Abstract
Dopamine transporters (DAT) are implicated in the pathogenesis and treatment of attention deficit hyperactivity disorder (ADHD) and are upregulated by chronic treatment with methylphenidate, commonly prescribed for ADHD. Methylation of the DAT1 gene in brain and blood has been associated with DAT expression in rodents’ brains. Here we tested the association between methylation of the DAT1 promoter derived from blood and DAT availability in the striatum of unmedicated ADHD adult participants and in that of healthy age-matched controls (HC) using Positron Emission Tomography (PET) and [11C]cocaine. Results showed no between-group differences in DAT1 promoter methylation or striatal DAT availability. However, the degree of methylation in the promoter region of DAT1 correlated negatively with DAT availability in caudate in ADHD participants only. DAT availability in VS correlated with inattention scores in ADHD participants. We verified in a post-mortem cohort with ADHD diagnosis and without, that DAT1 promoter methylation in peripheral blood correlated positively with DAT1 promoter methylation extracted from substantia nigra (SN) in both groups. In the cohort without ADHD diagnosis, DAT1 gene expression in SN further correlated positively with DAT protein expression in caudate; however, the sample size of the cohort with ADHD was insufficient to investigate DAT1 and DAT expression levels. Overall, these findings suggest that peripheral DAT1 promoter methylation may be predictive of striatal DAT availability in adults with ADHD. Due to the small sample size, more work is needed to validate whether DAT1 methylation in blood predicts DAT1 methylation in SN in ADHD and controls.
Keywords: DAT1/SLC6A3, Epigenetics, striatum, PET, postmortem
Graphical abstract
DAT1 promoter methylation in blood correlated negatively with dopamine transporter (DAT) availability in caudate in adults with ADHD. In a post-mortem cohort with and without ADHD diagnosis, DAT1 promoter methylation in blood correlated positively with DAT1 promoter methylation in substantia nigra (SN), and DAT1 gene expression in SN correlated positively with DAT protein expression in caudate. Peripheral DAT1 promoter methylation may thus be predictive of striatal DAT availability in ADHD.
Introduction
Attention deficit hyperactivity disorder (ADHD) is a neurodevelopmental disorder characterized by symptoms of inattention, hyperactivity, and impulsivity. These symptoms are thought to be caused, in part, by dysfunction in the striatal dopaminergic system. ADHD is associated with a blunted dopamine (DA) reward/motivation pathway, including changes in dopamine transporter (DAT) and dopamine D2/D3 receptor (DRD2) availability in the striatum (Dougherty et al., 1999; Spencer et al., 2005; Spencer et al., 2007; Volkow et al., 2007; Volkow et al., 2009b; Volkow et al., 2011). The stimulant medications methylphenidate and amphetamine are effective treatments for ADHD, and interact with the DAT either by blocking it and preventing DA reuptake or by facilitating the release of DA, respectively (Huang et al., 2011). We have shown that therapeutic doses of oral methylphenidate blocked DAT in the human brain and increased extracellular DA in the striatum, which in turn restored motivation and helped sustain attention (Volkow et al., 2002a; Volkow et al., 2009b; Volkow et al., 2011). Furthermore, chronic methylphenidate treatment of adults with ADHD was shown to upregulate DAT levels in the striatum (Wang et al., 2013), which might be a long lasting effect of the treatment since previously medicated adults with ADHD showed higher DAT availability than never medicated patients (Fusar-Poli et al., 2012). This may also explain why some studies have found higher DAT availability in adults with ADHD than in controls (Volkow et al., 2011).
ADHD has a strong genetic basis with a heritability estimate (h2) of 80% (Swanson et al., 2000). However, specific genes implicated in the disorder have remained elusive. The most extensively studied biomarker for ADHD is a polymorphism in the DAT gene (DAT1/SLC6A3), a 40-bp variable number tandem repeat (VNTR) located in the 3′-untranslated region of the gene (3′UTR), which produces two common alleles, designated as 9- and 10-repeats (9-R and 10-R) (Cornish et al., 2005). ADHD has been associated with the 10-R allele in some studies (Swanson et al., 2000; Cornish et al., 2005), but others have been unable to replicate this finding (Swanson et al., 2000; Langley et al., 2005; meta-analyzed by Costa et al., 2011). Similarly, there is evidence that the 7-R allele of the dopamine D4 receptor gene (DRD4) is associated with ADHD, but this has also not been universally replicated (LaHoste et al., 1996; Hawi et al., 2000; Swanson et al., 2000). Thus, these “risk allele” biomarkers commonly found in the general population lack validity and specificity for ADHD diagnosis (Swanson et al., 2000; Langley et al., 2005).
Genetic polymorphisms are static and may not sufficiently explain the gene/environment interactions for the development of ADHD. Epigenetic markers, such as methylation of DAT1, may account for underlying mechanisms of how gene and environment interact and how their interaction affects brain development (Wiers, 2012; Nikolova & Hariri, 2015; Lancaster et al., 2018). Recently, DAT1 methylation derived from blood was associated with responses to methylphenidate treatment in ADHD, including symptoms of hyperactivity and impulsivity (Ding et al., 2017). In preclinical studies in monkeys, DAT1 methylation in peripheral blood was associated with DAT availability in the basal ganglia and impulsivity (Rajala et al., 2014). Further, methylation of DAT1 extracted from striatum has been negatively correlated with DAT protein expression in rats with ADHD phenotype (Kim et al., 2014). It remains unknown, however, whether methylation of DAT1 in blood is associated with DAT availability or protein expression in the human brain and with symptoms of inattention or hyperactivity in ADHD patients.
Here we tested the association between DAT1 promoter methylation in peripheral blood and DAT availability in the brain of unmedicated adults with ADHD (n=13) and healthy volunteers (n=34). Participants underwent a scan with Positron Emission Tomography (PET) using [11C]Cocaine as a DAT radioligand (Volkow et al., 1998) to measure DAT availability (Wang et al., 2013). There were no group differences in DAT availability between cases and controls, confirming previous findings (Wang et al., 2013). Participants provided blood for analyses of DAT1 methylation and reported the presence and severity of ADHD symptoms assessed by the Conners’ Adult ADHD Rating Scale (CAARS). We further corroborated our findings in post-mortem brains of deceased individuals with a diagnosis of ADHD and of those without psychiatric diagnoses (controls). That is, we aimed to investigate in a post-mortem sample whether DAT1 methylation in blood is predictive for DAT1 methylation in substantia nigra (SN). Since dopaminergic neurons are projected from SN to striatum, we measured DAT1 methylation and gene expression (mRNA) transcribed from the DAT1 in SN and DAT protein expression in striatum. Based on the measurement, we tested the associations between the DAT1 methylation and DAT1 expression profiles in SN and striatum.
