Abstract
Follicular helper T (Tfh) cells are necessary for germinal center B cell maturation during primary immune responses; however, the T cells that promote humoral recall responses via memory B cells are less well defined. We herein characterize a human tonsillar CD4+ T cell subset with this function. These cells are similar to Tfh cells in terms of expression of the chemokine receptor CXCR5 and the inhibitory receptor PD-1, interleukin (IL)-21 secretion, and expression of the transcription factor BCL6; however, unlike Tfh cells that are located within the B cell follicle and germinal center, they reside at the border of the T cell zone and the B cell follicle in proximity to memory B cells, a position dictated by their unique chemokine receptor expression. They promote memory B cells to produce antibodies via CD40 ligand, IL-10, and IL-21. Our results reveal a unique extrafollicular CD4+ T cell subset in human tonsils, which specialize in promoting T cell-dependent humoral recall responses.
Introduction
Antigenic challenge by invading pathogens or protein vaccination induces a series of events leading to generation of T and B cell memory against the encountered antigen (Ag) (1–3). B cells mature in the germinal center (GC) within B cell follicles of secondary lymphoid organs (SLOs) (4–7). Maturation of GC B cells to memory B cells or long-lived plasma cells requires a specialized subset of CD4+ T cells, follicular helper T (Tfh) cells, which localize to GCs and provide critical survival and differentiation signals to GC B cells, including CD40 ligand (CD40L, CD154), and interleukins (IL)-21 and -4 (8–13).
Studies in mice reveal that GC responses begin in the days following initial Ag encounter, taking weeks for generation of memory B cells and long-lived plasma cells (14, 15). The latter secrete high levels of neutralizing immunoglobulin (Ig) for long periods after Ag clearance, providing the host with a first line of defense against re-infection (16–19), whereas memory B cells rapidly proliferate and differentiate to form secondary GC responses or into antibody-forming cells (AFCs) following re-encounter with the priming Ag (20, 21). Generation of memory B cells and maintenance of life-long protective antibodies produced by long-lived plasma cells are keys to the development of effective vaccines (22).
The pattern of antibody production in secondary immune responses differs remarkably from that of the primary response in terms of speed, magnitude, and specificity. Such differences are mainly a consequence of the intrinsic nature of memory B cells, including their robust proliferation and differentiation into AFCs (23, 24), their high-affinity B cell receptors (BCRs) acquired via the primary GC response (25–28), and their location at sites of Ag drainage in SLOs (29) including the splenic marginal zone (30), tonsillar mucosal epithelium (31), and bone marrow (32). While these features of memory B cells contribute to their accelerated recall responses upon Ag rechallenge, it is less clear if CD4+ T cells necessarily promote their secondary activation, analogous to the help provided by Tfh cells to GC B cells in the primary response. Yet, human memory B cells require CD4+ T cell help for their proper reactivation and differentiation (27, 33–36). Indeed, although human memory B cells proliferate and differentiate in vitro into AFCs in response to polyclonal signals including Toll-like receptor (TLR) stimuli and cytokines, they respond more robustly to additional signals including IL-10, IL-15, and IL-21, and/or CD40 engagement, which can be delivered by CD4+ T cells (30, 36, 37). The expression of BCRs with affinity higher than that of naïve B cells and constitutive display of CD80 and CD86 (31) suggest that human memory B cells capture antigen with their BCR, present the antigen along with costimulatory signals to stimulate CD4+ T cells, with receipt of appropriate help in return.
Vaccine studies demonstrate that isotype-switched immunoglobulin (Ig) with high affinity is detectable in the blood within 6-8 days upon re-vaccination (36, 38). If human CD4+ T cells provide help to memory B cells for their differentiation into AFCs for rapid recall responses, such help should be readily available. Rapid memory B cell differentiation is unlikely to be driven by Tfh cells, which would need to develop from naïve precursors, a process that takes days (39). Moreover, data from mice suggest that C-X-C motif chemokine receptor 5 (CXCR5)-expressing central memory CD4+ T cells, perhaps arising via the primary GC reaction, reside at the T cell zone during the resting stage and promote humoral recall responses by B cells (40–42). We therefore hypothesized that in humans a distinct subset of CD4+ T cells promote reactivation of memory B cells upon Ag recall.
We describe a subset of tonsillar CD4+ T cells that promotes maturation of memory B cells into AFCs. These cells display canonical Tfh markers indicative of the ability to provide B cell help, yet their expression of chemokine receptors and P-selectin glycoprotein ligand-1 (PSGL-1), a cell surface protein used for positioning of CD4+ T cells in SLOs (43–45), suggest that they reside in distinct locations from Tfh cells. Consistent with these findings, they colocalize with memory B cells extrafollicularly at the border of the T cell zone and B cell follicle (T-B border), a position resulting from their balanced migration response to the T cell zone chemokines, C-C chemokine ligands (CCL) 19 and 21, and the follicular C-X-C chemokine ligand (CXCL) 13, in comparison to T cell zone-resident CD45RA+ T and follicular-resident Tfh cells. These extrafollicular T helper cells promote memory B cell differentiation into AFCs via engagement of B cells by CD40 ligand, and through secretion of cytokines including IL-10 and IL-21. Our data provide evidence for a distinct population of extrafollicular CD4+ helper T cells which resides at the tonsillar T-B border, a unique cellular niche, well positioned to provide T cell-dependent help to memory B cells for humoral recall responses.
Materials and Methods
Human samples
Tonsils were obtained at time of routine pediatric tonsillectomy (ages 2-18) performed at Yale New Haven Children’s Hospital, New Haven, CT. The Institutional Review Board of the Yale University School of Medicine approved all experiments.
Surface staining
Cells were mechanically homogenized, followed by staining for surface molecules using LIVE/DEAD fixable Aqua (Invitrogen, 1:1000 dilution) and anti-CD4 PE-Cy5 (RPA-T4, BD, 1:50 dilution), anti-CD3 PE-Cy7 (UCHT1, eBioscience, 1:50 dilution), anti-CD45RA V450 (H100, eBioscience, 1:100 dilution), anti-CXCR5 conjugated with Alexa Fluor 488 (RF8B2, eBioscience, 1:50 dilution), Alexa Fluor 647 (RF8B2, BD, 1:50 dilution), or biotin (RF8B2, BD Biosciencese, 1:50 dilution)/streptavidin APC-Cy7 (1:400 dilution), anti-PSGL-1 PE (KPL-1, BD, 1:50 dilution) or APC (FLEG, eBioscience, 1:50 dilution), anti-ICOS FITC (C398.4A, eBioscience, 1:50 dilution), anti-CCR7 FITC (150503, R&D Systems, 1:50 dilution), anti-CD62L FITC (DREG56, eBioscience, 1:50 dilution), anti-CXCR4 PE-Cy5 (12G5, eBioscience, 1:50 dilution), anti-CD200 APC (OX104, eBioscience, 1:50 dilution), anti-OX40 PE-Cy5 (ACT35, BD, 1:50 dilution), anti-PD-1 PE-Cy7 (EH12.1, BD, 1:50 dilution), anti-CXCR3 BV421(1C6/CXCR3, BD, 1:50), anti-IL-2RA PE (M-A251, BD, 1:25), anti-CD19 APC-Cy7 (SJ25C1, eBioscience, 1:50 dilution), anti-IgD PE-Cy7 (IA6-2, BD, 1:50 dilution), anti-CD38 V450 (HIT2, BD, 1:50 dilution), and anti-IL-10R PE (3F9, Biolegend, 1:50 dilution).
Cell isolation
CD4+ T cells were enriched (Easysep Enrichment Kit; Stemcell Technologies) and sorted into CD45RA+, non-Tfh effectors (CD45RA− PSGL-1hi PD-1lo CXCR5lo), Tfh (CD45RA− PSGL-1lo PD-1hi CXCR5hi) and CD45RA− PSGL-1hi PD-1hi CXCR5hi T cells by FACSAria (BD Biosciences). Sorted subsets were used for intracellular staining, quantitative PCR (qPCR), measurement of cytokines, microarray experiments and T-B cell cocultures. For the latter, naïve B (CD3− CD19+ IgD+ CD38−), germinal center B (CD3− CD19+ IgD− CD38+) and memory B (CD3− CD19+ IgD− CD38−) cells were sorted and used with different T cell subsets.
Intracellular staining
For CD40L intracellular staining, sorted T cells were stimulated with PMA (100 ng/ml) and ionomycin (1 μg/ml) for an hour, fixed (BD CytoFix/CytoPerm™) and permeabilized (BD PERM/Wash™ solution). After permeabilization, cells were stained with anti-CD40L PE (24-31, Biolegend, 1:20 dilution). For IL-10 staining, total lymphocytes were stimulated with PMA (100 ng/ml) and ionomycin (1 μg/ml) for four hours and then for an additional two hours in the presence of Golgi plug (BD). Cells were stained for surface molecules prior to fixation. After fixation, cells were permeabilized and stained with anti-IL-10 Alexa Fluor 647 (JES3-9D7, Biolegend, 1:20 dilution). For BCL6, BLIMP-1, and Ki67 staining, total lymphocytes were stained for surface molecules prior to fixation. After fixation, cells were permeabilized and stained with anti-BCL6 PE (K112-91, BD, 1:50 dilution), anti-BLIMP-1 PE (6D3, BD, 1:50 dilution), or anti-Ki67 FITC (MOCP-21, BD, 1:100 dilution). FOXP3 staining was analogous to that of BCL6 except for changes in fixation and permeabilization (Biolegend FOXP3 FIX/PERM kit) and the use of anti-FOXP3 PE (206D, Biolegend, 1:10 dilution). Samples were loaded on LSR II (BD). Events were collected and analyzed with FlowJo software (TreeStar, CA).
Influenza-specific CD40L assay
PSGL-1hi PD-1hi CXCR5hi cells were isolated by flow cytometric sorting, and 5x104/well and the same number of autologous CD4-depleted cells were cocultured in the presence of Fluzone influenza virus vaccine (Sanofi Pasteur) in 96 well plates (U bottom) at 37°C for 16 hours and additional 2 hours with Golgi plug (BD). After stimulation, cells were collected and stained with Aqua Live/Dead (Invitrogen). After fixation/permeabilization, cells were stained with anti-CD40L PE (24-31, Biolegend, 1:20 dilution).
Confocal microscopy
Tonsils were placed in OCT medium, frozen with dry ice, then stored at −80 °C. 8-9 μm cryostat sections were fixed in acetone. After blocking with rat serum for 20 minutes, slides were stained with anti-CD19 FITC (HIB19, BD, 1:25 dilution), anti-IgD FITC (IA6-2, BD, 1:25 dilution) or PE (IgD26, Miltenyi Biotec, 1:25 dilution), anti-CD27 PE (O323, Biolegend, 1:25 dilution) or Alexa Fluor 647 (O323, Biolegend, 1:25 dilution), anti-CD38 Alexa Fluor 647 (HIT2, Biolegend, 1:10 dilution), anti-PSGL-1 PE (KPL-1, BD, 1:10 dilution), and anti-CD4 AL647 (OKT4, Biolegend, 1:25 dilution). For PD-1 staining, we used purified mouse anti-human PD-1 antibody (EH12.2H7, Biolegend, 1:40 dilution) followed by goat anti-mouse DyLight 649 (PoLy4053, Biolegend, 1:40 dilution). For secondary staining, anti-FITC Alexa Flour 488 antibody (Invitrogen, 1:100 dilution) was used to enhance the FITC signal. Slides were mounted (Prolong Gold Anti-Fade Mounting Medium; Invitrogen), and images acquired using 10X and 40X objectives (Olympus BX40), or a 20X objective (Zeiss LSM510 Meta Confocal Microscope). Within one image, areas of different anatomical compartments (T cell zone, follicular mantle, germinal center, T-B border, and subepithelium) were measured with ImageJ software. As defined previously in a murine study (39), the area of T-B border was determined as 80-100 μm on either side of the follicular mantle border. Cells of interests were quantified with ImageJ and their numbers per each anatomical compartment were calculated.
