Abstract
The papillary renal cell carcinoma (RCC)-associated (X;1)(p11;q21) translocation fuses the genes PRCC and TFE3 and leads to cancer by an unknown molecular mechanism. We here demonstrate that the mitotic checkpoint protein MAD2B interacts with PRCC. The PRCCTFE3 fusion protein retains the MAD2B interaction domain, but this interaction is impaired. In addition, we show that two t(X;1)-positive RCC tumor cell lines are defective in their mitotic checkpoint. Transfection of PRCCTFE3, but not the reciprocal product TFE3PRCC, disrupts the mitotic checkpoint in human embryonic kidney cells. Our results suggest a dominant-negative effect of the PRCCTFE3 fusion gene leading to a mitotic checkpoint defect as an early event in papillary RCCs.
Chromosomal translocations often occur as tumor-specific abnormalities, suggesting that the underlying molecular alterations are crucial for tumor development (1, 2). In a subset of renal cell carcinomas (RCCs) with chromophilic histology and a mainly papillary growth pattern, referred to as papillary RCCs, chromosomal translocations involving the Xp11 region, usually t(X;1)(p11;q21), are recurrently encountered (3–11). Positional cloning of the Xp11 breakpoint by us and others revealed that the t(X;1)(p11;q21) translocation results in an in-frame fusion of the transcription factor TFE3 gene on the X-chromosome to the PRCC gene on chromosome 1 (12–14). Consequently, two fusion genes are formed, TFE3PRCC and PRCCTFE3, both of which are expressed in t(X;1)-positive tumor cells (13).
TFE3 is a ubiquitously expressed transcription factor characterized by the presence of a basic region followed by helix-loop-helix and leucine zipper domains, both of which are needed for dimerization and DNA binding of the transcription factor (15–17). The fusion protein PRCCTFE3 retains all these domains. PRCC is also ubiquitously expressed and characterized by a relatively high proline content. We have shown that the N-terminal 156 amino acids of PRCC, when fused to TFE3, significantly elevate the transactivating capacity of this fusion protein as compared with wild-type TFE3 (18). Moreover, transfection studies with conditionally immortalized mouse renal proximal epithelial cells, from which chromophilic tumors are thought to arise, showed that PRCCTFE3 could bypass temperature-induced growth arrest and differentiation (19). On the basis of the limited functional information available, we chose to further characterize PRCC via the identification of interacting proteins through yeast two-hybrid screening. This resulted in the identification of MAD2B, member of a family of genes involved in processes of mitotic checkpoint control mechanisms (20–22). Our results indicate that PRCCTFE3 expression may contribute to RCC development through a mechanism that affects the PRCC–MAD2B interaction.
Materials and Methods
Yeast Two-Hybrid Analysis.
Yeast two-hybrid analysis and filter lift assays were basically performed as described by the manufacturer (Stratagene). In short, yeast cells (pJ69–4A), kindly provided by Philip James (University of Wisconsin, Madison, WI), were transfected with a bait plasmid carrying the PRCC coding sequence, and consecutively with DNA of the target plasmids containing cDNAs of a library of t(X;1)-positive tumor cells (13). Transfected yeast cells were first selected for the presence of both bait and target vector, after which colonies were scraped from the plates, titered, and replated on selective medium (without Leu, Trp, His, or Ade) at a density at least 10-fold more than was originally plated. These cells were then allowed to grow for at least 5 days at 30°C.
Deletion Constructs.
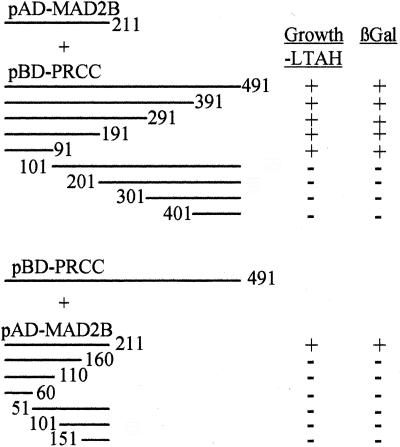
Deletion constructs were made by using primers 27–28 nt in length dispersed throughout the cDNA sequence. For the PRCC forward primers, the 5′ ends are located at positions 1, 101, 201, 301, and 401 (13). The 5′-end of the reverse primers start at positions 491, 391, 291, 191, and 91. For MAD2B, the 5′-end of the forward primers used are located at positions 1, 51, 101, 151, and 201, and at positions 211, 160, 110, and 60 for the reverse primers. The resulting PCR products were cloned into pGEM-T (Promega), isolated by appropriate restriction analysis for cloning in pBD-Gal4-T1c and pAD-Gal4-T3a, respectively, and checked by sequence analysis. The mentioned positions correspond to the amino acid positions in the protein.
Tissue Culture, Constructs, and Transfection.
Green monkey COS7 or COS1 cells were cultured and transiently transfected by electroporation as described before (18). CL89–12117 and CL89–17872 were cultured in RPMI-1640 medium (Life Technologies, Gaithersburg, MD) supplemented with 10% FCS, penicillin (100 units/ml) and streptomycin (100 μg/ml). CL89–12117 cells are diploid with t(X;1) as sole karyotypic abnormality. CL89–17827 cells exhibit several numerical anomalies next to t(X;1) (4). HeLa cells were cultured in DMEM with the same supplements.
