Significance
Cells require nuclear deformation to squeeze through tissue matrices. We have discovered that WD repeat domain 5 (WDR5; an epigenetic modulator of H3K4 methylation) is fundamental for cell polarity and migration in vitro and in vivo, independent of transcription. We have uncovered that the interactions between cells and the surrounding 3D confined conditions induce the upregulation of H3K4me3. Moreover, 3D environments control the deformability and the mechanical properties of the nucleus. We have identified that loss of WDR5 abrogates the H3K4 methylation and the nuclear changes induced by 3D conditions. Mechanistically, we found that myosin light chain kinase and myosin function were required for WDR5-mediated H3K4 methylation in 3D matrices. Our findings uncover functions of the epigenetic machinery when cells move through constricted conditions.
Keywords: WDR5, epigenetics, cell migration, H3K4me3, 3D matrix
Abstract
Cell migration through extracellular matrices requires nuclear deformation, which depends on nuclear stiffness. In turn, chromatin structure contributes to nuclear stiffness, but the mechanosensing pathways regulating chromatin during cell migration remain unclear. Here, we demonstrate that WD repeat domain 5 (WDR5), an essential component of H3K4 methyltransferase complexes, regulates cell polarity, nuclear deformability, and migration of lymphocytes in vitro and in vivo, independent of transcriptional activity, suggesting nongenomic functions for WDR5. Similarly, depletion of RbBP5 (another H3K4 methyltransferase subunit) promotes similar defects. We reveal that a 3D environment increases the H3K4 methylation dependent on WDR5 and results in a globally less compacted chromatin conformation. Further, using atomic force microscopy, nuclear particle tracking, and nuclear swelling experiments, we detect changes in nuclear mechanics that accompany the epigenetic changes induced in 3D conditions. Indeed, nuclei from cells in 3D environments were softer, and thereby more deformable, compared with cells in suspension or cultured in 2D conditions, again dependent on WDR5. Dissecting the underlying mechanism, we determined that actomyosin contractility, through the phosphorylation of myosin by MLCK (myosin light chain kinase), controls the interaction of WDR5 with other components of the methyltransferase complex, which in turn up-regulates H3K4 methylation activation in 3D conditions. Taken together, our findings reveal a nongenomic function for WDR5 in regulating H3K4 methylation induced by 3D environments, physical properties of the nucleus, cell polarity, and cell migratory capacity.
The tissue environment controls multiple cell functions including migration (1, 2). The extracellular matrix signals control the cytoskeleton and the mechanical properties of the nucleus, which has to be highly deformable to allow cells to migrate in confined spaces (3–5). Usually this nuclear deformability is controlled by lamin expression and the cytoskeleton, which act as mechanosensors (6). Chromatin contributes to nuclear stiffness (7–9), and alterations in chromatin associated with DNA repair machinery, including histone markers of DNA damage, have been linked to nuclear deformability of cancer cells when migrating through narrow spaces (10, 11). Epigenetic modifications, which include covalent changes in histones and DNA, regulate chromatin structure and gene expression (12). Some histone markers have been related to cancer progression and cell migration (13–15). The histone H3 is methylated at the residue K4 by specific methyltransferase complexes, which comprise a catalytic subunit (SET/MLL) and several core scaffolding subunits, including WDR5 (WD repeat-containing protein 5), ASH2L, RbBP5, and mDPY-30 (16). The cytoskeleton and actomyosin are critical actors during chromatin organization, nuclear positioning, and deformability (17, 18), and there is a link between WDR5 and myosin contractility in regulating cell reprogramming (19). However, the functional connections between WDR5 and H3K4 methylation in migrating cells remain unclear.
In this report, we identified that WDR5 critically regulates nuclear deformability, cell polarity, and cell migration in vitro, as well as cell invasion in a zebrafish transplantation model. This also occurred with RbBP5. In addition, we showed that cells cultured in 3D conditions up-regulate levels of H3K4me3 concomitant with altered nuclear mechanics. Further, depletion of WDR5 impairs both nuclear effects. We provide mechanistic insights into the influence of myosin IIA and MLC (myosin regulatory light chain) phosphorylation via myosin light chain kinase (MLCK) activity and cAMP levels on the WDR5-mediated H3K4 methylation. In addition, we identified that MLCK also localizes in the nucleus at the proximal euchromatin region together with H3K4me3 and WDR5. Together, our findings suggest a functional link between the MLCK/myosin pathway and WDR5, which is essential for cytoskeletal and nuclear changes of migrating cells in confined conditions.
Results
WDR5 Silencing Impairs Interstitial Cell Motility in Vitro and in Vivo.
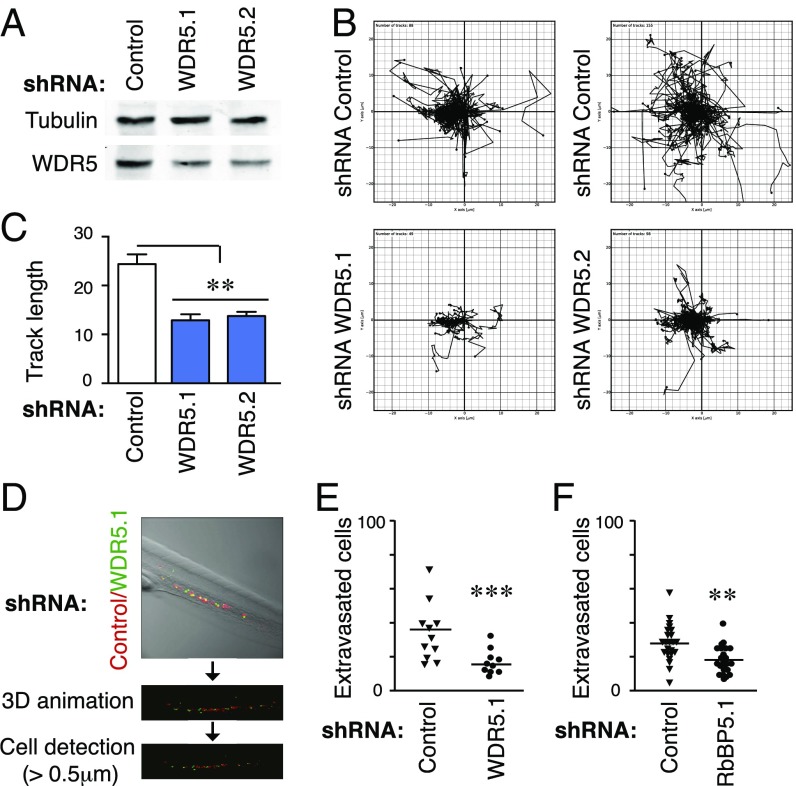
First, we stably depleted WDR5 expression in Jurkat cells (a human T cell acute lymphoblastic leukemia, T-ALL cell line; Fig. 1A). Then, we performed in vitro time-lapse cell migration to assess whether WDR5 depletion impaired the cell migration. Control and WDR5-depleted cells labeled with vital dyes mixed 1:1 were embedded in a 3D collagen type I matrix. WDR5 depletion significantly reduced cell movement measured as track length (Fig. 1 B and C and Movies S1 and S2). To test the importance of WDR5 in regulating cell migration through confined spaces in vivo, we injected a 1:1 mixture of labeled control and WDR5-depleted cells into the circulation of zebrafish embryos. We observed that although all cells dispersed immediately throughout the circulation with a plateau in the extravasation after 6 h (SI Appendix, Fig. S1A), WDR5 depletion greatly impaired extravasation (Fig. 1 D and E). Then, we expanded our interest to other H3K4 methyltransferase subunits and, by depletion of RbBP5 (SI Appendix, Fig. S1B), confirmed that RbBP5 silencing also impaired cell migration in vitro and in vivo (Fig. 1F, SI Appendix, Fig. S1 C and D, and Movies S3 and S4). Taken together, these results demonstrate that WDR5 is required for cell movement through confined spaces in vitro and in vivo.