Although the effects of gene methylation on gene expression are complex, gene methylation is generally seen as a ‘silencing’ epigenetic mark. That is, various studies have found that methylation of CpG islands in the promoter area have an inhibitory effect on transcription initiation, resulting in reduced gene expression (Brenet et al., 2011; Jones, 2012). Here, we therefore predicted (1) inverse correlations between methylation in DAT1 promoter regions derived from blood and brain and DAT availability measured with PET in the ADHD and control groups. We further expected (2) positive correlations between DAT1 methylation and ADHD symptoms in ADHD patients, given previous studies showing associations of DAT1 methylation derived from blood with symptoms of hyperactivity and impulsivity in ADHD (Ding et al., 2017), and with impulsivity in monkeys (Rajala et al., 2014). Few studies have explored whether methylation in peripheral tissues such as blood are predictors of methylation in brain. Here, we used the post-mortem cohort to verify that methylation levels in blood and brain are significantly related. Assuming the “mirror site model” that peripheral and neural tissue have similar methylation patterns (Aberg et al., 2013), we predicted that in the post-mortem cohort (3) DAT1 methylation in blood would be positively associated with DAT1 methylation in SN. Based on our previous assumption that methylation has an inhibitory effect on gene expression (Brenet et al., 2011; Jones, 2012), we also predicted that (4) DAT1 methylation in SN would be negatively correlated with DAT1 gene and DAT protein expression in SN and striatum, respectively.
Materials and Methods
Participants
Participants were selected from a PET scan database from Brookhaven National Laboratory (BNL). The sample consisted of 13 unmedicated adult ADHD subjects who participated in a previous PET study (Wang et al., 2013), and 34 healthy adults who served as healthy controls in PET imaging studies with [11C]Cocaine (Volkow et al., 2009a; Wang et al., 2013; Volkow et al., 2015). Participants provided written informed consent to the imaging and genetic studies and were paid for study participation. The studies were approved by the local Institutional Review Board (Committee on Research Involving Human Subjects, Stony Brook University, New York) and carried out at BNL. Table 1 summarizes demographic characteristics. Groups were matched for age, education, BMI, and socioeconomic status (all p>.33, ns). There were, however, more females in the ADHD group (n=7) than the healthy group (n=1) (χ2=17.7, p<.001). ADHD participants were recruited from a variety of sources, including clinical referral, and at least two clinicians interviewed the patients to ensure that they met DSM-IV diagnostic criteria for ADHD, as evidenced by the presence of at least 6 of 9 inattention symptoms (with or without 6 of 9 hyperactive/impulsive symptoms) as ascertained with a semi-structured interview using DSM-IV criteria. In addition, evidence was required from each subject’s history that some symptoms of ADHD were present in childhood (before age seven), even when the diagnosis was not made until adulthood. Participants were excluded if they had a prior history of more than three months of medication treatment for ADHD. They were also excluded it they had a present or history of substance use disorder (except tobacco). Smoking status was assessed with self-report and in our final sample there were 1 smoker and 1 ex-smoker in each group, thus no effect of smoking status on group (χ2=1.1, p=0.6). Exclusion criteria also included present or past history of psychiatric disease (axis I or II diagnosis other than ADHD), or neurological disease, medical conditions that may alter cerebral function (i.e., cardiovascular, endocrinological, oncological, or autoimmune diseases), current use of prescribed or over the counter medications, and/or head trauma with loss of consciousness of more than 30 minutes. Control participants were recruited from advertisements in the local newspapers; exclusion criteria other than allowance for ADHD were the same as for ADHD participants. Urine drug screens were obtained on all participants on the day of the PET scan to check for psychoactive drug use.
Table 1.
Demographics and clinical characteristics of ADHD participants and healthy controls
| ADHD (n=13) |
Healthy controls (n=34) |
||||
|---|---|---|---|---|---|
| Characteristic | Mean | SD | Mean | SD | p-value |
| Age, years | 34.1 | 8.2 | 35.2 | 6.0 | 0.61 |
| Years of education |
14.6 | 2.1 | 15.2 | 1.8 | 0.43 |
| Gender | 7 female | 54% | 1 female | 3% | <0.001 |
| BMI | 26.6 | 4.9 | 25.9 | 2.9 | 0.56 |
| VIQ | 96.1 | 4.6 | 93.4 | 6.5 | 0.24 |
| CAARS A inattention |
67.7 | 9.6 | 45.6 b | 7.7 | <0.001 |
| CAARS B hyperactivity |
63.5 | 10.9 | 44.2 b | 9.4 | <0.001 |
| CAARS C impulsivity |
56.2 | 13.4 | 44.8 b | 5.4 | 0.001 |
| BPND Caudate | 0.81 | 0.13 | 0.75 | 0.12 | 0.14 |
| BPND Putamen | 0.96 | 0.15 | 0.91 | 0.13 | 0.24 |
| BPND VS | 0.87 | 0.17 | 0.79 | 0.15 | 0.14 |
Abbreviations: ADHD attention deficit hyperactivity disorder, BMI body mass index, BPND nondisplaceable binding potential, CAARS Conners Adult Attention Rating Scale, VIQ verbal IQ, VS ventral striatum.
Clinical symptoms of ADHD were assessed using the Conners’ Adult Attention Rating Scale (CAARS) long version (Conners, 1998), which provides self-assessment of ADHD symptoms on a 0 (very minimal) to 3 (very much) point scale.
Positron Emission Tomography
Imaging data with [11C]Cocaine associated with each genetic sample were retrieved from the imaging dataset of the BNL Brain Imaging Center. All PET scans used in the current study were performed on a Siemens, HR+ scanner in 3D mode (resolution 4.5×4.5×4.5 mm). The procedures for subjects positioning and scanning protocols were described previously (Wang et al., 2013) along with the analytical approach for quantifying DAT availability (Fowler et al., 2001; Volkow et al., 2002b). In brief, dynamic scans were obtained after injection of 4–10 mCi of [11C]Cocaine (specific activity 0.5–1.5 Ci/mM at end of bombardment) for a total of 60 minutes. Arterial blood was obtained throughout the procedure to measure the concentration of unchanged [11C]Cocaine in plasma for quantification of DAT availability. Regions of interest (ROI) including caudate, putamen, and VS in both hemispheres were drawn directly on an emission image that represented the sum of images obtained during 10–54 min scans. Right and left cerebellar regions were drawn in three planes, 1.0, 1.4, and 1.8 cm above the canthomeatal line. ROIs were then projected onto the dynamic images to generate time activity curves for striatum and cerebellum. Average values for the striatal and cerebellar regions were computed from three slices, and values from the two hemispheres were also averaged. The cerebellum, where DAT expression is almost negligible, was used as reference region. Average values for the striatal and cerebellar regions were computed from the different slices where the regions were obtained. The time activity curves for tissue concentration along with the time activity curves for unchanged tracer in plasma were used to calculate the distribution volume (ml/gm) and the blood to tissue transport constant (K1) in striatum and cerebellum using a graphical analyses technique for reversible systems (Logan Plots) (Logan et al., 1996). The combination parameter Bmax/KD, which is a measure of free transporter concentration (binding potential; BPND), was calculated as the ratio of the distribution volume in striatum to that in cerebellum minus 1. This was used to quantify DAT availability, i.e., the number of transporters that are free to bind the radiotracer, for caudate, putamen and VS.