For the localization studies, the PRCC and MAD2B cDNAs were cloned into pECFP-N1 (CLONTECH) and pVSV2, respectively. For fluorescence resonance energy transfer (FRET) analysis, COS cells were transfected with 4 μg of the coding regions of MAD2B cloned into pECFP-N1, and 16 μg of the PRCC cDNA cloned into pEYFP-N1 (CLONTECH). For immunoprecipitation, 10 μg of both MAD2B-pSG8-VSV and PRCC-pEYFP-N1 were used.
T-REx-293 cells (Invitrogen) were cultured as described by the manufacturer. The cells were stably transfected by using Fugene according to the recommended protocol (Roche Diagnostics) with 5 μg of expression constructs carrying the coding sequences of PRCCTFE3 or TFE3PRCC cloned into a tetracyclin inducible expression vector (pcDNA/TO/myc-His; Invitrogen). The number of colonies was comparable between transfections with PRCCTFE3 and TFE3PRCC (15–35). Vector transfection yielded more colonies (200). Colonies were pooled for further experiments.
Polyclonal Antibodies, Immunolocalization, and Immunoprecipitation.
A polyclonal antibody directed against a PRCC synthetic peptide (GPPLGLPKPKKRKEP) was raised in rabbits (Genosys, The Woodlands, TX), according to standard procedures. The final bleed was used for purification using caprylic acid, followed by precipitation with ammonium sulfate (23, 24). A polyclonal antibody directed against recombinant MAD2B protein was raised using an expression construct encompassing the coding region of MAD2B coupled to the His6 tag of the pET28a vector (Novagen). The serum was used without further purification. A polyclonal antibody directed against green fluorescent protein (GFP), which recognizes cyano fluorescent protein (CFP), yellow fluorescent protein (YFP), and GFP, was kindly provided by W. Hendriks and E. Cuppen (Department of Cell Biology, University Medical Center, Nijmegen).
For the localization studies, cells were fixed [2% acid-free formaldehyde, 30 min, room temperature (rT)] 1–2 days after electroporation and stained directly with 0.5 μg/ml of 4′,6-diamidino-2-phenylindole (DAPI) (Serva) or incubated with the α-PRCC antibody (1:200) for 1 h (rT), followed by detection by using a goat α-rabbit secondary antibody (1 h, rT) coupled to the Alexis red fluorochrome (1:100; Alexis, Lausanne, Switzerland) and stained with DAPI, as described above. For immunoprecipitation, cells were incubated 24 h after transfection for 1 h in methionine-free DMEM before labeling with 50 μCi/ml [35S]methionine for 4 h. Cells were lysed by using RIPA buffer (150 mM NaCl/1% Nonidet P-40/0.5% sodiumdesoxycholate/0.1% SDS/50 mM Tris⋅HCl, pH 8). The supernatant was precleared by using 50 μl of preimmune serum (1 h, 4°C), after which 200 μl of protein A-agarose was added (50% in RIPA; 1 h, 4°C). Two microliters of the primary antibody or preimmune serum was added to the supernatant (2 h, 4°C), followed by addition of 100 μl of protein A-agarose (10%; 1 h, 4°C). After washes, the samples were boiled for 5 minutes in SDS sample buffer before loading onto SDS polyacrylamide gels (10%). The gels were fixed (15% HAc/10% methanol), fluorographed (Amplify, Amersham Pharmacia), and dried. Immunoprecipitated proteins were visualized by autoradiography.
FRET Microscopy.
FRET analysis was performed by using a Leitz orthoplan upright microscope (Leitz) equipped with an epi-illumination fluorescence detection system and a temperature-controlled specimen holder at 33°C. A Spex Fluorolog (Spex Industries, Metuchen, NJ) with two excitation monochromators was used as an excitation source, connected to the microscope via a UV-fiber-optic coupling. The emission monochromator was connected to the microscope via a fiberoptic cable (Ø12 mm). For spectral analysis, a filter cube fitted with a 455-nm dichroic mirror was used. Excitation and barrier filters were removed.
As excitation wavelength, 430 nm was chosen (suboptimal for CFP but optimal for separation from the tail of the YFP spectrum). Spectral data were collected with an integration time of 0.5 s/nm (8-nm slit). Data were sampled on line, stored, and displayed with the program dm3000 (Spex). The spectra were processed and normalized at 480 nm. For the image analysis, two other filter sets were used, the first to measure the free donor emission (excitation 440(8), 455-nm dichroic, 490 emitter) and the second to measure the total acceptor fluorescence by using a 515-nm dichroic mirror in combination with a 535(30) bandpass filter (Chroma Technology, Brattleboro, VT). Images were recorded (Leitz, ×50 n.a. 1.0 water immersion objective) by using an Image Intensifier (Delft Electronic Products, Roden, The Netherlands) connected to a JAI CV-M10 progressive scan charge-coupled device camera and digitized with an AG-5 framegrabber (Scion, Frederick, MD). Intensities for each pixel were corrected for illumination intensities at both wavelengths (Spex reference photomultiplier), quantum efficiencies, and molar extinction coefficients [ɛCFP(440) and ɛyFP(495), respectively 32.5 × 103 and 55.3 × 103 M−⋅cm−1 (25)]. The image processing (4 × 4 Gaussian filtering, arithmetic, false color overlay, and the intensity distribution histogram) was performed with Scion image for Windows NT, a modified version of NIH image. To obtain equal amounts of protein expression, different ratios of MAD2B-CFP and PRCC-YFP were used for the transfections. On the basis of Western blot analyses, the FRET analysis was performed with 4 μg of MAD2B-CFP/16 μg of PRCC-YFP.