Fig. 1.
WDR5-depleted cells present impaired cell migration through 3D environments and trafficking defects through blood vessel into tissues. (A) Jurkat cells were infected with specific shRNAs for WDR5. Cells were cultured for 3 wk, and after sorting, knockdown expression was confirmed by Western blotting. (B) Tracks of control or WDR5-depleted cells in a 3D collagen matrix. (C) Quantification of track length of cells in B. Mean n = 49–98 cells ± SEM. **P < 0.01. (D) Pipeline used for analyzing transplanted cell extravasation in zebrafish embryos. Representative micrograph showing the region of interest (caudal vein plexus and ventral fin fold) before and after 3D rendering of cells and size thresholding. (E) Graph shows the number of control and WDR5-depleted cells that had extravasated from D. n = 10 embryos. ***P < 0.001. (F) Graph shows extravasation of control and RbBP5 depleted cells in zebrafish embryos as in D. n = 23 embryos. **P < 0.01.
WDR5 Depletion Impairs Cell Polarity and Morphology Independent of Effects on Gene Transcription.
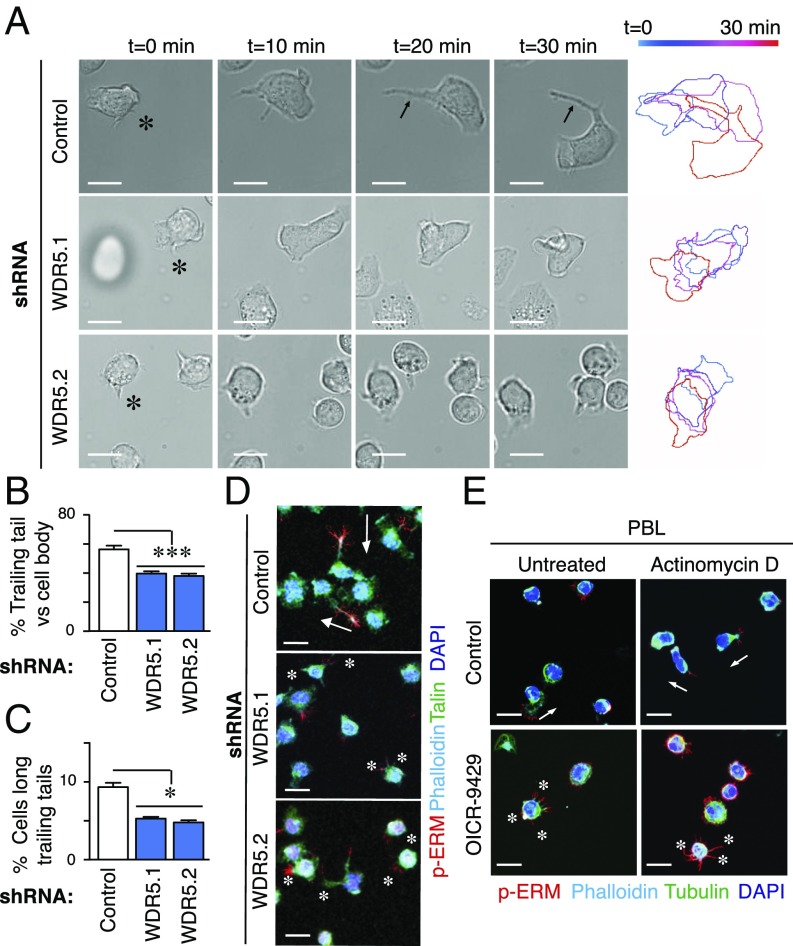
We observed that WDR5 localized mainly in the nucleus of Jurkat cells, whereas RbBP5 presented both cytoplasmic and nuclear distribution (SI Appendix, Fig. S2A). By time-lapse microscopy, we determined that WDR5-depleted cells adhered to VCAM1, but extended and retracted multiple transient protrusions and failed to form stable and elongated trailing tails to migrate (Fig. 2 A–C and Movies S5–S7). Human peripheral blood lymphocytes (PBL) treated with OICR-9429, a specific inhibitor of WDR5 (20), also present aberrant cell morphology (Movies S8 and S9). Interestingly, there were no significant changes in surface receptors expression on OICR-9429 treatment, and only minor differences between control and WDR5-depleted Jurkat cells (SI Appendix, Fig. S2 B and C). Moreover, we confirmed that WDR5 depletion did not affect cellular adhesion in 2D conditions to natural ligands such as collagen, VCAM1, and ICAM1 (SI Appendix, Fig. S2D). To further explore how WDR5 might control the cell polarity, we determined that the expression of cytoskeletal components was not affected in WDR5-depleted cells (SI Appendix, Fig. S3A); however, WDR5 silencing promoted loss of polarity and failure in trailing tail formation, as detected by phosphorylated ERM accumulation in the rear of polarized cells (Fig. 2D). To determine whether the aberrant polarity observed in WDR5-depleted cells was related to gene expression changes or a transcription-independent effect, we blocked mRNA transcription using actinomycin D (SI Appendix, Fig. S3B). Control cells treated with actinomycin D also presented long trailing tails, whereas WDR5-depleted cells showed aberrant cytoplasmic protrusions (SI Appendix, Fig. S3C). PBL formed short tails and less evident polarity than Jurkat cells, and treatment with OICR-9429 promoted defective polarity without any additional effect of actinomycin D treatment (Fig. 2E). Remarkably, RbBP5-depleted cells presented aberrant rounded cell morphologies (SI Appendix, Fig. S3D). These observations suggest that WDR5 is required for cell polarity and morphology independent of transcriptional activity.
