The arrangement of nucleosomes in chromatin plays a role in transcriptional regulation by restricting the accessibility of transcription factors and RNA polymerase II to cis-acting elements and promoters. For gene activation, the chromatin structure is altered to an open configuration.
KEYWORDS: histone chaperone, Tup family corepressor, chromatin repression, Schizosaccharomyces pombe, fbp1
ABSTRACT
The arrangement of nucleosomes in chromatin plays a role in transcriptional regulation by restricting the accessibility of transcription factors and RNA polymerase II to cis-acting elements and promoters. For gene activation, the chromatin structure is altered to an open configuration. The mechanism for this process has been extensively analyzed. However, the mechanism by which repressive chromatin is reconstituted to terminate transcription has not been fully elucidated. Here, we investigated the mechanisms by which chromatin is reconstituted in the fission yeast Schizosaccharomyces pombe fbp1 gene, which is robustly induced upon glucose starvation but tightly repressed under glucose-rich conditions. We found that the chromatin structure in the region upstream from fbp1 is closed by a two-step process. When cells are returned to glucose-rich medium following glucose starvation, changes in the nucleosome pattern alter the chromatin configuration at the transcription factor binding site to an inaccessible state, after which the nucleosome density upstream from fbp1 gradually increases via histone loading. Interestingly, this histone loading was observed in the absence of the Tup family corepressors Tup11 and Tup12. Analysis of strains carrying either gene disruptions or mutations affecting nine fission yeast histone chaperone genes demonstrated that the histone chaperone Asf1 induces nucleosome loading during glucose repression. These data establish a previously unappreciated chromatin reconstitution mechanism in fbp1 repression.
INTRODUCTION
Nucleosomes, which consist of core histone and DNA, are the fundamental structural unit of chromatin (1). The chromatin array is an important determinant for biochemical reactions, including transcription, since nucleosomes restrict the access of proteins to control sites within genomic DNA (2–4). For gene activation, the chromatin structure takes on an open configuration to allow access for trans-acting DNA-binding factors. Hence, transcriptional activation preferentially occurs in nucleosome-free, chromatin-accessible regions. Open chromatin regions are returned to a closed configuration for gene repression. While the mechanisms for the changes in chromatin configuration for gene activation have been extensively studied, the mechanisms for the changes in chromatin reconstitution for gene repression have not been well elucidated.
The fission yeast Schizosaccharomyces pombe fbp1 gene, encoding fructose-1,6-bisphosphatase, is robustly induced by glucose starvation (5, 6). fbp1 expression is strictly repressed by the Tup family transcriptional corepressors Tup11 and Tup12 (Tup11/12) and activated by transcriptional activators Atf1 and Rst2 (7–10). Atf1, a bZIP transcription factor, is regulated through phosphorylation by the mitogen-activated protein kinase (MAPK) pathway (11–13), while Rst2, a C2H2 Zn finger transcription factor, is regulated by the protein kinase A (PKA) pathway (7, 14). Two cis-acting elements required for fbp1 transcription have also been identified (15). Upstream activation sequence 1 (UAS1), containing a cyclic AMP (cAMP) response element (CRE), is the binding site for Atf1 (15), while UAS2, which resembles the Saccharomyces cerevisiae stress response element (STRE), is the critical binding site for Rst2 (8, 16).
In S. cerevisiae, the Tup corepressor Tup1 represses several genes, which are regulated by glucose, oxidative stress, DNA damage, and other cellular stress responses (17, 18), by establishing a repressive chromatin structure around the target gene promoter via the recruitment of histone deacetylases (HDACs) (19–22). The Drosophila and human Tup corepressors, the Groucho (Gro) and transducin-like enhancer of split (TLE) proteins, are also implicated in chromatin-regulated gene repression via the recruitment of HDACs (23, 24). We recently demonstrated that the S. pombe Tup corepressors Tup11/12 repress fbp1 transcription by two distinct mechanisms. First, Tup11/12 repress chromatin relaxation in the region upstream from the fbp1 promoter. Second, Tup11/12 interfere with the stable binding of RNA polymerase II (RNAPII) at the TATA box (25, 26). We also demonstrated that Tup11/12 repress the binding of Rst2 to the fbp1 upstream binding site (16). However, the role played by Tup11/12 in the reconstitution of repressive chromatin with regard to fbp1 repression has not been fully explained.
A group of histone chaperones facilitate and regulate the assembly and disassembly of nucleosomes for the control of transcription, replication, and DNA repair (27). However, the mechanisms by which a repressive chromatin state is established with the aid of these histone chaperones remain unclear. In fission yeast, there are 10 histone chaperones: the conserved histone chaperones CIA (CCG1-interacting factor A)/Asf1 (28), two Nap1 orthologs (Nap1/2) (29, 30), the FACT protein Pob3 (31), the CAF-1 complex protein Pcf1 (32), Spt6 (33), the Rtt106-like protein Mug183, the histone H2AZ chaperone Chz1 (34), the HIRA protein Hip1 (35), and the CENP-A nucleosome disassembly factor Ccp1 (36) homolog SPBC36B7.08c. Neither the relationships among these factors nor their division of labor has been fully elucidated.
We analyzed the mechanisms for chromatin reconstitution in the fission yeast fbp1 gene during glucose repression. Here, we demonstrate that the reconstitution of repressive chromatin takes place in two steps. First, the nucleosome-phasing pattern around the transcription factor binding sites changes in the absence of new nucleosome loading. Second, nucleosome loading upstream from the fbp1 promoter is gradually induced. We found that the histone chaperone Asf1, but not Tup11/12, is required for the reconstitution of repressive chromatin upstream from fbp1.
RESULTS
Two-step reconstitution of repressive chromatin upstream from fbp1.
