Abstract
Geological activities and climate oscillations during the Quaternary period profoundly impacted the distribution of species in Southwest China. Some plant species may be harbored in refugia, such as the dry-hot valleys of Southwest China. Cycas chenii X. Gong & W. Zhou, a critically endangered cycad species, which grows under the canopy in subtropical evergreen broad-leaved forests along the upstream drainage area of the Red River, is endemic to this refugium. In this study, 60 individuals of C. chenii collected from six populations were analyzed by sequencing two chloroplast intergenic spacers (cpDNA: psbA-trnH and trnL-trnF) and two nuclear genes (PHYP and RBP-1). Results showed high genetic diversity at the species level, but low within-population genetic diversity and high interpopulation genetic differentiation. A Bayesian phylogenetic tree based on cpDNA showed that five chloroplast haplotypes were clustered into two clades, which corresponds to the division of the western and eastern bank of the Red River. These data indicate a possible role for the Red River as a geographic barrier to gene flow in C. chenii. Based on our findings, we propose appropriate in situ and ex situ conservation strategies for C. chenii.
Keywords: Cycas chenii, Genetic variation, Phylogeography, Conservation
1. Introduction
Southwest China experienced glacial–interglacial cycles in the Pleistocene approximately 2.4–0.01 million years ago (MYA) (Zhou et al., 2006, Royden et al., 2008). Therefore, the dry-hot valleys of Southwest China, such as Red River valley, are recognized as potential refugia for some plant species (Guan and Zhou, 1996, Wang et al., 1996).
Extant cycads are composed of the two families (Cycadaceae and Zamiaceae) with ten genera, and are mainly distributed in Asia, Australia, South and Central America and Africa. Approximately 200 (62%) cycads are threatened with extinction (Jian et al., 2006, Hoffmann et al., 2010). There are about 25 Cycas species (21%) distributed in China (Calonje et al., 2016). The range of these species is often limited by habitat destruction and fragmentation, commonly attributed to planting economic crops along with over-collection for food, medicine and ornamentals (Wang et al., 1996). The Convention on International Trade in Endangered Species (CITES) of Wild Fauna and Flora (WFF) gave all cycads ‘First Grade’ conservation status in China (Xiao et al., 2004). China harbors abundant Cycas diversity, especially in the drainage areas of the Red River, which is considered a secondary diversification center of Cycas (Hill, 2008).
There are more than 14 Cycas species, of which 10 are endemic to the basin region of the Red River (Hill, 2008). Cycas chenii X. Gong & W. Zhou (Section Stangerioides) is a recently described species (Zhou et al., 2015). This species is distributed in Chuxiong and Honghe of Yunnan Province, China, along the upstream drainage areas of the Red River (also called Yuanjiang in China). It occurs on a range of substrates from limestone to shale or schist, which is characteristic of steep slopes at altitudes ranging from 500 m to 1300 m (Zhou et al., 2015). Only six populations have been discovered, four at the northeast side of the Red River and two at the southwest side. To date, the total population size of C. chenii is estimated at less than 500 individuals across its geographic distribution, with all the know populations of the species being far from any protected areas (Zhou et al., 2015). This species is threatened by ongoing land-clearing and over-collection. Narrow range, small population sizes and presence of a potential barrier to gene flow (the Red River) necessitates understanding extent and structure of genetic variation as a prerequisite for working out the appropriate conservation strategy for this species. The organelle DNA of cycads is maternally inherited and is transmitted only by seeds while their nuclear DNA is biparentally inherited and is transmitted via both seeds and pollens (Huang et al., 2004, Zhong et al., 2011, Feng et al., 2014). Therefore, using these two genetic markers together allows a greater understanding of the role that seed vs. pollen flow play in spatial structuring of genetic variation. Thus, we employed both maternally inherited chloroplast DNA (cpDNA) and biparentally inherited nuclear DNA (nDNA) markers to investigate (i) the extent and structure of genetic variation in C. chenii, and (ii) the role of the Red River in shaping this structure. Based on our results, we propose suitable conservation strategy for the species.
2. Materials and methods
2.1. Study species and population sampling
All C. chenii populations have less than 100 individuals. All currently known populations of C. chenii were sampled during August to September in 2012. Four populations (ZS, DT, WJ and SP) were sampled from the northeast of the Red River (NE group) and the other two (ML and LH) were from the southwest of the Red River (SW group) (Fig. 1). The distance between sampled individuals was at least 5 m, increasing the likelihood of sampling genetically unrelated individuals. Fresh leaves were dried in silica gel after collection and stored at room temperature until DNA extraction. Voucher specimens were stored in the herbarium of the Kunming Institute of Botany, Chinese Academy of Sciences (KUN).
