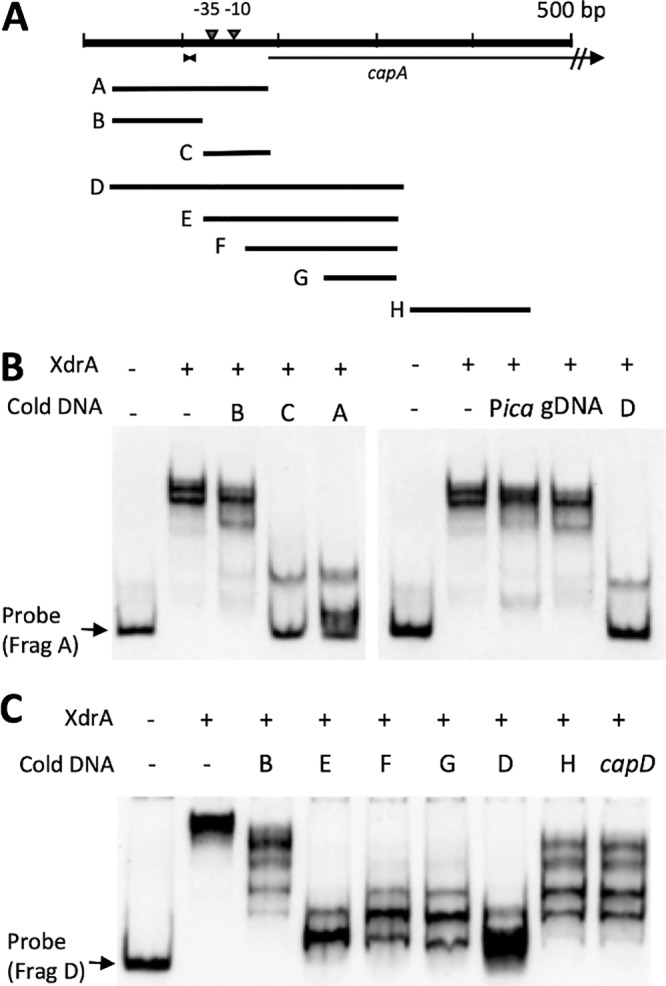
Mapping His6-XdrA binding to Pcap by EMSA. (A) Pcap-capA region used for EMSAs. The 10-bp inverted repeat is indicated by the facing arrowheads. DNA fragments used as probes or cold competitors are shown below. (B) EMSA was performed with 70.22 nM His6-XdrA using fragment A (0.078 nM) as a labeled probe. Cold DNA competitors were added at 500-fold excess. (C) EMSA performed with 28 nM His6-XdrA using fragment D (0.078 nM) as a labeled probe. Cold DNA competitors were added at 500-fold excess.
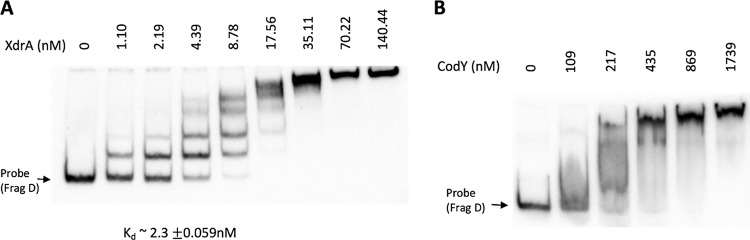
We previously showed that a 10-bp inverted repeat upstream of the −35 region of Pcap is essential for cap gene expression (8). However, XdrA bound to the Pcap is not affected by a mutation in the inverted repeat (data not shown), suggesting that the inverted repeat does not play an important role in XdrA binding. To determine the efficiency of binding, we used fragment D with increasing concentrations of His6-XdrA in an EMSA. The binding constant (Kd) was then determined to be ∼2.3 ± 0.059 nM (Fig. 3A), indicating that XdrA has a high affinity for binding.
FIG 3.
DNA-binding affinity of XdrA and CodY determined by EMSA with increasing amounts of protein. (A) His6-XdrA with constant amount of 0.078 nM probe. Dissociation constant Kd was determined by nonlinear regression; the standard deviation is from two replicates. (B) His6-CodY with constant amount of 0.62 nM probe in the presence of 2 mM GTP and 32 mM Ile.
XdrA binds Pcap and capA coding sequences.
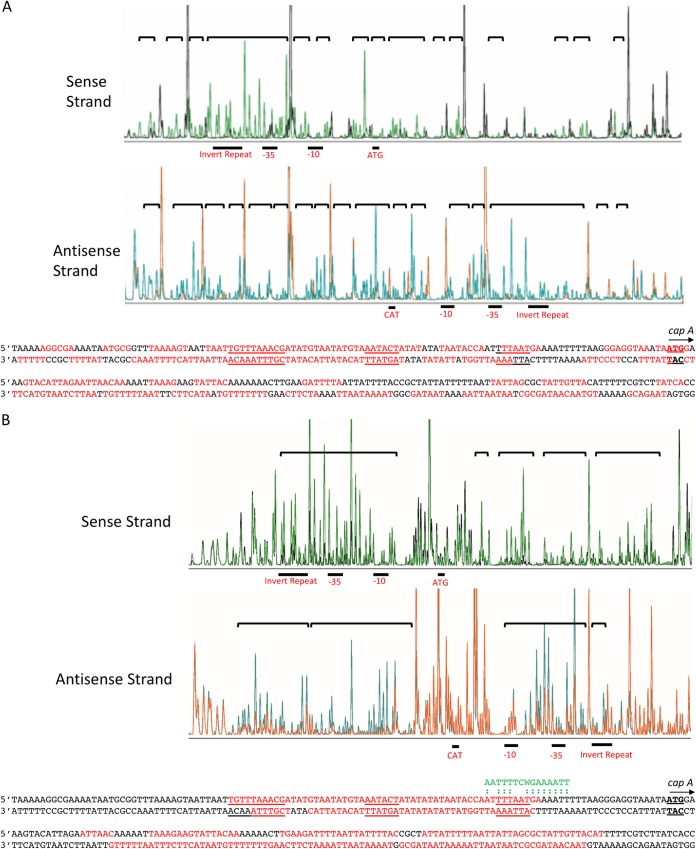
The above EMSA competition experiments revealed that XdrA specifically binds to a large region including the promoter of the cap operon and the capA gene. To further determine the binding site more precisely, we synthesized a 6-carboxyfluorescein (FAM)-VIC dual-labeled 312-bp probe (Fig. 2A, fragment D) and performed a fluorescence-based footprinting experiment on both the sense and antisense strands as described in Materials and Methods. As shown in Fig. 4A, XdrA protected almost continuously 237-bp regions on both the sense and antisense strands that centered around the capA ATG start codon. This region includes the −10 and −35 promoter regions, the upstream 10-bp inverted repeat, and a 123-bp capA coding region. The footprinting results are largely consistent with those of the EMSAs.