Fig. 2.
WDR5 expression affects the cell morphology and polarity. (A) Representative images of control or WDR5-depleted Jurkat cells on VCAM1 (2 μg/mL). Asterisks indicate the cell morphology tracked through time, and pseudocolor bar indicates time. (B) Graph shows the trailing edge length with respect to the cell body diameter from control or WDR5-depleted cells on VCAM1. Mean n = 15 cells ± SD. ***P < 0.001. (C) Graph shows the percentage of control or WDR5-depleted cells cultured on VCAM1 with extended trailing edge. Mean n = 3 ± SD. *P < 0.05. (D) Control or WDR5-depleted Jurkat cells were cultured on VCAM1, fixed and analyzed by confocal microscopy. White arrows indicate the presence of long trailing edges (identified by phospho-ERM) in control cells, whereas asterisks show multiple cytoplasmic protrusions induced in WDR5-depleted cells. (Scale bar, 10 μm.) (E) Primary T lymphocytes were cultured preincubated or not with actinomycin D and the WDR5 inhibitor (OICR-9429). Cell morphology was analyzed as in D. (Scale bar, 10 μm.)
Cell Confinement in 3D Environments Promotes WDR5-Mediated H3K4 Methylation.
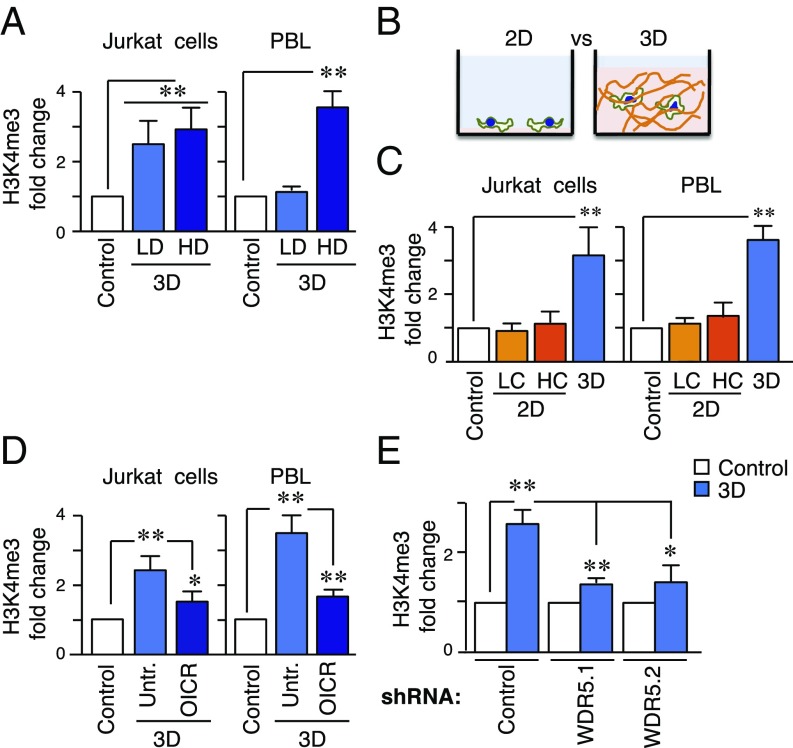
As WDR5 is critical for H3K4 methylation, we investigated the influence of 3D conditions on H3K4 trimethylation. We observed that H3K4me3 was elevated in PBL and Jurkat cells cultured in 3D collagen matrices (Fig. 3A and SI Appendix, Fig. S4A). Remarkably, Jurkat cells responded to low or high collagen densities, whereas PBL increased H3K4me3 only in a high-density matrix. As changes in the collagen density affect the pore size, stiffness, and ligand density, we cultured PBL in low-density collagen matrices with transglutaminase 2 (TGM2). TGM2 changes the microstructural properties without affecting collagen density or cell physiology directly (21). The addition of TGM2 to low collagen gel increased the levels of H3K4me3 in PBL (SI Appendix, Fig. S4B), suggesting that the properties of the extracellular space are critical for the up-regulation of H3K4me3. Then, we cultured PBL or Jurkat cells on thin-layer collagen (2D condition) or embedded in a collagen matrix (3D condition; Fig. 3B) and observed that the response to 3D conditions was responsible for inducing H3K4me3 (Fig. 3C and SI Appendix, Fig. S4C). RbBP5-depleted cells also presented a defective H3K4methylation induced by 3D environments, supporting the idea that H3K4 epigenetic modulators respond to confined conditions (SI Appendix, Fig. S4 D and E). 3D conditions also induced higher levels of H3K4me3 in a different cell type (MDA-MB-231, human breast adenocarcinoma) compared with these cells plated on plastic (Control) or 2D collagen plates (SI Appendix, Fig. S4F). Although cell migration through dense matrices requires matrix metalloprotease action (5), the up-regulation of H3K4me3 induced by 3D conditions was matrix metalloprotease-independent (SI Appendix, Fig. S4 G and H). We confirmed that WDR5 was critical for the effect of 3D environment on H3K4 methylation (Fig. 3 D and E). These data indicate that 3D constricted conditions promote H3K4 methylation, which is prevented by WDR5 inhibition or depletion.
Fig. 3.
WDR5 controls H3K4 methylation of cells in 3D conditions. (A) Levels of H3K4me3 in nuclear fractions isolated from Jurkat cells or PBL cultured in suspension (control), in a 0.4 mg/mL (LD) or 2 mg/mL (HD) 3D collagen matrix for 1 h. Graph shows mean n = 3 ± SD. **P < 0.01. (B) Schematic model of cells cultured on collagen-coated wells (2D) or in a collagen matrix (3D). (C) Levels of H3K4me3 from nuclear fractions from Jurkat cells cultured in suspension (Control), on 5 μg/mL (LC), 10 μg/mL (HC) collagen coated plates or embedded in a 3D matrix. Mean n = 3 ± SD. **P < 0.01. (D) Jurkat cells or PBL were incubated with the specific WDR5 inhibitor (OICR-9429 at 1 μM) and then culture in 3D. Nuclear fractions were isolated and H3K4me3 levels determined. Mean n = 3 ± SD. *P < 0.05; **P < 0.01. (E) Levels of H3K4me3 from nuclear fractions of control or stable WDR5-depleted cells in suspension or embedded in a 3D matrix. Mean n = 3 ± SD. *P < 0.05; **P < 0.01.
WDR5 Controls the Global Chromatin Structure and Nuclear Deformability in 3D Environments.