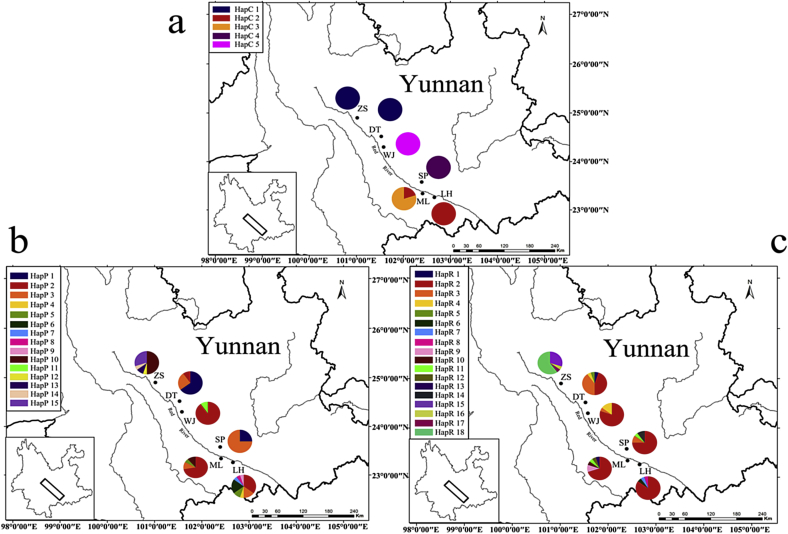
Fig. 1.
Geographic distribution of cpDNA (a) and nDNA (b. PHYP; c. RBP-1) haplotypes in C. chenii. ZS, Zhongshan; DT, Dutian; WJ, Wangjiadi; SP, Shiping; ML, Menglong; LH, Lianhua.
2.2. DNA extraction, PCR amplification and DNA sequencing
We extracted genomic DNA from dried leaves using the modified CTAB method (Doyle, 1991). After preliminary screening of DNA fragments from universal chloroplast and nuclear primers, we chose two cpDNA intergenic spacers, psbA-trnH and trnL-trnF (Taberlet et al., 1991, Shaw et al., 2005), and two nuclear genes, the phytochrome P gene PHYP (Zhou et al., 2015) and the gene that encodes the largest subunit of RNA polymerase II, RBP-1 (Liu et al., 2015). PCR amplification was carried out in 30 μL reactions. For cpDNA, the PCR reactions contained 10 ng of DNA, 3.0 μL of 10× PCR buffer, 1.5 μL of dNTPs (10 mM), 1.5 μL of MgCl2 (25 mM), 0.45 μL of Taq DNA Polymerase (5 U/μL), 0.45 μL of each primer, 1.5 μL of DMSO (20 mg/mL) and 19.65 μL of double-distilled water. For nDNA, the PCR reactions contained 20 ng DNA, 3.0 μL 10× PCR buffer, 1.5 μL dNTPs (10 mM), 2.25 μL MgCl2 (25 mM), 0.45 μL Taq DNA Polymerase (5 U/μL), 0.525 μL of each primer, 0.75 μL DMSO (20 mg/mL) and 18 μL double-distilled water. PCR amplification for cpDNA included an initial denaturation stage for 5 min at 80 °C, followed by 30 cycles of 1 min at 95 °C, 1 min annealing at 50 °C, extension for 1.5 min at 65 °C, and a final extension at 65 °C for 10 min. For nDNA: an initial denaturation stage at 94 °C for 5 min, was followed by 35 cycles at 95 °C for 1 min, annealing at 55 °C for 1 min, extension at 72 °C for 2 min, and a final extension for 10 min at 72 °C. All PCR products were sequenced in both directions with the same primers for the amplification reactions, using an ABI 3770 automated sequencer at Shanghai Majorbio Bio-pharm Technology Company Ltd.
2.3. Data analysis
We edited and assembled sequences using SeqMan (Swindell and Plasterer, 1997). Multiple alignments of the DNA sequences were performed in Clustal X, version 1.83 (Thompson et al., 1997), then the DNA sequences were adjusted by Bioedit, version 7.0.4.1 (Hall, 1999). Two cpDNA regions were combined by PAUP* 4.0b10 (Swofford, 2002). The concatenated sequence was used in the following analyses. For the two nuclear genes, heterozygous sites were resolved by applying the PHASE algorithm of DnaSP version 5.0 (Rozas et al., 2003). This program was also used for identification of haplotypes from the aligned DNA sequences and for calculation of Nei's nucleotide diversity (Pi) and haplotype diversity (Hd). Diversity and differentiation parameters (within-population diversity, Hs; total diversity, HT; differentiation for unordered and ordered alleles, GST and NST respectively), and a test whether NST is larger than GST, indicative of a phylogeographic structure (a situation when closely related haplotypes are more often found in the same area than less closely related haplotypes) were calculated with PERMUT (http://www.pierroton.intra.fr/genetics/labo/Software). Significance of the difference between NST and GST was assessed with 1000 random permutations following Burban et al. (1999). The hierarchical analysis of molecular variance (Excoffier et al., 1992) as implemented in Arlequin (Schneider et al., 2000) was used to estimate among-groups, among-populations within groups and within populations variance components. Isolation by distance (IBD) was tested between all pairs of populations as a correlation between genetic and geographic distance by computing Mantel test using GenAlEx version 6.3 (Peakall and Smouse, 2006). We calculated the ratio of pollen flow to seed flow following the formula pollen/seed migration ratio = [2(1/ΦSTc − 1) − (1/ΦSTn − 1)]/(1 − 1/ΦSTc), where ΦSTn and ΦSTc are levels of among-population differentiation calculated from nuclear and chloroplast markers, respectively (Mousadik and Petit, 1996).