FIG 4.
DNase I footprinting of the 5′-FAM-labeled sense strand and the 5′-VIC-labeled antisense strand of the Pcap probe. A reduction in intensity of DNase I-digested fragment in the presence of 674 nM XdrA (A) or 6.95 μM CodY (B) (black peaks for the sense strand panel and orange peaks for the antisense strand) compared to that in its absence (green peaks for the sense strand and blue peaks for the antisense strand) indicates protection. Protected regions are indicated by brackets. The protected sequences are indicated in red. CodY consensus sequences are shown in green above the matched sequences (indicated by colons) in the Pcap-capA region.
XdrA and CodY bind the capsule promoter with overlapping binding sites.
CodY has been shown to repress capsule gene expression (15–17). We previously employed EMSA and showed that CodY also bound to a Pcap-containing DNA fragment (Fig. 2, fragment A) used for the XdrA binding study above (15), suggesting that XdrA and CodY may physically interact when bound to Pcap or compete for binding. To investigate the potential interactions between these two repressors with regard to cap gene regulation, we first performed an EMSA to determine the approximate Kd of the purified His6-CodY protein using the 312-bp fragment D as a probe. We found that CodY bound with much less affinity than XdrA (Fig. 3B). However, the binding caused a smear when the CodY concentration was less than ∼400 nM, which prevented us from estimating an accurate Kd value. The smearing effect was reproducible. Nonetheless, these results suggest CodY binds much less efficiently than XdrA, with a Kd of more than ∼200 nM. We then determined the CodY binding site in the cap promoter region by DNase I footprinting using 6.95 μM His6-CodY. As shown in Fig. 4B, CodY protected two well-separated sites in the Pcap-capA region: one upstream of the ATG start codon and one downstream. The upstream site (∼55 bp) centers around the −35 promoter region in which the 10-bp inverted repeat was protected in the sense strand but partially protected in the antisense strand. The downstream site encompassing a 92-bp region is entirely within the capA coding sequence. Compared to the XdrA protected region, CodY protected a slightly narrower region, but both proteins protected sequences in the promoter region as well as the coding sequences. Interestingly, the protected regions of both proteins centered on the ATG start codon. These results revealed that the binding sites of XdrA and CodY overlapped substantially. Of note, we were unable to get meaningful CodY footprinting results in the presence of Ile and GTP or with a lower concentration of His6-CodY (3.48 μM).
Repression of capsule by XdrA requires the presence of codY in the chromosome.
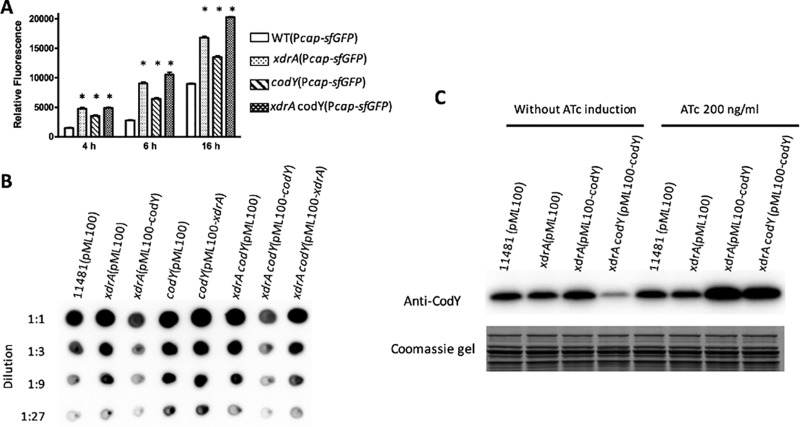
The extensive overlap of the XdrA and CodY binding sites in the Pcap-capA region suggests that the two regulators may interact functionally or physically to repress capsule gene expression. To test this possibility, we first measured the effect of cap gene expression by xdrA and/or codY mutations by using a plasmid carrying a Pcap-sfgfp reporter fusion at different growth phases. We found that the effect was observed in all growth phases but most profoundly at 4 h and 6 h (Fig. 5A). We then performed a genetic epistasis assay with capsule production at the 4-h time point as a readout. As shown in Fig. 5B, the increase in capsule in the xdrA mutant was complemented by the wild-type codY gene under the control of Pxyl-tetO in the presence of ATc, whereas the xdrA gene did not complement the codY mutation. Likewise, an xdrA-codY double mutant was complemented by the codY gene but not by xdrA. These results indicate that the codY mutation effect is epistatic to that of xdrA. Genetically, this suggests a hierarchical regulation of capsule in which CodY regulates capsule downstream of the XdrA. However, the xdrA mutation did not affect CodY production as shown by Western blotting (Fig. 5C), suggesting that XdrA does not regulate CodY. Furthermore, as both XdrA and CodY repress the cap operon by direct DNA binding, it is not likely that XdrA controls capsule genes indirectly through CodY.
FIG 5.