Nocodazole Treatment.
Pools of stably transfected T-REx 293 clones were used for the nocodazole assays. As a control, vector transfected cells were used. Twenty-four hours before nocodazole treatment, expression of the transfected constructs was induced by addition of tetracyclin (1 μg/ml) to the culture medium. T(X;1)-positive CL89–12117 and CL89–17872 cells, and HeLa cells were directly treated with nocodazole (100 ng/ml). Cells were fixed by using 3% paraformaldehyde in PBS at 0, 4, 8, 16, and 30 h after addition of nocodazole. Nuclei were identified by DAPI staining, after which mitotic nuclei were counted.
On the basis of the outcome of these experiments, nocodazole treatment was repeated and the distribution of 2N/4N cells was determined by FACS analysis (26) by using 106 cells/ml of pyrpidium iodide 24 h after nocodazole treatment. Similar experiments were performed by using 200 ng/ml of colcemid instead of nocodazole.
Results
Yeast Two-Hybrid Screening with PRCC.
A yeast two-hybrid screening by using the full coding sequences of PRCC as a bait and a RCC-derived cDNA library in yeast pJ69–4A cells resulted in 705 colonies that grew on selective medium, which were replated and screened in a lacZ filter lift assay. From the 173 clones that turned blue, 99 were chosen for further analysis. The corresponding plasmids were isolated and cloned into bacterial cells, after which the purified target and bait plasmids were reintroduced into yeast cells, plated on selective medium, and rescreened for their lacZ activity. Four of the target plasmids containing potential PRCC interactors remained positive. Sequence analysis revealed that these cDNAs, although not identical, were all derived from the same gene that was located on chromosome 1 (accession no. AL031731) and recently identified as MAD2B (20).
A Specific MAD2B-Binding Domain Within the PRCC Protein.
To determine which parts of the PRCC and MAD2B proteins are essential for the interaction, deletion constructs were made lacking increasing parts at the C- or N-terminal ends (Fig. 1) of their coding sequences. Each of the PRCC deletion constructs was transfected into yeast cells together with the full-length MAD2B construct and vice versa. Consequently, these transfectants were tested for their ability to grow on selective medium and lacZ activity. The results (Fig. 1) indicate that the N-terminal 100 amino acids of the PRCC protein, which are also present in the PRCCTFE3 fusion protein, are both essential and sufficient for binding to the MAD2B protein. In all cases, growth of colonies on selective medium was accompanied by lacZ activity. A further reduction of the critical interaction domain within the MAD2B protein could not be achieved, because both deletions of 50 amino acids at the N-terminal side or 51 amino acids at the C-terminal side resulted in a complete abolition of the interaction.
Figure 1.
Yeast two-hybrid analysis of PRCC and MAD2B deletion constructs. Schematic representation of the (deletion) constructs used and their capacity to grow on fully selective medium (without Leu, Trp, Ade, or His; indicated as −LTAH). In addition, the presence of β-galactosidase activity (indicated by βGal) as measured in a filter lift assay is indicated by +. The numbers correspond with the amino acid positions in the proteins.
Coexpression and Colocalization of MAD2B and PRCC.
The expression pattern of MAD2B was determined in normal tissues and renal tumor cell lines by using Northern blot analysis. Expression of MAD2B was present at comparable levels in all adult (heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas) and fetal (brain, liver, lung, muscle, and kidney) organs examined (not shown). MAD2B mRNA was also detected in human embryonic kidney 293 cells, in three t(X;1)-positive papillary RCCs, and in six nonpapillary RCC cell lines that were examined (not shown). Thus, similar to PRCC, MAD2B is ubiquitously expressed. These results indicate that the PRCC–MAD2B interaction, as demonstrated in yeast, is at least feasible in vivo.
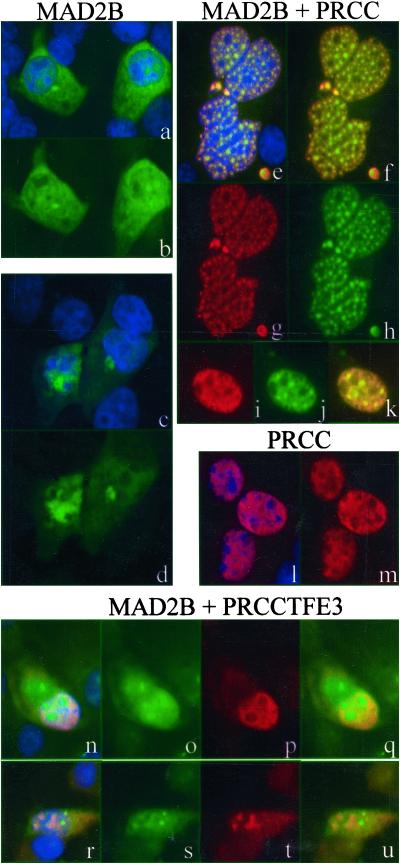
Because interaction between PRCC and MAD2B should also result in subcellular colocalization, we determined and compared the localization patterns of the PRCC and MAD2B proteins within cells. Expression constructs carrying the full coding sequences of VSV-tagged PRCC and/or CFP-tagged MAD2B were transiently transfected into COS cells, after which the subcellular localization was determined by direct visualization of the fluorescent CFP tag or through immunodetection by using a polyclonal anti-PRCC antibody (Fig. 2). Single PRCC transfectants showed fluorescence in the nucleus (l, m) as described before (18). In contrast, single MAD2B transfectants showed a ubiquitous cytoplasmic fluorescence (a–d). In some cases, intense fluorescence was observed near, although not overlapping with the nucleus (c, d). Cotransfection of MAD2B with PRCC revealed a completely different picture. MAD2B was now localized in the nucleus, mostly in nuclear patches or speckles. The overlay of both MAD2B and PRCC localization patterns revealed a (near) perfect colocalization of both proteins (e–k). On the basis of these results, we conclude that MAD2B is transferred to the nucleus through interaction with PRCC.