We inferred phylogenetic relationships among cpDNA and nDNA haplotypes using Bayesian approach implemented in MrBayes, version 3.2.1 (Ronquist et al., 2012), in which four simultaneous runs with four Markov chains each were run for 105 generations and trees were sampled every 100 generations, with the first 25% trees from each run being discarded. The nucleotide substitution model used was GTR. Phylogeographic relationships of haplotypes were inferred by statistical parsimony separately for cpDNA and nDNA data using NETWORK 4.2.0.1 software (Forster et al., 2007). Indels were treated as single mutational events in the Network analysis.
To estimate coalescent time between lineages, we used the evolutionary rates 1.01 × 10−9 and 5.1–7.1 × 10−9 mutation per site per year for synonymous sites for cpDNA and nDNA (Graur and Li, 2000). Estimation of the time of divergence was performed by BEAST, version 1.6.1 (Drummond and Rambaut, 2007) using the HKY + G and HKY model for cpDNA and two nDNA fragments, respectively, chosen by model-test in MEGA 6.06 (Tamura et al., 2013), and a strict molecular clock. The BEAST program was also used to perform a Bayesian skyline plot analysis to infer the historical demography of C. chenii. Posterior estimates of the mutation rate and time of divergence were obtained by Markov Chain Monte Carlo (MCMC) analysis. The analysis was run for 107 iterations with a burn-in of 104. Genealogies and model parameters were sampled every 104 iterations. Convergence of parameters and mixing of chains were followed by visual inspection of parameter trend lines and checking of effective sampling size (ESS) values in three pre-runs. The ESS parameter was found to surpass 200, which suggested acceptable mixing and sufficient sampling. Adequate sampling and convergence to the stationary distribution were checked using TRACER, version 1.5 (Drummond and Rambaut, 2004). The pairwise mismatch distributions were examined in DnaSP. The sum-of-squared deviations (SSD) between the observed and expected mismatch distributions were computed, and P-values were calculated as the proportion of simulations producing a larger SSD than the observed SSD. We also used Arlequin, version 3.11 (Excoffier et al., 2005) to calculate the raggedness index and its significance to quantify the smoothness of the observed mismatch distribution. DnaSP was used to examine neutrality tests, Tajima's D (Tajima, 1989) and Fu & Li's F* (Fu, 1997), for detecting departures from population equilibrium.
3. Results
3.1. Genetic diversity and differentiation
Combined, the two cpDNA fragments, psbA-trnH and trnL-trnF, comprised 1227 positions, of which four were nucleotide substitutions and four were indels, resulting in five chloroplast haplotypes (Table 1). Of those, three haplotypes (HapC3, HapC4 and HapC5) were population-specific (ML, SP and WJ), whereas haplotype HapC1 and HapC2 were detected in two populations each (Table 1, Fig. 1a). The nuclear gene PHYP had a length of 883 bp with 18 nucleotide substitutions and one indel. Fifteen haplotypes were detected, of which two (HapP2 and HapP3) were shared by four populations, and three (HapP1, HapP5 and HapP10) were shared by two populations. The remaining ten haplotypes were unique (Table 1, Fig. 1b). The nuclear gene RBP-1 had a length of 918 bp with 30 nucleotide substitutions and one indel. Of the 18 detected haplotypes, HapR2 was the most widely distributed haplotype (Table 1, Fig. 1c).
Table 1.
Population locations and distribution of cpDNA and nDNA haplotypes.
| Population code | Population location | Altitude (m) | Latitude (N°) | Longitude (E°) | n | cpDNA |
PHYP |
RBP-1 |
|---|---|---|---|---|---|---|---|---|
| Haplotypes (No.) | Haplotypes (No.) | Haplotypes (No.) | ||||||
| ZS | Zhongshan, Chuxiong | 1383 | 24.899 | 101.017 | 10 | HapC 1 (10) | HapP 10 (10),HapP 12 (1), HapP 13 (2),HapP 14 (1) HapP 15 (6) |
HapR 15 (6),HapR 16 (1), HapR 17 (1),HapR 18(12) |
| DT | Dutian, Chuxiong | 1078 | 24.521 | 101.532 | 10 | HapC 1 (10) | HapP 1 (13),HapP 2 (5) HapP 3 (2) |
HapR 1 (1),HapR 2 (9),HapR 3 (8), HapR 4 (1),HapR 5 (1) |
| WJ | Wangjiadi, Chuxiong | 1260 | 24.302 | 101.5817 | 10 | HapC 5 (10) | HapP 2 (18),HapP 11 (2) | HapR 2 (16),HapR 3 (1),HapR 4 (3) |
| SP | Shiping, Honghe | 1600 | 23.579 | 102.385 | 10 | HapC 4 (10) | HapP 1 (5),HapP 3 (15) | HapR 2 (15),HapR 3 (2),HapR 11 (1), HapR 14 (2) |
| ML | Menglong, Honghe | 489 | 23.339 | 102.413 | 10 | HapC 2 (2) HapC 3 (8) |
HapP 2 (13),HapP 3 (2), HapP 5 (1),HapP 10 (2) |
HapR 2 (14),HapR 9 (2),HapR 10 (1), HapR 11 (1),HapR 12 (1),HapR 13 (1) |
| LH | Lianhua, Honghe | 875 | 23.259 | 102.660 | 10 | HapC 2 (10) | HapP 2 (7),HapP 3(3), HapP 4 (1) HapP 5 (2),HapP 6(4), HapP 7 (1) HapP 8 (1),HapP 9 (1) |
HapR 2 (17),HapR 6 (1),HapR 7 (1), HapR 8 (1) |
| Total | 60 |
The within-population variation, Hs, was close to zero for cpDNA data, while exceeding 0.5 for both nuclear genes (Table 2). U tests showed that NST was not significantly greater than GST (GST = 0.936, NST = 0.984, P > 0.05), which indicated that there was no correlation between haplotype similarities and their geographic distribution in C. chenii.
