Abstract
Mutations at multiple sites in MEK1 occur in cancer, suggesting that their mechanisms of activation might be different. We analyzed 17 tumor-associated MEK1 mutants and found that they drove ERK signaling autonomously or in a RAS-RAF dependent manner. The latter are sensitive to feedback inhibition of RAF, which limits their functional output and often co-occur with RAS or RAF mutations. They act as amplifiers of RAF signaling. By contrast, another class of mutants delete a hitherto unrecognized negative regulatory segment of MEK1, is RAF- and phosphorylation-independent, unaffected by feedback inhibition of upstream signaling, and drives high ERK output and transformation in the absence of RAF activity. Moreover, these RAF-independent mutants are insensitive to allosteric MEK inhibitors, which preferentially bind to the inactivated form of MEK1. All the mutants were sensitive to an ATP-competitive MEK inhibitor. Thus, our study comprises a novel therapeutic strategy for tumors driven by RAF-independent MEK1 mutants.
Keywords: MEK1 mutants, RAF-dependency, ERK-dependent feedback, drug sensitivity
Introduction
RAS/RAF/MEK/ERK signaling is activated by upstream signaling pathways including those initiated by receptor tyrosine kinases and the strength and duration of the ERK signal is determined by negative feedback (1–3). Genetic lesions that deregulate the ERK pathway are important features of malignant transformation and are often drivers of tumor growth. Gain of function mutations of KRAS, NRAS, BRAF and loss of function lesions of NF1 occur commonly in human cancer. Activating RAF family fusion proteins and MEK1, MEK2, ARAF, HRAS, CRAF, and ERK, mutations also occur, but more rarely. The overall picture that emerges from these data is that mutations in different components of this pathway occur in many cancers and play a role in their development. What accounts for the differing frequencies of these mutations in cancer as a whole and in specific tumors and how the oncogenic consequences of mutations of the pathway may vary are poorly understood, if at all (4).
MEK1 and MEK2 are dual-specific serine/threonine and tyrosine kinases that become activated when phosphorylated by RAF kinases. They in turn phosphorylate and activate ERK1/2. Both MEK1 and MEK2 mutations occur in untreated human tumors (5–9) and have also been associated with the acquired resistance of tumors with BRAF V600E to RAF/MEK inhibitors (10,11). MEK1 mutations occur in multiple sites in the protein, without a single dominant hotspot, and the properties of the different mutants have not been well-defined.
Here, we have studied the mechanisms of activation of a panel of cancer associated MEK1 mutants and defined them into three different classes. The first is RAF-dependent, sensitive to ERK dependent feedback inhibition of RAS/RAF signaling, has low output and transforming activity in untransformed cells and, in human tumors, co-exists with other mutations that activate ERK. The second class of mutant has baseline RAF-independent activity but can be, to a greater or lesser degree, further activated when phosphorylated by RAF. These mutants are also sensitive to ERK dependent feedback and they are often but not always associated with other ERK-activating mutants in tumors. The third class of mutant is both RAF and phosphorylation independent, insensitive to feedback and constitutively active. It induces high levels of ERK output and transformation in untransformed cells and is unassociated with other ERK-activating mutations in tumors. These mutants are hyperactive and, unlike class 1 and 2 mutants, unresponsive to current MEK inhibitors, that bind to the inactive conformation of the enzyme. In contrast, an ATP-competitive MEK inhibitor suppresses the activity of all three classes of mutant. Thus, the work highlights the critical importance of functional characterization of different mutant alleles to understand their mechanism of activation, and their biologic effects, and to develop effective targeted therapies.
Results
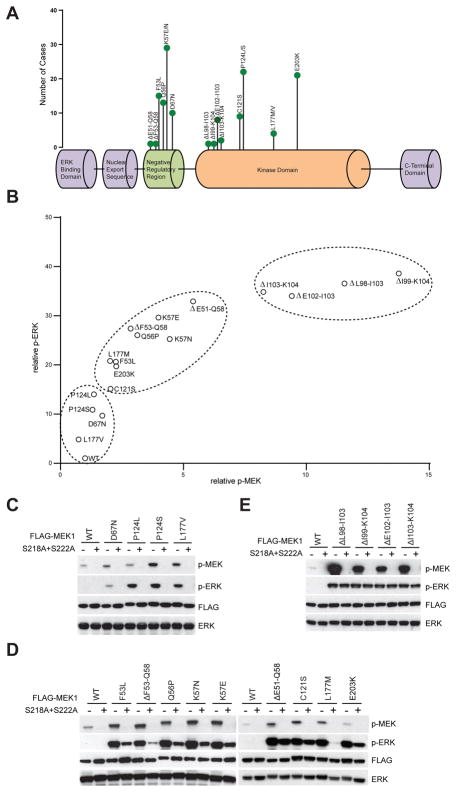
To characterize the functional consequences of MEK1 mutations, we cloned 17 MEK1 mutants that have been identified as recurrent in human tumors (Fig. 1A) and which, together, have been found in a wide variety of cancer types (cBioPortal(12)). When expressed in 293H cells, each mutant caused higher levels of p-ERK than WT MEK1 does (Fig. 1B and Fig. S1). They activated ERK signaling to varying degrees with different MEK/ERK phosphorylation patterns that can separate the mutations into different classes. Some mutants induced p-ERK more profoundly than pMEK (C121S, L177M, E203K, F53L, ΔF53–Q58, Q56P, K57E, K57N and ΔE51–Q58) (Fig. 1B). By contrast, others (e.g. ΔL98–I103, ΔE102–I103, ΔI99–K104 and ΔI103–K104) induced high levels of both pERK and pMEK. A third group (D67N, P124L, P124S and L177V) only weakly induced both pERK and pMEK. WT MEK1 is activated by its phosphorylation on S218 and S222 by RAF kinases (13,14). To test whether phosphorylation of these sites is similarly required for activation of the MEK1 mutants, we introduced both the S218A and S222A mutations into all of them. The double mutation completely abolished ERK activation in WT MEK1 and in four mutants (Fig. 1C), each of which was a weak activator of pERK in 293H cells (Fig. 1B). In contrast, the other 13 mutants all retained some degree of activation when S218 and S222 were replaced by alanine. This suggests some degree of phosphorylation-independent activation of catalysis by these MEK1 mutants. For nine of these, the double mutation clearly decreased, but did not abolish, the induction of pERK by the mutants (Fig. 1D). Thus, phosphorylation of these mutants enhances but is not required for induction of signaling. In four other mutants, those with in frame deletions in parts of the region between amino acids 98 and 104, signaling was unaffected by mutations of the two phosphorylation sites (Fig 1E). These were the mutants that, in Fig. 1B, caused hyperphosphorylation of both MEK and ERK. Thus, a set of MEK1 mutants signal in a phosphorylation-independent manner to varying degrees and the degree to which their activation is phosphorylation independent correlates directly with the level of their phosphorylation in cells.
Figure 1. MEK1 mutants are affected differently by S218/S222 phosphorylation.
A, Diagram of MEK1 mutations identified in human cancer from Cbioportal genomic database. B, 293H cells were transfected with vector, wild-type FLAG-MEK1, or 17 FLAG-MEK1 Mutants along with HA-ERK2. Western blot analysis was performed using antibodies against p-MEK, p-ERK, FLAG and total ERK. The relative p-ERK and p-MEK levels from western blot were determined by densitometry analysis using Image J. C–E, Ser218 and Ser222 of WT or mutant MEK1 were mutated into alanines (S218A+S222A), then WT or mutant MEK1 with or without S218A+S222A mutation along with HA-ERK2 were transfected into 293H cells. Western blot analysis was performed using antibodies against p-MEK, p-ERK, FLAG and total ERK.