To study the mechanism for the reconstitution of repressive chromatin, we analyzed transcription and the chromatin state of the fbp1 gene during glucose repression of the fbp1 gene in fission yeast. To this end, cells were subjected to glucose starvation for 3 h to induce fbp1 transcription and then shifted to glucose-rich medium for glucose repression. The abundance of fbp1 transcripts rapidly fell during glucose repression, and they disappeared at 60 min (Fig. 1A). To analyze the chromatin structure upstream from fbp1, we carried out an indirect end-labeling analysis using partially digested chromatin DNA with micrococcal nuclease (MNase) to map the nucleosomes and the nucleosome-free hypersensitive sites. Under glucose starvation conditions (0 min), MNase-sensitive bands were detected at UAS1 and at the TATA box (Fig. 1B, black and gray arrowheads, respectively), indicating that the chromatin state at these binding sites for transcription factors and RNA polymerase II had an accessible configuration. At 10 min after glucose repression, the MNase-sensitive sites at UAS1 disappeared (Fig. 1B and C), indicating that the chromatin state quickly closes following the initiation of glucose repression. In marked contrast, the MNase-sensitive bands at the TATA box only gradually weakened over time and disappeared 60 min after glucose repression (Fig. 1B and D). These results suggest that the chromatin state upstream from fbp1 changes into a repressive configuration upon glucose repression in two steps. In the first step, the chromatin state at transcription factor binding site UAS1 is altered within the first 10 min after glucose repression. In the second step, the chromatin state around the TATA box gradually changes to a closed configuration, and a repressive state is reached 60 min after glucose repression.
FIG 1.
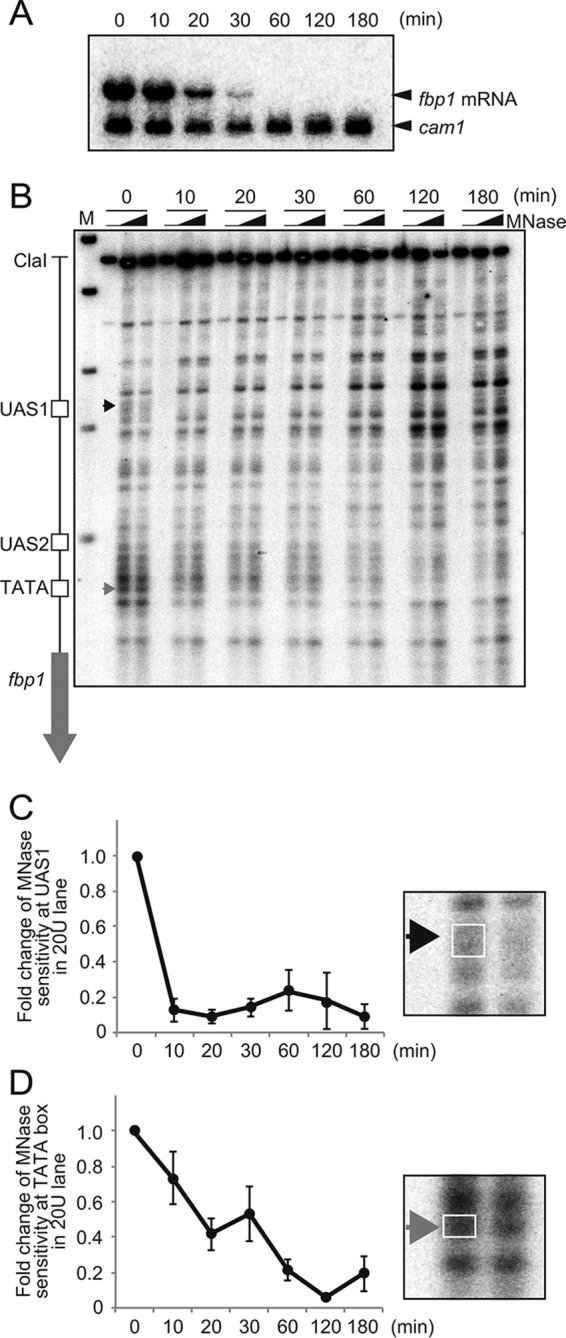
Two-step reconstitution of repressive chromatin upstream from fbp1. (A) Northern blot analysis to detect fbp1 transcripts. The indicated cells were cultured in YED medium containing glucose (0.1%) and glycerol (3%) for 3 h and transferred to YER medium containing glucose (6%). Cells were harvested at the indicated times. The cam1 transcript was used as an internal control (49). (B) Chromatin configurations around the fbp1 promoter in wild-type cells. The lanes represent chromatin from cells cultured in YER for the indicated times. The isolated chromatin was digested with MNase (0, 20, or 50 units/ml) at 37°C for 5 min. Purified DNA was digested with ClaI and analyzed by Southern blotting. The black arrowhead indicates regions with MNase-sensitive sites at UAS1 (positions −1162 to −1169 from the first A of the fbp1 open reading frame). The gray arrowhead indicates MNase-sensitive sites at the TATA box (positions −293 to −298). (C and D) Quantification of MNase-sensitive sites around UAS1 (C) and the TATA box (D). The intensities of the bands digested by MNase in the UAS1 and TATA regions (boxed on right) were quantified with FLA 7000, and the ratios of band intensities around the TATA box to the entire signal for each lane were calculated. The relative increases in the ratios at the indicated times after glucose starvation are indicated. The error bars represent the standard deviations from at least three independent experiments.
Nucleosome loading upstream from fbp1 in the second step of reconstitution of repressive fbp1 chromatin.
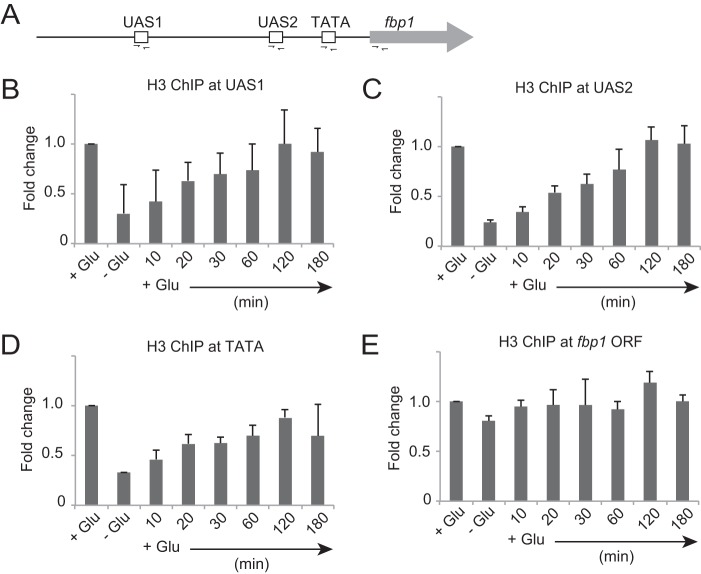
To understand the kinetics of nucleosome loading upstream from fbp1, we conducted a chromatin immunoprecipitation (ChIP) assay using anti-histone H3 antibody. We assessed histone H3 binding at UAS1, UAS2, the TATA box, and the fbp1 open reading frame (ORF) (Fig. 2A). During glucose repression, histone H3 binding gradually increased, plateauing 60 min after glucose repression for UAS1, UAS2, and the TATA box, while histone H3 at the fbp1 ORF was unchanged (Fig. 2B to E). Since nucleosome binding at the fbp1 ORF is also constant during transcriptional activation, nucleosomes upstream from fbp1, but not inside the ORF, may be subject to eviction or loading (37). These results suggest that nucleosome loading upstream from fbp1 is completed in the second step of fbp1 chromatin reconstitution. Interestingly, nucleosome loading at UAS1 is also completed 60 min after initiation of glucose repression, indicating that the observed prompt alteration of the MNase-sensitive band pattern at UAS1 (Fig. 1) is not associated with nucleosome loading.
