There are many different classes of proteins secreted from Phytophthora infestans that may influence or facilitate infection. Elucidating where and how they are secreted during infection is an important step toward developing methods to control their delivery processes. We used an inhibitor of conventional secretion to identify the following different classes of infection-associated extracellular proteins: cell wall-degrading and cell wall-modifying enzymes, microbe-associated molecular pattern-like proteins that may elicit immune responses, and apoplastic effectors that are predicted to suppress immunity. In contrast, secretion of a cytoplasmic effector that is translocated into host cells is nonconventional, as it is insensitive to inhibitor treatment. This evidence further supports the finding that proteins that are active in the apoplast and effector proteins that are active in the host cytoplasm are differentially secreted by P. infestans. Critically, it demonstrates that a disease-specific developmental structure, the haustorium, is a major secretion site for diverse protein classes during infection.
KEYWORDS: crop disease, filamentous plant pathogen, pathogenicity, secretome, virulence
ABSTRACT
The oomycete potato blight pathogen Phytophthora infestans secretes a diverse set of proteins to manipulate host plant immunity. However, there is limited knowledge about how and where they are secreted during infection. Here we used the endoplasmic reticulum (ER)-to-Golgi secretion pathway inhibitor brefeldin A (BFA) in combination with liquid chromatography-electrospray tandem mass spectrometry (LC-MS/MS) to identify extracellular proteins from P. infestans that were conventionally secreted from in vitro-cultured hyphae. We identified 19 proteins with predicted signal peptides that potentially influence plant interactions for which secretion was attenuated by BFA. In addition to inhibition by the apoplastic effector EPIC1, a cysteine protease inhibitor, we show that secretion of the cell wall-degrading pectinesterase enzyme PE1 and the microbe-associated molecular pattern (MAMP)-like elicitin INF4 was inhibited by BFA in vitro and in planta, demonstrating that these proteins are secreted by the conventional, Golgi-mediated pathway. For comparison, secretion of a cytoplasmic RXLR (Arg-[any amino acid]-Leu-Arg) effector, Pi22926, was not inhibited by BFA. During infection, whereas INF4 accumulated outside the plant cell, RXLR effector Pi22926 entered the plant cell and accumulated in the nucleus. The P. infestans effectors, the PE1 enzyme, and INF4 were all secreted from haustoria, pathogen structures that penetrate the plant cell wall to form an intimate interaction with the host plasma membrane. Our findings show the haustorium to be a major site of both conventional and nonconventional secretion of proteins with diverse functions during infection.
INTRODUCTION
Biotrophic pathogens rely on living host cells for growth, deploying a diverse set of secreted virulence determinants to facilitate successful infection (1–4). Over the last three decades, studies of pathogenic bacteria have revealed an arsenal of virulence factors (VFs) involved in pathogenesis, including secreted proteins classed as effectors, protein toxins, and enzymes and cell-surface structures such as capsular polysaccharides, lipopolysaccharides, and outer membrane proteins (5). Filamentous eukaryotic microbial plant pathogens such as fungi and oomycetes secrete a remarkable diversity of molecules at different stages of infection to manipulate the plant immune response (6). In the past 15 years, much research has focused on the activities of cytoplasmic effectors (acting inside host plant cells) and apoplastic effectors (acting in the extracellular space), particularly on where they localize and how they function to facilitate infection. Despite the intense research in this area, many other VFs remain to be characterized.
Phytophthora infestans, causing potato late blight, continues to pose a major threat to potato production worldwide, over 170 years after the Irish potato famine (7). As P. infestans is a model oomycete pathogen, many of its secreted proteins, including cytoplasmic and apoplastic effectors (previously summarized [8, 9]), have been characterized. However, there is still limited knowledge of the majority of secreted proteins representing the repertoire of the P. infestans infection secretome (10). Secreted carbohydrate-active enzymes (CAZymes), including cell wall-degrading enzymes (CWDEs), trypsin-like serine proteases, berberine bridge enzymes, carbonic anhydrases, small cysteine-rich proteins, and repeat-containing proteins, have been postulated to represent potential VFs (10).
Rust fungi and oomycetes produce intercellular hyphae with intracellular haustoria. These structures are formed by enzymatic digestion of the host cell wall and invagination of the host cell membrane. Haustoria create an intimate association with the host cell for molecular exchange, such as nutrient uptake in fungi (11–13). In addition, haustoria are sites for secretion of fungal and oomycete effectors. P. infestans haustorium formation was first examined by electron microscopy (EM) in potato leaf tissue as early as 1966 (14). It was not until 2007 that the first direct observation of effector secretion at haustoria was made (15). Since then, a small number of other cytoplasmic (16, 17) and apoplastic (9) effectors and the transmembrane protein HMP1 (18) have been found to accumulate at Phytophthora haustoria.
Conventional and nonconventional forms of secretion of different effector classes were first identified in the rice blast fungus Magnaporthe oryzae. The apoplastic effector “biotrophy-associated secreted protein 4” (Bas4) was conventionally secreted via the endoplasmic reticulum (ER)-to-Golgi secretion pathway from invasive hyphae (IH), as its secretion was sensitive to brefeldin A (BFA; an inhibitor of conventional ER-to-Golgi secretion). In contrast, the cytoplasmic effector Pwl2 (for pathogenicity toward weeping lovegrass) was nonconventionally secreted in a BFA-insensitive manner requiring the exocyst complex (19). In agreement with the study in M. oryzae, two distinct secretory pathways were also found in P. infestans, where the apoplastic effector EPIC1, a cysteine protease inhibitor, was conventionally secreted and the cytoplasmic effector Pi04314 was secreted in a BFA-insensitive manner to accumulate in the host nucleus (9).
We hypothesized that this might be a general rule for oomycete plant pathogens and sought to distinguish conventionally secreted proteins from nonconventionally secreted proteins using liquid chromatography-electrospray tandem mass spectrometry (LC-MS/MS). Although proteomic analysis of extracellular proteins was carried out for P. infestans previously (20), that study did not seek to discriminate between conventional and nonconventional secreted proteins. Here, we performed LC-MS/MS following growth of P. infestans in vitro in the presence or absence of BFA to specifically identify inhibitor-sensitive, conventionally secreted candidate VFs. We used a P. infestans transgenic line expressing EPIC1-mRFP (EPIC-monomeric red fluorescent protein), a BFA-sensitive secreted protein (9), to provide a marker for inhibitor activity. We also selected a further cytoplasmic RXLR effector, PITG_22926 (Pi22926), on the basis of its expression profile (21, 22) and its accumulation in the host nucleus to investigate whether translocation from P. infestans into host cells could be observed and whether it was sensitive or insensitive to BFA treatment. Using fusions of mRFP to a range of conventionally and unconventionally secreted proteins, we demonstrate that haustoria are general sites for protein secretion during infection.
RESULTS
Identification of a set of proteins that are conventionally secreted in vitro.
EPIC1 is a well-characterized apoplastic effector which has been shown to interact with and inhibit secreted plant defense proteases in the apoplast (23). It is secreted via the conventional ER-Golgi secretion pathway in P. infestans (9). To distinguish conventionally secreted proteins from those secreted nonconventionally, culture filtrate (CF) from an EPIC1 transformant constitutively expressing a C-terminally tagged mRFP fusion (EPIC1-mRFP) with a native signal peptide (SP) (9) was used for LC-MS/MS analysis with and without BFA treatment. Fusion protein expression and BFA sensitivity were confirmed by immunoblotting (24), showing that secretion of the EPIC1-mRFP fusion protein was attenuated by BFA treatment compared to the untreated control when grown in vitro (see Fig. S1 in the supplemental material).