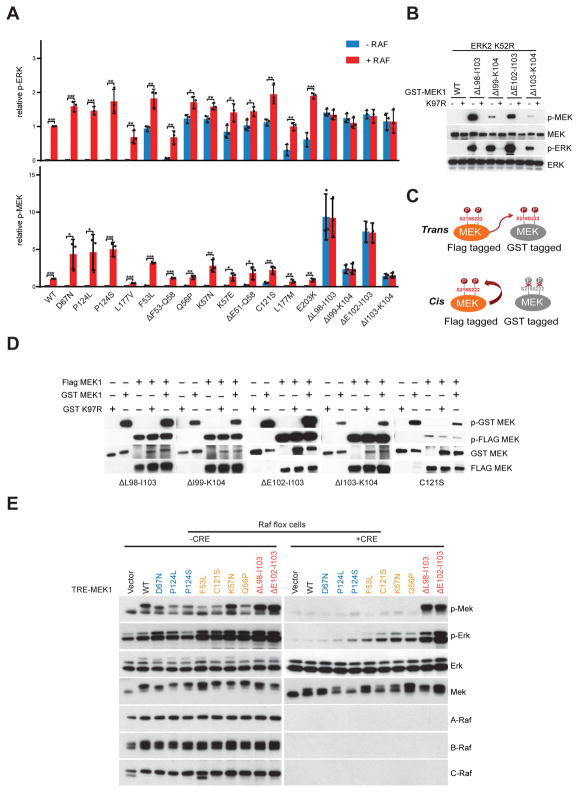
We therefore asked to what degree RAF kinase regulates the phosphorylation and kinase activity of these MEK1 mutants. We purified GST tagged MEK1 mutant proteins and performed in vitro kinase assays in the absence or presence of active BRAF kinase. Recombinant purified WT MEK1 was not phosphorylated in the absence of RAF kinase, nor were any of the four MEK1 mutant proteins whose kinase activity was abolished by S218A/S222A (Fig. 1C, Fig. 2A and S2A). Both the phosphorylation and kinase activities (with ERK2 as a substrate) of WT MEK1 and these mutants were potently induced by activated BRAF kinase (Fig. 2A). These are thus RAF and phosphorylation dependent MEK1 mutants. Presumably, their elevated activation in cells is due to enhanced activation by phosphorylation compared to WT. In contrast, the 9 mutants whose kinase activities were only partially decreased by S218A/S222A (Fig. 1D) all phosphorylated ERK2 in the absence of BRAF and their activity was further stimulated, marginally or significantly, by BRAF (Fig. 2A). These are thus RAF-regulated mutants. Phosphorylation of one of these (C121S) was detected in the absence of RAF and was further induced 4-fold by RAF (Fig. 2A). The other 8 mutants had no or barely detectable endogenous phosphorylation of S218 and S222 (Fig. 2A and S2B). These two groups together comprise the mutants that increase pERK to a much greater extent than pMEK in cells (Fig. 1B). In contrast, the four mutants with deletions in the aa 98–104 hyperactivate pMEK and pERK in cells and are completely insensitive to S218 and S222 mutations. In vitro, purified recombinant mutants of this class are phosphorylated in the absence of RAF kinases and addition of active BRAF affects neither their phosphorylation nor their kinase activity (Fig. 2A and S2C). These RAF-independent mutants are heavily phosphorylated in vitro and in cells, but their activity is phosphorylation independent.
Figure 2. Activity of MEK1 mutants is differently regulated by RAF kinase.
A, Purified GST fusion WT or mutant MEK1 proteins were incubated with recombinant inactive ERK2 K52R in the absence or presence of recombinant BRAF V600E at 30 °C for 15 min. Western blot analysis was performed using antibodies against p-ERK, ERK, p-MEK, MEK and BRAF. The relative p-ERK and p-MEK levels from western blot were determined by densitometry analysis using Image J. Representative data of western blot were shown from three independent experiments. Data are shown as mean(SD). T-test, *p<0.05, **P<0.01, ***p<0.001. B, K97R kinase dead mutation was introduced to WT or mutant MEK1. Purified GST fusion WT or mutant MEK1 proteins with or without kinase dead mutation were incubated with recombinant inactive ERK2 K52R as a substrate. Western blot analysis was performed using antibodies against p-MEK, p-ERK and total protein. C, Two possible models for auto-phosphorylation of MEK1 mutants. Active MEK1 mutant protein was labeled with FLAG tag, kinase dead MEK1 mutant protein was labeled with GST tag. Then MEK1 proteins labeled with two different tags were incubated together. If phosphorylation happens in trans, kinase dead one will be phosphorylated by th active one; if it’s in cis, only the active one will be phosphorylated. D, FLAG tagged active MEK1 mutant protein was purified through immunoprecipitation with an anti-FLAG antibody from 293H cells expressing mutant MEK1; active or kinase dead MEK1 mutant protein labeled with GST tag were purified from BL21(DE3); then MEK1 proteins labeled with two different tags were incubated either alone or together. Phosphorylation of different tagged MEK1 proteins were examined by immunoblotting. E, A-Raflox/lox; B-Raflox/lox;c-Raflox/lox;RERTert/ert MEF cells that inducibly express WT or mutant MEK1 were infected with Adeno-CRE virus and cultured in medium with 1 μM 4-Hydroxytamoxifen for a week. 300 ng/ml Doxycycline was added to the cells to induce the expression of those MEK1 proteins, cells were then collected after 24 hrs. Whole cell lysates were prepared and examined by Western blot.
We asked whether these mutants are phosphorylated by other kinases or whether they undergo autophosphorylation. To answer this question, we introduced a mutation that abrogates MEK kinase activity (K97R) into these MEK1 mutants. The activity of the K97R mutants was assessed with an in vitro kinase assay in which inactive ERK2 was used as the substrate. We found that the kinase dead mutation abolished ERK phosphorylation by the mutant MEK1 as expected. In addition, the phosphorylation of the K97R mutant was also abrogated in the absence of RAF kinases (Fig. 2B), supporting the hypothesis that these mutants autophosphorylate S218/S222. To determine whether this represents true autophosphoryation in cis or whether phosphorylation occurs in trans, we modified MEK1 mutant protein with a FLAG tag and the K97R mutant protein with a GST tag. The isolated FLAG tagged mutants and GST tagged kinase dead mutants were then co-incubated in a kinase assay and the phosphorylation of the tagged proteins was assessed. If phosphorylation occurs in trans, both FLAG tagged and GST tagged MEK1 mutants should be phosphorylated; if autophosphorylation is in cis, only the FLAG tagged protein will be phosphorylated (Fig. 2C). As shown in Figure 2D, FLAG tagged activating MEK1 mutants were phosphorylated, but they did not phosphorylate GST tagged K97R mutants. These data suggest that these MEK1 mutants are autophosphorylated in cis.
Our data show that MEK1 mutants may function in one of three different manners: RAF-dependent, -regulated or -independent activation. To test this idea, we utilized A-Raflox/lox; B-Raflox/lox; c-Raflox/lox; RERTert/ert MEF cells. The genes encoding ARAF, BRAF and CRAF can be deleted from this cell with an adenovirus encoding Cre recombinase administered together with 4-Hydroxytamoxifen. We used this system to assess the activity of MEK1 mutants in cells that express none of the WT RAF isoforms. As shown in Fig 2E, MEK1 mutants that function in a completely RAF independent manner (ΔL98–I103 and ΔE102–I103) are phosphorylated and potently stimulated ERK signaling despite the absence of expression of RAF kinases. In contrast, in cells expressing WT MEK or RAF-dependent MEK1 mutants (D67N, P124L and P124S), both MEK and ERK phosphorylation were decreased to trace levels in RAF-less cells. RAF-regulated MEK1 mutants (F53L, C121S, K57N and Q56P) have an intermediate effect on signaling when expressed in these cells; pMEK was almost completed eliminated, whereas a variable, but significant amount of ERK phosphorylation was retained. These data are consistent with our conclusion that MEK1 mutants can be placed into three functional classes based on their dependence on phosphorylation by RAF for activation (Table 1).