Quantitative PCR
Quantitative PCR was set up using Brilliant II SYBR® Green QPCR Master Mix (Agilent Technologies, Santa Clara, CA) according to manufacturer’s instructions. Primers of relevant genes were synthesized by Real Time Primers and the Keck Oligonucleotide Synthesis Facility at Yale University (GATA3, RORC, BCL6, PRDM1, PDCD1, CXCR5, SELPLG) or purchased from www.realtimeprimers.com (TBX21, GPR183). Primer sequences included: TBX21 forward primer: 5’-CAACAATGTGACCCAGATGA-3’, reverse primer: 5’-TGGCAAAGGGGTTATTATCA-3’; GATA3 forward primer: 5’- TCACAAAATGAACGGACAGAAC 3’,reverse primer: 5’-AGCTTGTAGTAGAGCCCACAGG 3’; RORC forward primer: 5’- CTGTAACGCGGCCTACTCCT -3, reverse primer: 5’- GTCTTGACCACTGGTTCCTGTT -3’; BCL6 forward primer: 5’- ATGCCAGTGATGTTCTTCTCAA 3, reverse primer: 5’- AAGGTTGCATTTCAACTGGTCT -3’; PRDM1 forward primer: 5’- GACTTTGCAGAAAGGCTTCACT 3, reverse primer: 5’- ACATTCTTTGGGCAGAGTTCAT -3’; PDCD1 forward primer: 5’-AAGGCGCAGATCAAAGAGAG-3, reverse primer: 5’-ACCCAGACTAGCAGCACCAG-3’; CXCR5 forward primer: 5’- CTGGAAATGGACCTCGAGAA reverse primer: 5’- GCAGGGCAGAGATGATTTTC 3’; SELPLG forward primer: 5’- GGCTGGGACCTTGTCACTAA-3, reverse primer: 5’- AGGCTTTCTCGGCTTCATCT -3’; and GPR183 forward primer: 5’-CTATGCAATGGGCTTTGACT-3’, reverse primer: 5’-TTGATGAGGAGTGGGAGTGT-3’. PCR reaction were performed on the Stratagene MX3005P™ (Agilent Technologies, Santa Clara, CA) with following conditions: step 1: Denaturation at 95°C for 10 minutes; Step 2: amplification (40 cycles) at 95°C for 30 seconds, 55°C for 1 minute, 72°C for 1 minute; and, Step3: termination (1 cycle) at 95°C for 1 minute, 55°C for 30 seconds, 95°C for 30 seconds. The expression of each transcript was standardized using expression of the housekeeping gene ACTIN.
T-B cell coculture
5 × 104/well of sorted T and B cells were cocultured in U-bottom 96 well plates in RPMI 1640 (GIBCO) supplemented with 1% L- glutamine, 1% penicillin/streptomycin, 1% sodium pyruvate, 10% Fetalplex and IL-2 (10 ng/ml). Supernatants were taken after 5 days for Ig analysis by ELISA. In blocking experiments, anti-CD3 + anti-CD28 (Dynabeads® Human T-Activator; Invitrogen, 1:1000 dilution) were added to the culture medium with the following: anti-IL-10 monoclonal antibody (PeproTech), IL-21R/Fc fusion protein (1 μg/ml, R&D Systems), anti-CD40L (5 μg/ml, eBioscience), and anti-ICOS ligand (5 μg/ml, eBioscience). Anti-IL-17 antibody (R&D system) and/or human IgG Fc fragment protein (Abcam) were used as isotype controls.
Cytokine measurement
Sorted T cells were stimulated with PMA (100 ng/ml) plus ionomycin (1 μg/ml) and cell supernatants obtained 6 hours later. Cytokines concentrations were determined using commercial kits, according to instructions of the manufacturers: IL-21, IL-4, IL-17, and interferon gamma (all from eBioscience), and IL-10 (Biolegend).
Migration assays
Enriched CD4+ T cells were washed and resuspended in migration medium (1640 RPMI with 0.5% fetal bovine albumin) at 1 × 106 cells/ml. 2 × 105 cells were loaded in a 3 μm pore transwell (BD Falcon), with 800 μl of migration medium placed in lower chambers. CCL19, CCL21, CXCL13 (all R&D Systems) and oxysterol (Avanti® Polar Lipids INC, Alabaster, Alabama) were added at a concentration of 1 μg/ml (CCL19, CCL21, CXCL13) or 5 μM (oxysterol) individually or collectively (Fig. 6). Cells were allowed to migrate at 37°C for 3 hours, followed by their collection from the lower chamber were collected and staining to quantify numbers of CD45RA+, Tfh, and PSGL-1hi PD-1hi CXCR5hi T cell populations. In some experiments, enriched CD4+ T cells were stained with either anti-CCR7 monoclonal antibody (16 μg/ml, R&D Systems) or anti-PSGL-1 biotin (16 μg/ml, Ancell) for 40 minutes either individually or collectively prior to migration. Before samples were loaded into for flow cytometry, the same numbers of beads (SpherotechINC) were added into each tube to standardize cell numbers between tubes. Events were analyzed with FlowJo software.
Gene expression array analysis
CD45RA+, effector memory, Tfh, and PSGL-1hi PD-1hi CXCR5hi T cells were sorted by flow cytometry. Independent samples were generated from three individual tonsils. mRNA from sorted cells was extracted using Trizol (Invitrogen, Carlsbad, CA) and purified (RNeasy Mini Kit; Qiagen), according to the manufacturer’s instructions. mRNA purity was verified by OD 260/280 and OD 260/230 ratio analysis (NanoDrop® ND-1000 Spectrophotometer; Thermo Scientific). Triplicates of Tfh and PSGL-1hi PD-1hi CXCR5hi T cells were made before RNA extraction. For one chip, 130-150 ng of mRNA per sample obtained from one individual tonsil were reverse transcribed to cDNA using a T7 oligo (dT) primer. Second-strand cDNA was synthesized, transcribed in vitro, and labeled via incorporation of biotin-16-UTP. Integrity of purified cDNA was assessed (Agilent 2100 Bioanalyzer) prior to hybridization. Labeled cDNA samples were hybridized to two Illumina HumanHT-12 v4 Expression Bead Chip genome-wide arrays using standardized Illumina reagents and protocols according to the manufacturer’s instructions. After washing and staining, Bead Chips were scanned on the Illumina Iscan. The Beadstudio software was used to export probe level expression values for each sample. The R v2.13.0 Bioconductor lumi v2.4.0 package was used for quality control analysis, variance stabilizing transformation and quantile normalization (46). Probes with less than two samples called present were excluded from analysis. The limma v3.8.1 package was used to identify differentially expressed genes (47). The GOstats v2.18.0 R package (48), Database for Annotation, Visualization and Integrated Discovery tool (49), and Ingenuity Pathways analysis systems were used for pathway and network analysis of differentially expressed gene sets. The gene expression array data is accessible in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nim.nih.giv/gds) with accession number GSE52156.
Statistics
The significance of the mean differences among three or more than three groups was determined by one-way ANOVA test with Bonferroni correction. For comparison of the mean differences between two groups, two-tailed Student’s or paired t-tests were performed. All statistical analyses were done with Prism Software.
Results
PSGL-1hi CD4+ T cells are geographically restricted to the tonsillar T cell zone
P-selectin glycoprotein ligand-1 (PSGL-1) is a T cell surface molecule that enables residence in the T cell zone by synergizing interaction between C-C chemokine receptor type 7 (CCR7) on T cells and CCL19 and CCL21, T cell zone chemokines (43). Murine experiments show that downregulation of Selplg (transcript for PSGL-1) by the transcription factors B-cell lymphoma 6 (Bcl6) and Achaete-scute complex homolog 2 (Ascl2) is required for Tfh cell differentiation (50, 51); thus, nearly all murine CD4+ T cells in B cell follicles and germinal centers (GCs) are PSGL-1lo (44), although PSGL-1lo cells also are found extrafollicularly (45), as they prepare for T zone exit to the follicle upon activation. To determine if the latter phenotype applies to humans, we stained human tonsil sections with antibodies to PSGL-1 (red), IgD (green), and CD4 (blue) (Fig. 1A, shown individually and merged). Consistent with murine studies, PSGL-1hi CD4+ T cells are in the T cell zone, and not in follicles and GCs (Fig. 1, A and B, with co-localization of purple PSGL-1+ CD4+ cells). Quantification of CD4+ cell numbers in T cell zones, follicular mantles, and GCs demonstrated that while both PSGL-1hi and PSGL-1lo populations reside in the T cell zone, as in mice (44), PSGL-1hi CD4+ T cells are not found within the follicular mantle and GCs (Fig. 1C). Thus, in humans, as in mice, PSGL-1 distinguishes extrafollicular CD4+ T cells from follicular CD4+ T cells: PSGL-1hi CD4+ cells reside only in the T cell zone and all GC CD4+ T cells are PSGL-1lo.
Fig. 1. PSGL-1hi CD4+ T cells are located in the T cell zone.
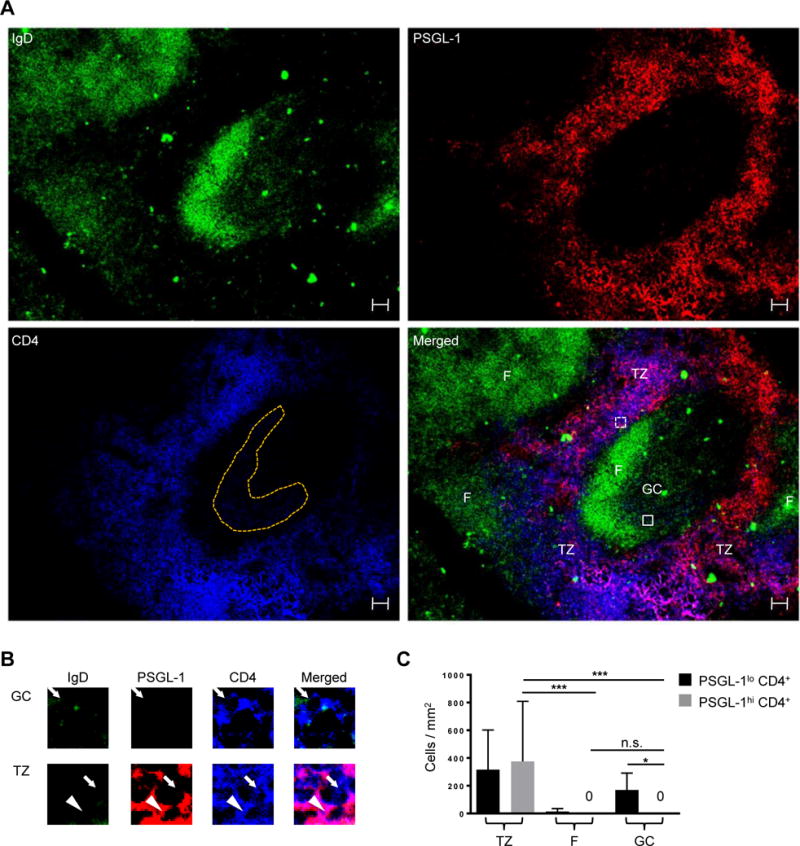
(A) PSGL-1 (red), IgD (green), and CD4 (blue) staining of human tonsils. TZ: T cell zone, F: follicular mantle, GC: germinal center. GC CD4+ T cells are shown in orange dots. Scale bar = 50 μm. (B) Cells in boxes are magnified. The PSGL-1hi CD4+ T cell is indicated by an arrowhead and PSGL-1lo CD4+ T cells are indicated by arrows. (C) PSGL-1hi CD4+ T and PSGL-1lo CD4+ T cells were quantified in the T cell zone, follicular mantle, and GCs. N = 10 different images from 7 tonsils. One-way ANOVA test. Bars indicate the mean and the SD. *P < 0.05, ***P < 0.001. TZ: T cell zone, F: follicular mantle, GC: germinal center.