Table 2.
Parameters of genetic diversity and differentiation, and results of neutrality tests and mismatch analysis for the combined cpDNA and two nDNA markers.
| Markers | Hd | Pi | Hs | HT | Tajima's D | Fu and Li’ F* | SSD | Raggedness index |
|---|---|---|---|---|---|---|---|---|
| cpDNA | 0.621 | 0.00143 | 0.059 | 0.920 | 1.987∗ | 1.519∗ | 0.029∗∗ | 0.111∗ |
| PHYP | 0.788 | 0.0027 | 0.518 | 0.850 | −0.819 | −0.961 | 0.036 | 0.222 |
| RBP-1 | 0.628 | 0.0024 | 0.471 | 0.662 | −1.894∗ | −3.056∗ | 0.113 | 0.309 |
∗P < 0.05; ∗∗P < 0.01.
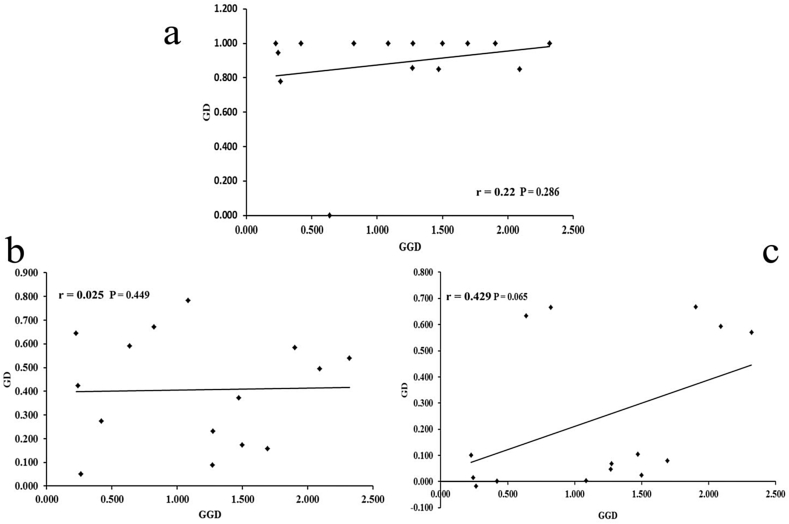
AMOVA revealed substantial differentiation between the two groups (NE and SW) for cpDNA (FCT = 0.76), but no group differentiation for either of the two nuclear genes (FCT = 0, for both nuclear genes). Mantel tests revealed no isolation by distance (IBD) (Fig. 2). The pollen/seed migration ratios were 111.7 and 197 for PHYP and RBP-1, respectively.
Fig. 2.
Plot of geographical distance (GGD) against genetic distance (GD) for six populations of C. chenii. (a. cpDNA; b. PHYP; c. RBP-1).
3.2. Phylogeny and divergent time of haplotypes
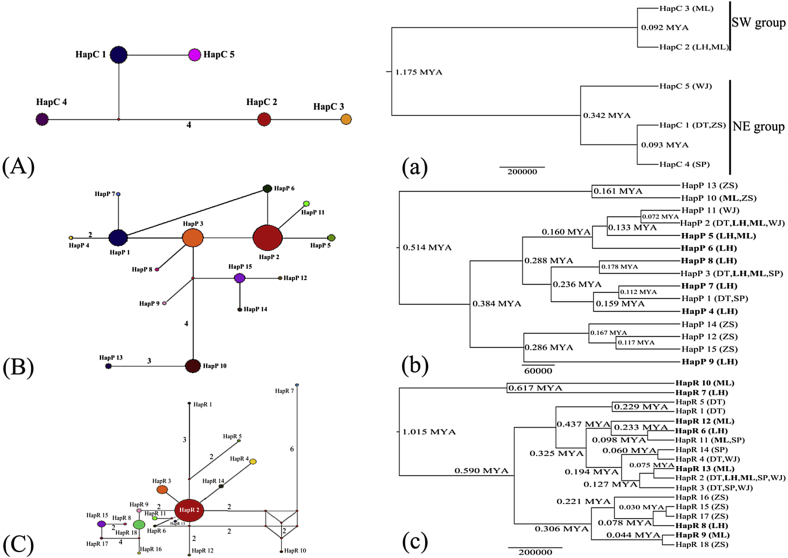
The cpDNA phylogenetic tree revealed two sub-clades corresponding to the two sides of the Red River, comprising three haplotypes (NE group), and two haplotypes (SW group), respectively (Fig. 3a). However, no clear clade structure was revealed from nDNA data.