FIG 2.
Kinetics of nucleosome loading upstream from fbp1 during chromatin reconstitution. (A) Schematic representation of the cis-regulatory elements in the fbp1 promoter region and amplification sites (determined by ChIP-qPCR) used in this study. (B to E) Histone H3 binding at UAS1 (B), UAS2 (C), the TATA box (D), and the fbp1 ORF (E) was determined by ChIP analysis. Cells were cultured to mid-log phase in YER (+Glu) and transferred to YED (−Glu) and cultured for 3 h. The cells were then transferred again to YER and cultured for the indicated times (+Glu). qPCR was performed using primer pairs to detect each segment indicated in panel A. The ChIP signal in the prp3 ORF was used for normalization. The error bars represent the standard deviations from three independent experiments.
Dissociation of transcription factor Atf1 in the first step of fbp1 chromatin repression.
Alteration of the nucleosome-phasing pattern can be caused by dissociation of DNA-binding factors, such as transcription factors. To determine whether the loss of MNase-sensitive sites at UAS1 10 min after glucose repression (Fig. 1) is associated with the removal of transcription factors, we examined Atf1 binding at UAS1. We detected the removal of Atf1 10 min after glucose repression (Fig. 3A). The Rst2 transcription activator, which binds to UAS2 upon transcriptional activation in an Atf1-dependent manner (16), also dissociated from the site 10 min after glucose repression (Fig. 3B). These data suggest that dissociation of transcriptional activators is associated with the alteration of the nucleosome-phasing pattern in the first step of chromatin reconstitution.
FIG 3.
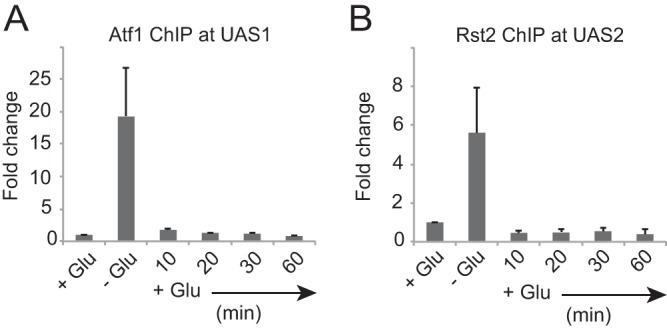
Dissociation kinetics of transcription factors from their binding sites upstream from fbp1 during glucose repression. Atf1 binding at UAS1 (A) and Rst2-flag binding at UAS2 (B) were determined by ChIP analysis using anti-Atf1 antibody and anti-DYKDDDDK antibody, respectively. Cells were cultured to mid-log phase in YER (+Glu) and then transferred to YED (−Glu) and cultured for 3 h. The cells were then transferred again to YER and cultured for the indicated times (+Glu). qPCR was performed using the primer pairs indicated in Fig. 2A. The ChIP signal in the prp3 ORF was used for normalization. The error bars represent the standard deviations from three independent experiments.
Tup11/12 corepressors are dispensable in nucleosome loading.
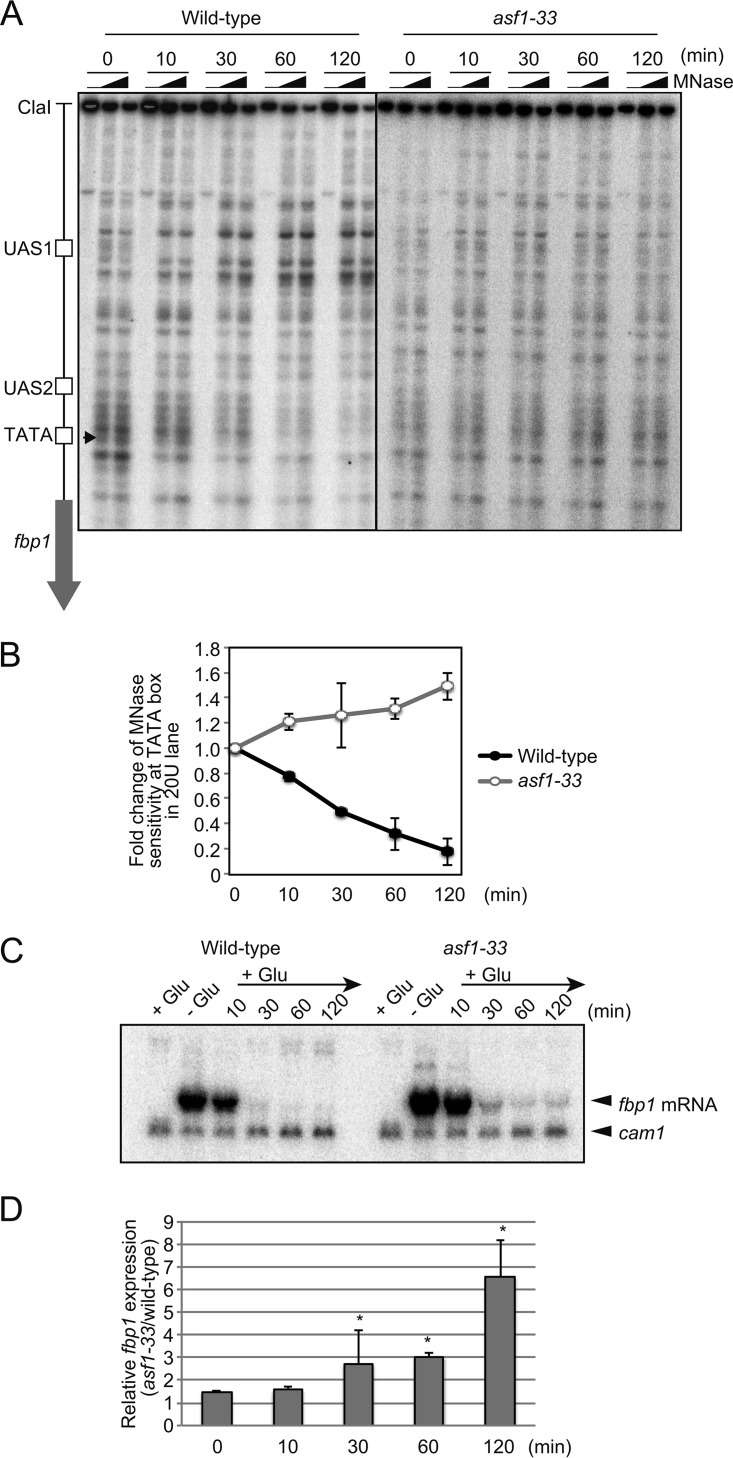
Tup1 family corepressors are believed to establish repressive chromatin through the recruitment of HDACs. We thus assumed that the fission yeast Tup1 orthologs Tup11/12 are required for the reconstitution of repressive chromatin upstream from fbp1. To examine this possibility, we analyzed the transcription and chromatin states of the fbp1 gene in a strain lacking Tup11/12. Under glucose starvation conditions, in tup11− tup12− (tupΔΔ) cells, expression of fbp1 was elevated 5-fold relative to that of wild-type cells, whereas fbp1 expression was gradually reduced over time and was barely detectable 60 min after glucose repression (Fig. 4A). The chromatin state upstream from fbp1 was altered to a repressive configuration in tupΔΔ cells after shifting the cells to a glucose-rich medium with slightly slower kinetics than in wild-type cells (Fig. 4B and C). To quantify nucleosome loading at the TATA box, we carried out a ChIP analysis to assess the binding kinetics of histone H3. In both wild-type and tupΔΔ cells, histone H3 binding at the TATA box gradually increased upon glucose refeeding (Fig. 4D), suggesting