Table 1.
Properties of WT and Mutant MEK1 Alleles
| MEK1 mutants | Phosphorylation dependency | RAF dependency | |
|---|---|---|---|
| Wild type | Dependent | Dependent | |
| Class I | D67N | Dependent | Dependent |
| P124L | Dependent | Dependent | |
| P124S | Dependent | Dependent | |
| L177V | Dependent | Dependent | |
| Class II | ΔE51–Q58 | Regulated | Regulated |
| ΔF53–Q58 | Regulated | Regulated | |
| E203K | Regulated | Regulated | |
| L177M | Regulated | Regulated | |
| C121S | Regulated | Regulated | |
| F53L | Regulated | Regulated | |
| K57E | Regulated | Regulated | |
| Q56P | Regulated | Regulated | |
| K57N | Regulated | Regulated | |
| Class III | ΔL98–I103 | Independent | Independent |
| ΔI99–K104 | Independent | Independent | |
| ΔE102–I103 | Independent | Independent | |
| ΔI103–K104 | Independent | Independent | |
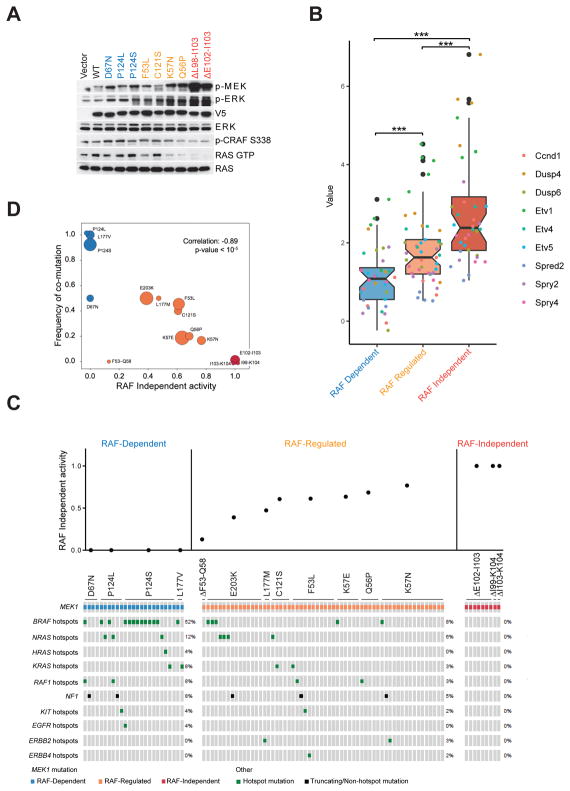
The mutants are thus categorized in terms of their dependence on upstream activation of RAF (Fig 2E): RAF-dependent mutants (blue), which are activated by RAF-dependent phosphorylation, those RAF-regulated mutants (orange) that are partially dependent on RAF but have varying amounts of RAF-independent activity and those RAF-independent mutants (red) that are completely RAF-independent. These categories in turn, predict whether signaling output will be sensitive to ERK-dependent feedback inhibition. In NIH3T3 cells, RAF-dependent MEK1 mutants activate pERK and ERK pathway output modestly (Fig. 3A, 3B and S3A) without affecting levels of RAS-GTP or CRAF phosphorylation (Fig. 3A). Increased ERK output is therefore not sufficient to feedback inhibit RAS/RAF. This is as expected if pathway output is self-limited by feedback inhibition of RAS or RAF. By contrast, RAF-independent mutants potently activate pERK while reducing CRAF S338 phosphorylation and RAS activation (Fig. 3A). In these cells, the ERK output was highest (Fig. 3B and S3A) and RAS-GTP and pCRAF were decreased compared to the levels in cells with WT MEK1. RAF-regulated MEK1 mutants have variable intermediate phenotypes: those with the highest levels of RAF-independent activation of ERK signaling (K57N, Q56P) cause higher levels of pERK and lower levels of RAS-GTP than the other tested mutants in this class (Fig 2, 3A and 3B). This result strengthens the model that the degree of RAF independence of MEK1 mutants is directly associated with their activation of the ERK pathway and inversely associated with their sensitivity to feedback inhibition of RAS-RAF. Degree of RAF independence is similarly associated with the ability of the MEK1 mutants to drive cell proliferation and transformation. The RAF-less cells are viable but do not proliferate (15). The expression of various MEK alleles was induced in the RAF-less cells and their ability to grow in soft agar was assessed (Fig. S3B). No growth in soft agar was noted in RAF-less cells in which expression of WT MEK1, RAF-dependent MEK1 mutants or RAF-regulated MEK1 mutants was induced. By contrast, RAF-independent mutants strongly promoted the anchorage-independent growth of the RAFless cells. Similar results were obtained when we examined the ability of the MEK1 mutant expressing RAF-less cells to form tumors in immunosuppressed (nude) mice (Fig. S3C). Tumors were observed as early as 14 days after implantation of cells expressing RAF-independent MEK1 mutants (red). In contrast, no palpable tumors were noted as late as 42 days after implantation of cells with RAF-dependent mutants (blue). Tumors did grow from cells in which RAF-regulated mutants (orange) were expressed, but more slowly than those expressing RAF-independent MEK1 mutants. The time of appearance of the former and their rate of growth were directly correlated with the degree to which the catalytic activity of the mutant was RAF-independent.
Figure 3. Loss of RAF dependency predicts enhanced ERK signaling output by MEK1 mutants.
A, NIH 3T3 cells stably expressing doxycycline-inducible WT or indicated mutant MEK1 were treated with doxycycline (300 ng/ml) for 24 hr. Expression and phosphorylation of the indicated proteins was assayed by western blot. B, ERK signaling ouptput was analyzed by real-time quantitative RT-PCR of nine validated ERK downstream target genes in NIH 3T3 cells inducibly expressing WT or mutant MEK1. The boxplot shows the log fold changes of gene expression (y axis) between 3 classes of MEK1 mutants (grouped in × axis and filled with three colors) and WT, which is overlaid with detailed changes of each gene plotted as colored points. Significant increasing changes were evaluated by T-tests among three groups. C, Co-mutation of MEK1 with RTK/RAS/RAF/NF1 in MEK1 mutant cancer patients. The data were collected from https://cbioportal.mskcc.org. Upper histogram showing RAF independent activity ratio (RAF independent in vitro kinase activity divided by kinase activity when RAF is added to the assay) RAF independent in vitro kinase activity was estimated by pERK in the absence of RAF, pERK in the presence of RAF was quantified as total MEK activity from Figure 2A. D, Weighted Pearson correlation between frequency of co-mutation and RAF independent activity ratio from c. The weights are based on the number of samples for each MEK1 mutation.