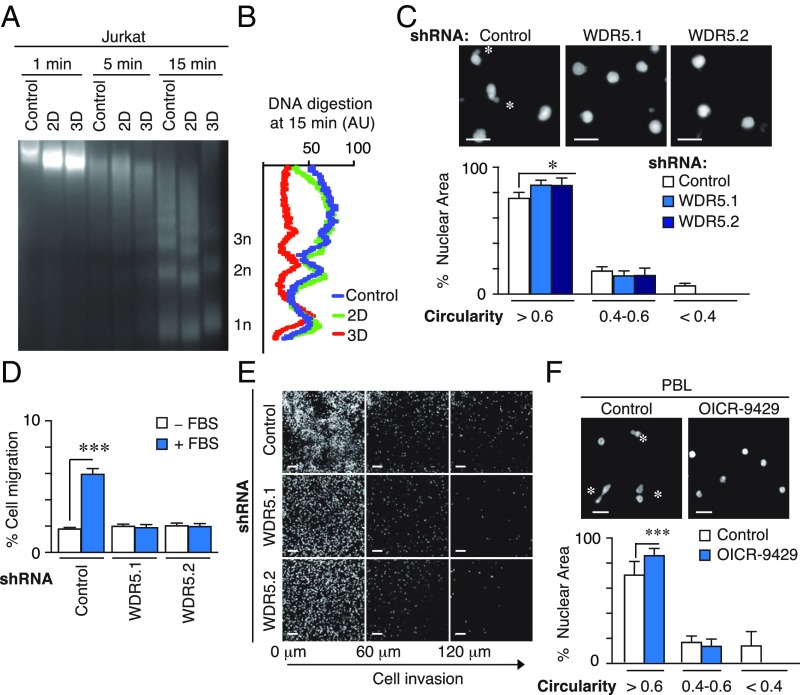
As global changes in H3K4 methylation might alter the overall structure of chromatin, we characterized the status of chromatin condensation by micrococcal nuclease (MNase) digestion. Jurkat (Fig. 4A) and PBL (SI Appendix, Fig. S5A) cultured in 3D collagen presented much greater sensitivity to DNA digestion than those cultured in suspension or on 2D conditions (Fig. 4B and SI Appendix, Fig. S5B), suggesting that high H3K4me3 levels correlated with less condensed chromatin structure when cells were embedded in 3D conditions. We confirmed that 3D environments did not sensitize WDR5-depleted cells for DNA digestion (SI Appendix, Fig. S5 C and D). The effect on the chromatin structure and cell migration might be related to high nuclear deformability when cells have to squeeze through narrow spaces. We determined that control cells embedded in 3D collagen matrices presented highly deformable nuclei (circularity index < 0.4) compared with the simple circular form observed in WDR5-depleted cells (Fig. 4C). Transwells of 3 µm pore size model physical barriers that require high nuclear deformability and active PBL migration (5). We observed that although control cells migrated across the 3D collagen matrix and the 3 µm pores, WDR5-depleted cells did not, although migration through nonrestrictive 8- and 5-µm pores was not affected (Fig. 4 D and E and SI Appendix, Fig. S5E). RbBP5 depletion abrogated nuclear deformability in 3D environments (SI Appendix, Fig. S5F), and OICR-9429 treatment also caused a dramatic change in nuclear morphology and invasion of PBL (Fig. 4F and SI Appendix, Fig. S5G). Together, these results confirm WDR5 requirement in nuclear deformability and 3D invasive migration.
Fig. 4.
WDR5 regulates chromatin conformation and nuclear deformability induced by 3D conditions. (A) Jurkat cells were cultured in suspension, 2D and 3D conditions. Then, cells were digested with micrococcal nuclease (MNase) at indicated times and DNA fragments resolved in agarose gel. (B) Signal profile of the mononucleosomes (1n), dinuclueosomes (2n), and trinucleosomes (3n) after micrococcal digestion (15 min) from cells in A. (C) Control or WDR5-depleted cells were embedded in 3D collagen, fixed and stained with DAPI. Asterisks mark the highly deformable nuclei. Graph shows the circularity index (rounded, >0.6; deformable, 0.4–0.6; or highly deformable, <0.4). Mean n = 3 ± SD. *P < 0.05. (D) Control or WDR5-depleted cells were seeded on collagen gel in the upper chamber of transwells and allowed to invade in response to 10% FBS for 24 h. Cells were collected from the bottom chamber, stained, and quantified. Mean n = 3 ± SD. ***P < 0.001. (E) Cells as in D were fixed after 6 h of cell migration in the collagen gel, stained with DAPI, and serial confocal sections were captured. (F) PBL cells were preincubated or not with OICR-9429, embedded in 3D collagen, fixed and stained with DAPI. Mean n = 3 ± SD. Asterisks mark the highly deformable nuclei. Graph shows the circularity index. Mean n = 3 ± SD. ***P < 0.01.
WDR5 Influences the Mechanical Properties of the Nucleus.
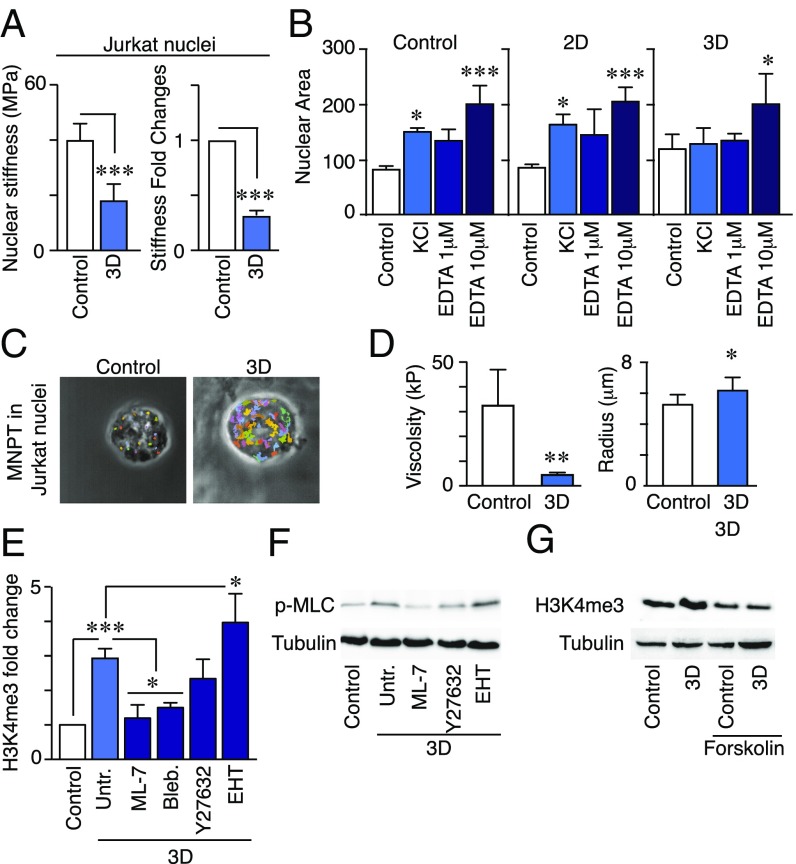
We isolated the nuclei from cells in suspension and embedded in 3D matrix to analyze their nuclear stiffness by AFM (atomic force microscopy). Isolation of the nuclei and their soft fixation onto coated coverslips allowed us to determine the nuclear surface and mechanical properties in the absence of any contribution of cytoskeleton and other cytosolic molecules. Nuclei from Jurkat cells in 3D conditions were softer than nuclei from cells in suspension (Fig. 5A) and had a flattened structure compared with nuclei from cells in suspension (SI Appendix, Fig. S6A). Nuclei from Jurkat cells cultured in 2D conditions were similar to suspension conditions (SI Appendix, Fig. S6B). To further verify how the nucleus responds to 3D environments, we analyzed the elasticity of isolated nuclei by osmotic swelling experiments. Isolated nuclei from cells in 3D and high H3K4me3 levels were bigger than those from cells in suspension or in 2D conditions and presented less capacity for nuclear area changes on KCl or EDTA addition (Fig. 5B, SI Appendix, Fig. S6C, and Movies S10–S15). WDR5 depletion also diminished the effect on the mechanical changes induced by 3D environment compared with control cells (SI Appendix, Fig. S6D). Moreover, PBL cultured in 3D had soft nuclei and WDR5 inhibition significantly diminished this 3D-mediated nuclear change (SI Appendix, Fig. S6E). As a complementary biophysical approach, we performed multiple-nuclear particle tracking in isolated nuclei from cells in suspension or in 3D conditions (Fig. 5C). High-contrast microscopy techniques with high temporal and spatial resolution allow particles to be tracked during their collision through Brownian motion with living cells (22). Nuclei from cells in 3D conditions presented a significant reduction in the nuclear viscosity compared with nuclei from cells in suspension (Fig. 5D), confirming that WDR5 was required for H3K4 methylation and nuclear mechanical changes induced by 3D environments.