that Tup11/12 are not pivotal for the reconstitution of the repressive chromatin state following fbp1 repression. Interestingly, the nucleosome occupancy in the tupΔΔ cells was lower than in the wild-type cells when the chromatin was transcriptionally active, and thus, it took longer for histone H3 to reappear at the TATA box in the tupΔΔ cells than in wild-type cells (Fig. 4D). These data suggest that Tup11/12 play an important role in the regulation of the chromatin state under transcriptionally active conditions. This is consistent with the finding that binding of Tup12 at UAS1-UAS2 increases under transcriptionally active conditions but quickly falls upon glucose repression, as described previously (8) (Fig. 4E).
FIG 4.
Tup11/12 corepressors are dispensable in nucleosome loading. (A) Northern blot analysis to detect fbp1 transcripts in wild-type and tupΔΔ cells. The indicated cells were cultured as for Fig. 1. (B) Chromatin structures around the fbp1 promoter in wild-type and tupΔΔ cells. Chromatin DNA was digested with MNase (0, 20, or 50 units/ml) at 37°C for 5 min. Purified DNA was digested with ClaI and analyzed by Southern blotting. (C) Quantification of MNase-sensitive sites around the TATA box. The intensities of the bands digested by MNase in the TATA region (white box in panel B) and UAS1 (black box in panel B) were quantified with FLA 7000 (Fuji Film, Japan), and the ratios of band intensities around the TATA box to the entire signal for each lane were calculated. (D) Histone H3 binding at the TATA box was determined by ChIP analysis, as for Fig. 2. (E) Binding of Tup12-flag at UAS1 and UAS2 was determined by ChIP analysis, as for Fig. 2. The error bars represent the standard deviations from at least three independent experiments.
Tup11/12 corepressors are dispensable in histone deacetylation in fbp1 gene repression.
Given the critical role the Tup repressors play in the recruitment of HDACs to establish repressive chromatin (19–24), we hypothesized that Tup11/12 are involved in the regulation of histone acetylation. To test this hypothesis, we compared histone deacetylation kinetics upstream from fbp1 in wild-type and tupΔΔ cells. As we previously demonstrated (37, 38), histone H3 acetylation at UAS1 and UAS2 increases under glucose starvation conditions in wild-type cells (see Fig. S1 in the supplemental material). The tupΔΔ cells exhibited a more pronounced increase in histone acetylation under transcriptionally active conditions, indicating that Tup11/12 are involved in the regulation of histone acetylation under active fbp1 expression conditions, presumably through modulating HDACs (see Fig. S1 in the supplemental material). This is consistent with the repressive function of Tup11/12 when the chromatin is transcriptionally active in fbp1 (Fig. 4). Wild-type cells exhibited rapid deacetylation, and acetylation levels returned to normal 20 min after glucose repression (see Fig. S1 in the supplemental material). This rapid response is in marked contrast to the slow demethylation of histone H3K4 in the 5′ region of the fbp1 ORF, where histone H3K4 trimethylation levels gradually decline and return to normal 120 min after initiation of glucose repression (see Fig. S2 in the supplemental material). More importantly, the tupΔΔ cells also exhibited deacetylation (see Fig. S1 in the supplemental material), indicating that the Tup11/12 corepressors are not pivotal in histone deacetylation in the glucose repression of the fbp1 gene.
Histone chaperone Asf1 is required for the establishment of repressive chromatin upstream from fbp1.