The results show that, unlike the RAF-independent MEK1 mutants, the RAF-dependent or regulated mutants drive transformation only weakly. We speculated that, in order to significantly activate ERK pathway, they must coexist with other lesions that activate RAF despite ERK-dependent feedback. This turned out to be the case. In Fig. 3C, MEK1 mutants are ordered on the X-axis by their degree of RAF independence. The Y-axis represents the fraction of MEK kinase activity that is RAF independent for each of these MEK1 mutants (RAF independent kinase activity divided by kinase activity when RAF is added to the in vitro kinase assay). Analysis of a large database (MSK-IMPACT) of human tumor-associated mutations showed that the RAF-dependent MEK1 mutants are almost always associated with activating mutations of RAS or RAF or RTK genes or NF1 inactivating lesions (Fig. 3C). In contrast, tumors that express any of the four RAF-independent mutants are unassociated with coexistent mutations that activate RAF signaling. MEK1 mutants with RAF-independent activity that is enhanced by RAF sometimes coexist with RAF activating mutations and sometimes do not. The data suggest that the frequency of coexistent RAF-activating mutations may be inversely correlated with the degree of RAF independence of the RAF regulated MEK1 mutants (Fig. 3D).
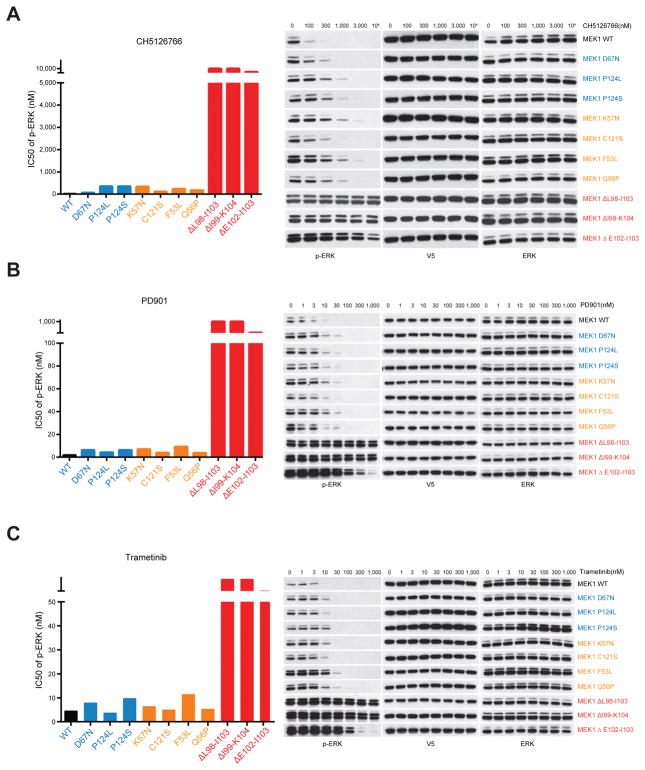
Our data suggest that the degree of RAF dependence of MEK1 mutants determines their ability to drive ERK pathway output and, consequently, whether they are co-selected with other mutations that drive RAF. The recurrence of these mutations in human cancer and their ability to activate ERK output suggested that they are tumor drivers, so we asked whether they were sensitive to MEK inhibitors. Several highly selective inhibitors of MEK are used in clinic as anticancer agents. These inhibitors bind to an allosteric site adjacent to the ATP pocket (16–19). They do not compete with ATP for binding and preferentially inhibit the inactive conformation of the enzyme. Here, we tested the effects of three different allosteric MEK inhibitors in NIH3T3 cells expressing WT or mutant MEK1 (Fig. 4). All three allosteric inhibitors lead to only small shifts in IC50 (~3–10 fold) in some of the RAF-dependent (blue) or RAF-regulated (orange) MEK1 mutants expressing cells relative to WT MEK1. In contrast, ERK activation in cells expressing RAF-independent (red) MEK1 mutants was only inhibited at ~10–100 fold higher doses (Fig. 4).
Figure 4. Sensitivity of MEK1 mutants to allosteric MEK inhibitors is associated with their RAF dependency.
A–C, WT or mutant MEK1 tagged with V5 were expressed in NIH3T3 cells upon cultured in medium containing doxycycline (300 ng/ml) for 24 hrs. Cells were then treated for 1 hr with increasing concentrations of three different allosteric MEK1 inhibitors CH5126766 (A) or PD901 (B) or Trametinib (C). IC50 of p-ERK inhibition to all three drugs were calculated on the basis of densitometry analysis of western blot results.
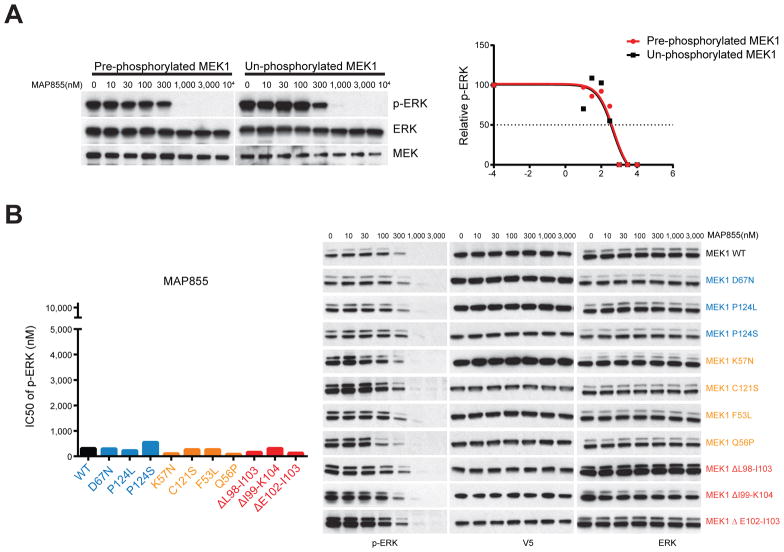
Previous studies suggest that the phosphorylation of MEK1 could affect its sensitivity to allosteric MEK inhibitors(17). Phosphorylation of purified MEK1 has been reported to cause a 20-fold decrease of its sensitivity to trametinib ((17) and table S1). We found that pre-phosphorylation of purified MEK1 kinase significantly reduced its sensitivity to the CH5126766 (100-fold) and PD901 (10-fold) (Fig. S4A and S4B) (Table S1). These data suggested that the constitutive RAF-independent hyperphosphorylation of RAF-independent MEK1 mutants (Fig. 2E, S2C) could cause their insensitivity to the allosteric MEK inhibitors. By contrast, the effects of a new selective ATP-competitive MEK inhibitor MAP855 are not reduced by phosphorylation of the enzyme. As shown in Figure 5A, the pre-phosphorylation of MEK1 doesn’t decrease its sensitivity to MAP855. As predicted, this drug effectively inhibited ERK signaling driven by each class of MEK1 mutants, including those with the aa 99–104 deletion (Fig. 5B).
Fig 5. MEK1 mutants are equally sensitive to a new ATP-competitive MEK inhibitor.
A, In the in vitro kinase assay, pre-phosphorylated MEK1 protein were treated with MAP855 at increasing concentrations. Alternatively, MEK1 were pre-treated with MAP855 before incubation with activated BRAF V600E kinase. The p-ERK level was quantified by densitometry, then normalized to the p-ERK level in untreated sample. The p-ERK response curves were generated using Prism 7.01. B, WT or mutant MEK1 tagged with V5 were expressed in NIH3T3 cells upon cultured in medium containing 300 ng/ml doxycycline for 24 hrs. Cells were then treated for 1 hr with increasing concentrations of MAP855. Expression and phosphorylation of the indicated proteins was assayed by western blot. IC50 values of p-ERK inhibition were calculated based on densitometry analysis of western blot result.