Analysis of the efficacy of BFA treatments of the samples of the transformant expressing EPIC1-mRFP (9) used for the LC-MS/MS assay. Download FIG S1, PDF file, 0.2 MB (216.9KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
LC/MS/MS of the CF ± BFA treatment samples in Fig. S1 identified a total of 421 proteins, peptides of which were detected in at least one biological replicate (see Table S1A in the supplemental material). Of these, 92 genes encoding proteins with predicted signal peptides (SP) were identified using a combination of Phobius version 1.01 (25) and SignalP4.1 (Table S1B). We focused on proteins that (i) contained a secretion SP; (ii) showed enrichment in CF in the absence of BFA (based on log2 intensity fold change values corresponding to absence of BFA/presence of BFA ratios of >1); and (iii) were detectable in two or more biological replicates in the absence of BFA. The total number of SP-containing proteins enriched in CF in the absence of BFA was 30 (Table S1C). Nineteen of these SP-containing proteins seen to be enriched in the absence of BFA are predicted to have roles in infection (Table 1). These comprised 6 apoplastic effectors, 6 carbohydrate-active enzymes (CAZymes), 6 candidate microbe-associated molecular patterns (MAMPs), and 1 in planta-induced protein (ipiB3) of unknown function (26) (Table 1).
TABLE 1 .
Infection-associated secreted proteins enriched in culture filtrate in the absence of BFAa
| Protein class | Protein ID | Protein name |
|---|---|---|
| Apoplastic effectors | D0NBV1 | Cystatin-like cysteine protease inhibitor EPIC1 |
| D0MSS2 | Cystatin-like cysteine protease inhibitor EPIC4 | |
| D0MVC9 | Kazal-like extracellular serine protease inhibitor EPI1 | |
| D0MVC6 | Kazal-like extracellular serine protease inhibitor EPI2 | |
| D0N2T6 | Kazal-like extracellular serine protease inhibitorEPI6 | |
| D0NCU6 | Small cysteine-rich protein SCR108 | |
| MAMP-like proteins | D0P3R6 | Elicitin-like protein INF4 |
| D0NUD3 | Elicitin-like protein | |
| D0NKU2 | Elicitin-like protein | |
| D0N3P2 | Secretory protein Opel | |
| D0P1U2 | NPP1-like protein | |
| D0N0V1 | NPP1-like protein | |
| Carbohydrate-active enzymes (CAZymes) | D0MS99 | Pectinesterase PE1 |
| D0MQ88 | Glucan 1,3-beta-glucosidase | |
| D0NCV1 | Glucan 1,3-beta-glucosidase | |
| D0NSK6 | Glycoside hydrolase | |
| D0N0L9 | Glycosyl transferase | |
| D0NNQ8 | 1,3-Beta-glucanosyltransferase | |
| In planta-induced protein | D0NIK4 | IpiB3-like protein |
The presence of a signal peptide was detected for all proteins listed in the table. ID, identifier.
(A) List of proteins identified from culture filtrate of Phytophthora infestans transformant expressing EPIC1-mRFP with or without brefeldin A treatment. (B) List of proteins with signal peptide or transmembrane domain. (C) List of proteins enriched in the absence of brefeldin A treatment fractions. Download TABLE S1, XLSX file, 0.3 MB (291.8KB, xlsx) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Among the apoplastic effectors were EPIC1 and EPIC4, which act in the apoplast to inhibit cysteine proteases (23, 27). Also detected were Kazal-like extracellular serine protease inhibitors EPI2, EPI6, and EPI1, which may play an important role in P. infestans colonization of the host apoplast. EPI1 has been shown to inhibit the tomato pathogenesis-related protease P69B (28). In addition, the small cysteine-rich protein SCR108, like SCR74 and SCR96 (29, 30), potentially influences host interactions.
Candidate MAMPs included 2 elicitin-like proteins that may be recognized by the pattern recognition receptor ELR (elicitin response) from the wild potato Solanum microdontum, which has been shown to detect INF1 (31). In addition, elicitin-like protein INF4, which evades detection by ELR (31), was also enriched in the CF in the absence of BFA. NPP1 (necrosis-inducing Phytophthora protein)-like proteins (NLPs) have been found in many plant pathogens, and two were enriched in CF in the absence of BFA treatment. Several members of this protein family can trigger cell death in plants (32, 33). NLPs contain a conserved peptide sequence that acts as a MAMP (34, 35) and is detected by the receptor-like protein RLP23 (36). In addition to elicitin-like proteins and NLPs, a P. infestans homologue of the Opel protein from P. parasitica, which elicits immune responses in Nicotiana spp. (37), was also enriched in the absence of BFA.
CAZymes enriched in the absence of BFA included glucan 1,3-beta-glucosidase, pectinesterase (PE), glycosyl transferase, and 1,3-beta-glucanosyltransferase, all of which have been predicted to be phytopathogen virulence factors (10, 38, 39). PE was first purified and characterized from P. infestans CF in 1985, and it was speculated that it contributes to pathogenicity during infection (39). Findings obtained here suggest that there are many conventionally secreted proteins potentially involved in infection that can be detected in CF from in vitro-grown P. infestans. We selected a cell wall-degrading enzyme (CWDE), PITG_01029, encoding a pectinesterase (which we renamed PE1), and elicitin-like INF4 for further study to verify their conventional secretion. INF4 was selected as it was reported to evade recognition by the elicitin receptor ELR (31) and may thus potentially be overexpressed in transgenic P. infestans without eliciting immune responses, allowing its secretion during infection to be investigated.
INF4 and PE1 are upregulated during infection and enhance colonization.
To determine whether INF4 and PE1 transcripts accumulated to the highest levels during infection, quantitative reverse transcriptase PCR (qRT-PCR) was performed on cDNA derived from mycelium (M), sporangia, and Nicotiana benthamiana infected with zoospores of P. infestans isolate 3928A. Infected samples from 12, 24, 33, and 48 h postinfection (hpi) were collected for RNA isolation and cDNA synthesis. The constitutively expressed actA gene from P. infestans was used as an endogenous control. Avr3a, an RXLR effector from P. infestans which is known to be upregulated during infection (15, 40), was used as a marker of successful infection. Results indicated that the transcript levels of INF4 and PE1 at 12 hpi were approximately 1,000- and 100-fold elevated, respectively, compared to expression in the M (normalized to a value of 1 [log value of 0]). INF4 showed increased transcript abundance relative to the M throughout infection, similarly to Avr3a (Fig. S2) (15, 40), whereas the level of expression of PE1 decreased after 12 hpi (Fig. S2), suggesting that it may particularly contribute to pathogenicity at an early stage of infection. Two further independent biological replicates yielded similar expression profiles (Fig. S2).