Epistasis assay. (A) Effect of single and double mutants of xdrA and codY on Pcap activity at different time points as indicated by a Pcap-sfgfp reporter (n = 4). (B) Capsule assay showing codY is epistatic to xdrA. Capsules were isolated from cultures grown in TSB-0G in the presence of 10 μg/ml chloramphenicol for 2 h and then induced with 200 ng/ml of ATc for 2 h and assayed by immunoblotting using an anticapsule antibody. (C) CodY production assayed by Western blotting. A portion of the Coomassie blue-stained gel was used as a loading control. Cell extracts were isolated from cultures grown in TSB-0G in the presence of 10 μg/ml chloramphenicol for 4 h or for 2 h and then induced with 200 ng/ml ATc for 2 h as indicated. *, P < 0.0001 versus the wild type.
The Western blotting results in Fig. 5C also showed that CodY, in the presence of the ATc inducer, was greatly overexpressed in the complemented strains compared to that in the wild type and the xdrA mutant. Without ATc, CodY was not overproduced and capsule production was not complemented (data not shown). Since the xdrA mutation resulted in an increased capsule phenotype in which codY is present as a single copy in the chromosome, the results of complementation in the presence of ATc suggest that a higher concentration of CodY is required for the repression of cap genes in the absence of XdrA. Taking these results altogether, we suggest that XdrA may play a secondary role by assisting in the binding of CodY to Pcap. This hypothesis might explain the epistatic effect of codY to xdrA in which only the overexpression of codY, but not the overexpression of xdrA, overcomes the effect of the double mutation. This might also explain how the loss of either codY or xdrA in the wild-type strain results in increased capsule production.
One possible mechanism by which XdrA promotes CodY binding is that the binding of XdrA to the Pcap DNA helps subsequent CodY binding. To test this possibility, we employed EMSA using an increasing amount of CodY in the presence of a constant amount of XdrA at a concentration causing ∼50% shift (Fig. 6). By comparing the intensity of the labeled fragments shifted by single protein to those shifted by both proteins, we found that the presence of XdrA did not augment the binding of CodY to Pcap, suggesting the binding of XdrA does not reduce the amount of CodY needed for binding. It should be noted here that the probe used in this cobinding experiment was labeled by FAM and VIC for direct observation of the shifted bands. As the method does not use an antibody for amplification of the probe signal, high concentrations of probes and proteins as indicated in Fig. 6 were needed in the reactions.
FIG 6.
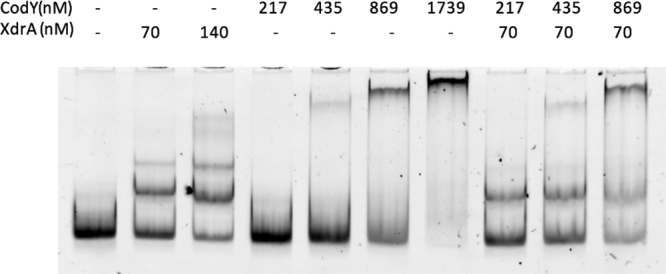
EMSA of Pcap with the His6-XdrA and His6-CodY proteins. A constant amount of His6-XdrA (70 nM) causing partial shift was incubated with FAM-VIC-labeled Pcap fragment (17.4 nM fragment D in Fig. 2A) in the presence of increasing amounts of His6-CodY. The EMSA gel was directly imaged with a fluorescence imager (Bio-Rad ChemiDoc MP imaging system using 530/28-nm emission filter).
Regulation of xdrA.
Many regulators have been identified that affect capsule expression at the transcriptional level in S. aureus. However, most of the regulators have been shown to indirectly regulate the cap genes. To exert their regulatory effect on capsule expression, these upstream regulators need to ultimately interact with those that directly interact with the Pcap region, most commonly, DNA-binding regulators. To determine whether XdrA serves as a downstream regulator for those indirect regulators, we tested the effect on xdrA expression of a group of known upstream regulators, including mgrA, agr, saeRS, clpC, sbcDC, and arlRS, by Northern blotting. As xdrA is an early gene (11), we isolated the mRNA of these regulatory mutants and the wild-type strain CYL11481 at an optical density at 600 nm (OD600) of ∼0.25. None of these regulators had an effect on xdrA gene expression. The use of cultures at a higher OD of ∼1.3 also did not show an effect (data not shown).
It has been reported that the xdrA and cap genes are affected by antibiotics targeting cell wall synthesis, such as fosfomycin or oxacillin (24, 25). To test whether oxacillin has an effect on xdrA, we used a red fluorescent protein, DsRed.T3, under the control of the xdrA promoter in the chromosome. The results in Fig. S2A in the supplemental material show that a subinhibitory concentration of oxacillin (65 ng/ml) slightly increased xdrA transcription (increased by only approximately 9%, P = 0.0249). At the same concentration, oxacillin reduced capsule promoter activity by approximately 40% as measured by using a capsule promoter fused to a green fluorescent protein reporter gene. However, oxacillin also similarly reduced capsule promoter activity in both the wild type and the xdrA mutant (Fig. S2B). These results suggest that a subinhibitory amount of oxacillin has a small but significant positive effect on xdrA transcription. However, the effect of oxacillin on Pcap promoter activity is not through XdrA, as the effect of oxacillin on capsule production is similar between the wild type and the xdrA mutant.
DISCUSSION
The expression of cap genes in S. aureus has been reported to be transcriptionally regulated by multiple regulators. Many of these regulators affect cap gene expression indirectly. In this study, we characterized the binding sites in the Pcap-capA region for two cap repressors, XdrA and CodY. Our results showed that both repressors bound to a broad region of sequence, including the 10-bp inverted repeat upstream of the promoter, the promoter region, and the coding region of the first gene of the cap operon. These results are in contrast to those for RbsR, an activator that we recently reported (9), which binds to a much shorter region (46 nucleotides [nt] of the sense strand and 16 nt of the antisense strand) encompassing the 10-bp inverted repeat and the −35 promoter region. Since the 10-bp inverted repeat is required for RbsR binding, it is possible that XdrA and CodY also repress cap gene expression by competing with RbsR binding at the inverted repeat, although the 10-bp inverted repeat was not essential for XdrA or CodY binding (data not shown).