Fig. 5.
Confined conditions promote changes in the mechanical properties of the nucleus and increase actomyosin contractility, which regulates H3K4 methylation. (A) Nuclei from cells cultured in suspension or 3D were isolated and left to sediment onto a coverslip. Nuclear stiffness was analyzed by AFM. Mean n = 30 nuclei ± SEM. (B) Isolated nuclei as in A were sedimented on poly-Lysine coated coverslips and their nuclear elasticity analyzed by adding KCl (10 μM) or different EDTA concentrations (1 and 10 μM). Graph shows the quantification of the nuclear area. Mean n = 3 ± SD. *P < 0.05; ***P < 0.001. (C) Representative examples of the multiple-nuclear particle tracking trajectories recorded in isolated nuclei from cells in suspension or in 3D. (Magnification: 337.5×.) (D) Graphs illustrate the quantification of the nuclear viscosity and the radius for the different cases. Mean n = 8–10 nuclei ± SEM. *P < 0.05; **P < 0.01. (E) Jurkat cells were incubated with Blebbistatin (50 μM), ML-7 (5 μM), Y27632 (25 μM), EHT 1864 (50 μM) for 30 min before culturing them in 3D. Nuclear fractions were isolated and H3K4me3 levels analyzed. Mean n = 3 ± SD. *P < 0.05; ***P < 0.001. (F) Cells as in E were cultured for 30 min, and phospho-MLC levels determined. (G) Jurkat cells were preincubated with forskolin (25 μM) and then cultured for 1 h embedded in 3D. H3K4 methylation was analyzed from nuclear fractions.
The Cytoskeleton Couples H3K4 Methylation Induced by 3D Matrix.
To elucidate the molecular mechanisms underlying the regulation of WDR5 and H3K4 methylation, we treated cells in 3D conditions with different chemical inhibitors against regulators of actin-based cell migration. We observed that blebbistatin (myosin II inhibitor) and ML-7 (MLCK inhibitor) blocked H3K4 methylation induced by 3D conditions. Moreover, Y27632 (Rho-associated protein kinase, ROCK inhibitor) did not affect H3K4 methylation, whereas EHT1864 (Rac inhibitor) enhanced H3K4me3 induction by the 3D matrix environment (Fig. 5E). Furthermore, using stable cell lines expressing Rac-WT, Rac-L61 (constitutively active), and Rac-N17 (dominant negative), we observed that the dominant negative mutant increased H3K4me3, whereas the active mutant reduced it (SI Appendix, Fig. S7A). Then, we verified that loss of myosin IIA reduced the levels of H3K4me3 in cells in a 3D matrix (SI Appendix, Fig. S7 B and C). Actomyosin contractility is regulated mainly by MLCK and ROCK, through MLC phosphorylation (23). We demonstrated that MLC is phosphorylated in Jurkat cells cultured in 3D matrix; MLCK inhibitor, but not ROCK inhibitor, prevented this effect, whereas Rac inhibition or the dominant negative mutant slightly increased it (Fig. 5F and SI Appendix, Fig. S7D). PKA and the intracellular levels of cAMP control MLCK activation and actomyosin contractility (24). We demonstrated that forskolin, a direct activator of adenylate cyclase, abrogated the up-regulation of H3K4me3 (Fig. 5G), confirming that the levels of cAMP regulate H3K4 methylation induced by 3D conditions. These results imply a fundamental role for the cytoskeleton, cAMP levels, and myosin activation on 3D-mediated H3K4 methylation.
MLCK Colocalizes with WDR5 at Euchromatin Sites and Is Critical for WDR5 Activity.