INF4 and pectinesterase (PE) are highly upregulated during infection. Download FIG S2, PDF file, 0.3 MB (266.8KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We investigated whether PE1 and INF4 could enhance P. infestans colonization under conditions of transient expression in planta, as has been shown for several RXLR effectors previously (see, for example, reference 41). We used Agrobacterium tumefaciens-mediated expression in N. benthamiana of PE1 and INF4 C-terminal fusions with mRFP, including the native signal peptides, followed by inoculation with P. infestans strain 88069. Significantly larger disease lesions were observed in areas expressing PE1-mRFP and INF4-mRFP than in those expressing secreted mRFP (control) at 6 days postinoculation (dpi) (Fig. S3A and S3B). This indicated that both PE1 and INF4 confer a benefit to P. infestans during infection. The stability of expressed fusion proteins was confirmed by immunoblotting (Fig. S3C). As anticipated, in contrast to INF1, INF4 did not elicit cell death when expressed in N. benthamiana (Fig. S3D).
INF4 and PE enhance P. infestans colonization. Download FIG S3, PDF file, 0.2 MB (221KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
INF4 and PE1 are conventionally secreted from haustoria.
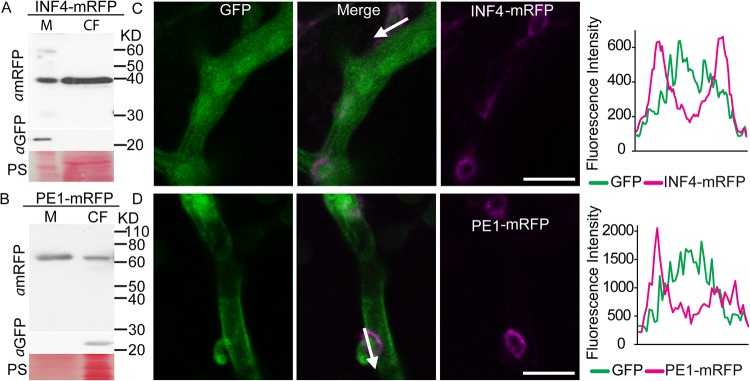
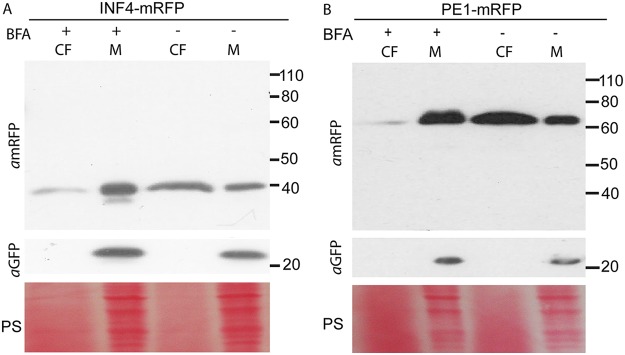
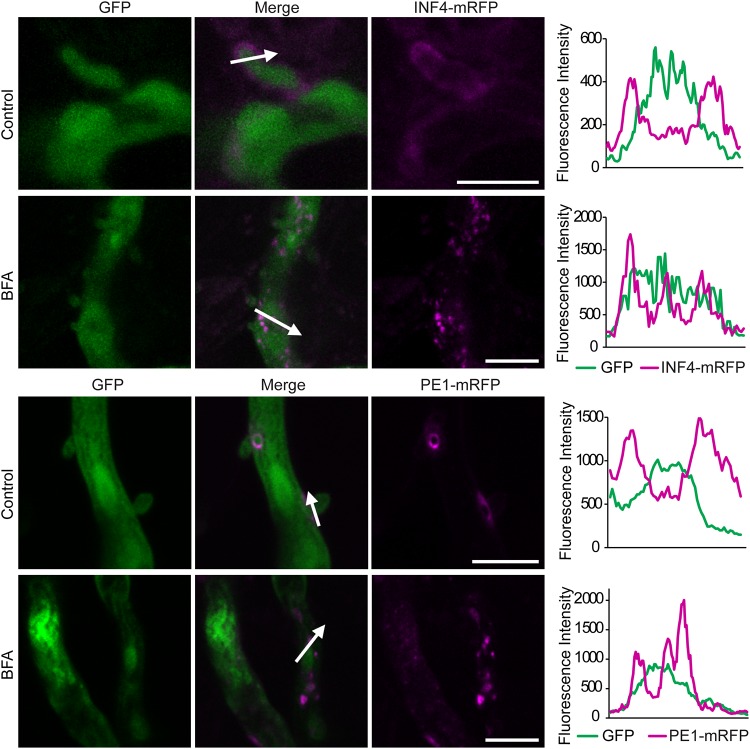
To localize PE1-mRFP and INF4-mRFP during infection, they were transformed into P. infestans. The native signal peptides for secretion were retained. Free green fluorescent protein (GFP) was expressed from the same vector to provide an intracellular control protein and to label hyphae (Fig. S4A). Two independent P. infestans transformants for each construct were used for in vitro analysis, showing that PE1-mRFP and INF4-mRFP were largely secreted from the mycelium into the CF. The absence of GFP in the CF indicated there was no detectable intracellular protein contamination of the CF (Fig. 1A and B). Confocal microscopy of N. benthamiana leaf tissue infected with the transformants revealed that secretion of both PE1-mRFP and INF4-mRFP occurred most strongly around the finger-like haustoria (Fig. 1C and D). Fluorescence from the INF4 fusion appeared to envelop the entire haustorium and extended to the surrounding hyphal surface, whereas PE1-mRFP accumulated particularly at the haustorial neck region, where it may help penetration (Fig. 1C and D). This is consistent with results from qRT-PCR, which showed that PE1 was most highly expressed at an early stage of infection (12 hpi). Thus, the haustorium is a site for secretion not only of the cytoplasmic RXLR effectors and apoplastic effectors (9, 15) but also of the MAMP-like protein INF4 and cell wall-degrading enzyme PE1.
FIG 1 .
Phytophthora infestans elicitor-like protein INF4 and cell wall-degrading enzyme pectinesterase PE1 are secreted from mycelium into the culture filtrate (CF) in vitro and at haustoria in planta. (A and B) Ham34 promoter-driven constitutive expression of the INF4 and PE1 fusion proteins tagged with monomeric red fluorescent protein (mRFP) in the mycelium (M) and secretion into the CF of in vitro-grown P. infestans were confirmed using an mRFP antibody. Enhanced green fluorescent protein (EGFP; detected with aGFP) was used as a marker of cytoplasmic protein, and the results showed that the CF preparation was not detectably contaminated with cellular protein. Size markers are indicated in kilodaltons, and protein loading is indicated by Ponceau staining (PS). (C and D) Confocal projections of P. infestans transformants expressing GFP in the hyphal cytoplasm and INF4-mRFP (C) or PE1-mRFP (D). Secretion of INF4 and PE fusions was observed particularly at haustoria in planta. The white arrows show the lines used to generate the fluorescence intensity profiles indicated in the graphs to the right of the images. The x-axis data in the graphs represent the distances (in micrometers) from one end of each white arrow in the images to the other end. Scale bars represent 10 µm.