Figure 2.
Subcellular (co)localization of MAD2B and PRCC after ectopic expression in COS cells. Cells were transiently transfected with MAD2B (a–d), PRCC and MAD2B (e–k), or PRCC (l, m). The localization of MAD2B is shown in green and that of PRCC in red. DAPI staining (blue) indicates the position of the nuclei. In f and k, the yellow overlay of the red and green signals indicates colocalization of the PRCC and MAD2B proteins. Localization of the MAD2B (green) and PRCCTFE3 (red) proteins after cotransfection is shown in n–u. The overlay of the red and green signals is shown in n, q, r, and u, indicating absence of colocalization for this combination. The blue DAPI staining in n and r again marks the positions of the nuclei. In a, c, e, l, n, and r, additional DAPI-positive nuclei can be seen that correspond to nontransfected cells.
Coimmunoprecipitation of MAD2B and PRCC.
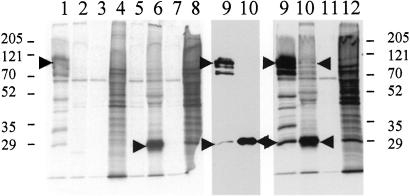
To confirm a direct physical interaction between MAD2B and PRCC, COS cells were transiently transfected with MAD2B-pSG8-VSV and/or PRCC-pEYFP-N1 expression constructs. Immunoprecipitations were performed on PRCC and MAD2B single (Fig. 3; lanes 1–4 and 5–8, respectively) and double (lanes 9–12) transfectants using a polyclonal anti-GFP antibody to precipitate PRCC (lanes 1 and 5) and a polyclonal anti-MAD2B antibody to precipitate MAD2B (lanes 2 and 6). In single MAD2B transfectants, the MAD2B protein (24.5 kDa, VSV tag included) could readily be detected (lane 6, arrow) but, as expected, not in single PRCC transfectants (lane 2). Single PRCC transfectants showed several bands corresponding to PRCC (lane 1), which we have consistently observed when using anti-PRCC or anti-GFP antibodies to precipitate this protein. These bands were absent in MAD2B transfected cells (lane 5). The correct PRCC band (84.5 kDa, YFP tag included) is marked by an arrow (lane 1). The lower molecular weight bands may correspond to specific degradation products or splice variants. In double transfected cells, the MAD2B protein could readily be detected after anti-GFP precipitation of the PRCC protein (Fig. 3 Middle; lane 9, arrow), indicating a physical interaction between the MAD2B and PRCC proteins. A longer exposure (Fig. 3 Right) also showed the opposite: PRCC can be detected in the anti-MAD2B immunoprecipitates (Fig. 3, lane 10, arrow), again confirming the physical interaction between these two proteins. Additional immunoprecipitations were performed on endogenous proteins by using HeLa cell extracts in conjunction with MAD2B and PRCC specific antibodies. Also in this case, coimmunoprecipitation of MAD2B with PRCC and vice versa was observed (not shown).
Figure 3.
Immunoprecipitation of the PRCC and MAD2B proteins. Lanes 1–8: immunoprecipations (IPs) performed on single transfectants by using an anti-GFP/YFP antibody (lanes 1 and 5) to precipitate PRCC and an anti-MAD2B antibody (lanes 2 and 6) to precipitate MAD2B. Lanes 3, 7, and 11 contain IP products obtained with preimmune sera instead of specific antibodies. Lanes 4, 8, and 12 contain 0, 5 μl of the total protein samples used for the IPs. Lanes 9–12 contain IP products of cotransfected cells by using anti-GFP/YFP (lane 9) or anti-MAD2B antibodies (lane 10), showing that MAD2B coimmunoprecipates with PRCC and vice versa (arrows). The middle section (lanes 9, 10) is a partial lighter exposure of the right section. Size markers (Bio-Rad) are indicated (Left and Right).
MAD2B and PRCC Interact in Vivo.