CD4+ T cells expressing canonical Tfh markers exist in the tonsillar T cell zone
Human tonsillar GC Tfh cells have been defined as either PD-1hi CXCR5hi or ICOShi CXCR5hi (52, 53). To further characterize PSGL-1hi cells, and to assess their relationship to Tfh cells, we, gated total CD4+ T cells into PD-1− CXCR5−, PD-1lo CXCR5lo, and PD-1hi CXCR5hi cells, using previously established gating strategy (52, 53). PD-1− CXCR5− cells were further gated into CD45RA+ naïve T cells and CD45RA− PSGL-1hi PD-1− CXCR5− cells. Based on their location and work by others (11, 12, 44, 53–55), we defined CD45RA− PSGL-1hi PD-1− CXCR5− CD4+ T cells as non-Tfh effectors. Of PD-1hi CXCR5hi CD4+ cells, 17.9 ± 5.8% (mean ± SD, n=12) expressed PSGL-1, gating based on the PSGL-1 expression on CD45RA+ naïve CD4+ T cells. Although PSGL-1hi PD-1hi CXCR5hi CD4+ T cells co-expressed the Tfh markers, PD-1 and CXCR5, they resided in the T cell zone according to microscopic findings (Fig. 1). The surface marker profile of PSGL-1hi PD-1hi CXCR5hi CD4+ T cells was similar to that of Tfh cells as both populations expressed ICOS, OX40, CXCR4, CD200, CD57, and CD40 ligand, with less expression of CD127, CCR7, and CD62L. In addition, both PSGL-1hi PD-1hi CXCR5hi CD4+ T cells and Tfh cells lacked FOXP3 expression characteristic of regulatory T cells and T follicular regulatory cells shown in mice and in humans (Fig. 2B) (56–60). PD-1lo CXCR5lo CD4+ cells, known to promote extrafollicular Ig production by naïve B cells (53), could be distinguished in flow cytometry from PSGL-1hi PD-1hi CXCR5hi CD4+ T cells and Tfh cells.
Fig. 2. Extrafollicular tonsillar CD4+ T cells express canonical markers of human Tfh cells.
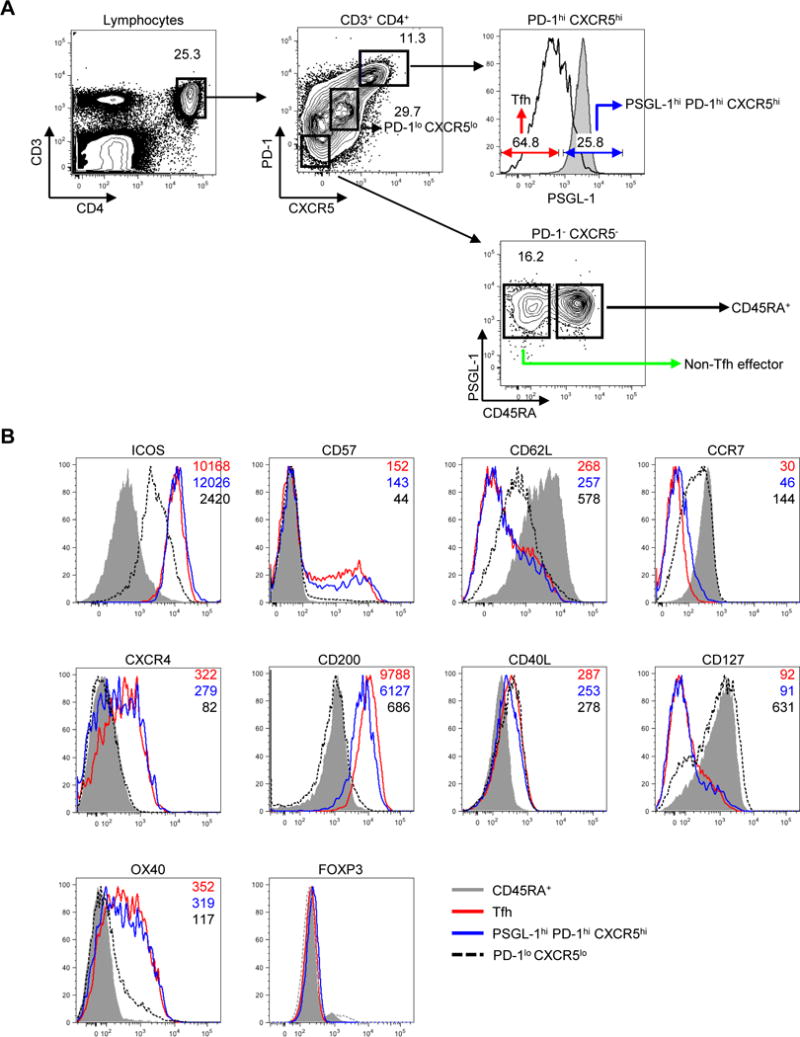
(A) Gating of tonsillar CD4+ T cells. Representative plots from twelve independent experiments. Shaded gray histograms indicate PSGL-1 expression on CD45RA+ naïve CD4+ T cells. (B) Expression of surface or intracellular proteins on CD45RA+ naïve, PD-1lo CXCR5lo CD4+ T cells, follicular helper T (Tfh), and PSGL-1hi PD-1hi CXCR5hi CD4+ T cells. Mean fluorescence intensity is shown on the right upper corner of each plot. All markers except CD40L were stained without cellular stimulation. Shaded gray histograms indicate expression of markers on CD45RA+ naïve CD4+ T cells as a control.
PSGL-1hi PD-1hi CXCR5hi T and Tfh cells are transcriptionally distinct
We next asked if PSGL-1hi PD-1hi CXCR5hi T cells had features distinct from those of Tfh cells in addition to surface marker expression and tonsillar location. We examined mRNA expression using microarrays comparing tonsillar Tfh and PSGL-1hi PD-1hi CXCR5hi T cells, alongside controls including naïve CD45RA+ and non-Tfh effector T cells (Fig. 3, A, B, and D; Supplemental Fig. 1, A, B, and C; Supplemental Tables 1-2). We did three independent sorts of Tfh and PSGL-1hi PD-1hi CXCR5hi cells, each sort from an individual tonsil, and then split each of these replicates into thirds for expression analysis by microarrays. The overall relatedness of the gene expression profiles of the 4 CD4+ T cell populations (Tfh, PSGL-1hi PD-1hi CXCR5hi, naïve, non-Tfh effector) was determined by hierarchical clustering of the 500 most variably expressed genes in the data set (Supplemental Fig. 1A). Expression patterns from naïve CD45RA+ T cells were most distinct from the other cell types. The expression profiles of the Tfh cells were most similar to those from PSGL-1hi PD-1hi CXCR5hi T cells (Fig. 3A, Supplemental Fig. 1A). Differences in gene expression between Tfh cells and PSGL-1hi PD-1hi CXCR5hi T cells were examined in the 9 replicates (false discovery rate [FDR] P < 0.05, log2 fold change > 0.5; Supplemental Table 1), and 320 differentially expressed genes were identified. Heatmap analysis revealed a high degree of uniformity and consistency in patterns of gene expression in the replicates of the two cell types (Supplemental Fig. 1B).
Fig. 3. PSGL-1hi PD-1hi CXCR5hi T cells exhibit unique patterns of gene expression and cytokine profiles in comparison to Tfh cells.
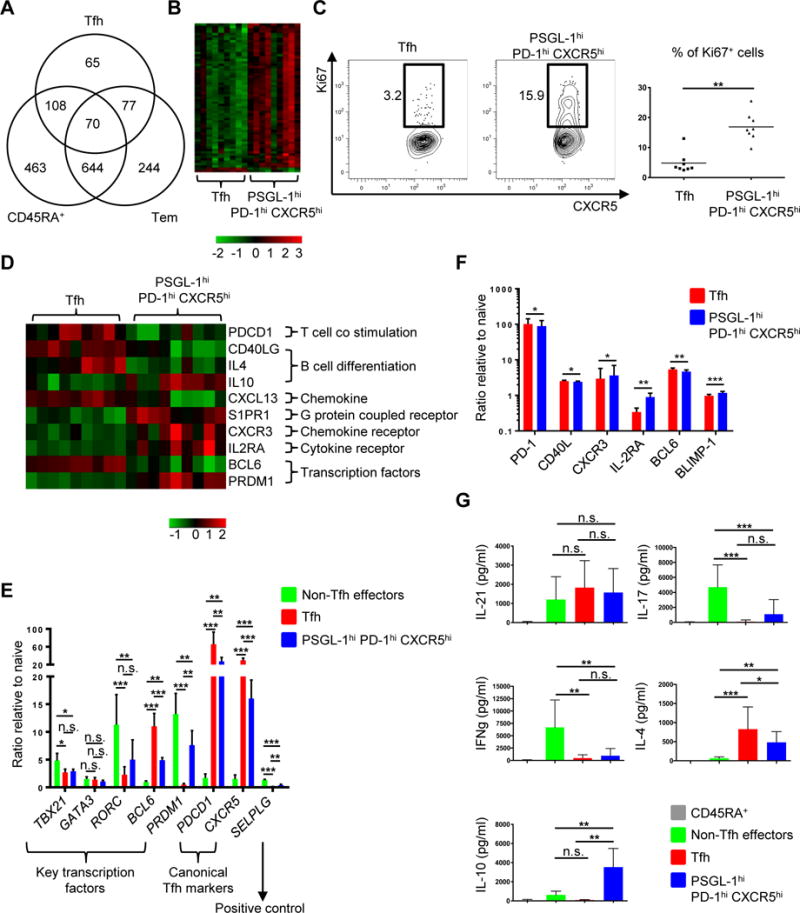
(A) Differentially expressed genes (FDR P < 0.05, log2 fold change > 0.5) between the 4 cell populations were identified in a series of pair wise analyses. The numbers of differentially expressed genes between the indicated cell population and PSGL-1hi PD-1hi CXCR5hi T cells and patterns of overlap are shown in Venn diagram format. Tfh: follicular helper T, Tem: effector memory cells. (B) Heat map of cell cycle genes (gene ontology (GO): 0022403). Columns represent 9 replicates of Tfh and PSGL-1hi PD-1hi CXCR5hi T cells. (C) Ki67 intracellular staining of Tfh and PSGL-1hi PD-1hi CXCR5hi T cells. Representative plots from eight independent experiments (left panel) and percentage of Ki67+ cells in Tfh and PSGL-1hi PD-1hi CXCR5hi T cell populations (right panel) are shown. Bars indicate the mean. n=8, paired t test, **P < 0.01. (D) Heat map of relevant genes differentially expressed between Tfh and PSGL-1hi PD-1hi CXCR5hi T cells (FDR P < 0.05, log2 fold change > 0.5). Columns represent 9 replicates of either Tfh or PSGL-1hi PD-1hi CXCR5hi T cells. (E) Quantitative PCR analysis of key transcription factors and canonical Tfh cell markers. Adjusted by expression of housekeeping gene (ACTIN). N = 4 or 5, One-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001. Bars indicate the mean and the SD. (F) Expression of PD-1, CD40L, CXCR3, IL-2RA, BCL6, and BLIMP-1 on Tfh and PSGL-1hi PD-1hi CXCR5hi T cells. The Y-axis represents the ratio of mean fluorescence intensities (MFI) to those of naïve CD45RA+ T cells. N = 5 or 6, paired t-test. *P < 0.05, **P < 0.01, ***P < 0.001. Bars indicate the mean and the SD. (G) Cytokine profile of CD4+ T cell subsets. Cytokines were measured after 6 hour-stimulation with PMA and ionomycin. N = 6 or more, One-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001. Bars indicate the mean and the SD. Tfh: follicular helper T cells.