An independent biological replicate of in vitro BFA treatment. Download FIG S4, PDF file, 0.2 MB (169.8KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
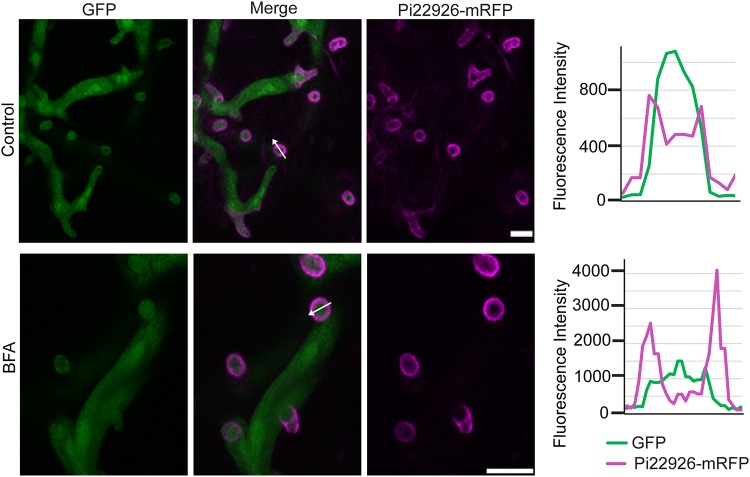
These P. infestans transformants were used to investigate secretion during in vitro growth, using the inhibitor BFA to confirm that PE1-mRFP and INF4-mRFP were conventionally secreted. In the absence of BFA, the intact fusion proteins were secreted into the CF in each case (Fig. 2). The cellular protein GFP was detected only in the mycelium, indicating negligible contamination of the CF with intracellular proteins. In BFA-treated transformants, secretion of PE1-mRFP and INF4-mRFP was effectively attenuated, with fusion proteins detected primarily in the CF samples not treated with BFA rather than in the CF samples treated with BFA (Fig. 2; see also Fig. S4B and C).
FIG 2 .
Brefeldin A (BFA) attenuates secretion of a MAMP-like protein and cell wall-degrading enzyme in vitro. BFA treatment of Phytophthora infestans transformants constitutively expressing mRFP-tagged INF4 or pectinesterase (PE1) was assayed by immunoblotting. (A) The secretion of INF4-mRFP was attenuated. (B) Secretion of the tagged cell wall-degrading enzyme PE1-mRFP was also sensitive to BFA treatment. αmRFP and αGFP were used as primary antibodies to detect mRFP and GFP, respectively. GFP expressed in the cytoplasm of the transformants was used as a cellular protein control to show that there was no detectable contamination of the filtered culture medium (CF) by mycelial (M) proteins. BFA-treated samples are indicated by a plus sign (+), and control samples are indicated by a minus sign (−). Protein size markers are indicated in kilodaltons, and protein loading is shown by Ponceau staining (PS).
In agreement with observations following the in vitro BFA sensitivity assay, confocal imaging of infected leaf tissue showed that mRFP fluorescence from P. infestans expressing PE1-mRFP or INF4-mRFP accumulated inside hyphae and haustoria after 24 h of exposure to BFA, suggesting that secretion had been attenuated in planta (Fig. 3). Moreover, in the presence of BFA, both PE1-mRFP and INF4-mRFP accumulated in vesicle-like structures (Fig. 3; seen also in the supplemental material). In total, 11 INF4-mRFP images each were obtained with and without BFA and 8 images each for PE1-mRFP. All images with BFA demonstrated fluorescence accumulation in vesicle-like structures.
FIG 3 .
Brefeldin A (BFA) attenuates secretion of INF4 and pectinesterase (PE1) from haustoria in planta. Confocal projections of the same transformants used in Fig. 2 in infected Nicotiana benthamiana revealed that INF4-mRFP and PE1-mRFP were secreted at haustoria. After BFA treatment, INF4-mRFP and PE1-mRFP were retained in the hyphae and haustoria, accumulating in vesicle-like structures, rather than outlining haustoria as in the water-treated control. White arrows represent the lines used for the fluorescence intensity profiles shown in the graphs to the right of each set of images. The x-axis data in the graphs represent the distances (in micrometers) from one end of each white arrow in the images to the other end. Scale bars = 10 µm. The BFA panel was imaged after 24 h of exposure to BFA (50 µg/ml). These images are representative of three independent biological replicates.
RXLR effector Pi22926 is nonconventionally secreted and translocated from Phytophthora infestans into host plant cells.
To further investigate whether cytoplasmic effectors from P. infestans are nonconventionally secreted from haustoria and delivered into host cells, and as a contrast to the conventionally secreted proteins examined in this work, an additional RXLR effector, Pi22926 (PITG_22926; Fig. S5A), was selected based on its induction during the biotrophic stage of infection (21). RXLR effector Pi22926, expressed transiently in N. benthamiana without SP-encoding sequences as a fusion protein (Pi22926-mRFP), accumulated in the nucleus and nucleolus (Fig. S5B), making it a suitable candidate effector for detecting translocation, as it is clearly detectable in a defined intracellular site in the host cell. The stability of the fusion protein was confirmed by immunoblotting (Fig. S5C).
Phytophthora infestans RXLR effector Pi22926 localizes to the nucleus and accumulates in the nucleolus. Download FIG S5, PDF file, 0.2 MB (166.9KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
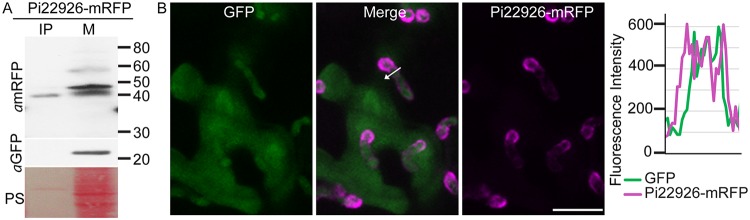
To observe whether Pi22926 was secreted at haustoria and delivered into host nuclei, we generated P. infestans transformants expressing SP-Pi22926-mRFP and coexpressing free GFP in the pathogen cytoplasm for labeling hyphae. Expression of SP-Pi22926-mRFP and GFP was driven by independent copies of the constitutive Ham34 promoter (Fig. S6A). Protein extraction and immunoblotting from in vitro-grown transgenic P. infestans showed that there were two bands from effector-mRFP fusions in the mycelium sample, the smaller of which was evident when immunoprecipitated (IP) from infected leaf material (Fig. 4A). No Pi22926-mRFP was detected in CF samples from in vitro growth, suggesting that the protein fusion was not stable under these conditions. GFP was detected in the mycelium fraction but was not detected in the IP from infected material, indicating that cytoplasmic proteins did not contaminate the mRFP_Trap beads.
FIG 4 .
Phytophthora infestans RXLR effector 22926 (Pi22926) is secreted from mycelium in vitro and localizes to haustoria during infection. (A) Expression of the Pi22926-monomeric red fluorescent protein (mRFP) fusion protein was confirmed using an mRFP antibody. Fusion protein was detected in in vitro-grown mycelium (M) and was IP from infected tissue by P. infestans transformants constitutively expressing Pi22926-mRFP fusions. Green fluorescent protein antibody (αGFP) was used as a cytoplasmic marker as described for Fig. 2. Size markers are indicated in kilodaltons, and protein loading is indicated by Ponceau staining (PS). (B) Confocal projection of intercellular hyphae and haustoria of the Pi22926-mRFP transformant in infected N. benthamiana leaf tissue, showing that the fusion protein was present at haustoria. The white arrow indicates the path used for the fluorescence intensity profile of mRFP and GFP fluorophores across the haustorium. The profile is shown at the right of the images. The x-axis data in the graphs represent the distance (in micrometers) from one end of the white arrow to the other end. Scale bar represents 10 µm.