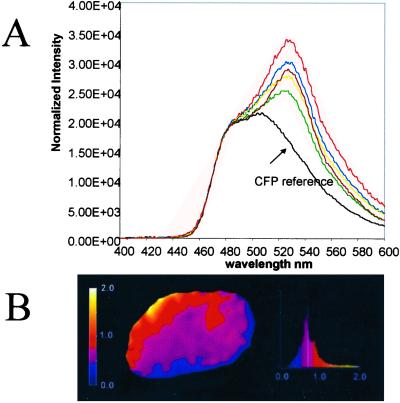
If a direct in vivo association between MAD2B and PRCC exists, we reasoned that we should be able to demonstrate this by FRET analysis. For this purpose, COS cells were transiently transfected with CFP- and YFP-labeled MAD2B and PRCC proteins, respectively. Fluorescence emission spectra were determined from nuclei of single living cells at 37°C at day one after transfection (Fig. 4A) by using a wavelength of 430 nm to excite CFP (MAD2B). Double transfected cells displayed an enhanced intensity at wavelengths between 500 and 600 nm, with a peak maximum at 525 nm, at the single PRCC-YFP emission maximum. Because spectral FRET analysis is sensitive to strong unequal expression of CFP and YFP proteins, we measured the free donor concentration and total acceptor concentration of both fusion proteins in the nucleus. The ratio of free donor (MAD2B-CFP) and total acceptor (PRCC-YFP) fluorescence is depicted in Fig. 4B showing a homogeneous distribution of the MAD2B and PRCC proteins with a mean ratio of 0.68. Because a fraction of the MAD2B-CFP fluorescence gives rise to sensitized emission (the PRCC-bound donor fluorescence), this ratio is a slight underestimate of the actual CFP/YFP concentration. Therefore, protein expression was also determined by Western blot analysis, which revealed that equal amounts of the MAD2B and PRCC proteins were expressed (not shown), thus validating the fluorescent measurements. The sensitized emission (Fig. 4A; sensitized emission intensity/CFP intensity at 525 nm: 69% +/−18%, mean of five cells) reflects a direct interaction between MAD2B and PRCC in the nucleus within the effective range of fluorescence energy transfer of 60 Å, the Föster radius for CFP-YFP.
Figure 4.
FRET analysis on single nuclei of cotransfected COS cells. (A) Normalized emission spectra of single nuclei of five different cells (each color represents a single nucleus) 1 day after electroporation. The CFP reference spectrum (of single transfected MAD2B-CFP cells) is indicated. Sensitized emission can be observed in all nuclei examined at 525 nm. (B) Ratio image of free donor fluorescence (MAD2B) and total acceptor fluorescence (PRCC) 1 day after electroporation and intensity distribution histogram of the ratio image (mean 0.68). The change in color represents a change in ratio (0.0–2.0), as indicated in the bar along the intensity distribution diagram.
Effect of the t(X;1) Translocation on the PRCC–MAD2B Interaction.
To evaluate the effect of the t(X;1) translocation on the interaction with MAD2B, PRCCTFE3 or TFE3PRCC cDNAs were introduced into yeast cells together with the MAD2B bait and analyzed in a yeast two-hybrid assay. The combination of PRCC and MAD2B was included for comparison. After transfection, 15 colonies that grew on medium without Leu and Trp and consequently contained both MAD2B and either one of the translocation products were screened for their ability to grow on full selective medium (without Leu, Trp, His, and Ade) and their capacity to drive the lacZ reporter. The combination of MAD2B with PRCC yielded 15/15 colonies that all turned blue. However, only 4 of 15 colonies containing the PRCCTFE3-MAD2B combination grew on full selective medium. These colonies turned weakly blue after prolonged incubation times. The combination with TFE3PRCC yielded 15/15 colonies that all turned blue in a filter lift assay, but this was due to the presence of transactivating regions in the N-terminal part of TFE3 because TFE3 alone with or without MAD2B gave the same results. PRCC alone did not give rise to any colonies on this selective medium (not shown).
When examining the subcellular (co)localization of the translocation products with MAD2B through transfection of the corresponding expression constructs into COS cells, patterns in cotransfectants remained unchanged compared with the patterns observed in the single transfectants. Despite the presence of the MAD2B-binding domain in PRCCTFE3, cotransfection with MAD2B did not lead to transfer of the MAD2B protein to the nucleus, as was seen in case of MAD2B and PRCC cotransfection (Fig. 2 n–u). In a small minority of the cells, partial colocalization was observed, whereas in the majority of the cells, MAD2B localization remained unaffected, which is completely in line with our observation in the yeast two-hybrid assay for this combination.
t(X;1)-Positive RCC Cells Exhibit a Mitotic Checkpoint Defect.
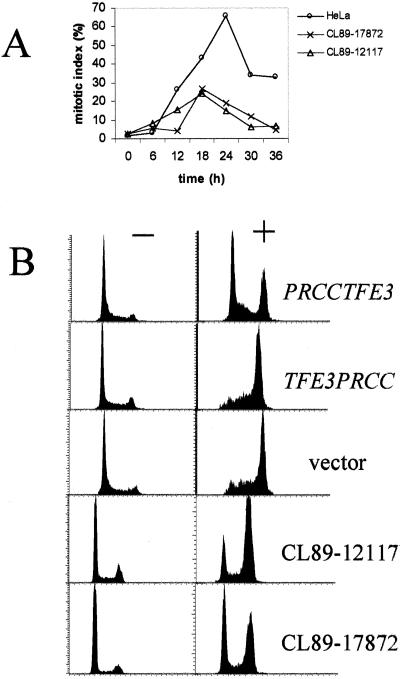
Because the PRCC interactor MAD2B is a member of the family of mitotic checkpoint proteins (20–22), we decided to investigate whether t(X;1)-positive tumor cells might have an impaired mitotic checkpoint. For this purpose, t(X;1)-positive tumor cell lines CL89–12117 and CL89–17872 were treated with nocodazole, which should result in accumulation of cells in mitosis if an intact mitotic checkpoint is present. As a control, HeLa cells were used that are known to respond normally to nocodazole treatment (27). Without nocodazole (time point 0 h), very few cells were mitotic. After addition of nocodazole, the percentage of mitotic cells increased significantly in HeLa cells. After 24 h of treatment, 66% of HeLa cells were mitotic, whereas this was the case for only 19% and 15% of CL89–12117 and CL89–17872 cells, respectively, indicating that the mitotic checkpoint that is challenged by nocodazole in the t(X;1)-positive tumor cells is impaired (Fig. 5A).