The unusually long DNA binding site for XdrA in the cap promoter and the capA coding region suggests that XdrA binds to the region multimerically as it wraps around the DNA for more than 200 bp. This might explain why multiple bands of XdrA-Pcap DNA complex were observed in the EMSAs (Fig. 3A). Bacterial DNA-binding regulators with long binding sites, such as E. coli Fur, have been shown to bind to a preferred site with a high affinity and then extend their binding at higher regulatory concentrations (26). As our results do not distinguish high-affinity from low-affinity sites, it is not known whether XdrA binds to a short preferential binding site first and then extends to the adjacent region. CodY also bound to the same region but with two distinct blocks totaling more than 100 bp in length. The finding that XdrA and CodY shared extensive overlapping binding sites and the subsequent genetic epistasis analyses (Fig. 5) led us to conclude that XdrA promotes the repression of the cap operon by CodY. The long binding site may be needed for XdrA to play an auxiliary role, such as DNA bending, in promoting CodY binding. Indeed, many members of Xre-HTH family cause bending upon DNA binding (27). However, the presence of XdrA did not increase the affinity of CodY, as shown by the in vitro cobinding experiments. Thus, the mechanism of the CodY-XdrA interaction remains to be elucidated.
The consensus CodY binding site, AATTTTCWGAAAATT (where W is A or T), in S. aureus has been defined, which closely resembles that in Bacillus and other low-GC Gram-positive organisms (15). A genome-wide search for in vivo binding in the S. aureus chromosome failed to identify a CodY binding site in the cap promoter region or in the capA gene (15). By examining the sequence, we found a very good 13-bp match with the 15-bp consensus sequence located at the −10 promoter region, but this is only partially within the CodY binding sequences as defined by our footprinting experiments. In addition, we found several matches with only 8 or 9 bp identical to the consensus sequence. These degenerate CodY boxes may provide weak binding sites for CodY, which may explain the high Kd value. The less than ideal binding sites may also explain the need for XdrA to promote CodY binding, though the mechanism involved is unknown. In Bacillus subtilis, CodY has been shown to bind as a dimer (28, 29). However, the long binding site we found here indicates that more than two molecules of CodY may bind to this region. It is possible that XdrA promotes multimerization of CodY.
XdrA was initially identified by its ability to bind the mecA promoter. However, xdrA mutation does not affect mecA expression, though it affects β-lactam resistance (10). A subsequent study by microarray analysis did not further reveal how XdrA affects β-lactam resistance (11). Instead, the gene profiling study suggests that XdrA is an activator of several genes, including spa, encoding protein A, and a repressor of several other genes, including the cap genes. By focusing on the regulation of spa, these authors found that XdrA was a direct activator of spa by acting on a cis element of approximately 100 bp in length just upstream of the spa −35 promoter region. However, they were unable to define the binding site either by EMSA or by footprinting (11). Our study showing XdrA repression of cap genes is consistent with the gene profiling study. A comparison between the XdrA binding site on the cap promoter and the putative binding region on the spa promoter, however, showed very little homology except that both are highly AT rich. Since XdrA activates spa but represses cap, it is possible that XdrA binds to different DNA sequences in two different regulatory modes. Most recently, XdrA was also found to affect biofilm formation and the expression of more than 100 genes (12). Surprisingly, cap genes were not found to be differentially regulated by XdrA in this study. The discrepancy could be due to the different strains used in our and their studies.
As XdrA is a regulator that directly binds to the cap promoter, it is possible that it serves as a downstream regulator in the cap regulatory network. However, our Northern analysis showed that none of the known indirect regulators of the cap genes we tested had an effect on xdrA transcription. This is consistent with the results from McCallum et al. showing that none of the regulators of spa (SarS, SarA, and Agr) had an effect on xdrA transcription (11). Thus, it is likely that XdrA does not serve as a downstream mediator for those indirect regulators affecting cap or spa, but rather, it may directly sense external signals. To identify a possible signal, we searched a database of microarray analyses (http://www.satmd.org) and found that a few cell wall-targeting antibiotics had an effect on xdrA and cap expression (24, 25). Although we showed that oxacillin had a small but significant effect on xdrA transcription, our genetic analyses suggest that the effect on capsule by oxacillin is not mediated by XdrA.
The cap operon has been shown to be transcribed primarily by a single promoter with a cis-acting element of a 10-bp inverted repeat 13 bp upstream of the −35 region of the promoter (8). Besides CodY and XdrA reported in this study, six other regulators, RbsR (9), KpdE (30), SpoVG (31), AirR (32), CcpE (33), and MsaB (34), have been shown to bind to this relatively simple promoter to affect capsule transcription. Furthermore, we also recently identified 4 putative Pcap-binding regulators yet to be characterized (9). Among these, the binding sites of RbsR, CodY, and XdrA have been mapped by footprinting, whereas EMSA was used to define the binding of other regulators. RbsR is an activator of the cap operon that binds to the 10-bp inverted repeat (9). This element is also likely the binding site for another activator, MsaB, as mutations within the repeat prevent MsaB from binding (34). Interestingly, as reported here, the inverted repeat is also part of the binding sites for CodY and XdrA repressors. Whether this element is involved in the binding of other Pcap-binding regulators remains to be determined. As Pcap is a simple promoter containing only one cis-acting element, the number of regulators capable of binding to this simple cap promoter is indeed remarkable. Besides these direct regulators, a large number of indirect regulators of capsule have been identified (9, 16, 30–39). The sheer number of direct and indirect regulators indicates that the regulatory network affecting capsule is extremely complex. How direct DNA-binding regulators interact with the upstream indirect regulators is largely unknown. This degree of complexity is surprising and further indicates that capsule is playing an intricate role in S. aureus infection that requires a large number of regulators to carefully control its production.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. NE1445 and NE1555 were obtained from the Nebraska transposon library collection (44) distributed by BEI Resources (https://www.beiresources.org/) through the Network of Antimicrobial Resistance in S. aureus (NARSA) program. The culture conditions were essentially as previously described (9).