The biologic function(s) of the genes that were differentially expressed between Tfh cells and PSGL-1hi PD-1hi CXCR5hi T cells were analyzed in a series of network and pathway analyses. Analysis of Gene Ontology (GO) biologic processes enriched in differentially expressed genes identified GO terms related to cell cycle, and with the exception of BCL6 and SMAD6, identified genes upregulated in PSGL-1hi PD-1hi CXCR5hi T cells (Fig. 3B and Supplemental Table 2). The top cluster of DAVID enriched terms and Ingenuity Pathway Analysis (IPA) also identified gene categories primarily related to cell cycle and the top functional network identified E2F proteins, transcription factors involved in cell cycle progression and DNA synthesis (61), as critical hubs, all with significant expression in PSGL-1hi PD-1hi CXCR5hi T cells and validated by quantitative PCR (Supplemental Fig. 1, C and D). Gene expression profiling suggested that PSGL-1hi PD-1hi CXCR5hi T cells proliferated more actively than Tfh cells via the E2F pathway. Indeed, PSGL-1hi PD-1hi CXCR5hi T cells contained more Ki67+ cells than did their Tfh counterparts (Fig. 3C).
Several other genes of interest were also differentially expressed between these two populations (Fig. 3D). As expected, Tfh cells more robustly expressed genes critical for Tfh cell function or differentiation, including PDCD1 (13), CD40 ligand (CD40LG) (12), interleukin-4 (IL4) (62, 63), BCL6 (64–66), and CXCL13, a ligand of CXCR5 and known to be produced by human Tfh cells (67, 68). In contrast, PSGL-1hi PD-1hi CXCR5hi T cells more significantly expressed genes involved in migration and B cell differentiation into AFCs including interleukin-10 (IL10) (69–71), sphingosine-1-phosphate receptors (S1PR), critical for T cell egress from either the thymus or peripheral lymphoid organs (72), C-X-C chemokine receptor type 3 (CXCR3), which enables T cells to migrate into inflamed tissues in response to C-X-C motif chemokine 10 (CXCL10) (73), as well as IL-2 receptor alpha (IL2RA), which is not robustly expressed in murine Tfh cells (55). The genes encoding Ascl2 (50), activin A (74), and Maf (68) which play critical roles in Tfh cell differentiation by upregulating CXCR5 and/or downregulating CCR7 and PSGL-1 were comparably expressed between PSGL-1hi PD-1hi CXCR5hi T cells.
We further dissected the RNA and protein differences between Tfh and PSGL-1hi PD-1hi CXCR5hi T cells using qPCR, assessing expression of key transcription factor genes and canonical Tfh markers (Fig. 3E). Non-Tfh effectors highly expressed the T-box transcription factor 21 (TBX21), the Th1 master regulator, RAR-related orphan receptor gamma (RORC), the Th17 master regulator, and PRDM1 (BLIMP-1, a BCL6 repressor), while Tfh cells highly expressed BCL6. As shown in microarray analysis, PSGL-1hi PD-1hi CXCR5hi T cells expressed less BCL6 and more PRDM1 than Tfh cells. The canonical Tfh markers, CXCR5 and PDCD1, were highly expressed in Tfh cells. We also compared protein expression of representative genes (Fig. 3D) between Tfh and PSGL-1hi PD-1hi CXCR5hi T cells by mean fluorescence intensity (MFI) and observed an identical tendency of expression of the genes at protein level (Fig. 3F). Regarding cytokine profiles, all activated CD4+ T subsets (non-Tfh effectors, Tfh, and PSGL-1hi PD-1hi CXCR5hi T cells) comparably produced IL-21 (Fig. 3G). Consistent with the qPCR data, non-Tfh effectors produced Th1 and Th17 cytokines, interferon-γ and IL-17, respectively. Both Tfh and PSGL-1hi PD-1hi CXCR5hi T cells produced IL-4, while IL-10 was made only by the latter (Fig. 3G). Thus, while Tfh and PSGL-1hi PD-1hi CXCR5hi T cells have similar gene expression profiles and BCL6 protein expression, the latter expressed significantly more transcripts related to cell cycle (alongside evidence of greater proliferation; Fig. 3C), migration (S1PR1 and CXCR3, low in CXCR5 and CXCL13), function (IL-10), and development (IL2RA and PRDM1). They also produced more IL-10 and less IL-4 than Tfh cells, while comparably producing IL-21.
PSGL-1hi CXCR5hi PD-1hi T cells promote differentiation of memory B cells into AFCs
We assessed the B helper function of human tonsillar CD4+ cells by measuring secreted immunoglobulin (Igs) during T and B cell cocultures (Fig. 4 and Supplemental Fig. 2). Memory B cells produced more IgG when cocultured with PSGL-1hi PD-1hi CXCR5hi T cells, compared to culture with Tfh cells (Fig. 4A). By contrast, GC B cells more robustly secreted IgG when cocultured with Tfh cells (Fig. 4A). Differential Ig secretion in co-cultures was not a consequence of distinct survival of T cell subsets, since we recovered equivalent numbers of Tfh and PSGL-1hi CXCR5hi PD-1hi T cells from the co-cultures when Igs were measured (Supplemental Fig. 2A). To elucidate the mechanism by which PSGL-1hi PD-1hi CXCR5hi T cells promoted memory B cells differentiation, we cocultured PSGL-1hi PD-1hi CXCR5hi T cells and memory B cells in the presence of anti-CD3 and anti-CD28 along with blockade of molecules known to contribute to T-B cell interactions. Anti-ICOSL antibody blockade of ICOS signaling did not affect Ig production by memory B cells, by contrast to the effects of CD40L blockade (Fig. 4B). Similarly, interruption of physical interactions between PSGL-1hi PD-1hi CXCR5hi T cells and memory B cells in transwell assays limited CD38++ IgG+ AFC formation (Supplemental Fig. 2B).
Fig. 4. PSGL-1hi PD-1hi CXCR5hi T cells promote memory B cell maturation to Ig production via contact and cytokines.
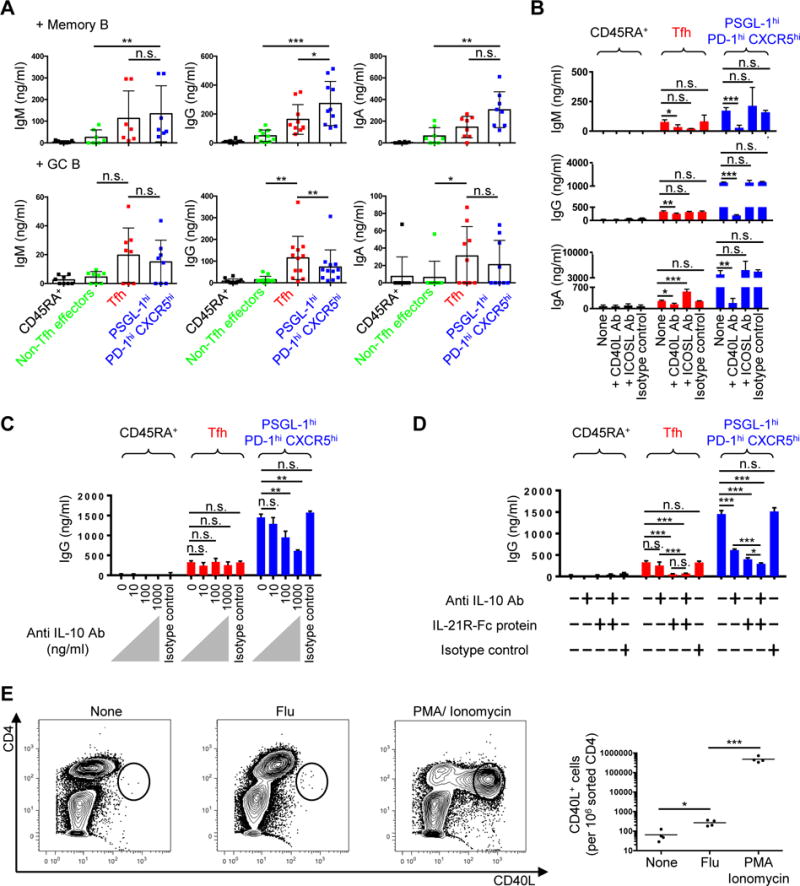
(A) Tonsillar CD45RA+ naïve, non-Tfh effectors, Tfh, and PSGL-1hi PD-1hi CXCR5hi T cells were cocultured with either autologous memory or GC B cells without exogenous stimulation. Immunoglobulins (Igs) were measured by ELISA 5 days after T-B coculture. N ≥ 7. One-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001. Bars indicate the mean and SD. (B) Anti-CD40L or anti-ICOS ligand antibodies were added to T cell plus memory B cell cocultures in the presence of anti-CD3 and anti-CD28 for 5 days. Representative data from three independent experiments. One-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001. Bars indicate the mean and the SD. (C) Indicated dose of anti-IL-10 antibody was added to T cell plus memory B cell cocultures in the presence of anti-CD3 and anti-CD28 for 5 days. Representative data from three independent experiments. One-way ANOVA test. **P < 0.01. Bars indicate the mean and the SD. (D) Anti-IL-10 (1 μg/ml) antibody and/or IL-21R-Fc (1 μg/ml) protein were added to T cell plus memory B cell cocultures in the presence of anti-CD3 and anti-CD28 for 5 days. IgG amounts in supernatants are shown. Representative data from two independent experiments. One-way ANOVA test. *P < 0.05, ***P < 0.001. Bars indicate the mean and the SD. (E) 5x104/well of sorted PSGL-1hi PD-1hi CXCR5hi T cells and autologous CD4-depleted cells were cocultured in the absence or presence of flu vaccine for 18 hours, or stimulated with PMA plus ionomycin for 4 hours. Golgi plug (BD) was added to each condition for last 2 hours of stimulation. CD40L intracellular staining was performed after stimulation. Far right panel showed CD40L+ cells/106 sorted CD4 cells. N = 4 different tonsils, paired t-test, *P < 0.05, ***P < 0.001. Bars indicate the mean.
We also asked if T-cell cytokines contributed to memory B cell differentiation in these co-culture assays, focusing initially upon IL-10 as it was exclusively produced by PSGL-1hi PD-1hi CXCR5hi T cells (Fig. 3F), and past studies demonstrating that this cytokine promotes B-cell production of isotype-switched Igs (69, 70). Accordingly, anti-IL-10 reduced in a dose-dependent manner the production of Ig upon PSGL-1hi PD-1hi CXCR5hi T cell and memory B cell cocultures (Fig. 4C and Supplemental Fig. 2C). As IL-10 and IL-21 work synergistically to promote Ig synthesis by B cells (9, 75), we went on to demonstrate that antibody blockade of both cytokines further inhibited Ig production in co-cultures, compared to single blockade, suggesting that these cytokines worked independently (Fig. 4D and Supplemental Fig. 2D). We also asked if PSGL-1hi PD-1hi CXCR5hi T cells contain antigen-specific memory cells, assessing CD40L expression in response to influenza antigen stimulation (75). PSGL-1hi PD-1hi CXCR5hi T cells upregulated CD40L upon stimulation, suggesting that this population contains antigen-specific memory cells (Fig. 4E). Collectively, these data reveal that PSGL-1hi PD-1hi CXCR5hi T cells help memory B cells differentiate into AFCs through contact (CD40L)- and cytokine (IL-10 and IL-21)-dependent signals.