DNA Extraction and Methylation Analysis
Genomic DNA was extracted from participants’ blood samples and from the substantia nigra in post-mortem brain samples. DNA methylation levels of 9 functional clusters (4 in promoter; assay IDs: ADS2165-RS1, ADS2165-RS2, ADS2795-FS1, ADS2795-FS2) across DAT1 were assessed using quantitative bisulfite pyrosequencing by EpigenDx following EpigenDx protocol. DNA methylation kits (Zymo Research, Inc., CA) were used to treat 500 ng of genomic DNA for each analysis and the manufacturer’s protocol was followed for DNA purification eluting samples to 46 μL. One μL of bisulfite treated DNA was combined with .2 μM of each primer for the polymerase chain reaction (PCR). One of the primers was labeled with biotin and purified with HPLC for the purpose of purifying the final PCR product with Sepharose beads. Next, the PCR product was bound to Streptavidin Sepharose HP (GE Healthcare Life Sciences) and the now immobilized PCR products were purified, washed, denatured with a .2 μM NaOH solution. Following the manufacturer’s protocol, the Pyrosequencing Vacuum Prep Tool (Pyrosequencing, Qiagen) was used for rewashing, .5 μM of sequencing primer was then annealed to the purified single stranded PCR products, and 10 μL of the PCR product were sequenced on the PSQ96 HS System (Pyrosequencing, Qiagen).
QCpG software (Pyrosequencing, Qiagen) was used to assess methylation at each CpG site. The percentage of methylated alleles was divided by the total number of alleles (methylated and unmethylated) to determine the level of methylation at each CpG site. Regional methylation was calculated by taking the average of all CpG site methylation levels measured within each cluster. Each experiment had non-CpG cytosines serving as internal controls to ensure complete bisulfite conversion of the input DNA. Furthermore, low, medium, and high methylated DNA was included as controls in each run. Finally, EpigenDx performed PCR bias testing by combining unmethylated control DNA with in vitro methylated DNA following different ratios (0%, 5%, 10%, 25%, 50%, 75%, 100%) and conducting bisulfite modification, PCR, and Pyrosequencing analysis (Muench et al., 2018).
The DAT1 assays analyzed 48 CpG dinucleotides across the gene including promoter, spanning from −886 to 50969 base pairs from the transcription start site (TSS), based on Ensembl Transcript ENST00000270349. We focused on methylation in the promoter and averaged methylation of 4 clusters in the promoter (28 CpG sites); ranging from −1001 to −321 base pairs from the TSS.
Statistical Analyses
All analyses were performed using SPSS 22 (IBM, Armonk, New York). There were no > 3SD statistical outliers for striatal DAT availability (BPND) measures (caudate, putamen and VS). Group comparisons in BPND measures, DAT1 methylation, and demographics were performed using two-tailed t-tests. Correlations analyses of DAT BPND with DAT1 promoter methylation were performed for four functional clusters in the promoter (28 CpG sites), for 3 striatal ROIs and for each group separately. Significance thresholds were corrected for multiple comparisons and set at alpha = 0.002 (i.e., 0.05/24). Uncorrected significance levels were set at alpha = 0.05, and results at p<0.1 were reported as trends. To explore directions of effects, post-hoc tests were performed for CpG sites within the averaged clusters that showed significance. We also explored whole DAT1 methylation effects on DAT BPND (Supplementary Table 1, Supplementary Figure 1). Because neither age nor gender correlated with either DAT1 methylation or DAT availability (all p>0.1), we did not include age or gender as covariates to main our analyses.
Post-mortem Analysis
Whole blood samples, and SN and caudate tissue of post-mortem human brains were obtained from the Human Brain Collection Core at the NIMH. The sample consisted of tissue from 8 individuals with a history of ADHD (2 female, mean age of death =27±13, 6 smokers) and 22 diagnosed as non-psychiatric subjects (12 female, mean age of death =41±15, 4 smokers). There were no gender differences between groups (χ2=2.1, p=0.2, ns), but ADHD subjects were younger than controls (t=2.4, p=0.02), and involved more active smokers (χ2=8.5, p=0.007). Methylation analyses of DNA from blood and SN was performed according to the before-mentioned methods.
Quantitative Polymerase Chain Reaction (qPCR) for DAT gene expression in substantia nigra
From 30 SN samples, RNA with good quality (RIN>5) was extracted from 16 samples (3 ADHD, 13 non-psychiatric). RNA samples were converted into cDNA, and gene expression levels were measured with Taqman qPCR assays (Thermo Fisher Scientific) for the target gene DAT1 (assay ID: Hs00997374_m1), and for housekeeping genes ACTB (assay ID: Hs99999903_m1), B2M (assay ID: Hs99999907_m1), and GUSB (assay ID: Hs99999908_m1). DAT1 gene expression was normalized to housekeeping gene expression profiles.
Western Blot for DAT protein expression in caudate
Caudate tissue samples from the same individuals (n=30) were lysed in RIPA buffer (50mM Tris pH 7.6, 150 mM NaCl, 1 mM EDTA, 0.5% deoxycholate, 1% NP40, 0.1% SDS) with protease and phosphatase inhibitors (Sigma). Proteins were diluted with Laemmli sample buffer, separated on 4–20% polyacrylamide gels (Biorad), and transferred onto nitrocellulose membranes. The blots were blocked with 5% nonfat dry milk in 1X TBS containing 0.1% Tween-20 and incubated with DAT (Goat Anti-SLC6A3, TA311170 OriGene, working concentration 3ug/ml) and β-actin primary antibodies (Monoclonal Anti-β-Actin antibody, A5316, dilution: 1:5000) overnight at 4°C. On the next day, the blots were incubated with horseradish peroxidase-conjugated secondary antibodies (Sigma) for 1 hour at room temperature. Signals were visualized with the enhanced chemiluminescence (ECL) detection system (Pierce).
Results
Behavioral and DAT1 Methylation Measures
While CAARS inattention, impulsivity, and hyperactivity scores were higher in ADHD participants compared to HC (all p≤ 0.001), there were no between-group differences in DAT1 promoter methylation or striatal DAT availability (Table 1).
DAT Brain Imaging Results
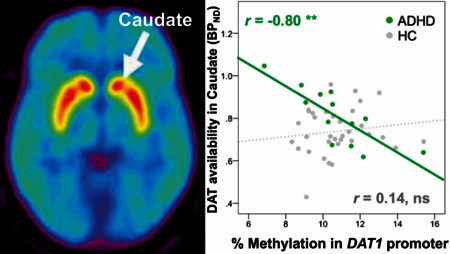
Degrees of methylation in the promoter cluster A (ADS2165) of DAT1 were negatively correlated with DAT availability (binding potential) in caudate in the ADHD participants only (r=−0.80, p=0.001, corrected for multiple comparisons); and in putamen: r=−0.66, p=0.014 and ventral striatum (VS) at trend level: r=−0.52, p=0.069, uncorrected for multiple comparisons (Figure 1) 1. DAT availability in VS correlated negatively with CAARS inattention scores in ADHD participants (r=−0.59, p=0.035). Post-hoc analysis showed that correlations of DAT1 methylation and DAT availability in ADHD participants were present for CpG sites 229, 230, and 231 (all p<0.001; see Supplementary Table 1 for all correlations of individual CpG sites).