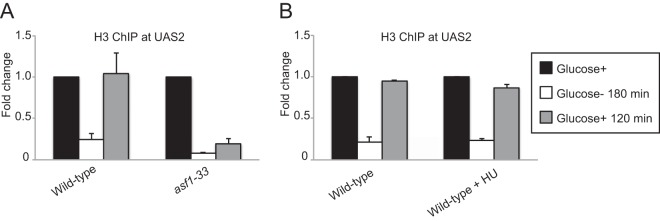
To explore the mechanisms that induce nucleosome loading to establish repressive chromatin, we screened strains with each of nine histone chaperone genes implicated in the loading of core histones deleted. We carried out a ChIP analysis to measure histone density before glucose starvation (Glucose+), at 180 min after glucose starvation (Glucose− 180 min), and after glucose repression for 120 min (Glucose+ 120 min). In wild-type cells, histone density at UAS2 was reduced upon glucose starvation and recovered after glucose repression (Fig. 5A). nap1Δ, nap2Δ, pob3Δ, pcf1Δ, mug183Δ, hip1Δ, chz1Δ, and spt6Δ cells showed histone occupancy levels at UAS2 indistinguishable from those of wild-type cells (see Fig. S3 in the supplemental material), indicating that these eight histone chaperones are not required for nucleosome reassembly in regions upstream from fbp1 following glucose repression. Since Asf1 is essential for cell proliferation, we used the temperature-sensitive allele asf1-33 (28). The asf1-33 cells showed critical defects in histone loading following glucose repression at the restrictive temperature, while wild-type cells exhibited normal assembly of nucleosomes after glucose repression under the same conditions (Fig. 5A). These results indicate that Asf1 is required for nucleosome reassembly upstream from fbp1 during glucose repression. Since Asf1 is known to be involved in chromatin reconstitution after replication (39, 40), we next questioned whether replication is required for chromatin repression in regions upstream from fbp1. We treated cells with hydroxyurea (HU) (20 mM) during glucose repression. While this treatment arrested cell proliferation, presumably due to replication arrest (see Fig. S4 in the supplemental material), it had little effect on chromatin reconstitution upstream from fbp1 during glucose repression (Fig. 5B). These results suggest that Asf1 induces loading of nucleosomes upstream from fbp1 during glucose repression independently of DNA replication. We next employed indirect end labeling using MNase digestion of chromatin DNA to examine the chromatin state in the fbp1 gene during glucose repression, as shown in Fig. 1. As expected, the asf1-33 cells showed defective chromatin repression upstream from the fbp1 ORF, including at the TATA box (Fig. 6A and B). However, the asf1-33 cells showed transcriptional repression of the fbp1 gene during glucose repression without reconstituting repressive chromatin around the TATA box (Fig. 6C). These results suggest that glucose repression of the fbp1 gene is not mediated solely by repressive chromatin formation in the promoter region. Since the asf1-33 cells show a significantly higher level of fbp1 transcription during the course of glucose refeeding than wild-type cells (Fig. 6D), the repressive chromatin established by Asf1-mediated nucleosome loading might contribute to the repression of transcriptional leakage of fbp1.
FIG 5.
Histone chaperone Asf1 is required for the formation of a condensed chromatin array upstream from fbp1 during glucose repression. (A) The indicated cells were cultured in YER at 25°C (Glucose+), transferred, and further cultured in YED for 3 h (Glucose− 180 min). Since the restrictive temperature for asf1-33 is 37°C, we incubated wild-type and asf1-33 cells at 37°C for 3 h and then transferred them again to YER and cultured them for 2 h at 37°C (Glucose+ 120 min). Histone H3 binding at UAS2 was determined by ChIP analysis, as for Fig. 2. (B) Cells were cultured at 30°C in YER (Glucose+), transferred, and further cultured in YED for 3 h (Glucose− 180 min). The cells were then transferred again to YER (containing HU [0 or 20 mM]) and cultured for 2 h (Glucose+ 120 min). Histone H3 binding at UAS2 was determined by ChIP analysis, as for Fig. 2. The error bars show the standard deviations from at least three independent experiments.
FIG 6.
Formation of a condensed chromatin array upstream from fbp1 is dispensable in the repression of fbp1 transcription. (A) Chromatin structures around the fbp1 promoter in wild-type and asf1-33 cells. The indicated cells were cultured as for Fig. 5A. Chromatin DNA was digested with MNase (0, 20, or 50 units/ml) at 37°C for 5 min. Purified DNA was digested with ClaI and analyzed by Southern blotting. (B) Quantification of MNase-sensitive sites around the TATA box, as for Fig. 4B. (C) Northern blot analysis to detect fbp1 transcripts in wild-type and asf1-33 cells. The indicated cells were cultured as for Fig. 5. (D) Quantification of fbp1 expression. The band intensities were quantified using Image J. The expression level of fbp1 was normalized to that of cam1. The fbp1 expression levels in asf1-33 cells relative to those in wild-type cells were calculated for each time point. The error bars show the standard deviations from at least two independent experiments. The significance of difference between wild-type and asf1-33 cells was calculated by Student's t test; *, P < 0.05.
DISCUSSION
In this study, we demonstrate that repressive chromatin is established upstream from fbp1 upon glucose repression in two steps. First, accessibility to the transcription factor binding site UAS1 is quickly reduced by an alteration of the nucleosome-phasing pattern. Second, a condensed chromatin array is reconstituted at the TATA box by 60 min after glucose repression. The first step is associated with the rapid dissociation of transcription factors from UAS1 and UAS2, while the second step is mediated by nucleosome loading by the histone chaperone Asf1. The dissociation of transcription factors may be a prerequisite for the later nucleosome loading, since ectopic recruitment of transcription factors results in constitutive fbp1 expression even under glucose-rich conditions (16).
We demonstrated that the Tup1 family corepressors Tup11/12 are not pivotal for the establishment of a condensed repressive chromatin array. This result was totally unexpected, given the role played by Gro and TLE in the establishment of repressive chromatin via the recruitment of HDACs (19–24). We previously demonstrated the critical role played by Tup11/12 in stress-specific chromatin regulation upstream from fbp1 and other genes (41). The role played by Tup11/12 in the determination of the transcription start site has been recently reported (25). These results suggest that the fission yeast Tup1 orthologs Tup11/12 might not act as simple corepressors but serve as transcriptional regulators via chromatin modulation. We found that Tup11/12 binding upstream from fbp1 is consistently enhanced when the chromatin is transcriptionally active. Thus, we propose that fission yeast Tup11/12 may play a role in limiting the level of transcriptional activation of fbp1.