Confocal projection of an additional transformant expressing SP-Pi22926-mRFP. Download FIG S6, PDF file, 0.3 MB (288.4KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
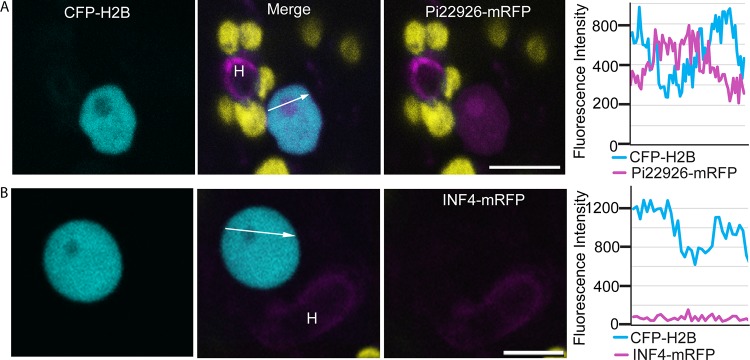
During infection, fluorescence from the Pi22926-mRFP fusion protein outlined the haustorium (Fig. 4B), further confirming that the haustorium is a major secretion site during infection. Importantly, confocal microscopy revealed that Pi22926-mRFP fusion protein from two independent P. infestans transformants was detectable in nuclei of haustoriated host cells, where it accumulated in the nucleolus in each case (Fig. 5A; see also Fig. S6B). From three independent biological replications for each transformant, a total of 42 haustoriated plant cells were examined, and mRFP fluorescence was observed in the nuclei in 40 of the 42 haustoriated host cells. This is in agreement with our previous observations obtained with RXLR effector Pi04314 (9). In contrast, no mRFP fluorescence was detected inside haustoriated host cells infected by transgenic lines expressing SP-INF4-mRFP (an example is shown in Fig. 5B).
FIG 5 .
Phytophthora infestans effector Pi22926 tagged with monomeric red fluorescent protein (Pi22926-mRFP) was translocated from the pathogen haustoria to the host nucleus and nucleolus. (A) Representative confocal projection of a haustoriated (H) N. benthamiana cell, in which the nucleus was labeled by CFP-NbH2B. Red fluorescence was observed in the nucleolus and nucleoplasm, indicating that Pi22926 fusion proteins have translocated from haustoria into the host cells. (B) Confocal projection of a typical nucleus from a leaf infected by the transformant expressing INF4-mRFP. No red fluorescence was detected in the nucleus. Chloroplast autofluorescence is indicated in yellow. The white arrows show the lines used for the fluorescence intensity profiles indicated in the graphs to the right of the image sets. The x-axis data in the graphs represent the distances (in micrometers) from one end of each white arrow in the images to the other end. These images are representative of 46 images of haustoriated cells from different independent biological replicates with four independent transformants. Bars, 10 µm.
To investigate whether secretion of Pi22926 was nonconventional, we treated infected leaf tissue with BFA. Confocal microscopy revealed that Pi22926-mRFP fluorescence still accumulated around haustoria following 24 h of exposure of infected N. benthamiana leaves to BFA (Fig. 6). This is consistent with the BFA-insensitive secretion of Pi04314 as reported previously (9) and further supports observations that cytoplasmic effectors and apoplast-active proteins are secreted from P. infestans haustoria via different pathways.
FIG 6 .
Secretion of the Phytophthora infestans cytoplasmic effector Pi22926 is insensitive to brefeldin A (BFA) during infection. Confocal projections of the Pi22926-mRFP-expressing transformant in infected N. benthamiana reveal that the fusion protein is preferentially secreted at haustoria even in the presence of BFA. White arrows indicate the lines used for the fluorescence intensity profiles shown in graphs to the right of each image. The x-axis data in the graphs represent the distances (in micrometers) from one end of each white arrow in the images to the other end. Scale bars represent 10 µm. The BFA panel was imaged after 24 h of exposure to BFA. These images are representative of more than 100 haustoria each either with or without BFA treatment from three independent biological replicates.
DISCUSSION
In this study, we identified candidate conventionally secreted proteins by LC-MS/MS analysis of the secreted proteome in the presence or absence of treatment with the ER-to-Golgi inhibitor BFA. This strategy revealed 19 P. infestans proteins with secretion signal peptides (SPs) that were enriched in extracellular medium in the absence of BFA and that are predicted to influence host plant interactions. These included apoplastic effectors, which suppress host defenses; elicitin-like proteins, NLPs, and Opel, which may elicit immune responses in the host; and CAZymes, which might change the biochemical characteristics of the host and pathogen cell walls to promote disease progression. We previously characterized the secretion of the typical apoplastic effector EPIC1, showing that it was sensitive to BFA (9). Therefore, we widened our study to a MAMP-like protein, INF4, and a cell wall-degrading enzyme, pectinesterase PE1, in this work. We showed that secretion of INF4 and PE1 is also sensitive to BFA and occurs at haustoria. This suggests the general rule that apoplast-active proteins are conventionally subject to ER-to-Golgi secretion. Moreover, and again in agreement with our earlier report (9), we show that a second cytoplasmic RXLR effector, Pi22926, is nonconventionally secreted from haustoria to be delivered to the host nucleus. These results are discussed below.
We observed that secreted PE1 localized specifically around the base of haustoria. As PE1 has a pectin-binding domain, this potentially reflects binding to the plant cell wall. PE proteins are major enzymes that facilitate plant cell wall modification and subsequent breakdown. Many pathogens use PE proteins to macerate the cell wall and gain access to plant cells (42). Overexpression of PE1 enhanced P. infestans colonization of N. benthamiana, suggesting that PEs play a critical role early in infection, softening cell walls to enable P. infestans to develop haustoria, leading to intimate contact with the host cell membrane.
Oomycetes are sterol auxotrophs, and elicitins have been shown to act as sterol carrier proteins (43), presumably to facilitate capture of sterols from the environment. Interestingly, some elicitins, such as INF1, are downregulated during infection (44, 45). This likely reflects the fact that they are targeted as MAMPs for recognition by host receptors (31). However, INF4 evades recognition by the elicitin receptor-like protein ELR (31). Here we confirmed that INF4 was not recognized in N. benthamiana (see Fig. S4D in the supplemental material) and so is unlikely to act as a MAMP. INF4 may have coevolved to escape host detection by receptors such as ELR but to retain its function in scavenging sterols, allowing this essential function to proceed during infection. These possibilities were not investigated further here but could be tested through a strategy such as mutagenesis of specific residues in INF4 to determine their impact on pathogenicity or sterol binding.