FIG. 1.
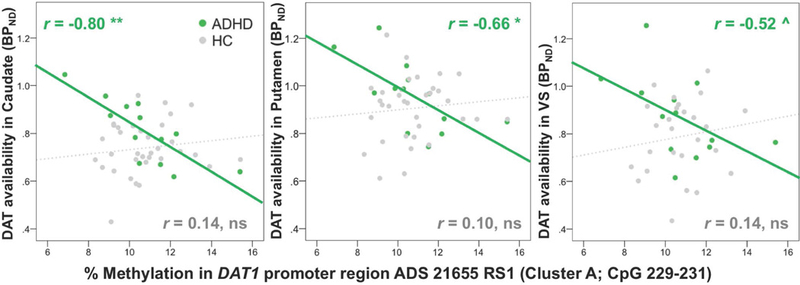
DAT1 methylation in promoter correlated negatively with DAT availability in caudate (corrected for multiple comparisons), and putamen, and at trend level in ventral striatum (VS) (uncorrected) in ADHD participants but not in healthy controls.
Post-mortem Results
DAT1 promoter methylation in blood and substantia nigra (SN)
The levels of DAT1 methylation in the promotor cluster A (ADS2165) in blood and in SN were positively correlated for ADHD participants (r=0.67, p=0.07, n=8) and controls (r=0.44, p=0.04, n=22); and when both groups were pooled together (r=0.45, p=0.01, n=30) (Figure 2). CpG site 230 showed the strongest correlation for controls (r=0.53, p=0.011) and groups combined (r=0.53, p=.003), which was also seen in the PET results.
FIG. 2.
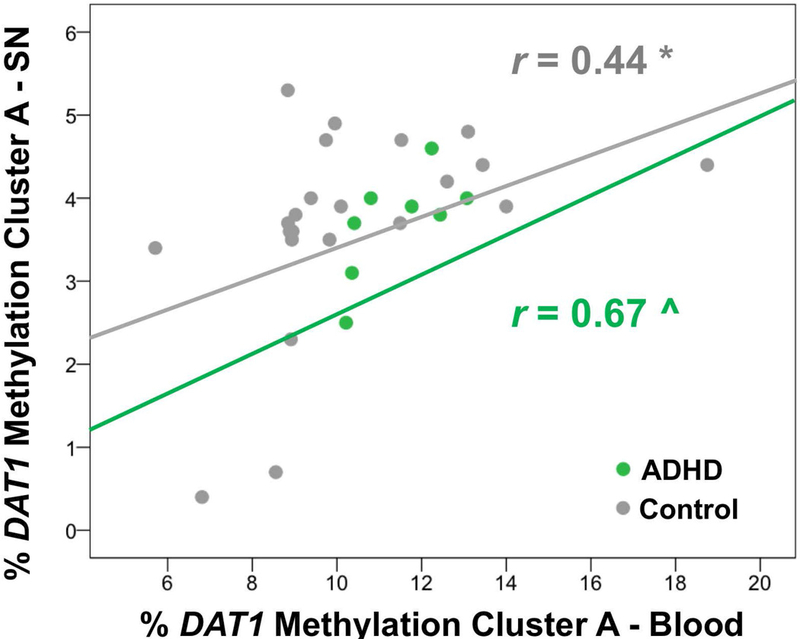
DAT1 promoter methylation (cluster A) in DNA extracted from blood correlated with DAT1 methylation in the same cluster extracted from Substantia Nigra (SN) both in ADHD participants (r=.67, p=.07) and in controls (r=.44, p=.04).
DAT1 promotor methylation in SN and DAT gene expression in SN and DAT protein expression levels in caudate.
Methylation of cluster A in SN was not correlated with DAT protein expression in caudate (p>0.05). After removal of outliers above 2 standard deviation (SD), only methylation at CpG site 231 in SN was negatively correlated with DAT protein expression in caudate (r=−0.61, p=0.006, n=19). The ADHD group had only 3 RNA samples for SN tissues (and one of them was a 3SD outlier for caudate protein expression), so any separate analyses for this group were precluded. In the non-psychiatric group, methylation of cluster A in SN was not correlated with DAT1 gene expression in SN. However, DAT1 gene expression in SN was correlated positively with DAT protein expression in caudate (r= 0.58, p =0.047, n=12) (Figure 3). Figure 4 summarizes overall findings on DAT1 methylation, DAT1 gene expression, and DAT protein expression in blood and brain.
FIG. 3.
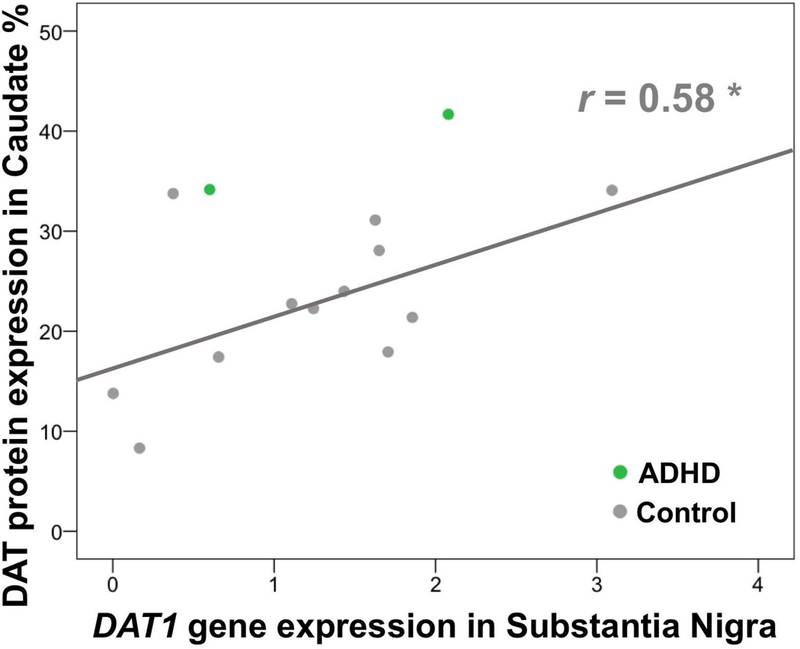
DAT1 gene expression (mRNA) in SN correlated positively with DAT protein expression in caudate in the control group (r=.58, p=.047). There were only 2 ADHD samples, which precluded analyses in this group.
FIG. 4.