Here, we demonstrate that only Asf1, among the nine histone chaperones tested, is required for the establishment of a condensed chromatin array upstream from fbp1 during glucose repression. Moreover, we found that Asf1-mediated nucleosome assembly takes place independently of replication. This result is consistent with the replication-independent nucleosome-loading activity of Asf1 detected in vitro using a budding yeast DEAE-CD fraction (42). A chromatin remodeler that harbors a chromodomain, Chd1, is also involved in this in vitro nucleosome-loading activity, which is counteracted by the Snf2 chromatin remodeler (42). Further investigation of the chromatin remodelers required to establish condensed nucleosome assays upstream from fbp1 during glucose repression is needed to better understand the mechanism for chromatin reconstitution in S. pombe cells upon exposure to glucose.
Unexpectedly, we found that fbp1 repression is accomplished even without the formation of a condensed chromatin array in asf1-deficient cells. This result is in marked contrast to the derepression of the budding yeast PHO5 gene in the absence of the Spt6-mediated chromatin assembly in the promoter region (43). It is possible that pleiotropic systems redundantly regulate transcription in the fission yeast fbp1 gene. The candidates for such systems might be the regulation of Atf1 and Rst2 via posttranslational modifications (8, 41). Further investigation of the pleiotropic gene repression mechanisms, including the interplay between chromatin modulations and transcriptional-regulatory factors, might reveal the mechanism underlying precise gene expression control.
MATERIALS AND METHODS
Fission yeast strains, genetic methods, and media.
Standard genetic procedures were carried out as described previously (44). Strain construction was carried out by mating haploids on sporulation medium (SPA), followed by tetrad dissection. The standard rich yeast extract medium, YEL (with 2% glucose), was used to culture cells. Yeast extract repressing (YER) medium (containing 6% glucose) and yeast extract derepressing (YED) medium (containing 0.1% glucose plus 3% glycerol) were used for glucose repression and derepression, respectively (9). Transformation was performed using the lithium-acetate method, as previously described (45). The S. pombe strains used in this study are listed in Table S1 in the supplemental material.
Deletion of the nap1+, nap2+, pob3+, pcf1+, mug183+, hip1+, and chz1+ genes.
The nap1+ sequence was amplified by PCR with Primstar GXL enzyme (TaKaRa Bio, Japan), using primers GGACGCTGTTTTATTTAGGACC and GTGTGCGAGCAAATTCCAG. The SalI-SacII fragment was eliminated from the cloned nap1+ sequence and replaced by the ura4+ marker gene to make nap1::ura4+. The AflII-AseI fragment carrying nap1::ura4+ was transformed into a wild-type fission yeast strain (SPH184). The mug183+ sequence was also amplified, using primers GGCAGAGTGCTTTTTACCAC and CGTTGATTACATCGGGAACAAC. The AatII-SnaBI fragment was eliminated from the cloned mug183+ sequence and replaced by the KanMX6+ marker gene to make mug183::KanMX6+. The PvuII-XbaI fragment carrying mug183::KanMX6+ was transformed into a wild-type fission yeast strain (SPH184). The pob3+ sequence was also amplified, using primers CTATCAGTTTAGAACGTTTCTAG and GTAGCAATTACAGGATAACGC. The ClaI-BglII fragment was eliminated from the cloned pob3+ sequence and replaced by the KanMX6+ marker gene to make pob3::KanMX6+. The SpeI-XbaI fragment carrying pob3::KanMX6+ was transformed into a wild-type fission yeast strain (SPH184). The pcf1+ sequence was also amplified, using primers CAATACTCATCAGTCTTTAAAACC and CCAACTCATACATAAGTTTCAC. The EcoNI-ScaI fragment was eliminated from the cloned pcf1+ sequence and replaced by the KanMX6+ marker gene to make pcf1::KanMX6+. The NdeI fragment carrying pcf1::KanMX6+ was transformed into a wild-type fission yeast strain (SPH184). To make the strains lacking hip1+, chz1+, and nap2+, flanking sequences (0.5 kbp) of hip1+, chz1+, and nap2+ were ligated with the ura4+ or KanMX6+ marker gene to construct hip1::ura4+, chz1::KanMX6+, and nap2::KanMX6+, which were transformed to make the disruptants.
Indirect end-labeling analysis using MNase-digested chromatin DNA.
Analysis of the chromatin structure by indirect end labeling using MNase-digested chromatin DNA was performed as previously reported (9, 46–48). The DNA samples were digested with ClaI, followed by Southern blotting using the probe, as described previously (9).
Northern blot analysis and ChIP.
Northern blot and ChIP analyses were performed as described previously (8). ChIP analysis was performed as described previously (25) using anti-Atf1 antibody (Abcam), anti-DYKDDDDK antibody (Wako; 018-22383), anti-H3 antibody (abcam; ab1791), anti-acetylated histone H3 antibody (Millipore; 06-599), and anti-trimethylated histone H3K4 antibody (abcam; ab8580).
Quantification of ChIP DNA.
DNA concentrations were quantified using a Thermal Cycler Dice real-time system TP800 (TaKaRa) and Thunderbird SYBR quantitative-PCR (qPCR) mix (Toyobo) with the following primer sets: fbp1-UAS1 (GGGATGAAAACAATCAACCTC and GGAATGCAGCAACGAAAATC), fbp1-UAS2 (GGGTGGAATGAGTCCGC and GTTCCGCGAATCATAAGCC), fbp1-TATA (CGCGGAACTAAACATAGCG and GCTAGAAACCGAGTGGTG), and fbp1-ORF (CGCCGATACAATCAGAAGC and CGATGAGTTTGCAGCATCC), and for the control site, prp3 (GCACAGTCGTTGTACAAATTCGTATTCCC and ACGATTCTAAACGCCTCTTGTTACGATCC).
Supplementary Material
ACKNOWLEDGMENTS
We thank Makoto Kawamukai and Hiroaki Kato for their interest in this research and for providing the asf1-33ts and spt6Δ S. pombe strains, respectively. We acknowledge the Radioisotope Research Center of Tokyo Metropolitan University for its support in the use of isotopes.
This work was supported by JSPS Kakenhi (25281021, 26116518, 24114509, 16K12598, and 16H02957 to K.H.; 26291018 to K.O.; and 16J02252 to R.A.) and the Takeda Science Foundation (to K.H.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00194-18.