The haustorium is emerging as a major site of secretion of cytoplasmic RXLR effectors and apoplastic effectors and is the location of the transmembrane protein Hmp1, which is essential for infection (9, 15–18). To demonstrate the importance of the haustorial interface as a battleground between pathogen and host, a wide range of secreted proteins need to be studied. Here, we showed that PE1 was secreted from haustoria (Fig. 1D), consistent with previous studies providing evidence for host cell wall degradation by P. infestans at sites of haustorium formation (46, 47). Interestingly, whereas INF4 was localized in the extrahaustorial matrix, PE1 was localized specifically as a ring toward the base of the haustorium. The localization of the MAMP-like INF4 to haustoria (Fig. 1C) suggests that this is a likely site of (MAMP)-triggered immunity (PTI) activation (48) and that it is, therefore, vital to suppress PTI responses using effectors also secreted at this location (9, 15). This is the first observation of the secretion and localization of a MAMP-like protein and a cell wall-degrading enzyme involved in P. infestans pathogenesis.
RXLR effector Pi22926 was secreted from haustoria (Fig. 4) and accumulated in plant cell nuclei (Fig. 5), indicating that it was translocated into host cells as anticipated. Two protein bands were detected in mycelium samples, the smaller of which was evident during infection. As indicated for Avr3a by Wawra et al. (49), the different sizes of fusion protein could be explained by proteolytic cleavage of the RXLR domain. The predicted size of Pi22926-mRFP after SP removal is 47.8 kDa, whereas cleavage at the RXLR domain would reduce the size of the fusion protein by 2.92 kDa to 44.88 kDa. Further investigation with transformants in which the RXLR motif has been mutated will be needed to investigate whether proteolytic cleavage occurs.
Conclusion.
PE1 and INF4 were enriched in the absence of BFA treatment in the P. infestans secretome. Indeed, secretion of both PE1-mRFP and INF4-mRFP in P. infestans transformants was attenuated by BFA treatment (Fig. 2 and 3). This confirms that they are conventionally secreted VFs. The observation of haustorial secretion of a MAMP-like protein and a cell wall-degrading enzyme together with cytoplasmic effector Pi22926 and previously identified secreted proteins (9, 15, 16, 18) provides strong evidence that the haustorium is a major site of general secretion during infection. This report brings new insight into virulence determinants in P. infestans infection and suggests that secretion processes may provide good targets for chemical control of this economically devastating crop disease.
MATERIALS AND METHODS
Phytophthora infestans culture and Nicotiana benthamiana inoculation.
Phytophthora infestans wild-type strains 88069 and 3928 A and transgenic lines were cultured as described by Grenville-Briggs et al. (50). Infection inoculum was prepared and placed in 10-µl droplets onto plant leaves as described by Whisson et al. (15). Samples for qRT-PCR were collected as 4.5-mm-diameter discs from inoculation sites at 12, 24, 36, and 48 hpi. Nonadhering and ungerminated spores were removed by rinsing the leaf discs with sterile water.
Phytophthora infestans transformation vector construction and transformation.
The Ham34 promoter (Ham34P) was cloned from oomycete expression vector pTOR (GenBank accession number EU257520). Gene-specific primers (see Table S2 in the supplemental material) modified to contain restriction enzyme recognition sites were used in PCRs. PCR products were purified and digested with KpnI restriction endonuclease and ligated into the same restriction site in plasmid pPLRAG (9) to yield Ham34P-pPLRAG (pHPRAG). P. infestans genes encoding INF4 (PITG_21410), pectinesterase (PITG_01029, PE1), and Pi22926 were cloned, retaining the native predicted signal peptides. Gene-specific primers (Table S2) with restriction enzyme recognition sites were used to amplify the genes from genomic DNA of isolate 3928A. PCR products were purified, digested with NotI restriction endonuclease, and ligated into the same restriction sites in plasmid pHPRAG to generate pHPRAG _INF4, pHPRAG _PE1, and pHPRAG _Pi22926. Transformation of P. infestans was carried out using a modified polyethylene glycol (PEG)-CaCl2-lipofectin protocol (51), modified as described by Avrova et al. (18). P. infestans transformants were maintained in the dark at 19°C on RyeA agar supplemented with 20 µg/ml geneticin antibiotic.
Oligonucleotide primers for qRT-PCR and constructs for P. infestans transformation. Download TABLE S2, DOCX file, 0.01 MB (13.1KB, docx) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Vector construction and Agrobacterium tumefaciens transient assays.
P. infestans genes encoding INF4, PE1, and Pi22926 were amplified, retaining the native predicted signal peptide and C-terminal mRFP from pHPRAG_INF4, pHPRAG_PE1, and pHPRAG_Pi22926, respectively. Gene-specific primers that included flanking Gateway recombination sites at both termini were used in nested PCR (Table S2). Purified PCR products were recombined into pDONR201 (Invitrogen) to generate entry clones. The entry clones with genes of interest were recombined with pB2GW7 and electroporated into Agrobacterium tumefaciens strain AGL1 for expression of fusion proteins in planta (52). A. tumefaciens transient-transformation assays (ATTAs) were performed as described by Wang et al. (9). Hypersensitive response (HR) analyses were carried out by infiltrating agrobacteria mediating expression of the INF1 construct as positive control or INF4-mRFP at an OD600 of 0.5. The degree of HR was assessed at 4 and 8 dpi as previously described (16).
Quantitative RT-PCR analysis.
RNA was extracted using a Qiagen RNeasy plant minikit (Qiagen, Crawley, United Kingdom), according to the manufacturer’s instructions. DNA contamination was removed using a Turbo DNA-free kit (Ambion) based on the manufacturer’s protocol. First-strand cDNA was synthesized from 5 µg of total RNA by oligo(dT) priming using Superscript II RNaseH reverse transcriptase (Invitrogen Life Technologies, Inc. Ltd., Paisley, United Kingdom) following the manufacturer’s recommendations. Power SYBR green master mix (Applied Biosystems, Foster City, CA) was used for real-time qRT-PCR, which was performed on a Chromo4 thermal cycler (MJ Research, United Kingdom) using Opticon Monitor 3 software with the following program: 95°C for 15 min followed by 40 cycles for quantitative PCR of 95°C for 15 s and 60°C for 1 min and, finally, melting curve analysis from 58°C to 95°C. The primer sequences used in the reactions are listed in Table S2. Data analysis was carried out as described by Avrova et al. (53). In each case, the constitutively expressed P. infestans actA gene was used as the internal control.
Western blotting and immunoprecipitation.