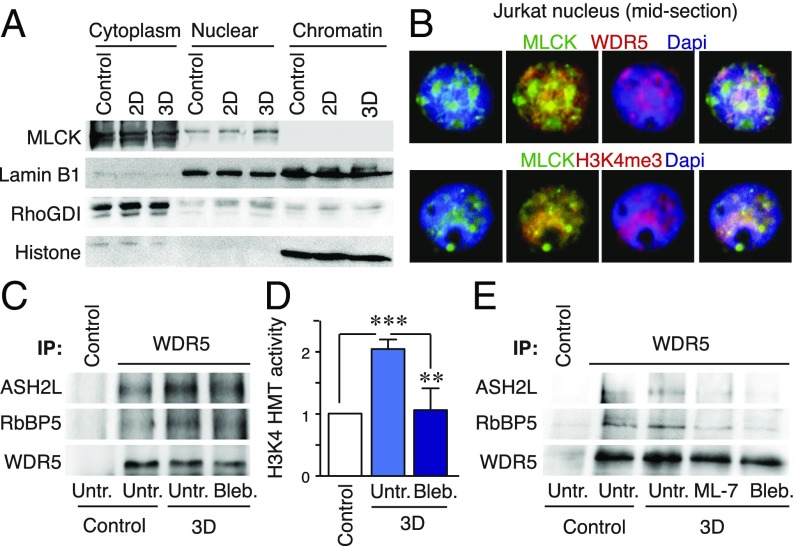
Then, we evaluated the localization of MLCK and its functional connections with WDR5. MLCK was present both in cytoplasmic and nuclear fractions of Jurkat cells (Fig. 6A and SI Appendix, Fig. S8A). We confirmed that phospho-MLC localized in the cytoplasm and the nucleus (SI Appendix, Fig. S8B), and that MLCK was localized in the nucleus of MDA-MB-231 cells (SI Appendix, Fig. S8C). Remarkably, nuclear MLCK localized at the euchromatin region, defined as the low-stained DAPI areas of the nucleus and was close to WDR5 and H3K4me3 (Fig. 6B), but not to H3K9me3 (SI Appendix, Fig. S8D). We confirmed that WDR5 and ASH2L did not alter their distribution in the nuclear soluble or chromatin bound fractions upon 3D conditions (SI Appendix, Fig. S8E). Therefore, we focused on analyzing how 3D environments might promote the catalytic activity of WDR5. We immunoprecipitated WDR5 and characterized that the methyltransferase activity bound to WDR5 fractions was up-regulated in 3D conditions, and blebbistatin treatment reduced this activity (Fig. 6 C and D). Finally, we evaluated whether MLCK and myosin might regulate the interactions between the H3K4 methyltransferase subunits, which in turn methylate H3K4. We demonstrated that MLCK and myosin activities control the interactions of WDR5 to RbBP5 and ASH2L (Fig. 6E). Together, these observations suggest that 3D environment up-regulated myosin contractility via MLCK, which controlled the methylation of H3K4 by WDR5.
Fig. 6.
MLCK localizes in the nucleus, at the vicinity of WDR5 and euchromatin regions, and regulates WDR5-mediated-H3K4 methylation. (A) Jurkat cells were cultured in suspension, 2D, and 3D conditions. Then, cells were lysed and cytoplasmic, nuclear, and chromatin-bound fractions analyzed by western blotting. RhoGDI, Lamin B1 and Histone H3 were used as internal control of cytoplasmic, nuclear/chromatin and chromatin fractions. (B) Midsection of a representative nucleus from isolated nuclei sedimented on poly-Lysine coated coverslips and stained to visualize MLCK, the chromatin, and WDR5 or H3K4me3. (C and D) Jurkat cells were incubated with Blebbistatin, for 30 min before culturing them in 3D. Then, WDR5 was immunoprecipitated (C), and its H3K4 methyltransferase activity (D) determined. Mean n = 4 ± SD. ***P < 0.001. (E) Jurkat cells were incubated with Blebbistatin or ML-7 inhibitors for 30 min before culturing them in 3D. Then WDR5 was immunoprecipitated and its interactions with ASH2L and RbBP5 were determined.
Discussion
Cell migration in 3D matrices is critical in physiological and pathological conditions, with nuclear deformability emerging as a critical determinant (5). Moreover, 3D elicits different intracellular signals from 2D conditions (25, 26). In this study, we have uncovered that WDR5 was critical for cell polarity, migration through confined spaces, the nuclear mechanics, and the up-regulation of H3K4me3 in 3D environments.
WDR5 is a common subunit of H3K4 methyltransferase complexes that plays an important role in multiple cell functions, including self-renewal, reprogramming, and abscission (27, 28). Here, we have defined that WDR5 regulated cell movement in vitro, and by using a zebrafish embryo transplantation model (29), we confirmed that WDR5 depletion abrogated cell migration in vivo. WDR5 might translocate to the mitochondria on viral infection (30). We found that WDR5 was mainly nuclear, but its silencing impaired cell morphology. Remarkably, these morphological defects in WDR5-depleted cells might be mediated independent of any transcriptional change, as actinomycin D treatment did not influence the cell polarity. Therefore, WDR5 had direct functional consequences in cell polarity through nontranscriptional functions, aligning with the idea that specific chromatin changes are uncoupled from gene activation (31).
We uncovered that the nuclei from Jurkat cells, normal lymphocytes, and a breast cancer cell line presented higher levels of H3K4me3 when in 3D conditions, suggesting that chromatin changes might be occurring because of restrictive 3D conditions. Mesenchymal cell migration depends on lamin expression, matrix metalloprotease activity, and DNA damage/repair machinery to allow cells to squeeze through narrow spaces (5, 10, 11, 17). Therefore, it is probable that chromatin changes might play a marginal role in the nuclear deformability of mesenchymal cells. Novel nongenomic functions have been proposed for changes in chromatin structure (32), including nuclear stiffness, morphology, and small deformations (33, 34). Our findings demonstrate that nuclei from cells in 3D conditions present more sensitivity to DNA digestion, concomitant with global chromatin decompaction, nuclear deformability, viscosity, and diminished nuclear stiffness, compared with cells in suspension. Recent data showing how chromatin decompaction by the nucleosomal binding protein HMGN5 decreases nuclear stiffness and nuclear elasticity on swelling conditions (9). Together, this aligns with the idea that euchromatin and chromatin plasticity contribute to nuclear mechanics and cell invasiveness (35, 36). In 3D conditions, the nucleus not only changes its position and stiffness (17, 37), but also its chromatin structure is highly complex and regulated and might contribute to deformability, thereby facilitating cell movement in confined spaces (33).
Myosin IIA is essential for cell protrusions and effective migration in dense collagen gels (38), and its deletion impairs the extravasation and migration through interstitial spaces of T-cells (39). The actomyosin cytoskeleton and Rho GTPases control nuclear deformation when cells migrate in confined spaces (40–42). Moreover, the cytoskeleton regulates chromatin and telomere dynamics related with nuclear changes (8), and it has been reported that nuclear Rac controls chromatin conformation and gene expression (43). MLCK and ROCK phosphorylate MLC differentially according to the composition and characteristics of the 3D matrix (44). Our data imply that 3D conditions promoted MLC phosphorylation through MLCK and that Rac inhibition enhanced this effect, consistent with the ability of Rac-stimulated PAK to antagonize MLCK (45). Remarkably, we have observed that H3K4 methylation induced by 3D conditions was dependent on cAMP levels. Mechanistically, elevation of cAMP by forskolin treatment inhibits MLCK and MLC phosphorylation (24) and activates Rac in several cell types (46, 47). MLCK localizes in the cytoplasm and inside the nucleus of several cell types (25, 48, 49). We have observed a significant amount of MLCK in the euchromatin region at the vicinity of WDR5 and H3K4me3. Our results show that 3D environments did not affect WDR5 localization at the chromatin, but promoted WDR5 interactions with ASH2L and RbBP5 in an MLCK/myosin-dependent manner. H3K4 methyltransferase assembly is critical for its activity (50) and suggests that MLCK and myosin contractility might regulate the complex formation between WDR5 and its molecular partners to catalyze H3K4 methylation. This aligns with previous reports indicating that 3D confinement controls H3K4 methylation and gene reprogramming (19), and transcriptional changes in cancer types where MLCK are also critical for cell invasion (51, 52).