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| Strains | ||
| S. aureus | ||
| RN4220 | Restriction-negative laboratory strain | J. Iandolo |
| Newman | Wild-type CP5 strain | T. Foster |
| CYL6401 | Strain Becker with 4-bp mutation in 10-bp invert-repeat | 8 |
| CYL11481 | Newman saeS(P18L) | 16 |
| NE1445 | USA300 FPR3757 xdrA::bursa | NARSA |
| NE1555 | USA300 FPR3757 codY::bursa | NARSA |
| CYL12837 | CYL11481 xdrA::bursa | This study |
| CYLA137 | CYL11481 xdrA::aphA-3 | This study |
| CYLA161 | CYL11481 codY::bursa | This study |
| CYLA163 | CYLA137 xdrA::aphA-3 codY::bursa | This study |
| CYL12968 | CYL11481 PxdrA::rfp | This study |
| E. coli | ||
| XL1-Blue | Host strain | Stratagene |
| CYL3259 | BL21(λDE3)(pLysS)(pTL3258) | This study |
| CYL4161 | Rosetta2(DE3)(pLysS)(pML4159) | This study |
| Plasmids | ||
| pLL31 | Shuttle vector with Pspac | 16 |
| pKAN | Vector for allele replacement | 40 |
| pRFP-F | RFP (DsRed.T3) fluorescent reporter | 40 |
| pML100 | Shuttle vector, with Pxyl-tetO | 41 |
| pLI50 | Shuttle vector | 42 |
| pML4155 | pML100 with xdrA | This study |
| pET28a(+) | E. coli His tag protein expression vector | Novagen |
| pET15b | E. coli His tag protein expression vector | Novagen |
| pML4159 | pET28a(+) with xdrA | This study |
| pTL3258 | pET15b with codY | This study |
| pCL11979 | pLL31 with codY | 16 |
| pMLA166 | pML100 with codY | This study |
| pCM11 | sfgfp expression vector | 43 |
| pMLE61 | pLI50 with Pcap::sfgfp | This study |
| pCL3169 | pGEM-T Easy with 614-bp Pcap fragment | 9 |
Strain and plasmid construction.
The primers used for strain and plasmid construction are listed in Table 2. The transposon mutants CYL12837 (xdrA::bursa) and CYLA161 (codY::bursa) were constructed by chromosomal phage transduction from NE1445 and NE1555, respectively, to CYL11481. To construct the xdrA-codY double mutant, ermB in CYL12837 was first replaced with aphA-3, coding for kanamycin resistance, using pKAN as described previously (40). The resultant strain, CYLA137, was transduced with codY::bursa from NE1555 to yield a double mutant xdrA::aphA-3 codY::bursa strain (CYLA163). All constructs were verified by PCR.
TABLE 2.
Oligonucleotide primers used in this study
| Name | Sequence |
|---|---|
| NM1739-2 | GATGATATCGAATTCCATTAATCATTTCACTTTCTGCTCA |
| NM1739-4 | GATGATATCGTCGACGGATCCCAGAAAGGTAGATAACAGAATGGATAGACA |
| NM1739-5 | CCTGGTGCCGCGCGGCAGCCATATGGATAGACAGAGTTTTACAGATTTAATTCAAACAA |
| NM1739-6 | TCGACGGAGCTCGAATTCGGATCCCTAGATATGTACTAATTCTTCTTTAGCATTTCTGTT |
| NM1739-7 | GGATAGACAGAGTTTTACAGATTTAATTCAAAC |
| NM1739-8 | GTTAAAGTATGTTTCAGCTTCTCTCATTTTAGAAG |
| codY7 | CATATGAGCTTATTATCTAAAAC |
| codY8 | GGATCCTTATTTACTTTTTTCT |
| cp8gs3 | CCATTATTTACCTCCCTTAAAAATTTTC |
| cp8gs3F | GAAAATTTTTAAGGGAGGTAAATAATGG |
| cp8gs4 | AACGATATGTAATATGTAAATAC |
| cp8gs5 | ACATATCGTTTAAACAATTAATTACTTT |
| cp8gs6 | CTACTTTAGAGTATAATTATTTTTAATTTC |
| cp8gs11 | GATATTTAGGTGATAAGACGA |
| cp8gs20 | GATTCACTAAAATTTGAGTGTTAGCTT |
| cp8gs21 | GATTTTAATTATTTTACCGCTATTATTTTTAATTATTAG |
| cap8A1 | ACTAAGGGTGACAATCCTCAG |
| cap8A2 | AAGTCCTTTGACACCTCATCTA |
| cap8D1 | TATGGATGGCGTTGAGTTATT |
| cap8D2 | CAACAGGATCTCTGCCTAGTA |
| icaR-P1 | CTGCAGGCAATTTCTTTACCTACCTTTCGTTAG |
| icaR-P2 | CTGCAGCTTATCCTTCAATTTTTATAACCCCCTAC |
| VIC-FP20 | VIC-5′GATTCACTAAAATTTGAGTGTTAGCTT |
| FAM-FP6 | 6FAM-5′CTACTTTAGAGTATAATTATTTTTAATTTC |
For xdrA complementation, pML4155 containing the CYL11481 xdrA gene under the control of Pxyl-tetO was constructed by ligating a 601-bp PCR fragment, amplified by primers NM1739-4 and NM1739-2, into the SalI and EcoRI sites of pML100. For codY complementation, pMLA166 containing the codY gene was constructed by ligating an 838-bp HindIII-SstI fragment in pCL11979 into similarly digested pML100. To express the recombinant His6-XdrA protein in E. coli, a 465-bp fragment containing the xdrA gene from CYL11481 was amplified using primers NM1739-5 and NM1739-6 and ligated to NheI-BamHI-digested pET-28a(+) (Novagen, Madison, WI). To express the recombinant His6-CodY protein in E. coli, a 773-bp fragment containing the codY gene from CYL11481 was amplified using primers codY7 and codY8 and ligated to NdeI-BamHI-digested pET-15b (Novagen). All clones were validated by restriction mapping and sequencing of the inserts. Superfolder green fluorescent protein gene (sfgfp) reporter plasmid pMLE61 containing the Pcap::sfgfp fusion was constructed by inserting the sfgfp fragment from pCM11 into the KpnI and EcoRI sites of pLI50, followed by ligating the Pcap fragment from pCL3169 into the HindIII and KpnI sites. The fusion of red fluorescent protein DsRed.T3 under the control of the xdrA promoter in the chromosome was accomplished by replacing the bursa insertion in CYL11481 xdrA::bursa with an rfp reporter using pRFP-F as described previously (40).