PSGL-1hi PD-1hi CXCR5hi T cells reside proximal to memory B cells at the T cell zone-B cell follicular border
We next investigated the location of PSGL-1hi PD-1hi CXCR5hi T cells, and for comparison, memory B cells, using confocal microscopy. For the latter, tonsil sections were stained with antibodies against CD27 and CD38, a memory B and GC B cell marker, respectively (76), in combination with CD19, to mark all B cells (Supplemental Fig. 2E). We used CD27 to demarcate the T cell zone from B cell follicles (Fig. 5A, red), as memory B cells, plasmablasts, and CD4+ T cells more strongly expressed CD27 in flow cytometry than did naïve and GC B cells (Supplemental Fig. 2E). Likewise, CD38 staining enabled differentiation of T cell zone and GCs from the follicular mantle (Fig 5A, blue), as GC B cells, plasmablasts, and some T cells expressed this protein (Supplemental Fig. 2E). When colors were merged, we observed that CD19+ CD27hi CD38− memory B cells were primarily found along the border of T cell zone-B cell follicle (T-B border) (Fig. 5A, far right panel; yellow cells as indicated by arrows). Quantification of microscopy data revealed that memory B cells were present mainly at the T-B border or B cell follicular mantles (Fig. 5C, left panel). This finding was consistent with work showing that memory B cells, especially those expressing CD27, are located mainly at the tonsillar T-B border (77).
Fig. 5. Memory B cells and PSGL-1hi PD-1hi CXCR5hi cells are enriched at the tonsillar T-B border.
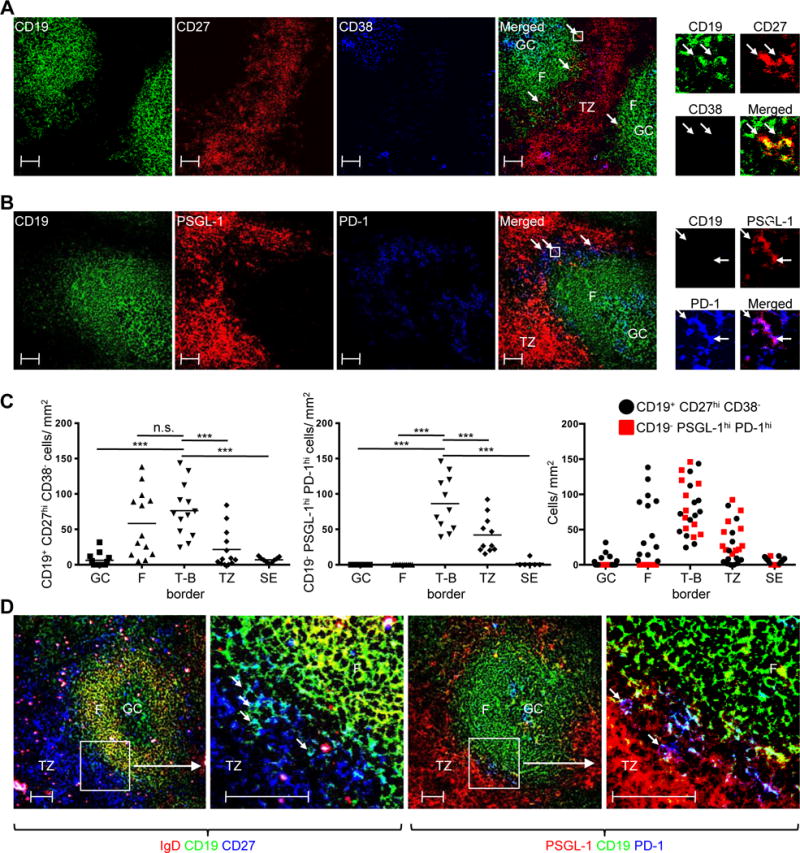
(A) CD27 (red), CD19 (green), and CD38 (blue) confocal staining. Arrows in merged picture indicate CD19+ CD27hi CD38− (yellow) cells. Cells in the box are magnified on top upper right. Scale bar = 50 μm. (B) PSGL-1 (red), CD19 (green), and PD-1 (blue) confocal staining. Arrows in merged picture indicates CD19− PSGL-1hi PD-1hi (purple) cells. Cells in the box are magnified on top upper right. Scale bar = 50 μm. (C) Numbers of CD19+ CD27hi CD38− cells (left panel) and CD19− PSGL-1hi PD-1hi (middle panel) cells per mm2 in indicated anatomic regions. Two graphs are merged (right panel). GC: germinal center, F: follicular mantle, T-B border: Border of the T cell zone and B cell follicle, TZ: T cell zone, SE: subepithelial area. Left panel: N = 13 images from 3 different tonsils, except the subepithelial area, in which N = 9. Middle panel; N = 11 images from 3 different tonsils, except subepithelial area, in which N = 6. One-way ANOVA test. ***P < 0.001. Bars indicate the mean. (D) Two left panels: IgD (red), CD19 (green), CD27 (blue) confocal staining. Boxed area in left panel magnified in next panel, where arrows indicate CD19+ CD27hi IgD− (light blue) cells. Two right panels: PSGL-1 (red), CD19 (green), PD-1 (blue) confocal staining. Boxed area magnified in next panel, where arrows indicate CD19− PSGL-1hi PD-1hi (purple) cells. Scale bar = 50 μm.
The PSGL-1 marked T cell zone (Fig. 5B, red) was adjacent to CD19 staining B cell follicles (Fig. 5B, green). As reported previously (78), PD-1hi cells were found either in GCs or at the T-B border (Fig. 5B, blue). When combined, CD19− PSGL-1lo PD-1hi cells (light blue) were present at the periphery of GCs while CD19− PSGL-1hi PD-1hi cells resided at the follicular outer edges (Fig. 5B, far right panel; purple cells as indicated by arrows), especially at the T-B border (Fig. 5C, middle panel). These CD19− PSGL-1hi PD-1hi cells were PSGL-1hi PD-1hi CXCR5hi T cells as determined by flow cytometry, since PD-1 was expressed only by CXCR5hi CD4+ T cells in human tonsils (Supplemental Fig. 2F). Quantification of memory B cells and CD19− PSGL-1hi PD-1hi T cells revealed that they were enriched at the T-B border (Fig. 5C, right panel). When identical areas were compared side-by-side, CD27 expressing memory B cells (Fig. 5D, left two panels; light blue cells as indicated by arrows) and CD19− PSGL-1hi PD-1hi T cells (Fig. 5D, right two panels; purple cells indicated by arrows) were colocalized at the T-B border.
Chemokine signaling and receptor expression by PSGL-1hi PD-1hi CXCR5hi T and memory B cells are consistent with their tonsillar co-localization
We next asked if chemokine signaling and receptor expression on PSGL-1hi PD-1hi CXCR5hi T and memory B cells were consistent with their co-localization at the tonsillar T-B border. We hypothesized that CCR7, PSGL-1, CXCR5, and Epstein-Barr virus-induced G-protein coupled receptor 2 (EBI2) collectively played a role in determining the location of both PSGL-1hi PD-1hi CXCR5hi T cells and memory B cells. To address this hypothesis, we first investigated their expression on T cells, using as EBI2’s transcript GPR183 as a surrogate for EBI2 (Fig. 6A). Compared to Tfh cells, PSGL-1hi PD-1hi CXCR5hi T cells expressed greater CCR7, PSGL-1 and GPR183, as markers of T zone homing, with less CXCR5, a follicular homing molecule. Consistent with these data, chemotaxis assays revealed that PSGL-1hi PD-1hi CXCR5hi cells migrated better in response to the T zone chemokines CCL19 and CCL21 than did Tfh cells, as assessed by the ratio of their migration to these two chemokines, compared to migration in their absence (PSGL-1hi PD-1hi CXCR5hi T cells = 3.06 ± 0.61, vs. Tfh cells = 1.14 ± 0.25; mean ± SD, n=10). The former also migrated less than did Tfh cells in response to the follicularly expressed CXCL13, a ligand for CXCR5, also as assessed by the ratio of their migration to the chemokine compared to migration in its absence (PSGL-1hi PD-1hi CXCR5hi cells = 2.69 ± 1.14, vs. Tfh cells = 5.26 ± 2.82; mean ± SD, n= 10) (Fig. 6B). In comparison, and as expected, naïve CD45RA+ T cells migrated well in response to CCL19 and CCL21 (9.97 ± 5.51, ratio relative to absence of CCL19 + 21; mean ± SD, n=10).
Fig. 6. Location of PSGL-1hi PD-1hi CXCR5hi T and memory B cells is dictated by signaling via CCR7, PSGL-1, EBI2, and CXCR5.
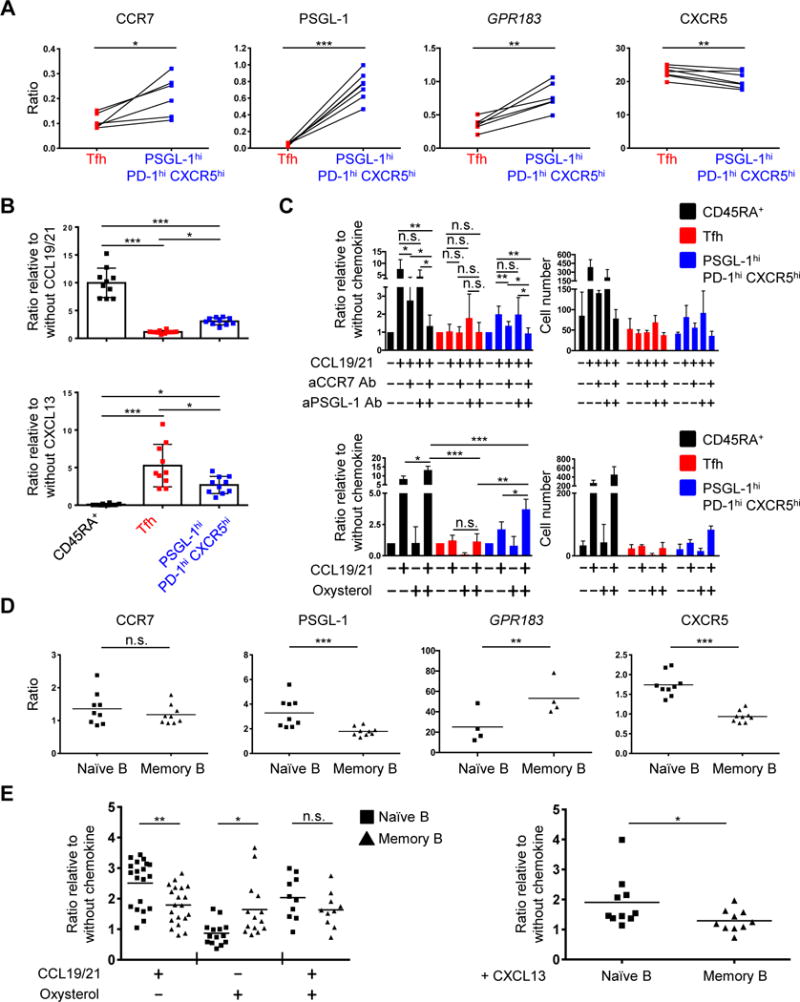
(A) Expression of CCR7, PSGL-1, GPR183, and CXCR5 on Tfh and PSGL-1hi PD-1hi CXCR5hi T cells. Y-axis represents ratio of mean fluorescence intensities (MFI; CCR7, PSGL-1, and CXCR5) or gene expression (GPR183) compared to naïve CD45RA+ T cells. N = 6 or 7, paired t-test. *P < 0.05, **P < 0.01, ***P < 0.001. Bars indicate the mean. Tfh: T follicular helper T cells. (B) Ratio of migrated cells in response to indicated chemokines compared to migration in absence of chemokines. N = 10 from 6 different tonsils (CCL19 and CCL21), or from 5 different tonsils (CXCL13). One-way ANOVA test. *P < 0.05, ***P < 0.001. Bars indicate the mean. Tfh: follicular helper T cells. (C) Upper panels: Ratio (left) and cell numbers (right) of migrated cells in response to CCL19 and CCL21 with or without pretreatment of anti-CCR7 and/or anti-PSGL-1 antibodies. Cell numbers are representative of six (upper panel) independent experiments. Lower panels: ratio (left) and cell number (right) of migrated cells in response to CCL19 and CCL21 and/or oxysterol. Cell numbers are representative of four independent experiments. One-way ANOVA test. *P < 0.05, **P < 0.01, ***P < 0.001. Bars indicate the mean and the SD. (D) Expression of CCR7, PSGL-1, GPR183, and CXCR5 by naïve and memory B cells. Y-axis represents ratio of MFI or gene expression to those of GC B cells. N = 9 for MFI, 4 for gene expression. Paired t-test. **P < 0.01, ***P < 0.001. Bars indicate the mean. (E) Ratio of migrated cells in response to CCL19 and CCL21 and/or oxysterol (left) or CXCL13 (right). N = 10 or more from 3 different tonsils, paired t-test (right). *P < 0.05, **P < 0.01. Bars indicate the mean.