Together, our findings revealed that WDR5 played a critical role in cell migration in vitro and in vivo by controlling dynamic changes in H3K4me3 levels, the physical properties of the nucleus, and the cell polarity and shape. Mechanistically, myosin phosphorylation via MLCK mediated WDR5 activity in 3D conditions, thereby enabling cell migration through confined spaces.
Materials and Methods
The human T cell line Jurkat was obtained from Christoph Ballestrem (The University of Manchester, United Kingdom). Primary human PBL were isolated from buffy coats of healthy anonymous donors (Blood Bank, Hospital Gregorio Marañón) after depletion of the monocyte fraction with CD14 microbeads. Jurkat and PBL were maintained in RPMI medium 1640 (Gibco) with Hepes (10 mM), l-glutamine (2 mM), 10% FCS, and 1% penicillin/streptomycin, in 5% CO2 at 37 °C. Human HEK293T and MDA-MB-231 cells were cultured in DMEM (Gibco), l-glutamine (2 mM), supplemented with 10% FCS and 1% penicillin/streptomycin. All cells were maintained in 5% CO2 at 37 °C.
Cells were cultured in suspension, on plates coated with matrix components or embedded in a collagen matrix. For 3D matrix, type I collagen solution from bovine were reconstituted at 0.4 or 2 mg/mL in RPMI and neutralized with 7.5% NaHCO3 and 25 mM Hepes. Cells were added to the collagen solution before polymerization at 37 °C for 45 min. In some cases, 1 μg TGM2 (R&D Systems) was added as crosslinking agent. For some experiments, cells were incubated with specific chemical inhibitors and the medium was also supplemented with these inhibitors.
SI Appendix includes supplementary material and methods, figures, and references.
Supplementary Material
Acknowledgments
We thank the Confocal Unit from the Instituto de Investigación Biomédica Gregorio Marañón and Bioimaging Facility from the University of Manchester (microscopes were purchased with grants from Wellcome Trust and the University of Manchester Strategic Fund) for their help with microscopy. We thank E. Zindy (Wellcome Trust Centre for Cell Matrix Research) for his assistance with image quantifications. E.M. was supported by a fellowship for Fondo de Garantía de Empleo Juvenil from Comunidad de Madrid and M.D. by a studentship from the Wellcome Trust. This work was supported by Spanish Ministry of Economy and Competitiveness Grants RYC-2015-18497 and SAF2017-86327-R (to J.R.-M.) and Grant FIS2015-70339-C2-1-R (to F.M.), Instituto de Salud Carlos III (ISCIII)/El Fondo Europeo de Desarrollo Regional (FEDER) Grant PI17/01324 (to R.S. and P.S.-M.), and European Research Council Grant StG 282059-PROMINENT (to A.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.E.D. is a guest editor invited by the Editorial Board.
See Commentary on page 8472.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719405115/-/DCSupplemental.
References
- 1.DuFort CC, Paszek MJ, Weaver VM. Balancing forces: Architectural control of mechanotransduction. Nat Rev Mol Cell Biol. 2011;12:308–319. doi: 10.1038/nrm3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charras G, Sahai E. Physical influences of the extracellular environment on cell migration. Nat Rev Mol Cell Biol. 2014;15:813–824. doi: 10.1038/nrm3897. [DOI] [PubMed] [Google Scholar]
- 3.Friedl P, Wolf K, Lammerding J. Nuclear mechanics during cell migration. Curr Opin Cell Biol. 2011;23:55–64. doi: 10.1016/j.ceb.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McGregor AL, Hsia CR, Lammerding J. Squish and squeeze-the nucleus as a physical barrier during migration in confined environments. Curr Opin Cell Biol. 2016;40:32–40. doi: 10.1016/j.ceb.2016.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf K, et al. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J Cell Biol. 2013;201:1069–1084. doi: 10.1083/jcb.201210152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buxboim A, et al. Matrix elasticity regulates lamin-A,C phosphorylation and turnover with feedback to actomyosin. Curr Biol. 2014;24:1909–1917. doi: 10.1016/j.cub.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahl KN, Engler AJ, Pajerowski JD, Discher DE. Power-law rheology of isolated nuclei with deformation mapping of nuclear substructures. Biophys J. 2005;89:2855–2864. doi: 10.1529/biophysj.105.062554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Makhija E, Jokhun DS, Shivashankar GV. Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc Natl Acad Sci USA. 2016;113:E32–E40. doi: 10.1073/pnas.1513189113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Furusawa T, et al. Chromatin decompaction by the nucleosomal binding protein HMGN5 impairs nuclear sturdiness. Nat Commun. 2015;6:6138. doi: 10.1038/ncomms7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denais CM, et al. Nuclear envelope rupture and repair during cancer cell migration. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raab M, et al. ESCRT III repairs nuclear envelope ruptures during cell migration to limit DNA damage and cell death. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- 12.Henikoff S, Greally JM. Epigenetics, cellular memory and gene regulation. Curr Biol. 2016;26:R644–R648. doi: 10.1016/j.cub.2016.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Gerlitz G, Bustin M. Efficient cell migration requires global chromatin condensation. J Cell Sci. 2010;123:2207–2217. doi: 10.1242/jcs.058271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yokoyama Y, et al. Cancer-associated upregulation of histone H3 lysine 9 trimethylation promotes cell motility in vitro and drives tumor formation in vivo. Cancer Sci. 2013;104:889–895. doi: 10.1111/cas.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, et al. Integrin α4β1 controls G9a activity that regulates epigenetic changes and nuclear properties required for lymphocyte migration. Nucleic Acids Res. 2016;44:3031–3044. doi: 10.1093/nar/gkv1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20:341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada T, et al. Nuclear lamin stiffness is a barrier to 3D migration, but softness can limit survival. J Cell Biol. 2014;204:669–682. doi: 10.1083/jcb.201308029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alam SG, et al. The mammalian LINC complex regulates genome transcriptional responses to substrate rigidity. Sci Rep. 2016;6:38063. doi: 10.1038/srep38063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Downing TL, et al. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater. 2013;12:1154–1162. doi: 10.1038/nmat3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grebien F, et al. Pharmacological targeting of the Wdr5-MLL interaction in C/EBPα N-terminal leukemia. Nat Chem Biol. 2015;11:571–578. doi: 10.1038/nchembio.