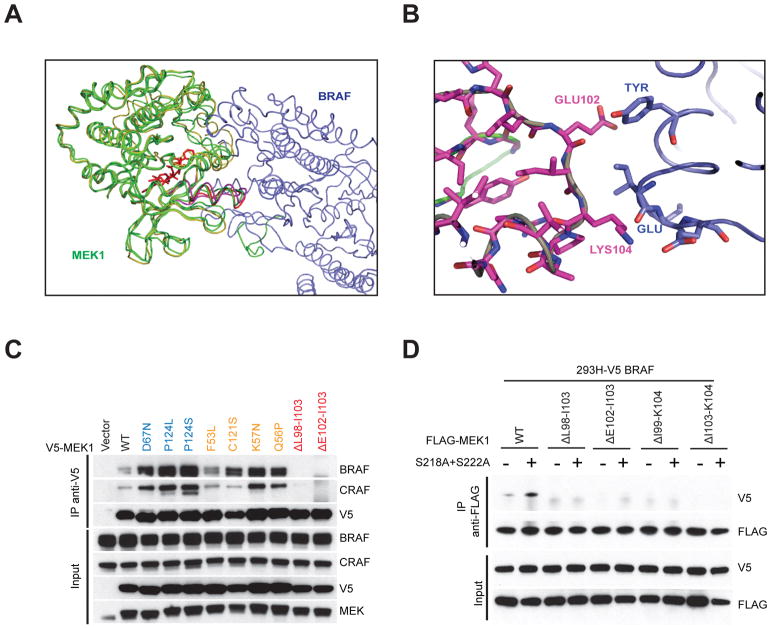
Thus, our work divides mutant MEK1 alleles into three groups, based on the degree of dependence of their catalytic activity on RAF-mediated phosphorylation of S218 and S222. The groups differ in the degree with which they can autonomously activate ERK signaling, their sensitivity to ERK-dependent feedback inhibition of RAS/RAF, whether they require cooperation with other mutant proteins that activate ERK signaling, and in their sensitivity to inhibitors that work by different mechanisms. The first type of mutant is completely RAF dependent and its activity is hyperstimulated by phosphorylation compared to that of WT. This type of mutant has little or no ability to drive signaling on its own and likely acts as an amplifier of the ERK-signaling driven by other mutants with which they coexist. The second group varies in its degree of RAF independence, which varies inversely with their coexistence with other ERK pathway mutants. The third group has unique properties and is comprised of four mutants with deletions in the region bounded by amino acids 98LIHLEI104K. The kinase activity of these mutants is constitutive and RAF and phosphorylation independent, although they are highly phosphorylated in cells due to their autophosphorylation in cis on S218 and S222. A computationally solved structure shows that this region of the protein interacts with BRAF (Fig. 6A, B and Fig. S5). This interaction is apparently required for the MEK1/BRAF interaction in cells since, unlike WT MEK1 or RAF-dependent or regulated MEK1 mutants, the RAF-independent mutants do not coimmunoprecipitate with BRAF or CRAF (Fig. 6C). This is not due to hyperphosphorylation of S218/S222 since the S218A/S222A double mutant does not restore binding to BRAF (Fig. 6D). The results are consistent with the structure and suggest that the interaction of 98–104 peptide in the β3-αC loop of MEK1 with BRAF is necessary for the binding of the two proteins. Since deletions in this region activate MEK1 in a RAF-independent manner, one might infer that RAF binding to this site inhibits MEK activity and that, for WT MEK1, this inhibition is relieved by RAF-mediated phosphorylation of S218/S222. However, this does not seem to be the case, since in RAF-less cells, WT MEK1 is also inactive. The data is more consistent with the hypothesis that aa 98–104 is a potent negative regulator of MEK1 kinase activity, deletion of which leads to constitutive activation. Deletions in the β3-αC loop have also been identified in EGFR, HER2 and BRAF as activating mutations in cancer patients (20). Displacement of helix-αC away from the β3–5 core to an inactive, αC-out conformation requires a minimum length of the loop for movement, implying that β3-αC deletions work by shifting the equilibrium to the active conformation. Foster et al. showed that kinase activity varies as a function of the insert length, with a maximum observed for a five amino acid deletion, also the most prevalent clinical deletion mutation(21). Analysis of all available kinase structures revealed that αC-out conformations preferentially occur in kinases with an intermediate β3-αC insert length(22). The deletions in MEK1 may result in a stabilization of the active conformation by a similar mechanism. Thus, it is likely that activation of WT MEK1 by RAF involves phosphorylation-dependent relief of the negative regulation by this domain. This is supported by the loss of any effect of phosphorylation on the activity of the deletion mutants, whereas activation of all the other mutants is enhanced by phosphorylation.
Fig 6. Deletions in aa 98–104 region of MEK1 reduced its binding to BRAF kinase.
A, Structure of MEK-BRAF binding complex. MEK-BRAF X-ray molecules are shown in green (MEK) and blue (BRAF), Molecular Dynamics (MD) calculated MEK is shown in gold. The allosteric MEK inhibitor G573 is shown in RED. The deletions region (magenta loop) can be seen in direct contact with BRAF. B, Close-up view of the interaction space between BRAF(blue) and the deletion region (magenta). Within this region GLU102 and LYS104 form polar interactions with an associated BRAF protein. LYS104 interacts with GLU side chain and a backbone carbonyl of an ILE in BRAF. GLU102 interacts with a TYR side chain in BRAF. C, NIH-3T3 cells inducibly expressing V5 tagged WT MEK1 or mutant MEK1 were exposed to 300ng/ml doxycycline for 24 hrs. Then cells were collected and subjected to immunoprecipitation with anti-V5 antibody. The input and pull-down proteins were assayed by Western blot. D, 293H cells stably expressing V5 tagged BRAF were transfected with FLAG tagged WT or mutant MEK1 with or without S218A+S222A mutation, 24 hrs after transfection, cells were collected and subjected to immunoprecipitation with anti-FLAG antibody. The input and pull-down proteins were assayed by Western blot.
Discussion
MEK1 mutants occur in more than 1% of human tumors with higher frequency in the histiocytosis, melanoma, colorectal, bladder, lung and other carcinomas. Mutations occur at many sites in the protein, with only 3 modest hotspots, K57, P124 and E203, the rest distributed at very low frequency at many sites. These frequencies hold for tumors that have not been treated with ERK pathway inhibitors, they may be higher in tumors with acquired resistance to RAF or MEK inhibitors. As is true for most proto-oncogenes, most of the low frequency mutants have not been studied and, until recently, there has been a general sense that they all function similarly and will respond to the same targeted drugs.
In this work, we have functionally characterized 17 oncogenic MEK1 mutants from human tumors and identified three different functional classes, based on their dependency on phosphorylation by RAF kinase or lack thereof (Table 1). Class 1 mutants are dependent on and hyperactivated by phosphorylaton of S218 and S222 by RAF, thus, they are sensitive to ERK-dependent feedback inhibition of upstream signaling. Generation of high ERK output in these cells, therefore, requires other molecular events that reduce the sensitivity of RAF to negative feedback. In support of this model, we found that these mutants always coexist with RAS, RAF or NF1 alterations in tumors. We believe these mutants serve as amplifiers of ERK signaling rather than autonomous activators. Melanomas with coexistent BRAF V600E and Class 1 MEK1 mutants have been identified in untreated tumors. Although their existence was shown to contribute to the ERK activation in tumors, they are unlikely to cause resistance to the RAF inhibitor treatment (23,24). In contrast, the Class 2 MEK1 mutant kinases have some level of basal, RAF-independent activity but are further activated by RAF. They sometimes coexist with RAS, RAF or NF1 and the degree to which they do varies inversely with fraction of their activity that is RAF-independent. This finding suggests that selection of these mutants, alone or together, is based on a requirement for a certain degree of ERK output, in a range that enhances growth but does not cause toxicity. In contrast to Class 1, Class 2 mutants have been found to cause acquired resistance to RAF inhibitors (24,25).