To investigate the role of PSGL-1 in responsiveness to CCL19 and CCL21, and further assess that of CCR7, we pretreated CD4+ T cells with either anti-CCR7 and/or PSGL-1 antibodies prior to migration assessment. We also assessed their migration to oxysterol, a ligand of EBI2, which is more abundant in T cell zones than follicles (79). Anti-CCR7 inhibited migration of PSGL-1hi PD-1hi CXCR5hi T cells (and as a control, naïve CD45RA+ T cells) toward CCL19 and CCL21, whereas blockade of PSGL-1 did not affect either population; however, dual antibody blockade more strongly inhibited migration of both populations compared to blocking CCR7 alone (Fig. 6C, upper panel). Both cell types also migrated more in response to CCL19 and CCL21 plus oxysterol than in response to either CCL19 and CCL21 or oxysterol alone. Collectively, these data indicated that PSGL-1hi PD-1hi CXCR5hi T cells migrate in response to extrafollicular and follicular chemokines, to a degree intermediate between T zone homing naïve T cells and follicular homing Tfh cells, suggesting they position themselves at a specific location, or perhaps niche, where the chemoattractive forces of extrafollicular and follicular chemokines are balanced. These data also supported the confocal microscopy images, in which PSGL-1hi PD-1hi CXCR5hi T cells were enriched at the tonsillar T-B border (Fig. 5, B and D).
We also investigated B cell chemokine expression and chemotaxis. Memory B cells, compared to their naïve counterparts, expressed more GPR183, with less CXCR5 and PSGL-1, with equivalent expression of CCR7 (Fig. 6D). Accordingly, memory B cells migrated more robustly in response to oxysterol (Fig. 6E, left), by contrast to naïve cells, which responded better to CXCL13 (Fig. 6E, right). The migratory capacity of naive and memory B cells in response to CCL19 and CCL21 plus oxysterol was equivalent, with the former also more responsive to CCL19 and CCL21 alone. Taken together, these data are consistent with the observation that memory B cells reside more distal from follicle than do naive B cells.
Discussion
Although multiple stimuli promote differentiation of human memory B cells into AFCs, including those delivered by cytokines and via Toll-like receptors, signals from CD4+ T cells more effectively promote such maturation (27, 33–36). Yet, the nature of the T cells that provide such help, and the mechanism by which they act, have not been well defined. We propose that these T helper cells are characterized by a unique cell surface markers, gene expression, response to chemokine signals, tonsillar location, and mechanism by which they drive memory B cells to mature into AFCs, distinguishing them from canonical B helper Tfh cells.
PSGL-1hi PD-1hi CXCR5hi T cells expressed surface molecules found on Tfh cells, including PD-1, CXCR5, and ICOS; however, they did not down-regulate PSGL-1, critical for emigration of nascent Tfh cells upon their activation in the T cell zone (43–45). This finding suggested these two T cell populations were distinct, while enabling a means for their separation microscopically and by flow cytometry, as well as their functional and molecular characterization. Tfh cells are located in follicles and GCs; by contrast, PSGL-1hi PD-1hi CXCR5hi T cells largely resided outside follicles, mainly at the T-B border. Such residence appeared to be mediated by chemokine receptor signaling, in that these cells were responsive to both T zone chemokines, CCL19 and CCL21 and to oxysterol, and to the B follicular chemokine, CXCL13 (79–83), suggesting that such signals balance them at the border of the T zone and follicle. In parallel, memory B cells migrated less well toward CXCL13 than did follicle-residing naïve B cells or Tfh cells, with co-residence with PSGL-1hi PD-1hi CXCR5hi T cells at the T-B border. We observed that, in functional terms, PSGL-1hi PD-1hi CXCR5hi T cells more robustly promoted Ig production upon co-culture with memory B cells than did Tfh cells, while the latter more strongly drove GC B cells toward Ig production. PSGL-1hi PD-1hi CXCR5hi T cells also had enhanced expression of genes comprising the E2F pathway, with Ki-67 staining revealing that they were more proliferative than Tfh cells. Tfh cells in general do not proliferate as well as other CD4+ T effector cells (84); since they expressed more PD-1 in our hands than PSGL-1hi PD-1hi CXCR5hi T cells (Fig. 3), the greater proliferative capacity of the latter cells might well be explained by the PD-1 inhibition of phosphorylation of retinoblastoma gene products, which suppress expression of E2F target genes (85). However, despite functional and geographical differences between PSGL-1hi PD-1hi CXCR5hi T cells and Tfh cells, we do not exclude the possibility that PSGL-1hi PD-1hi CXCR5hi T cells might be a transitory state in the Tfh differentiation program.
Human tonsillar CD4+ T cells that lack canonical markers of Tfh cells but that nonetheless provide help for B cell maturation to Ig production in vitro have been previously identified (11, 53). Are these cells distinct from the PSGL-1hi PD-1hi CXCR5hi T cells we describe? Ma and colleagues showed that tonsillar CD4+ CXCR5int T cells, stimulated with anti-CD3 and anti-CD28 antibodies for 8 days, promote naïve B cell maturation via IL-21 and ICOS (11). Although the gating strategy used in this work is distinct from that we employed, the described cells had low expression of PD-1 and CXCR5, along with modest production of IL-10 and IL-21, suggesting the CD4+ CXCR5int T cells are distinct from PSGL-1hi PD-1hi CXCR5hi T cells dissected herein. More recent work from Bentebibel and colleagues characterized tonsillar CD4+ ICOSlo CXCR5lo T cells that are specialized for B cell help at extrafollicular foci, driving naïve B cells to produce Ig to a greater degree than other CD4+ T cells subsets, help that was dependent upon ICOS, CD40L, IL-10, and IL-21 (53). These cells are located outside GCs and express BCL6 transcripts, albeit with minimal protein expression. Although these CD4+ ICOSlo CXCR5lo T cells share functional similarities with the PSGL-1hi PD-1hi CXCR5hi T cells we describe, the former appear specialized for promoting maturation of naïve B cells by contrast to the robust memory B cell help provided by PSGL-1hi PD-1hi CXCR5hi T cells in our hands. These differences might be attributable to differences between in vitro coculture systems - Bentebibel et al. cocultured T and B cells in the presence of staphylococcal enterotoxin for 8 days, whereas we cultured cells for only 5 days without addition of exogenous stimuli (84). Moreover, the PSGL-1hi PD-1hi CXCR5hi T cells we found are present alongside memory B cells at the tonsillar T-B border, while CD4+ ICOSlo CXCR5lo T cells are located mainly in the T cell zone (53). The latter also did not express BCL6 by flow cytometry in contrast to PSGL-1hi PD-1hi CXCR5hi T cells. Thus, it appears that the latter cells discussed herein are distinct from CD4+ ICOSlo CXCR5lo T cells phenotypically and geographically, as well as functionally. Nevertheless, because confocal images can distinguish only PD-1+ and PD-1−, the former cells at the T-B border might also include CD4+ ICOSlo CXCR5lo T cells. Further studies are needed to distinguish PSGL-1hi PD-1hi CXCR5hi T cells and CD4+ ICOSlo CXCR5lo T cells.
What is the relationship of tonsillar PSGL-1hi PD-1hi CXCR5hi T cells to circulating CD4+ CXCR5+ central memory T cells that promote maturation of naïve and memory B cells into AFCs via CD40L, IL-10, IL-21 and ICOS (86, 87)? Circulating CD4+ PD-1+ CXCR5+ central memory T cells from humans drive memory B cells to Ig secretion upon in vitro recall, leading to production of protective antibodies in the setting of HIV infection (52) and following influenza vaccination (88). Work in mice offers parallels, with in vivo studies describing emergence of CD4+ CXCR5+ central memory T cells after infection with lymphocytic choriomenigitis virus (LCMV) (42) or Listeria monocytogenes (40), which upon adoptive transfer locate to T cell zone in secondary lymphoid organs. After rechallenge, such cells preferentially assume a Tfh cell phenotype, gaining expression of PD-1, ICOS, and Bcl6, suggesting that PSGL-1hi PD-1hi CXCR5hi T cells arise from CD4+ CXCR5+ central memory T cells in vivo. Likewise, murine CD4+ CXCR5+ central memory T cells upregulate Bcl6 upon cognate interaction with memory B cells in a TCR:peptide-MHCII dependent manner at the T-B border, which in turn, provide potent help for differentiation into AFCs (89). Based upon these findings, we speculate that when CD4+ CXCR5+ central memory T cells are generated during the primary immune response, they enter secondary lymphoid organs within the T cell zone, migrating upon recall toward the T-B border for interaction with memory B cells.
An additional question raised by our studies is the fate of PSGL-1hi PD-1hi CXCR5hi T cells. He et al. (91) described circulating CD4+ CCR7lo PD-1hi CXCR5+ T cells in humans and mice that promoted antibody responses upon rechallenge. These cells also emerged in SLAM-associated protein (SAP)–deficient mice after immunization and patients with X-linked lymphoproliferative disease due to mutation of SH2D1A gene, suggesting an extrafollicular origin, given the critical role of SAP in Tfh differentiation and GC formation (90–93). Recently, Bentebibel et al. observed that influenza-specific CD4+ PD-1++ CXCR5+ CXCR3+ ICOS+ cells, capable of IL-10 and IL-21-dependent memory B cell help, emerge in the circulation after seasonal flu vaccination (75). Although these cells do not appear to represent bona fide PSGL-1hi PD-1hi CXCR5hi T cells as they are BCL6lo, they do share features with the latter, including surface marker expression, cytokine profiles, and the ability to promote memory B cell maturation toward AFCs, and therefore are candidates to represent the circulating progeny of PSGL-1hi PD-1hi CXCR5hi T cells that arise upon Ag stimulation (93).
In conclusion, we have identified a population of tonsillar B helper T cells that promote maturation of memory B cells in a cytokine- and contact-dependent manner, with localization in at the T-B border. They are distinct from classical follicular-resident Tfh cells, and likely play an important role in humoral recall responses in humans. Further characterization of these cells should expand our knowledge of human immune memory responses, and help guide development of more effective vaccines.
Supplementary Material
Supplemental Figure 1: PSGL-1hi PD-1hi CXCR5hi T cells proliferate via E2F pathway
Supplemental Figure 2: PSGL-1hi PD-1hi CXCR5hi T cells help memory B cells produce immunoglobulins (Igs) in a contact- and cytokine- (IL-10/21) dependent manner
Supplemental Table 1: Differentially expressed genes between Tfh cells and PSGL-1hi PD-1hi CXCR5hi T cells
Supplemental Table 2: Gene ontology terms from differentially expressed genes between Tfh cells and PSGL-1hi PD-1hi CXCR5hi T cells
Acknowledgments
We thank our patients for their participation. We also thank Drs. Jason Weinstein and Abhinav Seth, Department of Internal Medicine (Rheumatology) at the Yale University School of Medicine for critical reading of this manuscript.