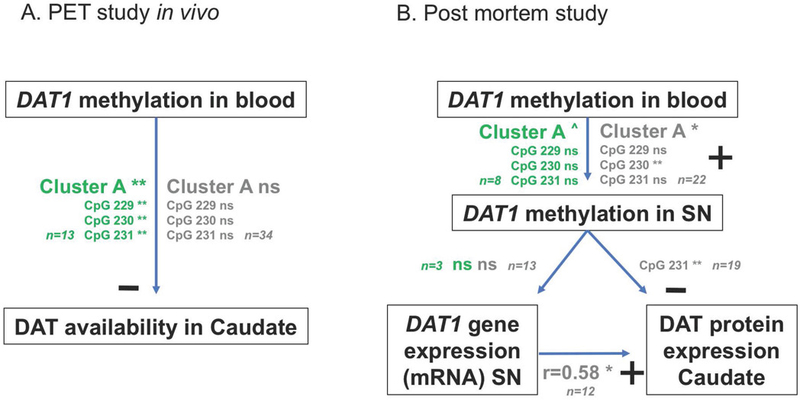
Summary of associations between DAT1 promoter methylation in blood and DAT availability in the in vivo human sample (Panel A), and with DAT1 methylation and DAT gene expression in substantia nigra (SN) and DAT protein expression in caudate in post-mortem sample. Results for ADHD patients are in green print, results for control groups in gray.
Last, DAT1 methylation of cluster A in blood did not correlate with DAT1 gene expression in SN or with DAT protein expression in Caudate (all p>0.05).
Discussion
DAT1 promoter methylation and striatal DAT availability
Here, we report an inverse association between methylation of a DAT1 promoter cluster in peripheral blood and striatal DAT availability measured with PET and [11C]cocaine in non-medicated adults with ADHD, but not in healthy volunteers. Results in caudate surived correction for multiple comparisons. The finding is partly in line with our hypothesis that methylation in DAT1 promoter regions derived from blood would inversely correlate with DAT availability measured with PET, which we based on preclinical studies associating peripheral DAT1 methylation with DAT availability in the basal ganglia (Rajala et al., 2014), and striatal DAT1 methylation with striatal DAT protein expression in rats with ADHD phenotype (Kim et al., 2014). The finding is corroborated by our post-mortem results from ADHD adults, since methylation of DAT1 in blood showed a positive association with DAT1 methylation in SN, where cell bodies of DA neurons are located (Giros et al., 1992; Giros & Caron, 1993). Therefore, we provide preliminary evidence that methylation of DAT1 in blood may be a relevant predictor of DAT expression in the human brain in ADHD, but not in healthy volunteers. Although we predicted negative associations between DAT1 methylation and striatal DAT availability in both groups, our findings of a negative association in subjects with ADHD is in line with previously reported associations of DAT1 methylation with DAT expression in preclinical models with ADHD phenotype (Kim et al., 2014; Rajala et al., 2014), and with improvements of ADHD symptoms in response to methylphenidate treatment in children with ADHD (Ding et al., 2017). The observed association between methylation of DAT1 and its expression might reflect the high concentration of CpG islands in the gene, which makes DAT expression particularly susceptible to modulation through epigenetic mechanisms, specifically DNA methylation (Shumay et al., 2010).
DAT1 promoter methylation and ADHD symptoms
Despite our second hypothesis that in the ADHD group DAT1 methylation would predict ADHD symptomatology, we did not find associations between these measures. Previous studies found that DAT1 methylation derived from blood correlated with symptoms of hyperactivity and impulsivity in children and adolescents with ADHD (Ding et al., 2017), and with impulsivity (and basal ganglia DAT availability) in monkeys (Rajala et al., 2014). In our study, however, the only significant association with ADHD symptomatology was a negative correlation of ventral striatal DAT availability with inattention symptoms in the ADHD group. The lack of an association may have several reasons, including that our sample size is small and consists of adults diagnosed with ADHD rather than children/adolescents. Moreover, studies on DAT availability and ADHD symptomatology have reported inconsistent findings (Del Campo et al., 2011), thus the relationship between DAT1 methylation and ADHD symptomatology, with DAT availability as a potential mediating endophenotype, remains to be explored in future research. We further did not find group differences in DAT1 methylation or striatal DAT availability. This was, however, expected as our PET data consisted of a subsample of a previous study that also did not find group differences in DAT availability between unmedicated ADHD participants and controls (Wang et al., 2013).
DAT1 promoter methylation in blood and DAT1 methylation and DAT expression in brain
Our third hypothesis was that DAT1 methylation in blood would positively associate with DAT1 methylation in SN. In the post-mortem samples, we did find a positive correlation between DAT1 promoter methylation from blood and from SN, both in the ADHD (at trend-level), and the non-psychiatric group. This is, to our knowledge, the first study showing an association of DAT1 promoter methylation in blood and brain in the same human subjects. The finding is in line with the “mirror site model” (Aberg et al., 2013) that predicts that peripheral and neural tissue have similar methylation patterns and are both associated with an endophenotype (here: DAT1 expression), which would be mediated by SN methylation. However, despite our fourth hypothesis that DAT1 methylation in SN would be predictive of striatal DAT1 gene expression, as was found in a preclinical model of ADHD by Kim el al. (2014), no associations were found between DAT1 methylation in SN and DAT1 gene expression in SN or protein expression in caudate in the cohort with ADHD. Also, DAT1 methylation in blood did not correlate with expression measures. This may be due to the small sample size in ADHD (n=3 and n=8 respectively), or the discrepancy might be explained by the fact that Kim et al. (2014) did not specify what part of the striatum was examined, and their rat model of ADHD phenotype was induced by alcohol exposure to the fathers of these rats, which is substantially different than the adults with ADHD in the current sample. In the non-psychiatric subjects, only a weak association of DAT1 methylation in SN (but not DAT1 methylation in blood) and caudate protein expression was found in CpG site 231. However, in the in vivo control sample there was no association between DAT1 methylation and DAT availability. Nevertheless, there was a clear association between DAT1 gene expression in the SN (but not DAT1 methylation in blood) and DAT protein expression in the caudate in the non-psychiatric subjects, which we could not assess in the ADHD group due to the restricted sample size (n=2 [and n=1 outlier in DAT protein expression]). This serves as preliminary evidence that DAT1 gene expression in SN regulates DAT protein expression in the caudate, further strengthening our rationale for examining DAT1 methylation associations between blood and SN where the DAT1 DNA and mRNA are located, whereas the DAT protein is enriched in striatum including the caudate. Nevertheless, the discrepancies between the in vivo (PET study) and ex vivo (postmortem study) findings in the non-psychiatric populations (controls) remain to be explored. That is, in controls the PET study did not show an association of DAT1 methylation in blood with striatal DAT availability whereas in the postmortem study DAT1 promoter methylation in blood was inversely correlated with DAT1 methylation in SN (opposite to what we expected) and only DAT1 methylation in CpG site 231 in SN showed a weak correlation with DAT protein expression in caudate whereas DAT1 gene expression in SN was associated with DAT protein expression in caudate. Some of these findings are contradictive to our hypothesized model that peripheral DAT1 methylation would predict striatal DAT1 expression/availability via DAT1 methylation and DAT1 gene expression in SN, in both control and ADHD groups. A possible interpretation to these findings is that DAT1 methylation is a predictor of striatal DAT expression in individuals with ADHD but not in controls.