Recombinant protein expression and purification.
To express His6-XdrA proteins, pML4159 was transformed into E. coli Rosetta 2(DE3)(pLysS) (Novagen). To express His6-CodY protein, pTL3258 was transformed into E. coli BL21(λDE3)(pLysS) (Novagen). Procedures for protein expression and purification were essentially as described previously (9). The purity of the His6-XdrA and His6-CodY proteins was examined by SDS-PAGE (results not shown).
Northern and Western blot analyses.
Total RNAs were isolated as described previously (9). For Northern blotting, the 430-bp xdrA-specific DNA probe was synthesized by using a PCR Dig Probe synthesis kit (Roche Applied Sciences) with primer pairs NM1739-7 and NM1739-8. Northern blotting was carried out as described previously (9). For Western blotting, bacterial cells were cultured for 4 h at 37°C with shaking at 225 rpm and normalized to an OD660 of 2. The cell extracts were used for Western blotting according to the protocol previously described and using anti-CodY antibody (39).
Nonradioactive DNase I footprinting.
A 312-bp probe, which corresponds to the −135 to +177 region of Pcap with respect to the transcriptional start site of the cap operon (Fig. 2A, fragment D), was synthesized by PCR using fluorescent dye 6-FAM-labeled primer 6-FAM-FP6 and VIC-labeled primer VIC-FP20. The PCR DNA fragments were purified using a NucleoSpin column (Clontech, Mountain View, CA) and used for DNase I footprinting according to the protocol of Zianni et al. (45) as previously described (9). Briefly, for XdrA-Pcap footprinting, the reaction mixtures (20 μl), consisting of 240 ng purified His6-XdrA, 86 ng of fluorescent dye-labeled DNA probe, 2 μg of bovine serum albumin (BSA), 0.1 μg of poly-l-lysine, and 1 μg of poly(dI-dC) in the binding buffer [20 mM HEPES (pH 7.6), 10 mM (NH4)2SO4, 1 mM dithiothreitol (DTT), 0.2% Tween 20, 30 mM KCl], were incubated at 23°C for 15 min. DNase I (0.06 U; New England BioLabs) was added to the reaction mixtures and incubated at 23°C for 3.5 min. The reactions were stopped by incubating at 78°C for 10 min. The CodY-Pcap footprinting reaction was similar to the XdrA-Pcap footprinting, except the 20-μl reaction mixtures contained 4 μg purified His6-CodY or 4 μg BSA for control, 88 ng of fluorescent dye-labeled DNA probe, and 0.04 U of DNase I. The DNA fragments were purified with a Qiagen Mini Elute PCR kit (Qiagen, Valencia, CA) and eluted in 25 μl of H2O. The experiments were repeated two times. Fifteen microliters of each purified DNA fragment, along with primers FAM-FP6 and VIC-FP20 and plasmid pCL3169, was submitted to Ohio State University Plant-Microbe Genomic Facility for fragment analysis on an Applied Biosystems 3730 DNA analyzer (45). The XdrA and CodY DNA binding sites were determined by aligning the sizes of the fragments and sequences of the probe.
Electrophoretic mobility shift assays.
EMSAs were performed as described previously (41). The DNA fragments (shown in Fig. 2A) were generated from strain Newman chromosome DNA by PCR (156-bp fragment A using primers cp8gs6 and cp8gs3 or 312-bp fragment D using primers cp8gs6 and cp8gs20) and labeled with digoxigenin-ddUTP using a Dig Gel Shift kit (Roche Applied Science, Indianapolis, IN). DNA fragments B, C, E, F, G, and H, capD, and Pica used for competition experiments were generated from strain Newman chromosome DNA by primer pairs cp8gs5/cp8gs6, cp8gs3/gp8gs4, cp8gs4/cp8gs11, cp8gs11/gp8gs3F, cp8gs11/cp8gs21, capA1/capA2, cap8D1/cap8D2, and icaR-P1/icaA-P1, respectively. A DNA fragment containing a 4-bp mutation within the 10-bp inverted repeat was amplified from CYL6401 chromosome using primers cp8gs6 and cp8gs3. An EMSA for testing the cobinding of XdrA and CodY was performed using FAM- and VIC-labeled fragment D amplified by primers 6-FAM-FP6 and VIC-FP20 as described in “Nonradioactive DNase I footprinting” above. Kd values, the protein concentrations at which 50% of the protein formed a complex with DNA, were determined by using Image Lab (Bio-Rad, Hercules, CA) software to quantitate the chemiluminescent signals in DNA bands (with subtraction of background signals). The data were fit with a binding equation by nonlinear regression using GraphPad Prism (San Diego, CA) software.
Fluorescence reporter assay.
S. aureus strains harboring the sfgfp reporter Pcap::sfgfp fusion (pMLE61) plasmid were grown in tryptic soy broth (TSB) without glucose (TSB-0G) with 10 μg/ml of chloramphenicol. The cultures were centrifuged and suspended in phosphate-buffered saline (PBS). Green florescence was measured in quadruplets in black 96-well microtiter plates (Costar 3720; Corning) using a Fluostar Omega plate reader (BMG Labtech) with an excitation wavelength of 485 nm and emission wavelength of 520 nm. The relative fluorescence values of promoter-reporter fusions were calculated by normalizing the average fluorescence from each sample to the corresponding absorbance at an OD600 and then subtracting the relative fluorescence value of the reporter plasmid control. For red fluorescence assays of the PxdrA-rfp fusion, the cultures were grown in TSB-0G for 2 h, and then 170 μg/ml chloramphenicol, 50 μg/ml kanamycin, and 50 μg/ml tetracycline were added and incubated for an additional 2.5 h for DsRed.T3 protein maturation (46, 47). Red fluorescence was measured as described above, using an excitation wavelength of 544 nm and an emission wavelength of 590 nm.