Figure 5.
Nocodazole treatment of t(X;1)-positive RCC and PRCCTFE3-transfected T-REx-293 cells. (A) Nocodazole assay of t(X;1)-positive RCC and HeLa cells. The mitotic index is given as the percentage of mitotic nuclei per 250–300 counted nuclei. (B) FACS analysis showing the result of nocodazole treatment of T-REx-293 cells transfected with PRCCTFE3, TFE3PRCC or empty vector negative controls (pcDNA/TO/myc-His), and t(X;1)-positive RCC CL89–12117 and CL89–17872 cells. The count of cells in arbitrary units is plotted as a function of the DNA content. Nocodazole treated cells are marked by +, nontreated cells by −.
PRCCTFE3-Transfected Cells Exhibit a Mitotic Checkpoint Defect.
To mimic the effects of the translocation, we transfected embryonal kidney 293 cells (T-R Ex-293) with a tetracyclin-inducible expression construct carrying the PRCCTFE3 or TFE3PRCC coding sequences and examined the response of these cells to nocodazole treatment 24 h after induction of TFE3PRCC or PRCCTFE3 expression by tetracyclin. On the basis of the experiments performed on the t(X;1)-positive tumor cells, cells were harvested after 24 h of nocodazole treatment and used for FACS analysis. The distribution of cells shifted markedly as a result of nocodazole treatment from 2N to 4N in vector transfected and TFE3PRCC transfected cells. Similar results were obtained with noninduced PRCCTFE3 transfected cells (not shown). In contrast, this shift was not observed in induced PRCCTFE3 transfected cells or the tumor cell lines CL89–12117 or CL89–17872 (Fig. 5B). Experiments in which we used colcemid instead of nocodazole yielded similar results (not shown).
To exclude the possibility that the observed difference was a result of differences in expression of the PRCCTFE3 and TFE3PRCC genes, we examined the respective mRNA and protein expression. Both mRNAs and proteins were found to be expressed at comparable levels (not shown).
Discussion
A yeast two-hybrid analysis to identify proteins that interact with PRCC yielded four independent partially overlapping clones, all corresponding to the same protein. Sequence analysis revealed that the corresponding gene was identical to MAD2B, a member of the family of mitotic checkpoint genes. This interaction was confirmed by using several in vitro and in vivo assays. Cotransfection of PRCC and MAD2B in COS cells showed that MAD2B, otherwise present in the cytoplasm, was transferred to the nucleus. Identical PRCC and MAD2B fluorescent patterns were observed indicating that, (i) the subcellular localization of MAD2B is affected by PRCC, and (ii) both proteins completely colocalize within the nucleus. In addition, the observed colocalization was shown to be the result of a direct physical association of these proteins, because sensitized emission as measured by FRET was observed, which is possible only when the fluorochromes coupled to the MAD2B and PRCC proteins are in close proximity to each other. Immunoprecipitation experiments also confirmed a physical interaction between PRCC and MAD2B.
MAD2B (also referred to as MAD2L2 or hRev7) was originally isolated by Cahill et al. (20) and is a member of the yeast family of MAD (mitotic-arrest-deficiency) and BUB (budding uninhibited by benzimidazole) genes, implied in the control of the spindle or mitotic checkpoint (20, 21, 27–34). Exit from mitosis requires the degradation of regulatory proteins, including mitotic cyclins and securin. This degradation is mediated by the anaphase-promoting complex (APC) through ubiquination. APC, in turn, is bound to the regulatory proteins CDC20 and/or CDH1. On activation of the spindle assembly checkpoint, MAD2 directly associates with CDC20-APC and inhibits activation of APC by CDC20 before anaphase (35). Very recently, it was found that the CDH1-APC-mediated mitotic checkpoint control is similarly regulated by MAD2B (21, 22). The MAD2 protein interacts with another checkpoint protein, MAD1, and the corresponding mitotic checkpoint control requires the formation of a MAD1–MAD2 complex (36). Here we show that MAD2B forms a similar functional complex with PRCC.
Several studies have indicated direct relationships between defective mitotic checkpoint genes and cancer (37). Decreased expression and mutation of the MAD2 (also referred to as MAD2L1) gene have been observed in primary human breast cancers and cell lines (38), and it has been suggested that such mutations may act in concert with BRCA2 deficiency in the pathogenesis of inherited breast cancers (39). MAD1L1 has been identified as a protein targeted by the HTLV-1 retroviral Tax protein, inactivating the spindle checkpoint in infected T cells, thus contributing to the development of T cell leukemias (40). Haploinsufficiency of Mad2 in mice led to a significant increase in the development of papillary lung adenocarcinomas, again implicating that mitotic checkpoint defects may contribute to tumor development (41). For another mitotic checkpoint gene, Bub1, it has been demonstrated that, when partially inactivated through expression of a dominant-negative form, it leads to loss of mitotic checkpoint control (37).