Fig. 3.
Network for C. chenii based on the cpDNA (A) and nDNA (B. PHYP; C. RBP-1) haplotypes. (The size of the circles corresponds to the frequency of each haplotype); BEAST-derived trees based on cpDNA (a) and nDNA (b. PHYP; c. RBP-1) haplotypes (Divergence times were shown on the nodes; haplotype group identity: Bold (SW group) and regular font (NE group), respectively).
The two clades (NE and SW groups) split at about 1.175 MYA (0.22–2.678 MYA, 95% HPD), while haplotypes within the two groups diverged much later (from 0.092 MYA to 0.342 MYA) (Fig. 3a). These results imply that haplotypes of C. chenii diverged in the Pleistocene.
3.3. Demographic analysis
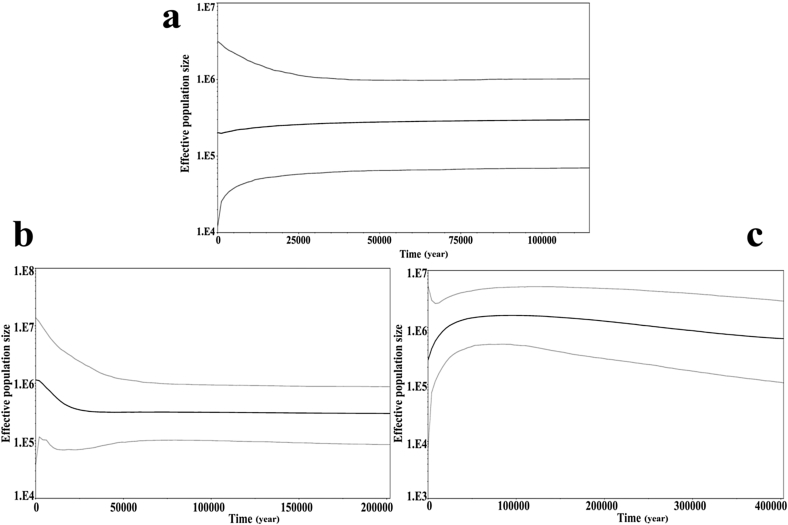
The Bayesian Skyline Plots produced from the cpDNA, PHYP and RBP-1 were in disagreement. For cpDNA, it showed a slow decline in the population size until approximate 1000 years ago, at which point a slight expansion occurred (Fig. 4a). For PHYP, C. chenii had a long history of constant population size, followed by a population expansion (about 25,000) (Fig. 4b). And for RBP-1, it showed that the species population size experienced a long period of continuous growth from 400,000–50,000 years ago, followed by a decline from about 50,000 years ago to the present (Fig. 4c).
Fig. 4.
Bayesian Skyline Plot based on cpDNA (a) and nDNA (b. PHYP; c. RBP-1) for the effective population size fluctuation throughout time. (Black line: median estimation; area between gray lines: 95% confidence interval).
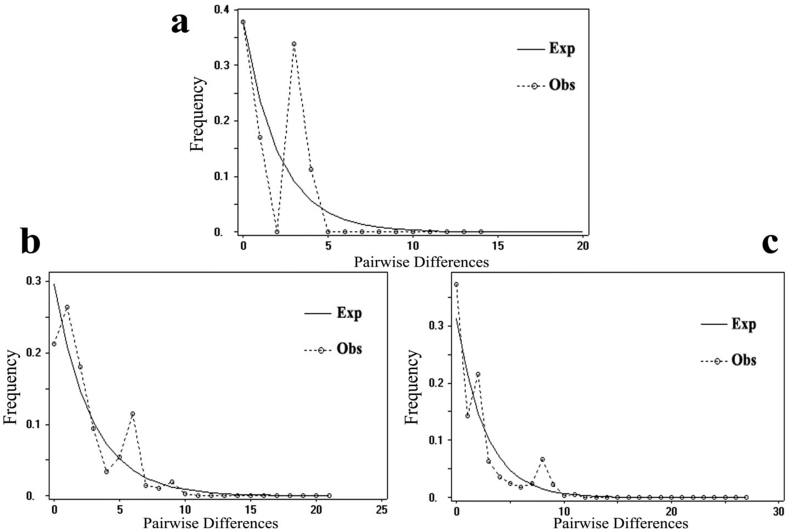
The mismatch analysis of cpDNA data revealed a multimodal pattern (Fig. 5a) with significantly positive SSD and raggedness index (Table 2), which indicated that C. chenii did not undergo a recent population expansion. This conclusion was also supported by the positive values of Tajima's D and Fu and Li’ F* (Table 2). The mismatch analysis of nDNA data also showed a multimodal pattern but non-significantly positive SSD and raggedness index. Tajima's D and Fu and Li’ F* values were negative for PHYP but not for the RBP-1 gene. Together, the results of mismatch analysis and the neutrality test applied to nDNA data indicate that C. chenii did not experience a recent population expansion (Fig. 5b–c; Table 2).