P. infestans transformants were cultured in amended lima bean (ALB) liquid medium containing 20 µg/ml geneticin antibiotic (54). Mycelium was separated after 2 dpi using a 70-µm-pore-size filter. To test the effects of BFA on protein secretion in vitro, the method described previously by Wang et al. (9) was followed. Briefly, mycelia of transformants expressing INF4-mRFP, PE1-mRFP, and Pi22926-mRFP were incubated for 24 h in the dark at 19°C in 1 ml of ALB liquid medium containing 20 µg/ml geneticin and 50 µg/ml BFA (5 mg/ml in a dimethyl sulfoxide [DMSO] stock solution of BFA was diluted in water to 50 µg/ml). To reduce background levels of plant proteins in the medium for LC-MS/MS analysis, EPIC1-mRFP was incubated in 1 ml of diluted ALB by the use of modified minimum medium (1:1 [vol/vol]) based on the method of Leal et al. (55) as follows. Briefly, 5 g d-glucose; 0.5 g KH2PO4; 0.5 g MgSO4; 0.5 mg FeSO4 ⋅ 7H2O and ZnSO4 ⋅ 7H2O; 0.02 mg CuSO4 ⋅ 5H2O, MnCl2 ⋅ 4H2O, and Na2MoO4 ⋅ 2H2O; 1 mg thiamine hydrochloride; 10 mg β-sitosterol; 4.52 g methionine; and 4.03 g aspartate were combined and double-distilled water (ddH2O) was added to reach a final volume of 1 liter. The PH was adjusted to 7 by addition of 1 M KOH, and then the reaction mixture was autoclaved for 20 min at 121°C. Amino acids were sterilized by the use of a 0.22-µm-pore-size filter and were added after autoclaving. The culture filtrate (CF) was retained separately after removing mycelia, and four times the sample volume of cold (−20°C) acetone (Thermo Fisher Scientific, Loughborough, United Kingdom) was used to precipitate proteins overnight. Protein was collected by centrifugation at 10,000 × g for 10 min at 4°C. One hundred milligrams of P. infestans mycelium was ground in liquid nitrogen (LN2) and resuspended in the same volume of 2% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample-loading buffer (100 mM Tris, 4% SDS, 20% glycerol, 0.2% bromophenol blue). To immunoprecipitate mRFP-tagged Pi22926 from N. benthamiana leaves that were infected by a transformant expressing Pi22926-mRFP, proteins were extracted using GTEN buffer (10% [vol/vol] glycerol, 25 mM Tris-HCl [pH 7.5], 1 mM EDTA, 150 mM NaCl) combined with 10 mM dithiothreitol (DTT), protease inhibitor cocktail, 1 mM phenylmethyl sulfonyl fluoride, and 0.2% Nonidet P-40 and were then incubated with mRFP-Trap_M beads (Chromtek) for 90 min at 4°C. Beads were washed three times in GTEN buffer containing protease inhibitor cocktail and 1 mM phenylmethyl sulfonyl fluoride and were resuspended with 2× SDS-PAGE sample-loading buffer (56). Samples were loaded onto a 12% bis-Tris NuPAGE Novex gel (Invitrogen). Gel electrophoresis and membrane blocking and washing steps were performed as described by McLellan et al. (57). Primary antibodies amRFP and aGFP (Sigma-Aldrich) were used at a 1:4,000 dilution. Secondary antibodies anti-rat immunoglobulin G (IgG) horseradish peroxidase (HRP) and anti-mouse IgG HRP (Sigma-Aldrich) were used at 1:5,000 dilutions. ECL substrate (Thermo Scientific Pierce, Rockford, IL) was used to detect protein bands on immunoblots, following the manufacturer’s protocol.
LC-MS/MS analysis.
Secreted protein mixtures were digested by the use of 1 µg/µl Trypsin Gold (reconstituted with 50 mM acetic acid) at a 1:100 enzyme-to-substrate (microgram) ratio. Digested peptides were run on an LTQ-Orbitrap XL instrument (Thermo Fisher, United Kingdom) coupled to a Dionex UltiMate 3000 high-performance liquid chromatography (HPLC) system (Thermo Fisher Scientific, United Kingdom). Eluent A (3% acetonitrile–0.05% formic acid) and eluent B (80% acetonitrile–0.04% formic acid) were used for a 2% to 40% gradient. The top 7 most intense peaks in each MS1 scan were then taken for MS2 analysis. The level of resolution for MS2 spectra was 6,000, and the spectra were fragmented using collision-induced dissociation (CID) and a mass range of 335 to 1,800 m/z. MS/MS data were analyzed using Maxquant version 1.6.0.16 against the P. infestans T30-4 database from Uniprot, the MS/MS–flow-injection mass spectrometry (FIMS) tolerance was set to 20 ppm, and the ion trap mass spectrometer (ITMS) match tolerance was 0.15. Fixed modifications included carbamidomethyl (C), and variable modifications included oxidation (M); a maximum value of 2 missed cleavages was used. The protein false-discovery rate (FDR) was set to 0.01. The minimum peptide length was 7. Output data from Maxquant were filtered by the use of Perseus_1.5.6.0, peptide intensity data were transformed and used for P value analysis, and a two-way t test was used for analyzing differentially expressed proteins in the two samples being compared. Signal peptide (SP) and transmembrane prediction was performed using Phobius version 1.01 (25) and SignalP 4.1 (58) on amino acid sequences. hmmsearch version 3.1b1 (59) (PFAM release 31.0 [24 February 2017]) was used to identify PFAM domains within amino acid sequences. Gathering cutoffs (--cut_ga) was applied to hmmsearch to reduce levels of false positives.
Confocal microscopy.
N. benthamiana leaf pieces infected with biotrophy-stage P. infestans were mounted on slides and imaged using a Nikon A1R confocal microscope with a 40×/0.8 water dipping lens. Enhanced green fluorescent protein (EGFP) was imaged with 488-nm excitation, and emissions were collected at wavelengths between 500 and 530 nm. Cyan fluorescent protein (CFP) was imaged with 456-nm excitation, and emissions were collected at 480 nm. Imaging of mRFP was conducted using 561-nm excitation, and emissions were collected at wavelengths between 600 and 630 nm. The pinhole was set to 1 Airy unit for the fluorophore with the longest wavelength. Single optical-section images and z-series were collected from leaf tissue that was not heavily colonized by P. infestans to minimize the potential for interference from autofluorescence from the cell damage and death caused by infection. For BFA treatments in planta, 50 µg/ml BFA–water was infiltrated into leaf tissue infected by transformants (9). Treated leaves were imaged after 24 h.
Data availability.
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (60) partner repository with the data set identifier PXD009802.
Accession number(s).
UniProt protein accession numbers are listed in Table 1.
INF4-mRFP fluorescence during infection following treatment with BFA. Download MOVIE S1, AVI file, 14 MB (14.4MB, avi) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PE1-mRFP fluorescence during infection following treatment with BFA. Download MOVIE S2, AVI file, 14 MB (14.4MB, avi) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We thank Piers Hemsley for helpful discussions and for advice with protein analyses.
We are grateful to the China Scholarship Council for funds to support S.W. and to the Biotechnology and Biological Sciences Research Council (BBSRC) (grants BB/N009967/1, BB/L026880/1, and BB/J016500/1) and the Scottish Government Rural and Environment Science and Analytical Services Division (RESAS) for funding provided to P.R.J.B., P.C.B., L.W., and S.C.W.
Footnotes
Citation Wang S, Welsh L, Thorpe P, Whisson SC, Boevink PC, Birch PRJ. 2018. The Phytophthora infestans haustorium is a site for secretion of diverse classes of infection-associated proteins. mBio 9:e01216-18. https://doi.org/10.1128/mBio.01216-18.