In t(X;1)-positive renal cell carcinomas, PRCC is expressed together with TFE3PRCC and PRCCTFE3 (13). The MAD2B interaction domain of PRCC is retained in the PRCCTFE3 fusion protein. However, in contrast to PRCC, PRCCTFE3 interacts only weakly with MAD2B. Thus, as a result of the translocation, the PRCC-MAD2B interaction appears to be compromised. In t(X;1)-positive tumor cells, we indeed found an impaired mitotic checkpoint as assessed by nocodazole and colcemid assays. The same effect was achieved after transfection of PRCCTFE3 into 293 embryonic kidney cells, whereas this effect was not seen after transfection of TFE3PRCC or empty vector controls. Therefore, we suggest that expression of PRCCTFE3 may act as a dominant-negative PRCC mutant. Because of its lack of binding to MAD2B, transfer of MAD2B to the nucleus is impaired. Expression of PRCCTFE3 leads to a bypass of the mitotic checkpoint even when disrupted spindles are present. This defect may represent a crucial step in the development of t(X;1)-positive renal cell carcinomas and is in line with our previous observation that conditionally immortalized mouse renal proximal epithelial cells transfected with PRCCTFE3 could bypass temperature-induced growth arrest and differentiation (19).
Acknowledgments
We thank Wiljan Hendriks en Edwin Cuppen [Department of Cell Biology, University Medical Center (UMC), Nijmegen] for providing the anti-GFP antiserum, Egbert Oosterwijk (Department of Urology, UMC) for the gift of the nonpapillary renal cell carcinoma cell lines, and Arie Pennings (Department Hematology, UMC) for expert help with the FACs analyses. Jose Thijssen, Diederik de Bruijn, and Helma van den Hurk (Department Human Genetics, UMC) are acknowledged for advice and support. This work was supported by the Dutch Cancer Society, Grant KWF98–1804.
Abbreviations
- RCC
renal cell carcinoma
- GFP
green fluorescent protein
- DAPI
4′,6-diamidino-2-phenylindole
- FRET
fluorescence resonance energy transfer
- CFP
cyano fluorescent protein
- YFP
yellow fluorescent protein
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Rabbitts T H. Nature (London) 1994;372:143–149. doi: 10.1038/372143a0. [DOI] [PubMed] [Google Scholar]
- 2.Dos Santos N R, Geurts van Kessel A. Anticancer Res. 1999;19:4697–4714. [PubMed] [Google Scholar]
- 3.Thoenes W, Stoerkel S, Rumpelt H J. Pathol Res Pract. 1986;181:125–143. doi: 10.1016/S0344-0338(86)80001-2. [DOI] [PubMed] [Google Scholar]
- 4.Meloni A M, Dobbs R M, Jr, Pontes J E, Sandberg A A. Cancer Genet Cytogenet. 1993;65:1–6. doi: 10.1016/0165-4608(93)90050-v. [DOI] [PubMed] [Google Scholar]
- 5.Tonk V, Wilson K S, Timmons C F, Schneider N R, Tomlinson G E. Cancer Genet Cytogenet. 1995;81:72–75. doi: 10.1016/s0165-4608(94)00195-2. [DOI] [PubMed] [Google Scholar]
- 6.Dijkhuizen T, van den Berg E, Wilbrink M, Weterman M A J, Geurts van Kessel A, Storkel S, Folkers R P, Braam A, de Jong B. Genes Chromosomes Cancer. 1995;14:43–50. doi: 10.1002/gcc.2870140108. [DOI] [PubMed] [Google Scholar]
- 7.Hernandez-Marti M J, Orellana-Alonso C, Badia-Garrabou L, Verdeguer Miralles A, Paradis-Alos A. Cancer Genet Cytogenet. 1995;83:82–83. doi: 10.1016/0165-4608(94)00184-7. [DOI] [PubMed] [Google Scholar]
- 8.Carcao M D, Taylor G P, Greenberg M L, Bernstein M L, Champagne M, Hershon L, Baruchel S. Med Pediat Oncol. 1998;31:153–158. doi: 10.1002/(sici)1096-911x(199809)31:3<153::aid-mpo5>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 9.Desangles F, Camparo P, Fouet C, Houlgatte A, Arborio M. Cancer Genet Cytogenet. 1999;113:141–144. doi: 10.1016/s0165-4608(98)00261-1. [DOI] [PubMed] [Google Scholar]
- 10.Perot C, Bougaran J, Boccon G L, Storkel S, Leverger G, van den Akker J, Taillemite J L, Couturier J. Cancer Genet Cytogenet. 1999;110:54–56. doi: 10.1016/s0165-4608(98)00181-2. [DOI] [PubMed] [Google Scholar]
- 11.de Jong B, Molenaar I M, Leeuw J A, Idenburg V I S, Oosterhuis J W. Cancer Genet Cytogenet. 1986;21:165–169. doi: 10.1016/0165-4608(86)90042-7. [DOI] [PubMed] [Google Scholar]
- 12.Weterman M A J, Wilbrink M, Janssen I, Janssen H A P, van den Berg E, Fisher S E, Craig I, Geurts van Kessel A. Cytogenet Cell Genet. 1996;75:2–6. doi: 10.1159/000134444. [DOI] [PubMed] [Google Scholar]