Fig. 5.
Mismatch distribution of cpDNA (a) and nDNA (b. PHYP; c. RBP-1) haplotypes based on pairwise sequence difference against the frequency of occurrence for C. chenii.
4. Discussion
4.1. Genetic variation
The high level of total genetic diversity in C. chenii based on cpDNA data (HT = 0.92) was comparable with what is commonly observed in other Cycas species: mean HT of 0.67 deduced from 170 Cycas species (Petit et al., 2005), HT = 1.000 for Cycas simplicipinna (Feng et al., 2014), HT = 0.896 for Cycas multipinnata (Gong et al., 2015) and HT = 0.56 for Cycas debaoensis (Zhan et al., 2011). The total genetic diversity of C. chenii was also high for both nuclear markers (PHYP and RBP-1) (HT = 0.85 and 0.662, respectively) (Table 2). The within-population diversity of cpDNA was low for the organelle markers (Hs = 0.059) and high for nuclear markers (Hs = 0.52 and 0.47). The organelle DNA is maternally inherited in Cycas and dispersed only by seeds, whereas nuclear DNA is biparentally inherited and dispersed by seeds and pollens (Huang et al., 2004). The seeds of Cycas are usually large and heavy, falling near the mother plant. Therefore, the limited seed dispersal capacity of Cycas may be the main cause of high population genetic structuring as shown by cpDNA (Hamrick and Godt, 1990). Our study conforms to generally observed low within-population variation and high variation in genetic differentiation among populations of cycads (Walters and Decker-Walters, 1991), e.g. Cycas pectinata (HS = 0.077, GST = 0.387), C. debaoensis (HS = 0.179, GST = 0.684), C. multipinnata (HS = 0.225, GST = 0.749), Cycas dolichophylla (HS = 0.32, GST = 0.678) (Yang and Meerow, 1996, Zhan et al., 2011, Gong et al., 2015, Zheng et al., 2016).
Analysis of cpDNA data revealed two distinct genetic clades in C. chenii that correspond to NE and SW sides of the Red River, but also no difference in structuring of genetic diversity for ordered vs. unordered alleles (i.e. NST ≈ GST), and no correlation between geographic and genetic distances. The latter indicate a limited gene flow among the last remaining and highly isolated populations of C. chenii. The physiographic pattern of mountains dissected by deep valleys in the Red River Fault (RRF) zone (Li et al., 2008) could limit seed dispersal in a northeast-southwest direction, thus promoting the population isolation and differentiation of the NE group and SW group in C. chenii. Another factor contributing to the observed structure of genetic variation in C. chenii appears to be high fragmentation of the species to a large extent due to destroyed habitat. In sum, the present study suggests limited gene flow in C. chenii that result from i) vicariance (the Red River) and ii) habitat destruction and fragmentation. These two processes are responsible for the existence of the two genetic clusters, which, however, display a mosaic-like genetic structure within each of the two parts of the species range (NE and SW groups), high genetic diversity at the species level and low genetic diversity within populations.
The nDNA data revealed lower differentiation between the NE and SW groups than the cpDNA (Fig. 3b–c), and the migration ratio of pollen/seed was higher than 100 for both nuclear genes. These results suggest much higher importance of pollen than seed flow in C. chenii, indicating an existing pollen flow not only among the populations within, but also between the NE and SW groups.
4.2. Demographic history
Some Gymnosperm species experienced population expansion during the recent glacial periods, such as Cycas revoluta (Chiang et al., 2009) and Taxus wallichiana (Liu et al., 2013). The mismatch analysis and neutrality tests showed that C. chenii did not experience a recent population expansion, and the Bayesian Skyline Plot based on cpDNA showed that C. chenii had experienced a slow range reduction since approximately 50,000 years ago, which is similar to the population dynamics of C. debaoensis, C. simplicipinna and C. multipinnata (Zhan et al., 2011, Feng et al., 2014, Gong et al., 2015). The cpDNA data revealed a long period of slow decline in C. chenii until approximately 1000 years ago, at which point a slight population expansion occurred (Fig. 4a). During the recent 1000 years, the established warmer climate was able to promote population growth in C. chenii. The population dynamics revealed by the two nuclear genes differed. The gene RBP-1 showed a population expansion during the Quaternary, followed by a decline over the Last Glacial Maximum (LGM) (Fig. 4c); in contrast, the PHYP gene showed an expansion in C. chenii population about 25,000 years ago (Fig. 4b). This contradiction in population dynamics revealed by the cpDNA and nDNA genes could possibly be explained by the different inheritance of the two genomes.
Although details are not clear, C. chenii appears to have experienced slow population contraction during the glacial period. The latter can explain the observed low within-population genetic variation in C. chenii because the larger genetic loss results from the slower range contraction or shift (Arenas et al., 2012). More widely distributed in the basin of the Red River before the glacial epoch, this species was probably forced into several isolated dry-hot Red River valley refugia during glaciation.