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fraley SI, et al. Three-dimensional matrix fiber alignment modulates cell migration and MT1-MMP utility by spatially and temporally directing protrusions. Sci Rep. 2015;5:14580. doi: 10.1038/srep14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tseng Y, Kole TP, Wirtz D. Micromechanical mapping of live cells by multiple-particle-tracking microrheology. Biophys J. 2002;83:3162–3176. doi: 10.1016/S0006-3495(02)75319-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vicente-Manzanares M, Ma X, Adelstein RS, Horwitz AR. Non-muscle myosin II takes centre stage in cell adhesion and migration. Nat Rev Mol Cell Biol. 2009;10:778–790. doi: 10.1038/nrm2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leitman EM, et al. MLCK regulates Schwann cell cytoskeletal organization, differentiation and myelination. J Cell Sci. 2011;124:3784–3796. doi: 10.1242/jcs.080200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–1712. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 26.Shin JW, Mooney DJ. Extracellular matrix stiffness causes systematic variations in proliferation and chemosensitivity in myeloid leukemias. Proc Natl Acad Sci USA. 2016;113:12126–12131. doi: 10.1073/pnas.1611338113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- 28.Ang Y-S, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell. 2011;145:183–197. doi: 10.1016/j.cell.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley VL, et al. Focused chemical genomics using zebrafish xenotransplantation as a pre-clinical therapeutic platform for T-cell acute lymphoblastic leukemia. Haematologica. 2015;100:70–76. doi: 10.3324/haematol.2014.110742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang YY, et al. WDR5 is essential for assembly of the VISA-associated signaling complex and virus-triggered IRF3 and NF-kappaB activation. Proc Natl Acad Sci USA. 2010;107:815–820. doi: 10.1073/pnas.0908967107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shachar S, Voss TC, Pegoraro G, Sciascia N, Misteli T. A high-throughput imaging-based mapping platform for the systematic identification of gene positioning factors. Cell. 2015;162:911–923. doi: 10.1016/j.cell.2015.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bustin M, Misteli T. Nongenetic functions of the genome. Science. 2016;352:aad6933. doi: 10.1126/science.aad6933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irianto J, et al. Nuclear constriction segregates mobile nuclear proteins away from chromatin. Mol Biol Cell. 2016;27:4011–4020. doi: 10.1091/mbc.E16-06-0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stephens AD, Banigan EJ, Adam SA, Goldman RD, Marko JF. Chromatin and lamin A determine two different mechanical response regimes of the cell nucleus. Mol Biol Cell. 2017;28:1984–1996. doi: 10.1091/mbc.E16-09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pajerowski JD, Dahl KN, Zhong FL, Sammak PJ, Discher DE. Physical plasticity of the nucleus in stem cell differentiation. Proc Natl Acad Sci USA. 2007;104:15619–15624. doi: 10.1073/pnas.0702576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimamoto Y, Tamura S, Masumoto H, Maeshima K. Nucleosome-nucleosome interactions via histone tails and linker DNA regulate nuclear rigidity. Mol Biol Cell. 2017;28:1580–1589. doi: 10.1091/mbc.E16-11-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petrie RJ, Koo H, Yamada KM. Generation of compartmentalized pressure by a nuclear piston governs cell motility in a 3D matrix. Science. 2014;345:1062–1065. doi: 10.1126/science.1256965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rai V, Thomas DG, Beach JR, Egelhoff TT. Myosin IIA heavy chain phosphorylation mediates adhesion maturation and protrusion in three dimensions. J Biol Chem. 2017;292:3099–3111. doi: 10.1074/jbc.M116.733402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobelli J, et al. Confinement-optimized three-dimensional T cell amoeboid motility is modulated via myosin IIA-regulated adhesions. Nat Immunol. 2010;11:953–961. doi: 10.1038/ni.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kubow KE, Conrad SK, Horwitz AR. Matrix microarchitecture and myosin II determine adhesion in 3D matrices. Curr Biol. 2013;23:1607–1619. doi: 10.1016/j.cub.2013.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiam HR, et al. Perinuclear Arp2/3-driven actin polymerization enables nuclear deformation to facilitate cell migration through complex environments. Nat Commun. 2016;7:10997. doi: 10.1038/ncomms10997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobelli J, Estin Matthews M, Chen S, Krummel MF. Activated T cell trans-endothelial migration relies on myosin-IIA contractility for squeezing the cell nucleus through endothelial cell barriers. PLoS One. 2013;8:e75151. doi: 10.1371/journal.pone.0075151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Navarro-Lérida I, et al. Rac1 nucleocytoplasmic shuttling drives nuclear shape changes and tumor invasion. Dev Cell. 2015;32:318–334. doi: 10.1016/j.devcel.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 44.Ávila-Rodríguez D, et al. The shift in GH3 cell shape and cell motility is dependent on MLCK and ROCK. Exp Cell Res. 2017;354:1–17. doi: 10.1016/j.yexcr.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 45.Sanders LC, Matsumura F, Bokoch GM, de Lanerolle P. Inhibition of myosin light chain kinase by p21-activated kinase. Science. 1999;283:2083–2085. doi: 10.1126/science.283.5410.2083. [DOI] [PubMed] [Google Scholar]
- 46.Aslam M, et al. cAMP controls the restoration of endothelial barrier function after thrombin-induced hyperpermeability via Rac1 activation. Physiol Rep. 2014;2:e12175. doi: 10.14814/phy2.12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schlegel N, Waschke J. VASP is involved in cAMP-mediated Rac 1 activation in microvascular endothelial cells. Am J Physiol Cell Physiol. 2009;296:C453–C462. doi: 10.1152/ajpcell.00360.2008. [DOI] [PubMed] [Google Scholar]
- 48.Smith A, Bracke M, Leitinger B, Porter JC, Hogg N. LFA-1-induced T cell migration on ICAM-1 involves regulation of MLCK-mediated attachment and ROCK-dependent detachment. J Cell Sci. 2003;116:3123–3133. doi: 10.1242/jcs.00606. [DOI] [PubMed] [Google Scholar]
- 49.Poperechnaya A, Varlamova O, Lin PJ, Stull JT, Bresnick AR. Localization and activity of myosin light chain kinase isoforms during the cell cycle. J Cell Biol. 2000;151:697–708. doi: 10.1083/jcb.151.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dou Y, et al. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol. 2006;13:713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 51.Velez DO, et al. 3D collagen architecture induces a conserved migratory and transcriptional response linked to vasculogenic mimicry. Nat Commun. 2017;8:1651. doi: 10.1038/s41467-017-01556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mierke CT, Frey B, Fellner M, Herrmann M, Fabry B. Integrin α5β1 facilitates cancer cell invasion through enhanced contractile forces. J Cell Sci. 2011;124:369–383. doi: 10.1242/jcs.071985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.