- 13.Weterman M A J, Wilbrink M, Geurts van Kessel A. Proc Natl Acad Sci USA. 1996;93:15294–15298. doi: 10.1073/pnas.93.26.15294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sidhar S K, Clark J, Gill S, Hamoudi R, Crew A J, Gwilliam R, Ross M, Linehan W M, Birdsall S, Shipley J, Cooper C S. Hum Mol Genet. 1996;5:1333–1338. doi: 10.1093/hmg/5.9.1333. [DOI] [PubMed] [Google Scholar]
- 15.Beckmann H, Su L K, Kadesch T. Genes Dev. 1990;4:167–179. doi: 10.1101/gad.4.2.167. [DOI] [PubMed] [Google Scholar]
- 16.Beckmann H, Kadesch T. Genes Dev. 1991;5:1057–1066. doi: 10.1101/gad.5.6.1057. [DOI] [PubMed] [Google Scholar]
- 17.Roman C, Matera A G, Cooper C, Artandi S, Blain S, Ward D C, Calame K. Mol Cell Biol. 1992;12:817–827. doi: 10.1128/mcb.12.2.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weterman M A J, van Groningen J J M, Jansen A, Geurts van Kessel A. Oncogene. 2000;19:69–74. doi: 10.1038/sj.onc.1203255. [DOI] [PubMed] [Google Scholar]
- 19.Weterman M A J, van Groningen J J M, Hartog A, Geurts van Kessel A. Oncogene. 2001;20:1414–1424. doi: 10.1038/sj.onc.1204213. [DOI] [PubMed] [Google Scholar]
- 20.Cahill D P, Dacosta L T, Carson-Walter E B, Kinzler K W, Vogelstein B, Lengauer C. Genomics. 1999;58:181–187. doi: 10.1006/geno.1999.5831. [DOI] [PubMed] [Google Scholar]
- 21.Pfleger M C, Salic A, Lee E, Kirschner W. Genes Dev. 2001;15:1759–1764. doi: 10.1101/gad.897901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Fang G. Genes Dev. 2001;15:1765–1770. doi: 10.1101/gad.898701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Page M, Thorpe R. In: The Protein Protocols Handbook. Walker J M, editor. Totawa, NJ: Humana; 1996. pp. 721–722. [Google Scholar]
- 24.Page M, Thorpe R. In: The Protein Protocols Handbook. Walker J M, editor. Totawa, NJ: Humana; 1996. p. 723. [Google Scholar]
- 25.Tsien R Y. Annu Rev Biochem. 1998;67:509–544. doi: 10.1146/annurev.biochem.67.1.509. [DOI] [PubMed] [Google Scholar]
- 26.Brons P P, Pennings A H, Haanen C, Wessels H M, Boezeman J B. Cytometry. 1990;11:837–844. doi: 10.1002/cyto.990110710. [DOI] [PubMed] [Google Scholar]
- 27.Hoyt M A, Totis L, Roberts B T. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- 28.Li R, Murray A W. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- 29.Roberts B T, Farr K A, Hoyt M A. Mol Cell Biol. 1994;14:8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hardwick K G, Murray A W. J Cell Biol. 1995;131:709–720. doi: 10.1083/jcb.131.3.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor S S, McKeon F. Cell. 1997;89:727–735. doi: 10.1016/s0092-8674(00)80255-x. [DOI] [PubMed] [Google Scholar]
- 32.Taylor S S, Ha E, McKeon F. J Cell Biol. 1998;142:1–11. doi: 10.1083/jcb.142.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu J, Bousbaa H, Logarinho E, Li Z, Williams B C, Lopes C, Sunkel C E, Goldberg M L. J Cell Biol. 1999;146:13–28. doi: 10.1083/jcb.146.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dobles M, Liberal V, Scott M L, Benezra R, Sorger P K. Cell. 2000;101:635–645. doi: 10.1016/s0092-8674(00)80875-2. [DOI] [PubMed] [Google Scholar]
- 35.Fang G, Yu H, Kirschner M W. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen R H, Brady D M, Smith D, Murray A W, Hardwick K G. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wassmann K, Benezra R. Curr Opin Genet Dev. 2001;11:83–90. doi: 10.1016/s0959-437x(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 38.Percey M J, Myre K A, Neely C K, Azim J N, Ethier S P, Petty E M. Genes Chrom Cancer. 2000;29:356–362. doi: 10.1002/1098-2264(2000)9999:9999<::aid-gcc1044>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 39.Lee H, Trainer A H, Friedman L S, Thristlewaite F C, Evans M J, Ponder B A J, Venkitaraman A R. Mol Cell. 1999;4:1–10. doi: 10.1016/s1097-2765(00)80182-3. [DOI] [PubMed] [Google Scholar]
- 40.Jin D Y, Spencer F, Jeang K T. Cell. 1998;93:81–91. doi: 10.1016/s0092-8674(00)81148-4. [DOI] [PubMed] [Google Scholar]
- 41.Michel L S, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger P K, Murty V V, Benezra R. Nature (London) 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]