Uniquely, Class 3 mutants are completely RAF-independent. In cells, they drive high levels of ERK output and are insensitive to the ERK-dependent feedback inhibition of RAS and RAF. Thus, unlike Class 1 mutants, they autonomously drive signaling. Tumors in which Class 3 mutants coexist with RAS, RAF or NF1 mutants have not so far been identified.
The currently available MEK inhibitors are all allosteric and bind to the inactive form of the enzyme. Class 1 and 2 mutants were also sensitive to these drugs, suggesting that a significant fraction of these enzymes is in the un-phosphorylated, inactive state. Our data suggests that the aa 98–104 region of MEK1 is auto-inhibitory and its deletion causes Class 3 enzymes to permanently adopt the active conformation. In support of this idea, Class 3 mutants are resistant to allosteric MEK inhibitors. The other option to inhibit the activity of this class of mutant MEK1 would be using an ATP-competitive MEK inhibitor. Although there is one such compound E6201 which is currently under clinical investigation, it also has many non-MEK targets (26–28). Here, we examined a newly developed selective MEK inhibitor MAP855 which functions by competing ATP binding to MEK protein. In contrast to the cases with allosteric MEK inhibitors, all three classes of MEK1 mutants are sensitive to this new ATP-competitive inhibitor. Such drugs may be useful in treating tumors dependent on any classes of MEK1 mutants and in preventing Class 2 or Class 3 MEK1 mutants-dependent acquired resistance.
Taken together, our work suggests that detailed functional analysis of multiple oncogenic alleles in a given protein is a powerful means for understanding different biochemical strategies for oncogenic activation and is necessary if we are to successfully use the genomics of the tumor to develop effective therapies.
Methods
Cell culture
293H, NIH-3T3 and Phoenix AMPHO cells were purchased from ATCC between 2013 and 2015. A-Raflox/lox; B-Raflox/lox; c-Raflox/lox; RERTert/ert MEF cells were provided by Dr. Mariano Barbacid in Feb 2016 and was confirmed by MSK-IMPACT. These cell lines and all other cells with inducible expression of MEK1 mutants were tested negative for mycoplasma contamination. All the cells were grown in Dulbecco’s modified Eagle’s medium with glutamine, antibiotics, and 10% FBS. Cells engineered to inducible express a protein were maintained in standard medium with 100 μg/ml hygromycin and 2 μg/ml puromycin.
NIH-3T3 cells were used to construct the stable lines with inducible expression of mutant MEK1s to study MEK1 mutant-driven signaling output and their response to different types of MEK inhibitors. Conditional RAF knockout MEF cells were employed to determine the RAF dependency of mutant MEK1 driven ERK pathway activation. Co-expression and co-immunoprecipitation experiments were performed with the 293H cells, in which more than two exogenous proteins were transiently expressed. Cell lines were used after less than 3 months of passages post receipt for the above experiments.
Antibodies
Western blot, immunoprecipitation and in vitro kinase assays were performed as previously described. The following antibodies were used: anti-p217/p221-MEK1/2 (p-MEK1/2)(#9154), anti-p202/p204-ERK1/2 (p-ERK1/2) (#4370), anti-MEK1/2 (#4694), anti-ERK1/2 (#4696) from Cell Signaling, anti-V5 (R960-25) from Thermo Fisher Scientific, anti-ARAF (sc-408) and anti-BRAF (sc-5284) from Santa Cruz Biotechnology, anti- FLAG (F3165) from Sigma, anti-CRAF (610152) from BD Transduction Laboratories, anti-p-CRAF S338 (05-538) from Millipore, and anti-RAS antibody from the active RAS pull-down and detection kit (Thermo Fisher Scientific, #16117). For immunoprecipitations of tagged proteins, the following reagents were used: anti-V5 agarose affinity gel (Invitrogen), anti-FLAG M2 affinity gel (Sigma), protein G agarose gel (Invitrogen).
Plasmids
The pcDNA3-FLAG-MEK1 was obtained from Biomyx. The pcDNA3-BRAF-V5 was constructed as previously described(29). The pGEX6P1 was obtained from Addgene. The MEK1 gene was sub-cloned into pGEX6P1 for in vitro protein purification. Plasmids TTIGFP-MLUEX and pMSCV-rtTA3-PGK-Hygro for inducible gene expression were provided by Scott Lowe’s lab at MSKCC. The MEK1 gene was sub-cloned into TTIGFP-MLUEX vector harboring the Tet-responsive promoter. Mutations were introduced by using the site-directed Mutagenesis Kit (Stratagene).
Compounds
Trametinib and CH5126766 were obtained from GlaxoSmithKline and Chugai Pharmaceuticals, respectively. PD0325901 was synthesized in the MSKCC Organic Synthesis Core Facility by O. Ouerfelli. MAP855((1-((3S,4S)-4-(8-(2-chloro-4-(pyrimidin-2-yloxy)phenyl)-7-fluoro-2-methyl-1H-imidazo[4,5-c]quinolin-1-yl)-3-fluoropiperidin-1-yl)-2-hydroxyethanone)) was obtained from Novartis (compound No. 1, WO2015022662). The referenced patent provides detailed structure and methods for synthesis. Doxycycline and 4-Hydroxytamoxifen from Sigma Aldrich; Puromycin and Hygromycin stock solution from invitrogen. 4-Hydroxytamoxifen was dissolved in methanol to yield 5 mM stock, all other drugs were dissolved in DMSO to yield 10 mM stock and stored at −20°C.
Generation of Rafless cells
A-Raflox/lox; B-Raflox/lox; c-Raflox/lox; RERTert/ert MEF cells were generously provided by Mariano Barbacid. MEF cells were isolated from embryonic day 13.5 embryos and immortalized as previously reported(15). For generation of Rafless cells, A-Raflox/lox; B-Raflox/lox; c-Raflox/lox; RERTert/ert MEF cells were infected with Adeno-Cre particles (multiplicity of infection = 100) for 24 hrs, then cells were cultured in medium with 1 μM 4-Hydroxytamoxifen (Sigma) for a week.
Inducible gene expression in cells
Retroviruses encoding rtTA or MEK1 genes were packaged in Phoenix-AMPHO cells obtained from ATCC. The supernatant-containing virus was filtered with 0.45 μM PVDF membrane. The target cells were infected with virus for 8 hrs. 48 hrs later, cells were selected in medium containing Puromycin (2 μg/ml) or Hygromycin (100 μg/ml) for 3 days. The positive infected cell populations were further sorted using GFP as a marker after overnight exposure to 1μg/ml doxycycline. GFP positive cells were then cultured and expanded in medium with doxycycline along with antibiotics. For the conditional RAF knockout cells, the inducible expression cells were created in the absence of 4-Hydroxytamoxifen.
Transfections
Cells were seeded in 60 mm or 100 mm plates and transfected the following day using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. A ratio of DNA to lipofectamine of 1 μg DNA / 3 μl lipofectamine was employed.
Quantitative RT-PCR
Total RNA was collected from cultured cells using the SV Total RNA Isolation System (promega). cDNA was synthesized with ImProm-II™ Reverse Transcription System using Oligo(dT) Primer (Promega), and RT-PCR was performed on a ViiA™ 7 Real-Time PCR instrument with SYBR® Green PCR Master Mix (Applied Biosystems™), using the following program: holding at 50 °C for 2 min and polymerase activation at 95 °C for 10 min, followed by 40 cycles of amplification (95 °C for 15 s, 60 °C for 1 min). GAPDH expression was used as an internal reference to normalize input cDNA. Fold changes of the gene expression between the mutants and WT were averaged from the 3 replicates and logarithm transformed before clustering analysis. The heatmap was plotted by R package with genes in the row were clustered based on similarity of Pearson Correlation Coefficient. One tail t-test were performed using R package for comparisons among 3 different groups.