Implications of findings
Previous studies reported that treatment naïve ADHD patients with higher striatal DAT availability respond more reliably to methylphenidate treatment (Krause, 2008). Therefore, if peripheral DAT1 methylation can accurately predict striatal DAT availability in ADHD, the question remains whether DAT1 methylation is also associated with treatment responses in ADHD. Genetic biomarkers, such as the 3′UTR VNTR have previously been proposed in the diagnosis of ADHD (Gold et al., 2014) and for their association with striatal DAT availability in healthy volunteers (Shumay et al., 2011). Because DNA methylation is a dynamic measure, it might, however, better reflect the expression of proteins sensitive to modification by environmental exposures. The expression of DAT1 is dynamic and sensitive to circadian rhythms (Ferris et al., 2014), age (Volkow et al., 1996), addictive drugs including tobacco (Newberg et al., 2007; Volkow et al., 2015; Ashok et al., 2017), medication exposures (Wang et al., 2013), among others. Although multiple factors regulate DAT expression, methylation of DAT1 is of particular interest as it changes dynamically in response to various environmental influences (Shumay et al., 2010). Thus, to the extent that DAT1 methylation in peripheral blood predicts DAT expression in the brain in ADHD, it may have potential as a biomarker to monitor treatment progress and to predict response to medication in this population (Adriani et al., 2017; Ding et al., 2017).
Methodological considerations
The present study is limited by the small sample sizes, in both the in vivo and post-mortem ADHD groups. Therefore, we consider the main finding on the association between DAT1 promoter methylation and DAT availability in 13 ADHD patients preliminary and in need of replication. A larger sample may have provided the power to detect associations between DAT availability and ADHD symptom severity, particularly impulsive behavior. An adequate sample size in the post-mortem ADHD group may have provided insight on whether the observed effect of DAT1 methylation in blood on DAT availability in caudate, was mediated through DAT1 methylation in SN and gene and protein expression profiles; which we were unable to test. Our study is also limited in its ability to infer implications for treatment, as we only have one-time point. Future studies on the relation between DAT1 methylation and ADHD should use a longitudinal approach to assess whether peripheral DAT1 methylation changes with treatment. Indeed, Wang et al. (2013) showed that striatal DAT availability upregulated after 1 year of treatment with methylphenidate. However, we currently lack corresponding evidence of changes in DAT1 methylation. Furthermore, despite previous findings that DAT1 promoter methylation and age were positively correlated in alcohol-dependent patients (Nieratschker et al., 2014; Muench et al., 2018), in our sample we did not corroborate an association between age and methylation of DAT1 whether we analyzed each group separately (p>0.38) or when we combined them (p=0.48). When correcting our main analyses for age, associations between DAT1 methylation in blood and striatal DAT availability in ADHD remained at an uncorrected level. Further, the results of our study may have been affected by the unbalanced gender distribution, which was due to an unbalanced gender distribution in a previous ADHD study (Wang et al., 2013) (see Supplemental Material 1 for sample selection from BNL protocols). Previous research found evidence for slightly higher striatal DAT availability in women than in men (Koch et al., 2007), but we did not find an effect of gender on DAT1 methylation or DAT availability in our sample. Finally, we did not a priori hypothesize that the association between DAT methylation and striatal DAT availability would be specific to ADHD participants and have no explanation of why such an association would not be present in controls other than speculating that a greater sensitivity for epigenetic regulation of DAT1 via DNA methylation might underlie vulnerability for ADHD.
Summary
In sum, DAT1 promoter methylation in blood may be a novel biomarker for striatal DAT availability in adults with ADHD as evidenced from our PET and post-mortem findings but more work is required to validate this. Imaging epigenetics is a novel and promising research field, and only few studies have evaluated epigenetic markers as predictors of neuroimaging measures. In healthy volunteers, serotonin transporter methylation predicted threat-related reactivity in the amygdala (Nikolova et al., 2014) and resting state functional connectivity in salience networks (Muehlhan et al., 2015); methylation of the Monoamine oxidase A (MAO-A) gene predicted MAO-A expression in brain (Shumay et al., 2012), and Catechol-O-methyltransferase (COMT) methylation predicted prefrontal cortex activation on a working memory task (Ursini et al., 2011). In alcohol use disorder, DAT1 promoter methylation was associated with neural alcohol cue reactivity (Wiers et al., 2015). Furthermore, while DAT1 promoter methylation predicted striatal responses during reward processing in healthy controls, this effect was not present in the alcohol-dependent group. Together, these findings further support the relevance of peripheral methylomic variation in the DAT1 promoter for dopamine-related neuronal phenotypes (Muench et al., 2018). Despite challenges in integrating methods of epigenomics and neuroimaging data (Bogdan et al., 2017; Lancaster et al., 2018), imaging epigenetics has a strong potential for investigating dynamic brain-based pathways relevant to behavior and neuropsychiatry.
Supplementary Material
Acknowledgements
The work was supported by the National Institutes of Health Intramural Research Program and grant number Y1AA-3009 to NDV. Postmortem tissues on blood and brain, and qPCR data generation and analysis were obtained from DIRP, NIMH, HBCC. We thank Elena Shumay, Karen Torres, and Chris Wong for their contributions.
Abbreviations
- ADHD
attention deficit hyperactivity disorder
- BPND
nondisplaceable binding potential
- CAARS
Conners’ Adult ADHD Rating Scale
- COMT
Catechol-O-methyltransferase
- DRD2
Dopamine D2/D3 receptor
- DRD4
Dopamine D4 receptor gene
- DAT
Dopamine transporter
- ECL
Enhanced chemiluminescence
- MAO-A
Monoamine oxidase A
- PCR
Polymerase chain reaction.
- PET
Positron Emission Tomography
- ROI
Regions of interest
- SD
Standard deviation
- SN
substantia nigra
- TSS
transcription start site
- VS
ventral striatum
- VIQ
verbal IQ
- VNTR
variable number tandem repeat
Footnotes
When correcting these correlations for age, DAT1 methylation in cluster A correlated negatively with DAT availability in caudate (r=−0.73, p=0.003); putamen (r=−0.62, p=0.016) and at trend level in VS (r=−0.46, p=0.069) in the ADHD participants (uncorrected for multiple comparisons).
Conflict of Interest Statements
The authors declare no conflicts of interest.
Data Accessibility Statement
Data is archived at NIH/NIAAA/LNI. Please contact corresponding author for access of data.
Data will become accessible by corresponding author for whoever requires.