The deduced divergence time of the two cpDNA lineages (NE and SW) mainly fall into the Calabrian (1.175 MYA, Fig. 3a). In the Quaternary, the climate oscillated repeatedly from 2.4 MYA to the present, and the dry-hot valley of southwest China could serve at that time as refugia for many plant species suffering range contraction due to glaciations (Guan and Zhou, 1996, Wang et al., 1996).
4.3. Implications for conservation
The present decline of C. chenii throughout its distribution range is known to be largely caused by over-collection and habitat destruction. Designing a suitable conservation plan requires knowledge of the extent and structure of species genetic variation and demographic history. The results show that C. chenii has relatively high genetic diversity at the species level, low genetic diversity within populations and high genetic differentiation among populations. The last two features appear to be the result of range contraction during the species’ evolution rather than recent habitat loss. Presence of a clear phylogeographic structure, i.e. two haplotype clades separated by the Red River, implies that conservation efforts cannot focus on one part of the distribution range. In order to preserve present genetic diversity, at least two protected areas must be established, representing NE and SW groups. Because of high population genetic differentiation and the presence of many unique haplotypes, every existing population is important. Populations which can-not be protected in situ, and those harboring the highest diversity and unique haplotypes (such as ML, WJ and SP) must be the highest priority for ex situ conservation. Representative seed collection must be done in all existing populations.
Acknowledgments
This research was supported by the United Fund of the NSFC and the Yunnan Natural Science Foundation (Grant No. U1136602 to X. G.). We thank Wei Zhou, Jian Liu and Meng-meng Guan for their assistance with field sampling.
(Editor: Weibang Sun)
Footnotes
Peer review under responsibility of Editorial Office of Plant Diversity.
References
- Arenas M., Ray N., Currat M. Consequences of range contractions and range shifts on molecular diversity. Mol. Biol. Evol. 2012;29(1):207–218. doi: 10.1093/molbev/msr187. [DOI] [PubMed] [Google Scholar]
- Burban C., Petit R.J., Carcreff E. Rangewide variation of the maritime pine bast scale Matsucoccus feytaudi Duc. (Homoptera: Matsucoccidae) in relation to the genetic structure of its host. Mol. Ecol. 1999;8(10):1593–1602. doi: 10.1046/j.1365-294x.1999.00739.x. [DOI] [PubMed] [Google Scholar]
- Calonje M., Stevenson D.W., Stanberg L. 2016. The World List of Cycads, online edition.http://www.cycadlist.org Available from: [Google Scholar]
- Chiang Y., Hung K., Moore S. Paraphyly of organelle DNAs in Cycas Sect. Asiorientales due to ancient ancestral polymorphisms. BMC Evol. Biol. 2009;9(1):1–19. doi: 10.1186/1471-2148-9-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle J. Springer; Berlin: 1991. DNA Protocols for Plants–CTAB Total DNA Isolation. Molecular Techniques in Taxonomy. [Google Scholar]
- Drummond A.J., Rambaut A. University of Oxford; UK: 2004. Tracer-MCMC Trace Analysis Tool. [Google Scholar]
- Drummond A.J., Rambaut A. BEAST: bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007;7(1):1–8. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131(2):479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Laval G., Schneider S. Arlequin version. 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Feng X.Y., Wang Y.H., Gong X. Genetic diversity, genetic structure and demographic history of Cycas simplicipinna (Cycadaceae) assessed by DNA sequences and SSR markers. BMC Plant Biol. 2014;14(1):1–16. doi: 10.1186/1471-2229-14-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster M., Forster P., Watson J. Fluxus Technology Ltd; Clare: 2007. NETWORK (version 4.2. 0.1): A Software for Population Genetics Data Analysis. [Google Scholar]
- Fu Y.X. Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics. 1997;147(2):915–925. doi: 10.1093/genetics/147.2.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y.Q., Zhan Q.Q., Nguyen K.S. The Historical Demography and Genetic Variation of the Endangered Cycas multipinnata (Cycadaceae) in the Red River Region, Examined by Chloroplast DNA Sequences and Microsatellite Markers. PLoS One. 2015;10(2):e0117719. doi: 10.1371/journal.pone.0117719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graur D., Li W.H. Dynamics of genes in populations. Fundam. Mol. Evol. 2000;58 [Google Scholar]
- Guan Z.T., Zhou L. Sichuan Science and Technology Press; Chengdu: 1996. Cycads of China. [Google Scholar]
- Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hamrick J.L., Godt M.J.W. Sinnaur Associates Inc; Sunderland: 1990. Allozyme Diversity in Plant Species. Plant Population Genetics, Breeding and Genetic Resources. [Google Scholar]