Active RAS pull-down assay
Cells were cultured in 10cm dishes until 70–80% confluence. GTP-bound Ras was quantitated using the RAF1 Ras-binding domain (RBD) pull-down from Detection Kit (Thermo Scientific), as instructed by the manufacturers.
Anchorage-independent cell growth
Rafless cells were seeded in 0.4% agar on six-well plates, incubated at 37°C for 4 weeks, and stained with 0.005% crystal violet (Sigma-Aldrich) for 1 hour at room temperature. Soft agar colonies were quantified using Gen5 (BioTek). Captured images were analyzed using uniform size and shape parameters.
Immunoprecipitation
Cells were collected on ice and lysed in lysis buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 0.1%NP40, 1 mM EDTA) supplemented with protease and phosphatase inhibitor cocktail tablets (Roche). Immunoprecipitations were carried out at 4 °C for 1 hr, followed by five washes with lysis buffer. The pull-down complexes were eluted with 1XSDS loading buffer and assayed by Western blotting.
Expression and purification of recombinant MEK1
Human wild type MEK1, as well as all the mutants used in this study, were subcloned into pGEX6P1, expressed as glutathione-S-transferase fusions and purified by Pierce™ Glutathione Agarose (Thermo Fisher Scientific).
In vitro kinase assay
In vitro kinase assays were conducted in the presence of 200 μM ATP, at 30 °C for 15 min. Briefly, GST-MEK1 or mutants were incubated in the absence or presence of active BRaf (V600E) Protein (Upstate). Changes in MEK1 phosphorylation were estimated by immunoblotting for p-MEK. To test the kinase activity of WT or mutant MEK1 protein, recombinant inactive ERK2 protein (GenWay Biotech) was used as a substrate and the reaction was terminated with the addition of 1XSDS loading buffer and boiling. Kinase activity was estimated by immunoblotting for pERK. When the subsequent in vitro kinase assay was performed after immunoprecipitation, 0.02% SDS was added to the wash buffer and one extra wash with kinase buffer was required.
Structural analysis of MEK1-RAF Complex
All structure models of MEK1 or MEK1/BRAF complex were generated from 10ns AMBER15 constant pressure simulations using the PARM99(30) force field for the proteins and PARM@FROSST force field for the small molecules at 1atm and 300K. Conformationally and topologically averaged AM1-BCC(31) charges were generated for the small molecules. WATMD(32) was used to generate time averaged structures for evaluation of the PPI’s.
Mutational data and enrichment analysis
Mutational data were obtained from three sources: 1) The Cancer Genome Atlas (TCGA); 2) various published studies in which mutational data was publicly available, and 3) and an ongoing prospective clinical sequencing initiative at Memorial Sloan Kettering that utilizes a targeted capture-based sequencing of protein-coding exons and select introns of 314 or 410 cancer-associated genes (MSK-IMPACT). As mutation calling algorithms and mutation filtering and reporting practices varied from study to study, somatic mutational data review and correction were undertaken where possible as previously described. Mutation calls were uniformly re-annotated to gene transcripts in Ensembl release 75 (Gencode release 19), and a single canonical effect per mutation was reported using Variant Effect Predictor (VEP) version 81 and vcf2maf version 1.6.2. To exclude putative germline variants misattributed as somatic mutations, we exclude any variant identified in ExAC r0.3 with a minor population allele frequency greater than 0.06%. We sought to determine the enrichment of co-occurring RTK/RAS/RAF lesions in tumors possessing MEK1 mutants in all cancer types.
Xenograft Model Studies
Nu/nu athymic mice were obtained from Harlan Laboratories and maintained in compliance with Institutional Animal Care and Use Committee (IACUC) guidelines. Rafless cells with WT or mutant MEK1 expression were implanted subcutaneously into the left and right posterior flanks of 4–6 week old Nu/nu athymic female mice (n=5 mice per group). Tumor formation was measured twice a week. All studies were performed in compliance with institutional guidelines under an IACUC approved protocol. Investigators were not blinded when assessing the outcome of the in vivo experiments.
Statistical analysis
Results are mean values ± s.d. Two tailed t-test were performed using GraphPad Prism 7.01 for comparisons among different groups. Investigators were not blinded when assessing the outcome of the in vivo experiments. All cellular experiments were repeated at least three times.
Supplementary Material
Significance.
Mutants with which MEK1 mutants coexist and their sensitivity to inhibitors are determined by allele-specific properties. This study shows the importance of functional characterization of mutant alleles in single oncogenes and identifies a new class of MEK1 mutants, insensitive to current MEK1 inhibitors but treatable with a new ATP-competitive inhibitor.
Acknowledgments
Financial support: This research is supported by grants from the National Institutes of Health (R01 CA169351, P01 CA129243, R35 CA210085 and U54 OD202355), and from Mr. William H. Goodwin and Mrs. Alice Goodwin and the Commonwealth Foundation for Cancer Research and The Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center. This work is also supported by Functional Genomics Initiative of MSKCC (O. A-W.) and the NIH/NCI Cancer Center Support Grant P30 CA008748.
We are grateful to Piro Lito and Vanessa S. Rodrik-Outmezguine for useful discussions.
Footnotes
Conflict of interest: N.R. is on the scientific advisory board of and receives research funding from Chugai, and is on the scientific advisory board of and owns stock in Beigene, Wellspring and Kura. N.R. is also on the scientific advisory board of Daiichi-Sankyo, Astra-Zeneca and Takeda, and is a consultant to Novartis. D. M., G. C., D. S. and H. M. are employees of Novartis Institutes for Biomedical Research.