REFERENCES
- 1.Kornberg RD. 1974. Chromatin structure: a repeating unit of histones and DNA. Science 184:868–871. doi: 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg RD, Lorch Y. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98:285–294. doi: 10.1016/S0092-8674(00)81958-3. [DOI] [PubMed] [Google Scholar]
- 3.Wolffe AP. 1997. Histones, nucleosomes and the roles of chromatin structure in transcriptional control. Biochem Soc Trans 25:354–358. doi: 10.1042/bst0250354. [DOI] [PubMed] [Google Scholar]
- 4.Wolffe AP. 1994. Nucleosome positioning and modification: chromatin structures that potentiate transcription. Trends Biochem Sci 19:240–244. doi: 10.1016/0968-0004(94)90148-1. [DOI] [PubMed] [Google Scholar]
- 5.Hoffman CS, Winston F. 1991. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes Dev 5:561–571. doi: 10.1101/gad.5.4.561. [DOI] [PubMed] [Google Scholar]
- 6.Hoffman CS, Winston F. 1989. A transcriptionally regulated expression vector for the fission yeast Schizosaccharomyces pombe. Gene 84:473–479. doi: 10.1016/0378-1119(89)90523-4. [DOI] [PubMed] [Google Scholar]
- 7.Higuchi T, Watanabe Y, Yamamoto M. 2002. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol Cell Biol 22:1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirota K, Hoffman CS, Ohta K. 2006. Reciprocal nuclear shuttling of two antagonizing Zn finger proteins modulates Tup family corepressor function to repress chromatin remodeling. Eukaryot Cell 5:1980–1989. doi: 10.1128/EC.00272-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hirota K, Hoffman CS, Shibata T, Ohta K. 2003. Fission yeast Tup1-like repressors repress chromatin remodeling at the fbp1+ promoter and the ade6-M26 recombination hotspot. Genetics 165:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janoo RT, Neely LA, Braun BR, Whitehall SK, Hoffman CS. 2001. Transcriptional regulators of the Schizosaccharomyces pombe fbp1 gene include two redundant Tup1p-like corepressors and the CCAAT binding factor activation complex. Genetics 157:1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanoh J, Watanabe Y, Ohsugi M, Iino Y, Yamamoto M. 1996. Schizosaccharomyces pombe gad7+ encodes a phosphoprotein with a bZIP domain, which is required for proper G1 arrest and gene expression under nitrogen starvation. Genes Cells 1:391–408. doi: 10.1046/j.1365-2443.1996.d01-247.x. [DOI] [PubMed] [Google Scholar]
- 12.Shiozaki K, Russell P. 1996. Conjugation, meiosis, and the osmotic stress response are regulated by Spc1 kinase through Atf1 transcription factor in fission yeast. Genes Dev 10:2276–2288. doi: 10.1101/gad.10.18.2276. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson MG, Samuels M, Takeda T, Toone WM, Shieh JC, Toda T, Millar JB, Jones N. 1996. The Atf1 transcription factor is a target for the Sty1 stress-activated MAP kinase pathway in fission yeast. Genes Dev 10:2289–2301. doi: 10.1101/gad.10.18.2289. [DOI] [PubMed] [Google Scholar]
- 14.Kunitomo H, Higuchi T, Iino Y, Yamamoto M. 2000. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11(+) gene, which encodes a pivotal transcription factor for sexual development. Mol Biol Cell 11:3205–3217. doi: 10.1091/mbc.11.9.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neely LA, Hoffman CS. 2000. Protein kinase A and mitogen-activated protein kinase pathways antagonistically regulate fission yeast fbp1 transcription by employing different modes of action at two upstream activation sites. Mol Cell Biol 20:6426–6434. doi: 10.1128/MCB.20.17.6426-6434.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asada R, Umeda M, Adachi A, Senmatsu S, Abe T, Iwasaki H, Ohta K, Hoffman CS, Hirota K. 2017. Recruitment and delivery of the fission yeast Rst2 transcription factor via a local genome structure counteracts repression by Tup1-family corepressors. Nucleic Acids Res 45:9361–9371. doi: 10.1093/nar/gkx555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roth SY. 1995. Chromatin-mediated transcriptional repression in yeast. Curr Opin Genet Dev 5:168–173. doi: 10.1016/0959-437X(95)80004-2. [DOI] [PubMed] [Google Scholar]
- 18.Wahi M, Komachi K, Johnson AD. 1998. Gene regulation by the yeast Ssn6-Tup1 corepressor. Cold Spring Harbor Symp Quant Biol 63:447–457. doi: 10.1101/sqb.1998.63.447. [DOI] [PubMed] [Google Scholar]
- 19.Davie JK, Edmondson DG, Coco CB, Dent SY. 2003. Tup1-Ssn6 interacts with multiple class I histone deacetylases in vivo. J Biol Chem 278:50158–50162. doi: 10.1074/jbc.M309753200. [DOI] [PubMed] [Google Scholar]
- 20.Gromoller A, Lehming N. 2000. Srb7p is a physical and physiological target of Tup1p. EMBO J 19:6845–6852. doi: 10.1093/emboj/19.24.6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Z, Reese JC. 2004. Redundant mechanisms are used by Ssn6-Tup1 in repressing chromosomal gene transcription in Saccharomyces cerevisiae. J Biol Chem 279:39240–39250. doi: 10.1074/jbc.M407159200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Z, Reese JC. 2004. Ssn6-Tup1 requires the ISW2 complex to position nucleosomes in Saccharomyces cerevisiae. EMBO J 23:2246–2257. doi: 10.1038/sj.emboj.7600227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Courey AJ. 2000. Groucho/TLE family proteins and transcriptional repression. Gene 249:1–16. doi: 10.1016/S0378-1119(00)00161-X. [DOI] [PubMed] [Google Scholar]
- 24.Malave TM, Dent SY. 2006. Transcriptional repression by Tup1-Ssn6. Biochem Cell Biol 84:437–443. doi: 10.1139/o06-073. [DOI] [PubMed] [Google Scholar]