This study was supported in part by American College of Rheumatology Research and Education Foundation Scientist Development Award (S.T.K.), and National Institutes of Health Grants R37 AR40072, P30 AR053495, and the Lupus Research Alliance (J.C.), and National Institutes of Health Grant R01 AR068994 (J.C. and P.G.G.).
Footnotes
The authors declare no competing financial interests.
References
- 1.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357:1903–1915. doi: 10.1056/NEJMoa066092. [DOI] [PubMed] [Google Scholar]
- 2.Naniche D, Garenne M, Rae C, Manchester M, Buchta R, Brodine SK, Oldstone MB. Decrease in measles virus-specific CD4 T cell memory in vaccinated subjects. J Infect Dis. 2004;190:1387–1395. doi: 10.1086/424571. [DOI] [PubMed] [Google Scholar]
- 3.Amanna IJ, Slifka MK. Contributions of humoral and cellular immunity to vaccine-induced protection in humans. Virology. 2011;411:206–215. doi: 10.1016/j.virol.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allen CD, Okada T, Cyster JG. Germinal-center organization and cellular dynamics. Immunity. 2007;27:190–202. doi: 10.1016/j.immuni.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 6.Good-Jacobson KL, Shlomchik MJ. Plasticity and heterogeneity in the generation of memory B cells and long-lived plasma cells: the influence of germinal center interactions and dynamics. Journal of immunology. 2010;185:3117–3125. doi: 10.4049/jimmunol.1001155. [DOI] [PubMed] [Google Scholar]
- 7.Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunological reviews. 2012;247:52–63. doi: 10.1111/j.1600-065X.2012.01124.x. [DOI] [PubMed] [Google Scholar]
- 8.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, Forster R. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J Exp Med. 2000;192:1545–1552. doi: 10.1084/jem.192.11.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bryant VL, Ma CS, Avery DT, Li Y, Good KL, Corcoran LM, de Waal Malefyt R, Tangye SG. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. Journal of immunology. 2007;179:8180–8190. doi: 10.4049/jimmunol.179.12.8180. [DOI] [PubMed] [Google Scholar]
- 10.Schmitt N, Morita R, Bourdery L, Bentebibel SE, Zurawski SM, Banchereau J, Ueno H. Human dendritic cells induce the differentiation of interleukin-21-producing T follicular helper-like cells through interleukin-12. Immunity. 2009;31:158–169. doi: 10.1016/j.immuni.2009.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma CS, Suryani S, Avery DT, Chan A, Nanan R, Santner-Nanan B, Deenick EK, Tangye SG. Early commitment of naive human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol Cell Biol. 2009;87:590–600. doi: 10.1038/icb.2009.64. [DOI] [PubMed] [Google Scholar]
- 12.Craft JE. Follicular helper T cells in immunity and systemic autoimmunity. Nature reviews Rheumatology. 2012;8:337–347. doi: 10.1038/nrrheum.2012.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Good-Jacobson KL, Szumilas CG, Chen L, Sharpe AH, Tomayko MM, Shlomchik MJ. PD-1 regulates germinal center B cell survival and the formation and affinity of long-lived plasma cells. Nature immunology. 2010;11:535–542. doi: 10.1038/ni.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi Y, Dutta PR, Cerasoli DM, Kelsoe G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl)acetyl. V. Affinity maturation develops in two stages of clonal selection. J Exp Med. 1998;187:885–895. doi: 10.1084/jem.187.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blink EJ, Light A, Kallies A, Nutt SL, Hodgkin PD, Tarlinton DM. Early appearance of germinal center-derived memory B cells and plasma cells in blood after primary immunization. J Exp Med. 2005;201:545–554. doi: 10.1084/jem.20042060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manz RA, Thiel A, Radbruch A. Lifetime of plasma cells in the bone marrow. Nature. 1997;388:133–134. doi: 10.1038/40540. [DOI] [PubMed] [Google Scholar]
- 17.DiLillo DJ, Hamaguchi Y, Ueda Y, Yang K, Uchida J, Haas KM, Kelsoe G, Tedder TF. Maintenance of long-lived plasma cells and serological memory despite mature and memory B cell depletion during CD20 immunotherapy in mice. Journal of immunology. 2008;180:361–371. doi: 10.4049/jimmunol.180.1.361. [DOI] [PubMed] [Google Scholar]
- 18.Slifka MK, Antia R, Whitmire JK, Ahmed R. Humoral immunity due to long-lived plasma cells. Immunity. 1998;8:363–372. doi: 10.1016/s1074-7613(00)80541-5. [DOI] [PubMed] [Google Scholar]
- 19.Slifka MK, Ahmed R. Long-lived plasma cells: a mechanism for maintaining persistent antibody production. Curr Opin Immunol. 1998;10:252–258. doi: 10.1016/s0952-7915(98)80162-3. [DOI] [PubMed] [Google Scholar]
- 20.Tangye SG, Tarlinton DM. Memory B cells: effectors of long-lived immune responses. Eur J Immunol. 2009;39:2065–2075. doi: 10.1002/eji.200939531. [DOI] [PubMed] [Google Scholar]
- 21.Dogan I, Bertocci B, Vilmont V, Delbos F, Megret J, Storck S, Reynaud CA, Weill JC. Multiple layers of B cell memory with different effector functions. Nature immunology. 2009;10:1292–1299. doi: 10.1038/ni.1814. [DOI] [PubMed] [Google Scholar]
- 22.Sallusto F, Lanzavecchia A, Araki K, Ahmed R. From vaccines to memory and back. Immunity. 2010;33:451–463. doi: 10.1016/j.immuni.2010.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Crotty S, Felgner P, Davies H, Glidewell J, Villarreal L, Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. Journal of immunology. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 24.Pinna D, Corti D, Jarrossay D, Sallusto F, Lanzavecchia A. Clonal dissection of the human memory B-cell repertoire following infection and vaccination. Eur J Immunol. 2009;39:1260–1270. doi: 10.1002/eji.200839129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernasconi NL, Onai N, Lanzavecchia A. A role for Toll-like receptors in acquired immunity: up-regulation of TLR9 by BCR triggering in naive B cells and constitutive expression in memory B cells. Blood. 2003;101:4500–4504. doi: 10.1182/blood-2002-11-3569. [DOI] [PubMed] [Google Scholar]
- 26.Good KL, Avery DT, Tangye SG. Resting human memory B cells are intrinsically programmed for enhanced survival and responsiveness to diverse stimuli compared to naive B cells. Journal of immunology. 2009;182:890–901. doi: 10.4049/jimmunol.182.2.890. [DOI] [PubMed] [Google Scholar]
- 27.Tangye SG, Avery DT, Deenick EK, Hodgkin PD. Intrinsic differences in the proliferation of naive and memory human B cells as a mechanism for enhanced secondary immune responses. Journal of immunology. 2003;170:686–694. doi: 10.4049/jimmunol.170.2.686. [DOI] [PubMed] [Google Scholar]
- 28.McHeyzer-Williams LJ, McHeyzer-Williams MG. Antigen-specific memory B cell development. Annu Rev Immunol. 2005;23:487–513. doi: 10.1146/annurev.immunol.23.021704.115732. [DOI] [PubMed] [Google Scholar]
- 29.Mamani-Matsuda M, Cosma A, Weller S, Faili A, Staib C, Garcon L, Hermine O, Beyne-Rauzy O, Fieschi C, Pers JO, Arakelyan N, Varet B, Sauvanet A, Berger A, Paye F, Andrieu JM, Michel M, Godeau B, Buffet P, Reynaud CA, Weill JC. The human spleen is a major reservoir for long-lived vaccinia virus-specific memory B cells. Blood. 2008;111:4653–4659. doi: 10.1182/blood-2007-11-123844. [DOI] [PubMed] [Google Scholar]
- 30.Ettinger R, Sims GP, Robbins R, Withers D, Fischer RT, Grammer AC, Kuchen S, Lipsky PE. IL-21 and BAFF/BLyS synergize in stimulating plasma cell differentiation from a unique population of human splenic memory B cells. Journal of immunology. 2007;178:2872–2882. doi: 10.4049/jimmunol.178.5.2872. [DOI] [PubMed] [Google Scholar]
- 31.Liu YJ, Barthelemy C, de Bouteiller O, Arpin C, Durand I, Banchereau J. Memory B cells from human tonsils colonize mucosal epithelium and directly present antigen to T cells by rapid up-regulation of B7-1 and B7-2. Immunity. 1995;2:239–248. doi: 10.1016/1074-7613(95)90048-9. [DOI] [PubMed] [Google Scholar]
- 32.Ellyard JI, Avery DT, Phan TG, Hare NJ, Hodgkin PD, Tangye SG. Antigen-selected, immunoglobulin-secreting cells persist in human spleen and bone marrow. Blood. 2004;103:3805–3812. doi: 10.1182/blood-2003-09-3109. [DOI] [PubMed] [Google Scholar]
- 33.Lanzavecchia A, Parodi B, Celada F. Activation of human B lymphocytes: frequency of antigen-specific B cells triggered by alloreactive or by antigen-specific T cell clones. Eur J Immunol. 1983;13:733–738. doi: 10.1002/eji.1830130908. [DOI] [PubMed] [Google Scholar]
- 34.Lanzavecchia A, Parodi B. In vitro stimulation of IgE production at a single precursor level by human alloreactive T helper clones. Clin Exp Immunol. 1984;55:197–203. [PMC free article] [PubMed] [Google Scholar]
- 35.Kindler V, Zubler RH. Memory, but not naive, peripheral blood B lymphocytes differentiate into Ig-secreting cells after CD40 ligation and costimulation with IL-4 and the differentiation factors IL-2, IL-10, and IL-3. Journal of immunology. 1997;159:2085–2090. [PubMed] [Google Scholar]
- 36.Bernasconi NL, Traggiai E, Lanzavecchia A. Maintenance of serological memory by polyclonal activation of human memory B cells. Science. 2002;298:2199–2202. doi: 10.1126/science.1076071. [DOI] [PubMed] [Google Scholar]
- 37.Good KL, Bryant VL, Tangye SG. Kinetics of human B cell behavior and amplification of proliferative responses following stimulation with IL-21. Journal of immunology. 2006;177:5236–5247. doi: 10.4049/jimmunol.177.8.5236. [DOI] [PubMed] [Google Scholar]
- 38.Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, James J, Air GM, Capra JD, Ahmed R, Wilson PC. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kerfoot SM, Yaari G, Patel JR, Johnson KL, Gonzalez DG, Kleinstein SH, Haberman AM. Germinal center B cell and T follicular helper cell development initiates in the interfollicular zone. Immunity. 2011;34:947–960. doi: 10.1016/j.immuni.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, Jenkins MK. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 2011;35:583–595. doi: 10.1016/j.immuni.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tubo NJ, Pagan AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, Jenkins MK. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 2013;153:785–796. doi: 10.1016/j.cell.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hale JS, Youngblood B, Latner DR, Mohammed AU, Ye L, Akondy RS, Wu T, Iyer SS, Ahmed R. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 2013;38:805–817. doi: 10.1016/j.immuni.2013.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Veerman KM, Williams MJ, Uchimura K, Singer MS, Merzaban JS, Naus S, Carlow DA, Owen P, Rivera-Nieves J, Rosen SD, Ziltener HJ. Interaction of the selectin ligand PSGL-1 with chemokines CCL21 and CCL19 facilitates efficient homing of T cells to secondary lymphoid organs. Nature immunology. 2007;8:532–539. doi: 10.1038/ni1456. [DOI] [PubMed] [Google Scholar]