- Hill K. The genus Cycas (Cycadaceae) in China. Telopea. 2008;12(1):71–118. [Google Scholar]
- Hoffmann M., Hilton-Taylor C., Angulo A. The impact of conservation on the status of the world's vertebrates. Science. 2010;330(6010):1503–1509. doi: 10.1126/science.1194442. [DOI] [PubMed] [Google Scholar]
- Huang S., Hsieh H.-T., Fang K. Patterns of genetic variation and demography of Cycas taitungensis in Taiwan. Botanical Rev. 2004;70(1):86–92. [Google Scholar]
- Jian S.G., Zhong Y., Liu N. Genetic variation in the endangered endemic species Cycas fairylakea (Cycadaceae) in China and implications for conservation. Biodivers. Conserv. 2006;15(5):1681–1694. [Google Scholar]
- Li Y.G., He D.M., Ye C.Q. Spatial and temporal variation of runoff of Red River Basin in Yunnan. J. Geogr. Sci. 2008;18(3):308–318. [Google Scholar]
- Liu J., Moeller M., Provan J. Geological and ecological factors drive cryptic speciation of yews in a biodiversity hotspot. New Phytol. 2013;199(4):1093–1108. doi: 10.1111/nph.12336. [DOI] [PubMed] [Google Scholar]
- Liu J., Zhou W., Gong X. Species delimitation, genetic diversity and population historical dynamics of Cycas diannanensis (Cycadaceae) occurring sympatrically in the Red River region of China. Front. Plant Sci. 2015;6:696. doi: 10.3389/fpls.2015.00696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousadik A., Petit R.J. Chloroplast DNA phylogeography of the argan tree of Morocco. Mol. Ecol. 1996;5(4):547–555. doi: 10.1111/j.1365-294x.1996.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Peakall R.O.D., Smouse P.E. GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes. 2006;6(1):288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit R.J., Duminil J., Fineschi S. Invited review: comparative organization of chloroplast, mitochondrial and nuclear diversity in plant populations. Mol. Ecol. 2005;14(3):689–701. doi: 10.1111/j.1365-294X.2004.02410.x. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Mark v.d.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royden L.H., Burchfiel B.C., van der Hilst R.D. The geological evolution of the Tibetan Plateau. Science. 2008;321(5892):1054–1058. doi: 10.1126/science.1155371. [DOI] [PubMed] [Google Scholar]
- Rozas J., Sánchez-DelBarrio J.C., Messeguer X. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19(18):2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Schneider S., Roessli D., Excoffier L. Arlequin: a software for population genetics data analysis. User Man. Ver. 2000;2:2496–2497. [Google Scholar]
- Shaw J., Lickey E.B., Beck J.T. The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am. J. Bot. 2005;92(1):142–166. doi: 10.3732/ajb.92.1.142. [DOI] [PubMed] [Google Scholar]
- Swindell S.R., Plasterer T.N. 1997. SEQMAN. Sequence Data Analysis Guidebook; pp. 75–89. [PubMed] [Google Scholar]
- Swofford D.L. Sinauer Associates, Inc; Sunderland: 2002. PAUP*: Phylogenetic Analysis Using Parsimony (and other methods), Version 4.0b10. Massachusetts. [Google Scholar]
- Taberlet P., Gielly L., Pautou G. Universal primers for amplification of three non-coding regions of chloroplast DNA. Plant Mol. Biol. 1991;17(5):1105–1109. doi: 10.1007/BF00037152. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123(3):585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters T.W., Decker-Walters D.S. Patterns of allozyme diversity in the West Indies cycad Zamia pumila (Zamiaceae) Am. J. Bot. 1991;78(3):436–445. [Google Scholar]
- Wang F.X., Liang H.B., Chen T.Q. Guangdong Science and Technology Press; Guangzhou: 1996. Cycads in China. [Google Scholar]
- Xiao L.Q., Ge X.J., Gong X. ISSR variation in the endemic and endangered plant Cycas guizhouensis (Cycadaceae) Ann. Bot. 2004;94(1):133–138. doi: 10.1093/aob/mch119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S.L., Meerow A.W. The Cycas pectinata (Cycadaceae) complex: genetic structure and gene flow. Int. J. Plant Sci. 1996;157(4):468–483. [Google Scholar]
- Zhan Q.Q., Wang J.F., Gong X. Patterns of chloroplast DNA variation in Cycas debaoensis (Cycadaceae): conservation implications. Conserv. Genet. 2011;12(4):959–970. [Google Scholar]
- Zheng Y., Liu J., Gong X. Tectonic and climatic impacts on the biota within the Red River Fault, evidence from phylogeography of Cycas dolichophylla (Cycadaceae) Sci. Rep. 2016;6:33540. doi: 10.1038/srep33540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z.R., Li N., Qian D. Maternal inheritance of plastids and mitochondria in Cycas L. (Cycadaceae) Mol. Genet. Genomics. 2011;286(5–6):411–416. doi: 10.1007/s00438-011-0653-9. [DOI] [PubMed] [Google Scholar]
- Zhou S.Z., Wang X.L., Wang J. A preliminary study on timing of the oldest Pleistocene glaciation in Qinghai–Tibetan Plateau. Quat. Int. 2006;154:44–51. [Google Scholar]
- Zhou W., Guan M.M., Gong X. Cycas chenii (Cycadaceae), a new species from China, and its phylogenetic position. J. Syst. Evol. 2015;53(6):489–498. [Google Scholar]