References
- 1.Dong C, Waters SB, Holt KH, Pessin JE. SOS phosphorylation and disassociation of the Grb2-SOS complex by the ERK and JNK signaling pathways. J Biol Chem. 1996;271:6328–32. doi: 10.1074/jbc.271.11.6328. [DOI] [PubMed] [Google Scholar]
- 2.Dougherty MK, Muller J, Ritt DA, Zhou M, Zhou XZ, Copeland TD, et al. Regulation of Raf-1 by direct feedback phosphorylation. Mol Cell. 2005;17:215–24. doi: 10.1016/j.molcel.2004.11.055. [DOI] [PubMed] [Google Scholar]
- 3.Pratilas CA, Taylor BS, Ye Q, Viale A, Sander C, Solit DB, et al. (V600E)BRAF is associated with disabled feedback inhibition of RAF-MEK signaling and elevated transcriptional output of the pathway. Proc Natl Acad Sci U S A. 2009;106:4519–24. doi: 10.1073/pnas.0900780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang MT, Asthana S, Gao SP, Lee BH, Chapman JS, Kandoth C, et al. Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nat Biotechnol. 2016;34:155–63. doi: 10.1038/nbt.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arcila ME, Drilon A, Sylvester BE, Lovly CM, Borsu L, Reva B, et al. MAP2K1 (MEK1) Mutations Define a Distinct Subset of Lung Adenocarcinoma Associated with Smoking. Clin Cancer Res. 2015;21:1935–43. doi: 10.1158/1078-0432.CCR-14-2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond EL, Durham BH, Haroche J, Yao Z, Ma J, Parikh SA, et al. Diverse and Targetable Kinase Alterations Drive Histiocytic Neoplasms. Cancer Discov. 2016;6:154–65. doi: 10.1158/2159-8290.CD-15-0913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grisham RN, Sylvester BE, Won H, McDermott G, DeLair D, Ramirez R, et al. Extreme Outlier Analysis Identifies Occult Mitogen-Activated Protein Kinase Pathway Mutations in Patients With Low-Grade Serous Ovarian Cancer. J Clin Oncol. 2015;33:4099–105. doi: 10.1200/JCO.2015.62.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, et al. A landscape of driver mutations in melanoma. Cell. 2012;150:251–63. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikolaev SI, Rimoldi D, Iseli C, Valsesia A, Robyr D, Gehrig C, et al. Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat Genet. 2011;44:133–9. doi: 10.1038/ng.1026. [DOI] [PubMed] [Google Scholar]
- 10.Emery CM, Vijayendran KG, Zipser MC, Sawyer AM, Niu L, Kim JJ, et al. MEK1 mutations confer resistance to MEK and B-RAF inhibition. Proc Natl Acad Sci U S A. 2009;106:20411–6. doi: 10.1073/pnas.0905833106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moriceau G, Hugo W, Hong A, Shi H, Kong X, Yu CC, et al. Tunable-combinatorial mechanisms of acquired resistance limit the efficacy of BRAF/MEK cotargeting but result in melanoma drug addiction. Cancer Cell. 2015;27:240–56. doi: 10.1016/j.ccell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer discov. 2012;2:401–4. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan M, Templeton DJ. Identification of 2 serine residues of MEK-1 that are differentially phosphorylated during activation by raf and MEK kinase. J Biol Chem. 1994;269:19067–73. [PubMed] [Google Scholar]
- 14.Zheng CF, Guan KL. Activation of MEK family kinases requires phosphorylation of two conserved Ser/Thr residues. EMBO J. 1994;13:1123–31. doi: 10.1002/j.1460-2075.1994.tb06361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drosten M, Sum EY, Lechuga CG, Simon-Carrasco L, Jacob HK, Garcia-Medina R, et al. Loss of p53 induces cell proliferation via Ras-independent activation of the Raf/Mek/Erk signaling pathway. Proc Natl Acad Sci U S A. 2014;111:15155–60. doi: 10.1073/pnas.1417549111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caunt CJ, Sale MJ, Smith PD, Cook SJ. MEK1 and MEK2 inhibitors and cancer therapy: the long and winding road. Nat Rev Cancer. 2015;15:577–92. doi: 10.1038/nrc4000. [DOI] [PubMed] [Google Scholar]
- 17.Gilmartin AG, Bleam MR, Groy A, Moss KG, Minthorn EA, Kulkarni SG, et al. GSK1120212 (JTP-74057) is an inhibitor of MEK activity and activation with favorable pharmacokinetic properties for sustained in vivo pathway inhibition. Clin Cancer Res. 2011;17:989–1000. doi: 10.1158/1078-0432.CCR-10-2200. [DOI] [PubMed] [Google Scholar]
- 18.Ishii N, Harada N, Joseph EW, Ohara K, Miura T, Sakamoto H, et al. Enhanced inhibition of ERK signaling by a novel allosteric MEK inhibitor, CH5126766, that suppresses feedback reactivation of RAF activity. Cancer Res. 2013;73:4050–60. doi: 10.1158/0008-5472.CAN-12-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lito P, Saborowski A, Yue J, Solomon M, Joseph E, Gadal S, et al. Disruption of CRAF-mediated MEK activation is required for effective MEK inhibition in KRAS mutant tumors. Cancer Cell. 2014;25:697–710. doi: 10.1016/j.ccr.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eck MJ, Yun CH. Structural and mechanistic underpinnings of the differential drug sensitivity of EGFR mutations in non-small cell lung cancer. Biochim Biophys Acta. 2010;1804:559–66. doi: 10.1016/j.bbapap.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster SA, Whalen DM, Ozen A, Wongchenko MJ, Yin J, Yen I, et al. Activation Mechanism of Oncogenic Deletion Mutations in BRAF, EGFR, and HER2. Cancer Cell. 2016;29:477–93. doi: 10.1016/j.ccell.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 22.Mobitz H. The ABC of protein kinase conformations. Biochim Biophys Acta. 2015;1854:1555–66. doi: 10.1016/j.bbapap.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Shi H, Moriceau G, Kong X, Koya RC, Nazarian R, Pupo GM, et al. Preexisting MEK1 exon 3 mutations in V600E/KBRAF melanomas do not confer resistance to BRAF inhibitors. Cancer discov. 2012;2:414–24. doi: 10.1158/2159-8290.CD-12-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trunzer K, Pavlick AC, Schuchter L, Gonzalez R, McArthur GA, Hutson TE, et al. Pharmacodynamic effects and mechanisms of resistance to vemurafenib in patients with metastatic melanoma. J Clin Oncol. 2013;31:1767–74. doi: 10.1200/JCO.2012.44.7888. [DOI] [PubMed] [Google Scholar]
- 25.Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goto M, Chow J, Muramoto K, Chiba K, Yamamoto S, Fujita M, et al. E6201 [(3S,4R,5Z,8S,9S,11E)-14-(ethylamino)-8, 9,16-trihydroxy-3,4-dimethyl-3,4,9,19-tetrahydro-1H-2-benzoxacyclotetradecine-1,7 (8H)-dione], a novel kinase inhibitor of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase (MEK)-1 and MEK kinase-1: in vitro characterization of its anti-inflammatory and antihyperproliferative activities. J Pharmacol Exp Ther. 2009;331:485–95. doi: 10.1124/jpet.109.156554. [DOI] [PubMed] [Google Scholar]
- 27.Narita Y, Okamoto K, Kawada MI, Takase K, Minoshima Y, Kodama K, et al. Novel ATP-competitive MEK inhibitor E6201 is effective against vemurafenib-resistant melanoma harboring the MEK1-C121S mutation in a preclinical model. Mol Cancer Ther. 2014;13:823–32. doi: 10.1158/1535-7163.MCT-13-0667. [DOI] [PubMed] [Google Scholar]
- 28.Zhang W, Borthakur G, Gao C, Chen Y, Mu H, Ruvolo VR, et al. The Dual MEK/FLT3 Inhibitor E6201 Exerts Cytotoxic Activity against Acute Myeloid Leukemia Cells Harboring Resistance-Conferring FLT3 Mutations. Cancer Res. 2016;76:1528–37. doi: 10.1158/0008-5472.CAN-15-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao Z, Torres NM, Tao A, Gao Y, Luo L, Li Q, et al. BRAF Mutants Evade ERK-Dependent Feedback by Different Mechanisms that Determine Their Sensitivity to Pharmacologic Inhibition. Cancer Cell. 2015;28:370–83. doi: 10.1016/j.ccell.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zgarbova M, Sponer J, Otyepka M, Cheatham TE, 3rd, Galindo-Murillo R, Jurecka P. Refinement of the Sugar-Phosphate Backbone Torsion Beta for AMBER Force Fields Improves the Description of Z- and B-DNA. J Chem Theory Comput. 2015;11:5723–36. doi: 10.1021/acs.jctc.5b00716. [DOI] [PubMed] [Google Scholar]
- 31.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem. 2002;23:1623–41. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 32.Velez-Vega C, McKay DJ, Aravamuthan V, Pearlstein R, Duca JS. Time-averaged distributions of solute and solvent motions: exploring proton wires of GFP and PfM2DH. J Chem Inf Model. 2014;54:3344–61. doi: 10.1021/ci500571h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.