- 44.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, Dong X, Odegard JM, Kaech SM, Dent AL, Crotty S, Craft J. In vivo regulation of Bcl6 and T follicular helper cell development. Journal of immunology. 2010;185:313–326. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odegard JM, Marks BR, DiPlacido LD, Poholek AC, Kono DH, Dong C, Flavell RA, Craft J. ICOS-dependent extrafollicular helper T cells elicit IgG production via IL-21 in systemic autoimmunity. J Exp Med. 2008;205:2873–2886. doi: 10.1084/jem.20080840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 47.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic acids research. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–258. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 49.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 50.Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, Nakatsukasa H, Neelapu SS, Chen W, Clevers H, Tian Q, Qi H, Wei L, Dong C. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014 doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hatzi K, Nance JP, Kroenke MA, Bothwell M, Haddad EK, Melnick A, Crotty S. BCL6 orchestrates Tfh cell differentiation via multiple distinct mechanisms. J Exp Med. 2015;212:539–553. doi: 10.1084/jem.20141380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A, Haddad EK, Poignard P, Crotty S. Human Circulating PD-1CXCR3CXCR5 Memory Tfh Cells Are Highly Functional and Correlate with Broadly Neutralizing HIV Antibody Responses. Immunity. 2013 doi: 10.1016/j.immuni.2013.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bentebibel SE, Schmitt N, Banchereau J, Ueno H. Human tonsil B-cell lymphoma 6 (BCL6)-expressing CD4+ T-cell subset specialized for B-cell help outside germinal centers. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:E488–497. doi: 10.1073/pnas.1100898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–663. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 55.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, Crotty S. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34:932–946. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hori S, Nomura T, Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. Science. 2003;299:1057–1061. doi: 10.1126/science.1079490. [DOI] [PubMed] [Google Scholar]
- 57.Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, Fan HM, Liu ZM, Neelapu SS, Dong C. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, Fagarasan S, Liston A, Smith KG, Vinuesa CG. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sage PT, Tan CL, Freeman GJ, Haigis M, Sharpe AH. Defective TFH Cell Function and Increased TFR Cells Contribute to Defective Antibody Production in Aging. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallin EF, Jolly EC, Suchanek O, Bradley JA, Espeli M, Jayne DR, Linterman MA, Smith KG. Human T-follicular helper and T-follicular regulatory cell maintenance is independent of germinal centers. Blood. 2014;124:2666–2674. doi: 10.1182/blood-2014-07-585976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 62.King IL, Mohrs M. IL-4-producing CD4+ T cells in reactive lymph nodes during helminth infection are T follicular helper cells. J Exp Med. 2009;206:1001–1007. doi: 10.1084/jem.20090313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reinhardt RL, Liang HE, Locksley RM. Cytokine-secreting follicular T cells shape the antibody repertoire. Nature immunology. 2009;10:385–393. doi: 10.1038/ni.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, Barnett B, Dent AL, Craft J, Crotty S. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang YH, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu D, Rao S, Tsai LM, Lee SK, He Y, Sutcliffe EL, Srivastava M, Linterman M, Zheng L, Simpson N, Ellyard JI, Parish IA, Ma CS, Li QJ, Parish CR, Mackay CR, Vinuesa CG. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 67.Kim CH, Rott LS, Clark-Lewis I, Campbell DJ, Wu L, Butcher EC. Subspecialization of CXCR5+ T cells: B helper activity is focused in a germinal center-localized subset of CXCR5+ T cells. J Exp Med. 2001;193:1373–1381. doi: 10.1084/jem.193.12.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kroenke MA, Eto D, Locci M, Cho M, Davidson T, Haddad EK, Crotty S. Bcl6 and Maf cooperate to instruct human follicular helper CD4 T cell differentiation. Journal of immunology. 2012;188:3734–3744. doi: 10.4049/jimmunol.1103246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choe J, Choi YS. IL-10 interrupts memory B cell expansion in the germinal center by inducing differentiation into plasma cells. Eur J Immunol. 1998;28:508–515. doi: 10.1002/(SICI)1521-4141(199802)28:02<508::AID-IMMU508>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 70.Conti P, Kempuraj D, Kandere K, Di Gioacchino M, Barbacane RC, Castellani ML, Felaco M, Boucher W, Letourneau R, Theoharides TC. IL-10, an inflammatory/inhibitory cytokine, but not always. Immunol Lett. 2003;86:123–129. doi: 10.1016/s0165-2478(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 71.Yoon SO, Zhang X, Berner P, Choi YS. IL-21 and IL-10 have redundant roles but differential capacities at different stages of Plasma Cell generation from human Germinal Center B cells. J Leukoc Biol. 2009;86:1311–1318. doi: 10.1189/jlb.0409268. [DOI] [PubMed] [Google Scholar]
- 72.Cyster JG. Chemokines, sphingosine-1-phosphate, and cell migration in secondary lymphoid organs. Annu Rev Immunol. 2005;23:127–159. doi: 10.1146/annurev.immunol.23.021704.115628. [DOI] [PubMed] [Google Scholar]
- 73.Segerer S, Banas B, Wornle M, Schmid H, Cohen CD, Kretzler M, Mack M, Kiss E, Nelson PJ, Schlondorff D, Grone HJ. CXCR3 is involved in tubulointerstitial injury in human glomerulonephritis. Am J Pathol. 2004;164:635–649. doi: 10.1016/S0002-9440(10)63152-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Locci M, Wu JE, Arumemi F, Mikulski Z, Dahlberg C, Miller AT, Crotty S. Activin A programs the differentiation of human TFH cells. Nature immunology. 2016;17:976–984. doi: 10.1038/ni.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bentebibel SE, Lopez S, Obermoser G, Schmitt N, Mueller C, Harrod C, Flano E, Mejias A, Albrecht RA, Blankenship D, Xu H, Pascual V, Banchereau J, Garcia-Sastre A, Palucka AK, Ramilo O, Ueno H. Induction of ICOS+CXCR3+CXCR5+ TH cells correlates with antibody responses to influenza vaccination. Sci Transl Med. 2013;5:176ra132. doi: 10.1126/scitranslmed.3005191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Agematsu K. Memory B cells and CD27. Histol Histopathol. 2000;15:573–576. doi: 10.14670/HH-15.573. [DOI] [PubMed] [Google Scholar]
- 77.Steiniger B, Timphus EM, Jacob R, Barth PJ. CD27+ B cells in human lymphatic organs: re-evaluating the splenic marginal zone. Immunology. 2005;116:429–442. doi: 10.1111/j.1365-2567.2005.02242.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang C, Thudium KB, Han M, Wang XT, Huang H, Feingersh D, Garcia C, Wu Y, Kuhne M, Srinivasan M, Singh S, Wong S, Garner N, Leblanc H, Bunch RT, Blanset D, Selby MJ, Korman AJ. In vitro characterization of the anti-PD-1 antibody nivolumab, BMS-936558, and in vivo toxicology in non-human primates. Cancer Immunol Res. 2014;2:846–856. doi: 10.1158/2326-6066.CIR-14-0040. [DOI] [PubMed] [Google Scholar]
- 79.Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, Guerini D, Baumgarten BU, Roggo S, Wen B, Knochenmuss R, Noel S, Gessier F, Kelly LM, Vanek M, Laurent S, Preuss I, Miault C, Christen I, Karuna R, Li W, Koo DI, Suply T, Schmedt C, Peters EC, Falchetto R, Katopodis A, Spanka C, Roy MO, Detheux M, Chen YA, Schultz PG, Cho CY, Seuwen K, Cyster JG, Sailer AW. Oxysterols direct immune cell migration via EBI2. Nature. 2011;475:524–527. doi: 10.1038/nature10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pereira JP, Kelly LM, Xu Y, Cyster JG. EBI2 mediates B cell segregation between the outer and centre follicle. Nature. 2009;460:1122–1126. doi: 10.1038/nature08226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401:708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- 82.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, Moser B. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J Exp Med. 2000;192:1553–1562. doi: 10.1084/jem.192.11.1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Haynes NM, Allen CD, Lesley R, Ansel KM, Killeen N, Cyster JG. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. Journal of immunology. 2007;179:5099–5108. doi: 10.4049/jimmunol.179.8.5099. [DOI] [PubMed] [Google Scholar]
- 84.Rasheed AU, Rahn HP, Sallusto F, Lipp M, Muller G. Follicular B helper T cell activity is confined to CXCR5(hi)ICOS(hi) CD4 T cells and is independent of CD57 expression. Eur J Immunol. 2006;36:1892–1903. doi: 10.1002/eji.200636136. [DOI] [PubMed] [Google Scholar]
- 85.Patsoukis N, Brown J, Petkova V, Liu F, Li L, Boussiotis VA. Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci Signal. 2012;5:ra46. doi: 10.1126/scisignal.2002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N, Lavecchio EM, Punaro M, Pascual V, Banchereau J, Ueno H. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chevalier N, Jarrossay D, Ho E, Avery DT, Ma CS, Yu D, Sallusto F, Tangye SG, Mackay CR. CXCR5 expressing human central memory CD4 T cells and their relevance for humoral immune responses. Journal of immunology. 2011;186:5556–5568. doi: 10.4049/jimmunol.1002828. [DOI] [PubMed] [Google Scholar]
- 88.Bentebibel SE, Khurana S, Schmitt N, Kurup P, Mueller C, Obermoser G, Palucka AK, Albrecht RA, Garcia-Sastre A, Golding H, Ueno H. ICOS(+)PD-1(+)CXCR3(+) T follicular helper cells contribute to the generation of high-avidity antibodies following influenza vaccination. Scientific reports. 2016;6:26494. doi: 10.1038/srep26494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ise W, Inoue T, McLachlan JB, Kometani K, Kubo M, Okada T, Kurosaki T. Memory B cells contribute to rapid Bcl6 expression by memory follicular helper T cells. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11792–11797. doi: 10.1073/pnas.1404671111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McCausland MM, Yusuf I, Tran H, Ono N, Yanagi Y, Crotty S. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. Journal of immunology. 2007;178:817–828. doi: 10.4049/jimmunol.178.2.817. [DOI] [PubMed] [Google Scholar]
- 91.He J, Tsai LM, Leong YA, Hu X, Ma CS, Chevalier N, Sun X, Vandenberg K, Rockman S, Ding Y, Zhu L, Wei W, Wang C, Karnowski A, Belz GT, Ghali JR, Cook MC, Riminton DS, Veillette A, Schwartzberg PL, Mackay F, Brink R, Tangye SG, Vinuesa CG, Mackay CR, Li Z, Yu D. Circulating Precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T Cells Indicate Tfh Cell Activity and Promote Antibody Responses upon Antigen Reexposure. Immunity. 2013;39:770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 92.Tsai LM, Yu D. Follicular helper T-cell memory: establishing new frontiers during antibody response. Immunol Cell Biol. 2014;92:57–63. doi: 10.1038/icb.2013.68. [DOI] [PubMed] [Google Scholar]
- 93.Schmitt N, Bentebibel SE, Ueno H. Phenotype and functions of memory Tfh cells in human blood. Trends Immunol. 2014;35:436–442. doi: 10.1016/j.it.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: PSGL-1hi PD-1hi CXCR5hi T cells proliferate via E2F pathway
Supplemental Figure 2: PSGL-1hi PD-1hi CXCR5hi T cells help memory B cells produce immunoglobulins (Igs) in a contact- and cytokine- (IL-10/21) dependent manner
Supplemental Table 1: Differentially expressed genes between Tfh cells and PSGL-1hi PD-1hi CXCR5hi T cells
Supplemental Table 2: Gene ontology terms from differentially expressed genes between Tfh cells and PSGL-1hi PD-1hi CXCR5hi T cells