- 25.Asada R, Takemata N, Hoffman CS, Ohta K, Hirota K. 2015. Antagonistic controls of chromatin and mRNA start site selection by Tup family corepressors and the CCAAT-binding factor. Mol Cell Biol 35:847–855. doi: 10.1128/MCB.00924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. 2008. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature 456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- 27.Loyola A, Almouzni G. 2004. Histone chaperones, a supporting role in the limelight. Biochim Biophys Acta 1677:3–11. doi: 10.1016/j.bbaexp.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Tanae K, Horiuchi T, Matsuo Y, Katayama S, Kawamukai M. 2012. Histone chaperone Asf1 plays an essential role in maintaining genomic stability in fission yeast. PLoS One 7:e30472. doi: 10.1371/journal.pone.0030472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grande M, Lambea E, Fajardo A, Lopez-Aviles S, Kellogg D, Aligue R. 2008. Crosstalk between Nap1 protein and Cds1 checkpoint kinase to maintain chromatin integrity. Biochim Biophys Acta 1783:1595–1604. doi: 10.1016/j.bbamcr.2008.03.019. [DOI] [PubMed] [Google Scholar]
- 30.Walfridsson J, Khorosjutina O, Matikainen P, Gustafsson CM, Ekwall K. 2007. A genome-wide role for CHD remodelling factors and Nap1 in nucleosome disassembly. EMBO J 26:2868–2879. doi: 10.1038/sj.emboj.7601728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lejeune E, Bortfeld M, White SA, Pidoux AL, Ekwall K, Allshire RC, Ladurner AG. 2007. The chromatin-remodeling factor FACT contributes to centromeric heterochromatin independently of RNAi. Curr Biol 17:1219–1224. doi: 10.1016/j.cub.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kunoh T, Habu T. 2014. Pcf1, a large subunit of CAF-1, required for maintenance of checkpoint kinase Cds1 activity. Springerplus 3:30. doi: 10.1186/2193-1801-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kiely CM, Marguerat S, Garcia JF, Madhani HD, Bahler J, Winston F. 2011. Spt6 is required for heterochromatic silencing in the fission yeast Schizosaccharomyces pombe. Mol Cell Biol 31:4193–4204. doi: 10.1128/MCB.05568-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luk E, Vu ND, Patteson K, Mizuguchi G, Wu WH, Ranjan A, Backus J, Sen S, Lewis M, Bai Y, Wu C. 2007. Chz1, a nuclear chaperone for histone H2AZ. Mol Cell 25:357–368. doi: 10.1016/j.molcel.2006.12.015. [DOI] [PubMed] [Google Scholar]
- 35.Blackwell C, Martin KA, Greenall A, Pidoux A, Allshire RC, Whitehall SK. 2004. The Schizosaccharomyces pombe HIRA-like protein Hip1 is required for the periodic expression of histone genes and contributes to the function of complex centromeres. Mol Cell Biol 24:4309–4320. doi: 10.1128/MCB.24.10.4309-4320.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong Q, Yin FX, Gao F, Shen Y, Zhang F, Li Y, He H, Gonzalez M, Yang J, Zhang S, Su M, Chen YH, Li F. 2016. Ccp1 homodimer mediates chromatin integrity by antagonizing CENP-A loading. Mol Cell 64:79–91. doi: 10.1016/j.molcel.2016.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Adachi A, Senmatsu S, Asada R, Abe T, Hoffman CS, Ohta K, Hirota K. 2018. Interplay between chromatin modulators and histone acetylation regulates the formation of accessible chromatin in the fission yeast fbp1 upstream regulatory region. Genes Genet Syst 92:267–276. doi: 10.1266/ggs.17-00018. [DOI] [PubMed] [Google Scholar]
- 38.Takemata N, Oda A, Yamada T, Galipon J, Miyoshi T, Suzuki Y, Sugano S, Hoffman CS, Hirota K, Ohta K. 2016. Local potentiation of stress-responsive genes by upstream noncoding transcription. Nucleic Acids Res 44:5174–5189. doi: 10.1093/nar/gkw142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanematsu F, Takami Y, Barman HK, Fukagawa T, Ono T, Shibahara K, Nakayama T. 2006. Asf1 is required for viability and chromatin assembly during DNA replication in vertebrate cells. J Biol Chem 281:13817–13827. doi: 10.1074/jbc.M511590200. [DOI] [PubMed] [Google Scholar]
- 40.Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT. 1999. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 41.Hirota K, Hasemi T, Yamada T, Mizuno KI, Hoffman CS, Shibata T, Ohta K. 2004. Fission yeast global repressors regulate the specificity of chromatin alteration in response to distinct environmental stresses. Nucleic Acids Res 32:855–862. doi: 10.1093/nar/gkh251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robinson KM, Schultz MC. 2003. Replication-independent assembly of nucleosome arrays in a novel yeast chromatin reconstitution system involves antisilencing factor Asf1p and chromodomain protein Chd1p. Mol Cell Biol 23:7937–7946. doi: 10.1128/MCB.23.22.7937-7946.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adkins MW, Tyler JK. 2006. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell 21:405–416. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Gutz H, Heslot H, Leupold U, Loprieno N. 1974. Schizosaccharomyces pombe, p 395–446. In King RD. (ed), Handbook of genetics, vol 1 Plenum, New York, NY. [Google Scholar]
- 45.Hirota K, Tanaka K, Watanabe Y, Yamamoto M. 2001. Functional analysis of the C-terminal cytoplasmic region of the M-factor receptor in fission yeast. Genes Cells 6:201–214. doi: 10.1046/j.1365-2443.2001.00415.x. [DOI] [PubMed] [Google Scholar]
- 46.Hirota K, Fukuda T, Yamada T, Ohta K. 2009. Analysis of chromatin structure at meiotic DSB sites in yeasts. Methods Mol Biol 557:253–266. doi: 10.1007/978-1-59745-527-5_16. [DOI] [PubMed] [Google Scholar]
- 47.Hirota K, Mizuno K, Shibata T, Ohta K. 2008. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26. Mol Biol Cell 19:1162–1173. doi: 10.1091/mbc.e07-04-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hirota K, Steiner WW, Shibata T, Ohta K. 2007. Multiple modes of chromatin configuration at natural meiotic recombination hot spots in fission yeast. Eukaryot Cell 6:2072–2080. doi: 10.1128/EC.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeda T, Yamamoto M. 1987. Analysis and in vivo disruption of the gene coding for calmodulin in Schizosaccharomyces pombe. Proc Natl Acad Sci U S A 84:3580–3584. doi: 10.1073/pnas.84.11.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.