References
- Aberg KA, Xie LY, McClay JL, Nerella S, Vunck S, Snider S, Beardsley PM & van den Oord EJ (2013) Testing two models describing how methylome-wide studies in blood are informative for psychiatric conditions. Epigenomics, 5, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriani W, Romano E, Pucci M, Pascale E, Cerniglia L, Cimino S, Tambelli R, Curatolo P, Granstrem O, Maccarrone M, Laviola G & D’Addario C (2017) Potential for diagnosis versus therapy monitoring of attention deficit hyperactivity disorder: a new epigenetic biomarker interacting with both genotype and auto-immunity. European child & adolescent psychiatry [DOI] [PubMed]
- Ashok AH, Mizuno Y, Volkow ND & Howes OD (2017) Association of Stimulant Use With Dopaminergic Alterations in Users of Cocaine, Amphetamine, or Methamphetamine: A Systematic Review and Meta-analysis. JAMA psychiatry, 74, 511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdan R, Salmeron BJ, Carey CE, Agrawal A, Calhoun VD, Garavan H, Hariri AR, Heinz A, Hill MN, Holmes A, Kalin NH & Goldman D (2017) Imaging Genetics and Genomics in Psychiatry: A Critical Review of Progress and Potential. Biological psychiatry, 82, 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenet F, Moh M, Funk P, Feierstein E, Viale AJ, Socci ND & Scandura JM (2011) DNA methylation of the first exon is tightly linked to transcriptional silencing. PloS one, 6, e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conners CK (1998) Rating scales in attention-deficit/hyperactivity disorder: use in assessment and treatment monitoring. The Journal of clinical psychiatry, 59 Suppl 7, 24–30. [PubMed] [Google Scholar]
- Cornish KM, Manly T, Savage R, Swanson J, Morisano D, Butler N, Grant C, Cross G, Bentley L & Hollis CP (2005) Association of the dopamine transporter (DAT1) 10/10-repeat genotype with ADHD symptoms and response inhibition in a general population sample. Mol Psychiatry, 10, 686–698. [DOI] [PubMed] [Google Scholar]
- Costa A, Riedel M, Muller U, Moller HJ & Ettinger U (2011) Relationship between SLC6A3 genotype and striatal dopamine transporter availability: a meta-analysis of human single photon emission computed tomography studies. Synapse, 65, 998–1005. [DOI] [PubMed] [Google Scholar]
- Del Campo N, Chamberlain SR, Sahakian BJ & Robbins TW (2011) The roles of dopamine and noradrenaline in the pathophysiology and treatment of attention-deficit/hyperactivity disorder. Biological psychiatry, 69, e145–157. [DOI] [PubMed] [Google Scholar]
- Ding K, Yang J, Reynolds GP, Chen B, Shao J, Liu R, Qian Q, Liu H, Yang R, Wen J & Kang C (2017) DAT1 methylation is associated with methylphenidate response on oppositional and hyperactive-impulsive symptoms in children and adolescents with ADHD. The world journal of biological psychiatry : the official journal of the World Federation of Societies of Biological Psychiatry, 18, 291–299. [DOI] [PubMed] [Google Scholar]
- Dougherty DD, Bonab AA, Spencer TJ, Rauch SL, Madras BK & Fischman AJ (1999) Dopamine transporter density in patients with attention deficit hyperactivity disorder. Lancet, 354, 2132–2133. [DOI] [PubMed] [Google Scholar]
- Ferris MJ, Espana RA, Locke JL, Konstantopoulos JK, Rose JH, Chen R & Jones SR (2014) Dopamine transporters govern diurnal variation in extracellular dopamine tone. Proceedings of the National Academy of Sciences of the United States of America, 111, E2751–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler JS, Volkow ND, Wang GJ, Gatley SJ & Logan J (2001) [(11)]Cocaine: PET studies of cocaine pharmacokinetics, dopamine transporter availability and dopamine transporter occupancy. Nuclear medicine and biology, 28, 561–572. [DOI] [PubMed] [Google Scholar]
- Fusar-Poli P, Rubia K, Rossi G, Sartori G & Balottin U (2012) Striatal dopamine transporter alterations in ADHD: pathophysiology or adaptation to psychostimulants? A meta-analysis. The American journal of psychiatry, 169, 264–272. [DOI] [PubMed] [Google Scholar]
- Giros B & Caron MG (1993) Molecular characterization of the dopamine transporter. Trends in pharmacological sciences, 14, 43–49. [DOI] [PubMed] [Google Scholar]
- Giros B, el Mestikawy S, Godinot N, Zheng K, Han H, Yang-Feng T & Caron MG (1992) Cloning, pharmacological characterization, and chromosome assignment of the human dopamine transporter. Molecular pharmacology, 42, 383–390. [PubMed] [Google Scholar]
- Gold MS, Blum K, Oscar–Berman M & Braverman ER (2014) Low Dopamine Function in Attention Deficit/Hyperactivity Disorder: Should Genotyping Signify Early Diagnosis in Children? Postgraduate Medicine, 126, 153–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawi Z, McCarron M, Kirley A, Daly G, Fitzgerald M & Gill M (2000) No association of the dopamine DRD4 receptor (DRD4) gene polymorphism with attention deficit hyperactivity disorder (ADHD) in the Irish population. Am J Med Genet, 96, 268–272. [DOI] [PubMed] [Google Scholar]
- Huang YS, Tsai MH & Guilleminault C (2011) Pharmacological treatment of ADHD and the short and long term effects on sleep. Current pharmaceutical design, 17, 1450–1458. [DOI] [PubMed] [Google Scholar]
- Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nature reviews. Genetics, 13, 484–492. [DOI] [PubMed] [Google Scholar]
- Kim P, Choi CS, Park JH, Joo SH, Kim SY, Ko HM, Kim KC, Jeon SJ, Park SH, Han SH, Ryu JH, Cheong JH, Han JY, Ko KN & Shin CY (2014) Chronic exposure to ethanol of male mice before mating produces attention deficit hyperactivity disorder-like phenotype along with epigenetic dysregulation of dopamine transporter expression in mouse offspring. Journal of neuroscience research, 92, 658–670. [DOI] [PubMed] [Google Scholar]
- Koch W, Pogarell O, Popperl G, Hornung J, Hamann C, Gildehaus FJ, Seelos K, Lewis D, Favit A & Tatsch K (2007) Extended studies of the striatal uptake of 99mTc-NC100697 in healthy volunteers. Journal of nuclear medicine : official publication, Society of Nuclear Medicine, 48, 27–34. [PubMed] [Google Scholar]
- Krause J (2008) SPECT and PET of the dopamine transporter in attention-deficit/hyperactivity disorder. Expert review of neurotherapeutics, 8, 611–625. [DOI] [PubMed] [Google Scholar]
- LaHoste GJ, Swanson JM, Wigal SB, Glabe C, Wigal T, King N & Kennedy JL (1996) Dopamine D4 receptor gene polymorphism is associated with attention deficit hyperactivity disorder. Mol Psychiatry, 1, 121–124. [PubMed] [Google Scholar]
- Lancaster K, Morris JP & Connelly JJ (2018) Neuroimaging Epigenetics: Challenges and Recommendations for Best Practices. Neuroscience, 370, 88–100. [DOI] [PubMed] [Google Scholar]
- Langley K, Turic D, Peirce TR, Mills S, Van Den Bree MB, Owen MJ, O’Donovan MC & Thapar A (2005) No support for association between the dopamine transporter (DAT1) gene and ADHD. Am J Med Genet B Neuropsychiatr Genet, 139B, 7–10. [DOI] [PubMed] [Google Scholar]
- Logan J, Fowler JS, Volkow ND, Wang GJ, Ding YS & Alexoff DL (1996) Distribution volume ratios without blood sampling from graphical analysis of PET data. Journal of cerebral blood flow and metabolism : official journal of the International Society of Cerebral Blood Flow and Metabolism, 16, 834–840. [DOI] [PubMed] [Google Scholar]