Capsule immunoblotting.
To measure capsule production, capsules were prepared as described previously (16) from cultures grown in TSB-0G. Serially diluted samples (1.5 μl each) were applied with a pipette directly to nitrocellulose membranes. The membranes were treated with a specific anticapsule antibody for detection as described previously (16).
Statistics.
Comparisons between means were analyzed by paired Student t tests with GraphPad Prism (San Diego, CA).
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grant AI113766 from the National Institute of Allergy and Infectious diseases. Nebraska transposon mutants were obtained through the Network of Antimicrobial Resistance in S. aureus (NARSA) program supported by the NIAID/NIH. We also acknowledge the UAMS sequencing core and proteomic core supported in part by National Institutes of Health grants P20GM103420, P30GM103450, and P20GM103625.
We thank Ravi Gupta and Dereje Gudeta for critical reading of the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00203-18.
REFERENCES
- 1.Novick RP. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol 48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 2.Bronner S, Monteil H, Prévost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol Rev 28:183–200. doi: 10.1016/j.femsre.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 3.Junecko J, Zielinska AK, Mrak LN, Ryan DC, Graham JW, Smeltzer M, Lee CY. 2012. Transcribing virulence in Staphylococcus aureus. World J Clin Infect Dis 2:63–76. doi: 10.5495/wjcid.v2.i4.63. [DOI] [Google Scholar]
- 4.O'Riordan K, Lee JC. 2004. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17:218–234. doi: 10.1128/CMR.17.1.218-234.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee CY, Lee JC. 2006. Staphylococcal capsules, p 456–463. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-positive pathogens, 2nd ed. American Society for Microbiology, Washington, DC. [Google Scholar]
- 6.Sau S, Bhasin N, Wann ER, Lee JC, Foster TJ, Lee CY. 1997. The Staphylococcus aureus allelic genetic loci for serotype 5 and 8 capsule expression contain the type-specific genes flanked by common genes. Microbiology 143:2395–2405. doi: 10.1099/00221287-143-7-2395. [DOI] [PubMed] [Google Scholar]
- 7.Sau S, Sun J, Lee CY. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol 179:1614–1621. doi: 10.1128/jb.179.5.1614-1621.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ouyang S, Sau S, Lee CY. 1999. Promoter analysis of the cap8 operon, involved in type 8 capsular polysaccharide production in Staphylococcus aureus. J Bacteriol 181:2492–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lei MG, Lee CY. 2015. RbsR activates capsule but represses the rbsUDK operon in Staphylococcus aureus. J Bacteriol 197:3666–3675. doi: 10.1128/JB.00640-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ender M, Berger-Bächi B, McCallum N. 2009. A novel DNA-binding protein modulating methicillin resistance in Staphylococcus aureus. BMC Microbiol 9:15. doi: 10.1186/1471-2180-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCallum N, Hinds J, Ender M, Berger-Bächi B, Meier PS. 2010. Transcriptional profiling of XdrA, a new regulator of spa transcription in Staphylococcus aureus. J Bacteriol 192:5151–5164. doi: 10.1128/JB.00491-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeFrancesco AS, Masloboeva N, Syed AK, DeLoughery A, Bradshaw N, Li GW, Gilmore MS, Walker S, Losick R. 2017. Genome-wide screen for genes involved in eDNA release during biofilm formation by Staphylococcus aureus. Proc Natl Acad Sci U S A 114:E5969–E5978. doi: 10.1073/pnas.1704544114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonenshein AL. 2005. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr Opin Microbiol 8:203–207. doi: 10.1016/j.mib.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Brinsmade SR. 2017. CodY, a master integrator of metabolism and virulence in Gram-positive bacteria. Curr Genet 63:417–425. doi: 10.1007/s00294-016-0656-5. [DOI] [PubMed] [Google Scholar]
- 15.Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. 2010. Direct targets of CodY in Staphylococcus aureus. J Bacteriol 192:2861–2877. doi: 10.1128/JB.00220-10 (Erratum, 192:4258, 2010.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luong TT, Sau K, Roux C, Sau S, Dunman PM, Lee CY. 2011. Staphylococcus aureus ClpC divergently regulates capsule via sae and codY in strain Newman but activates capsule via codY in strain UAMS-1 and in strain Newman with repaired saeS. J Bacteriol 193:686–694. doi: 10.1128/JB.00987-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. 2009. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol 191:2953–2963. doi: 10.1128/JB.01492-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montgomery CP, Boyle-Vavra S, Roux A, Ebine K, Sonenshein AL, Daum RS. 2012. CodY deletion enhances in vivo virulence of community-associated methicillin-resistant Staphylococcus aureus clone USA300. Infect Immun 80:2382–2389. doi: 10.1128/IAI.06172-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rivera FE, Miller HK, Kolar SL, Stevens SM Jr, Shaw LN. 2012. The impact of CodY on virulence determinant production in community-associated methicillin-resistant Staphylococcus aureus. Proteomics 12:263–268. doi: 10.1002/pmic.201100298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roux A, Todd DA, Velázquez JV, Cech NB, Sonenshein AL. 2014. CodY-mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J Bacteriol 196:1184–1196. doi: 10.1128/JB.00128-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ibberson CB, Jones CL, Singh S, Wise MC, Hart ME, Zurawski DV, Horswill AR. 2014. Staphylococcus aureus hyaluronidase is a CodY-regulated virulence factor. Infect Immun 82:4253–4264. doi: 10.1128/IAI.01710-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waters NR, Samuels DJ, Behera RK, Livny J, Rhee KY, Sadykov MR, Brinsmade SR. 