- Muehlhan M, Kirschbaum C, Wittchen HU & Alexander N (2015) Epigenetic variation in the serotonin transporter gene predicts resting state functional connectivity strength within the salience-network. Human brain mapping, 36, 4361–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muench C, Wiers CE, Cortes CR, Momenan R & Lohoff FW (2018) Dopamine Transporter Gene Methylation is Associated with Nucleus Accumbens Activation During Reward Processing in Healthy but not Alcohol-Dependent Individuals. Alcoholism, clinical and experimental research, 42, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newberg A, Lerman C, Wintering N, Ploessl K & Mozley PD (2007) Dopamine transporter binding in smokers and nonsmokers. Clinical nuclear medicine, 32, 452–455. [DOI] [PubMed] [Google Scholar]
- Nieratschker V, Grosshans M, Frank J, Strohmaier J, von der Goltz C, El-Maarri O, Witt SH, Cichon S, Nothen MM, Kiefer F & Rietschel M (2014) Epigenetic alteration of the dopamine transporter gene in alcohol-dependent patients is associated with age. Addiction biology, 19, 305–311. [DOI] [PubMed] [Google Scholar]
- Nikolova YS & Hariri AR (2015) Can we observe epigenetic effects on human brain function? Trends in cognitive sciences, 19, 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolova YS, Koenen KC, Galea S, Wang CM, Seney ML, Sibille E, Williamson DE & Hariri AR (2014) Beyond genotype: serotonin transporter epigenetic modification predicts human brain function. Nature neuroscience, 17, 1153–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajala AZ, Zaitoun I, Henriques JB, Converse AK, Murali D, Epstein ML & Populin LC (2014) Dopamine transporter gene susceptibility to methylation is associated with impulsivity in nonhuman primates. Journal of neurophysiology, 112, 2138–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumay E, Chen J, Fowler JS & Volkow ND (2011) Genotype and ancestry modulate brain’s DAT availability in healthy humans. PloS one, 6, e22754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumay E, Fowler JS & Volkow ND (2010) Genomic features of the human dopamine transporter gene and its potential epigenetic States: implications for phenotypic diversity. PloS one, 5, e11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumay E, Logan J, Volkow ND & Fowler JS (2012) Evidence that the methylation state of the monoamine oxidase A (MAOA) gene predicts brain activity of MAO A enzyme in healthy men. Epigenetics : official journal of the DNA Methylation Society, 7, 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TJ, Biederman J, Madras BK, Dougherty DD, Bonab AA, Livni E, Meltzer PC, Martin J, Rauch S & Fischman AJ (2007) Further evidence of dopamine transporter dysregulation in ADHD: a controlled PET imaging study using altropane. Biological psychiatry, 62, 1059–1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TJ, Biederman J, Madras BK, Faraone SV, Dougherty DD, Bonab AA & Fischman AJ (2005) In vivo neuroreceptor imaging in attention-deficit/hyperactivity disorder: a focus on the dopamine transporter. Biological psychiatry, 57, 1293–1300. [DOI] [PubMed] [Google Scholar]
- Swanson JM, Flodman P, Kennedy J, Spence MA, Moyzis R, Schuck S, Murias M, Moriarity J, Barr C, Smith M & Posner M (2000) Dopamine genes and ADHD. Neurosci Biobehav Rev, 24, 21–25. [DOI] [PubMed] [Google Scholar]
- Ursini G, Bollati V, Fazio L, Porcelli A, Iacovelli L, Catalani A, Sinibaldi L, Gelao B, Romano R, Rampino A, Taurisano P, Mancini M, Di Giorgio A, Popolizio T, Baccarelli A, De Blasi A, Blasi G & Bertolino A (2011) Stress-related methylation of the catechol-O-methyltransferase Val 158 allele predicts human prefrontal cognition and activity. The Journal of neuroscience : the official journal of the Society for Neuroscience, 31, 6692–6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Ding YS, Fowler JS, Wang GJ, Logan J, Gatley SJ, Hitzemann R, Smith G, Fields SD & Gur R (1996) Dopamine transporters decrease with age. Journal of nuclear medicine : official publication, Society of Nuclear Medicine, 37, 554–559. [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Ding YS, Wang GJ & Gatley SJ (1998) Positron emission tomography radioligands for dopamine transporters and studies in human and nonhuman primates. Advances in pharmacology, 42, 211–214. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Logan J, Alexoff D, Zhu W, Telang F, Wang GJ, Jayne M, Hooker JM, Wong C, Hubbard B, Carter P, Warner D, King P, Shea C, Xu Y, Muench L & Apelskog-Torres K (2009. a) Effects of modafinil on dopamine and dopamine transporters in the male human brain: clinical implications. JAMA : the journal of the American Medical Association, 301, 1148–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang G, Ding Y & Gatley SJ (2002. a) Mechanism of action of methylphenidate: insights from PET imaging studies. Journal of attention disorders, 6 Suppl 1, S31–43. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Logan J, Franceschi D, Maynard L, Ding YS, Gatley SJ, Gifford A, Zhu W & Swanson JM (2002. b) Relationship between blockade of dopamine transporters by oral methylphenidate and the increases in extracellular dopamine: therapeutic implications. Synapse, 43, 181–187. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Kollins SH, Wigal TL, Newcorn JH, Telang F, Fowler JS, Zhu W, Logan J, Ma Y, Pradhan K, Wong C & Swanson JM (2009. b) Evaluating dopamine reward pathway in ADHD: clinical implications. JAMA, 302, 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Newcorn J, Fowler JS, Telang F, Solanto MV, Logan J, Wong C, Ma Y, Swanson JM, Schulz K & Pradhan K (2007) Brain dopamine transporter levels in treatment and drug naive adults with ADHD. NeuroImage, 34, 1182–1190. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Newcorn JH, Kollins SH, Wigal TL, Telang F, Fowler JS, Goldstein RZ, Klein N, Logan J, Wong C & Swanson JM (2011) Motivation deficit in ADHD is associated with dysfunction of the dopamine reward pathway. Molecular psychiatry, 16, 1147–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Smith L, Fowler JS, Telang F, Logan J & Tomasi D (2015) Recovery of dopamine transporters with methamphetamine detoxification is not linked to changes in dopamine release. NeuroImage, 121, 20–28. [DOI] [PubMed] [Google Scholar]
- Wang GJ, Volkow ND, Wigal T, Kollins SH, Newcorn JH, Telang F, Logan J, Jayne M, Wong CT, Han H, Fowler JS, Zhu W & Swanson JM (2013) Long-term stimulant treatment affects brain dopamine transporter level in patients with attention deficit hyperactive disorder. PloS one, 8, e63023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiers CE (2012) Methylation and the human brain: towards a new discipline of imaging epigenetics. European archives of psychiatry and clinical neuroscience, 262, 271–273. [DOI] [PubMed] [Google Scholar]
- Wiers CE, Shumay E, Volkow ND, Frieling H, Kotsiari A, Lindenmeyer J, Walter H & Bermpohl F (2015) Effects of depressive symptoms and peripheral DAT methylation on neural reactivity to alcohol cues in alcoholism. Translational psychiatry, 5, e648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