2016. A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol Microbiol 101:495–514. doi: 10.1111/mmi.13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prax M, Lee CY, Bertram R. 2013. An update on the molecular genetics toolbox for staphylococci. Microbiology 159:421–435. doi: 10.1099/mic.0.061705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petek M, Baebler S, Kuzman D, Rotter A, Podlesek Z, Gruden K, Ravnikar M, Urleb U. 2010. Revealing fosfomycin primary effect on Staphylococcus aureus transcriptome: modulation of cell envelope biosynthesis and phosphoenolpyruvate induced starvation. BMC Microbiol 10:159. doi: 10.1186/1471-2180-10-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyle-Vavra S, Yin S, Jo DS, Montgomery CP, Daum RS. 2013. VraT/YvqF is required for methicillin resistance and activation of the VraSR regulon in Staphylococcus aureus. Antimicrob Agents Chemother 57:83–95. doi: 10.1128/AAC.01651-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Escolar L, Pérez-Martín J, de Lorenzo V. 2000. Evidence of an unusually long operator for the fur repressor in the aerobactin promoter of Escherichia coli. J Biol Chem 275:24709–24714. doi: 10.1074/jbc.M002839200. [DOI] [PubMed] [Google Scholar]
- 27.Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. 2009. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science 323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blagova EV, Levdikov VM, Tachikawa K, Sonenshein AL, Wilkinson AJ. 2003. Crystallization of the GTP-dependent transcriptional regulator CodY from Bacillus subtilis. Acta Crystallogr D Biol Crystallogr 59:155–157. doi: 10.1107/S0907444902018358. [DOI] [PubMed] [Google Scholar]
- 29.Levdikov VM, Blagova E, Joseph P, Sonenshein AL, Wilkinson AJ. 2006. The structure of CodY, a GTP- and isoleucine-responsive regulator of stationary phase and virulence in Gram-positive bacteria. J Biol Chem 281:11366–11373. doi: 10.1074/jbc.M513015200. [DOI] [PubMed] [Google Scholar]
- 30.Zhao L, Xue T, Shang F, Sun H, Sun B. 2010. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect Immun 78:3506–3515. doi: 10.1128/IAI.00131-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jutras BL, Chenail AM, Rowland CL, Carroll D, Miller MC, Bykowski T, Stevenson B. 2013. Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins. PLoS One 8:e66683. doi: 10.1371/journal.pone.0066683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun F, Ji Q, Jones MB, Deng X, Liang H, Frank B, Telser J, Peterson SN, Bae T, He C. 2012. AirSR, a [2Fe-2S] cluster-containing two-component system, mediates global oxygen sensing and redox signaling in Staphylococcus aureus. J Am Chem Soc 134:305–314. doi: 10.1021/ja2071835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding Y, Liu X, Chen F, Di H, Xu B, Zhou L, Deng X, Wu M, Yang CG, Lan L. 2014. Metabolic sensor governing bacterial virulence in Staphylococcus aureus. Proc Natl Acad Sci U S A 111:E4981–E4990. doi: 10.1073/pnas.1411077111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Batte JL, Samanta D, Elasri MO. 2016. MsaB activates capsule production at the transcription level in Staphylococcus aureus. Microbiology, 162:575–589. doi: 10.1099/mic.0.000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupta RK, Luong TT, Lee CY. 2015. RNAIII of the Staphylococcus aureus agr system activates global regulator MgrA by stabilizing mRNA. Proc Natl Acad Sci U S A 112:14036–14041. doi: 10.1073/pnas.1509251112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luong TT, Newell SW, Lee CY. 2003. Mgr, a novel global regulator in Staphylococcus aureus. J Bacteriol 185:3703–3710. doi: 10.1128/JB.185.13.3703-3710.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luong TT, Lee CY. 2006. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology 152:3123–3131. doi: 10.1099/mic.0.29177-0. [DOI] [PubMed] [Google Scholar]
- 38.Chen Z, Luong TT, Lee CY. 2007. The sbcDC locus mediates repression of type 5 capsule production as part of the SOS response in Staphylococcus aureus. J Bacteriol 189:7343–7350. doi: 10.1128/JB.01079-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham JW, Lei MG, Lee CY. 2013. Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J Bacteriol 195:4506–4516. doi: 10.1128/JB.00758-13 (Erratum, 196:2324, 2014.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bose JL, Fey PD, Bayles KW. 2013. Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl Environ Microbiol 79:2218–2224. doi: 10.1128/AEM.00136-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lei MG, Cue D, Roux CM, Dunman PM, Lee CY. 2011. Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol 193:5231–5241. doi: 10.1128/JB.05454-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee CY, Buranen SL, Zhi-Hai Y. 1991. Construction of single-copy integration vectors for Staphylococcus aureus. Gene 103:101–105. doi: 10.1016/0378-1119(91)90399-V. [DOI] [PubMed] [Google Scholar]
- 43.Lauderdale KJ, Malone CL, Boles BR, Morcuende J, Horswill AR. 2010. Biofilm dispersal of community-associated methicillin-resistant Staphylococcus aureus on orthopedic implant material. J Orthop Res 28:55–61. doi: 10.1002/jor.20943. [DOI] [PubMed] [Google Scholar]
- 44.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zianni M, Tessanne K, Merighi M, Laguna R, Tabita FR. 2006. Identification of the DNA bases of a DNase I footprint by the use of dye primer sequencing on an automated capillary DNA analysis instrument. J Biomol Tech 17:103–113. [PMC free article] [PubMed] [Google Scholar]
- 46.Bevis BJ, Glick BS. 2002. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat Biotechnol 20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 47.Sörensen M, Lippuner C, Kaiser T, Mißlitz A, Aebischer T, Bumann D. 2003. Rapidly maturing red fluorescent protein variants with strongly enhanced brightness in bacteria. FEBS Lett 552:110–114. doi: 10.1016/S0014-5793(03)00856-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.