REFERENCES
- 1.Goldberg DE, Cowman AF. 2010. Moving in and renovating: exporting proteins from Plasmodium into host erythrocytes. Nat Rev Microbiol 8:617–621. doi: 10.1038/nrmicro2420. [DOI] [PubMed] [Google Scholar]
- 2.Koeck M, Hardham AR, Dodds PN. 2011. The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cell Microbiol 13:1849–1857. doi: 10.1111/j.1462-5822.2011.01665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozkurt TO, Schornack S, Banfield MJ, Kamoun S. 2012. Oomycetes, effectors, and all that jazz. Curr Opin Plant Biol 15:483–492. doi: 10.1016/j.pbi.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Rafiqi M, Ellis JG, Ludowici VA, Hardham AR, Dodds PN. 2012. Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr Opin Plant Biol 15:477–482. doi: 10.1016/j.pbi.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Yang J, Yu J, Yao Z, Sun L, Shen Y, Jin Q. 2005. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res 33:D325–D328. doi: 10.1093/nar/gki008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Asai S, Shirasu K. 2015. Plant cells under siege: plant immune system versus pathogen effectors. Curr Opin Plant Biol 28:1–8. doi: 10.1016/j.pbi.2015.08.008. [DOI] [PubMed] [Google Scholar]
- 7.Fry WE, Birch PR, Judelson HS, Grünwald NJ, Danies G, Everts KL, Gevens AJ, Gugino BK, Johnson DA, Johnson SB, McGrath MT, Myers KL, Ristaino JB, Roberts PD, Secor G, Smart CD. 2015. Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology 105:966–981. doi: 10.1094/PHYTO-01-15-0005-FI. [DOI] [PubMed] [Google Scholar]
- 8.Whisson SC, Boevink PC, Wang S, Birch PR. 2016. The cell biology of late blight disease. Curr Opin Microbiol 34:127–135. doi: 10.1016/j.mib.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S, Boevink PC, Welsh L, Zhang R, Whisson SC, Birch PRJ. 2017. Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol 216:205–215. doi: 10.1111/nph.14696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raffaele S, Win J, Cano LM, Kamoun S. 2010. Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics 11:637. doi: 10.1186/1471-2164-11-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hahn M, Neef U, Struck C, Göttfert M, Mendgen K. 1997. A putative amino acid transporter is specifically expressed in haustoria of the rust fungus Uromyces fabae. Mol Plant Microbe Interact 10:438–445. doi: 10.1094/MPMI.1997.10.4.438. [DOI] [PubMed] [Google Scholar]
- 12.Struck C, Siebels C, Rommel O, Wernitz M, Hahn M. 1998. The plasma membrane H(+)-ATPase from the biotrophic rust fungus Uromyces fabae: molecular characterization of the gene (PMA1) and functional expression of the enzyme in yeast. Mol Plant Microbe Interact 11:458–465. doi: 10.1094/MPMI.1998.11.6.458. [DOI] [PubMed] [Google Scholar]
- 13.Voegele RT, Struck C, Hahn M, Mendgen K. 2001. The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc Natl Acad Sci U S A 98:8133–8138. doi: 10.1073/pnas.131186798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehrlich MA, Ehrlich HG. 1966. Ultrastructure of the hyphae and haustoria of Phytophthora infestans and hyphae of P. parasitica. Can J Bot 44:1495–1503. doi: 10.1139/b66-164. [DOI] [Google Scholar]
- 15.Whisson SC, Boevink PC, Moleleki L, Avrova AO, Morales JG, Gilroy EM, Armstrong MR, Grouffaud S, van West P, Chapman S, Hein I, Toth IK, Pritchard L, Birch PR. 2007. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 16.Gilroy EM, Breen S, Whisson SC, Squires J, Hein I, Kaczmarek M, Turnbull D, Boevink PC, Lokossou A, Cano LM, Morales J, Avrova AO, Pritchard L, Randall E, Lees A, Govers F, van West P, Kamoun S, Vleeshouwers VG, Cooke DE, Birch PR. 2011. Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol 191:763–776. doi: 10.1111/j.1469-8137.2011.03736.x. [DOI] [PubMed] [Google Scholar]
- 17.Liu T, Song T, Zhang X, Yuan H, Su L, Li W, Xu J, Liu S, Chen L, Chen T, Zhang M, Gu L, Zhang B, Dou D. 2014. Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun 5:4686. doi: 10.1038/ncomms5686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avrova AO, Boevink PC, Young V, Grenville-Briggs LJ, van West P, Birch PR, Whisson SC. 2008. A novel Phytophthora infestans haustorium-specific membrane protein is required for infection of potato. Cell Microbiol 10:2271–2284. doi: 10.1111/j.1462-5822.2008.01206.x. [DOI] [PubMed] [Google Scholar]
- 19.Giraldo MC, Dagdas YF, Gupta YK, Mentlak TA, Yi M, Martinez-Rocha AL, Saitoh H, Terauchi R, Talbot NJ, Valent B. 2013. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat Commun 4:1996. doi: 10.1038/ncomms2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meijer HJ, Hua C, Kots K, Ketelaar T, Govers F. 2014. Actin dynamics in Phytophthora infestans; rapidly reorganizing cables and immobile, long-lived plaques. Cell Microbiol 16:948–961. doi: 10.1111/cmi.12254. [DOI] [PubMed] [Google Scholar]
- 21.Cooke DE, Cano LM, Raffaele S, Bain RA, Cooke LR, Etherington GJ, Deahl KL, Farrer RA, Gilroy EM, Goss EM, Grünwald NJ, Hein I, MacLean D, McNicol JW, Randall E, Oliva RF, Pel MA, Shaw DS, Squires JN, Taylor MC, Vleeshouwers VG, Birch PR, Lees AK, Kamoun S. 2012. Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLoS Pathog 8:e1002940. doi: 10.1371/journal.ppat.1002940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AMV, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JIB, Bulone V, Cai G, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, Fischbach MA, Fugelstad J, Gilroy EM, Gnerre S, Green PJ, Grenville-Briggs LJ, Griffith J, Grünwald NJ, Horn K, Horner NR, Hu CH, Huitema E, Jeong DH, Jones AME, Jones JDG, Jones RW, Karlsson EK, Kunjeti SG, Lamour K, Liu Z, et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 23.Song J, Win J, Tian M, Schornack S, Kaschani F, Ilyas M, van der Hoorn RA, Kamoun S. 2009. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc Natl Acad Sci U S A 106:1654–1659. doi: 10.1073/pnas.0809201106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chardin P, McCormick F. 1999. Brefeldin A: the advantage of being uncompetitive. Cell 97:153–155. doi: 10.1016/S0092-8674(00)80724-2. [DOI] [PubMed] [Google Scholar]
- 25.Käll L, Krogh A, Sonnhammer ELL. 2007. Advantages of combined transmembrane topology and signal peptide prediction—the Phobius web server. Nucleic Acids Res 35:W429–W432. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieterse CM, van West P, Verbakel HM, Brassé PW, van den Berg-Velthuis GC, Govers F. 1994. Structure and genomic organization of the ipiB and ipiO gene clusters of Phytophthora infestans. Gene 138:67–77. doi: 10.1016/0378-1119(94)90784-6. [DOI] [PubMed] [Google Scholar]
- 27.Tian M, Win J, Song J, van der Hoorn R, van der Knaap E, Kamoun S. 2007. A Phytophthora infestans cystatin-like protein targets a novel tomato papain-like apoplastic protease. Plant Physiol 143:364–377. doi: 10.1104/pp.106.090050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tian M, Huitema E, Da Cunha L, Torto-Alalibo T, Kamoun S. 2004. A Kazal-like extracellular serine protease inhibitor from Phytophthora infestans targets the tomato pathogenesis-related protease P69B. J Biol Chem 279:26370–26377. doi: 10.1074/jbc.M400941200. [DOI] [PubMed] [Google Scholar]
- 29.Liu Z, Bos JI, Armstrong M, Whisson SC, da Cunha L, Torto-Alalibo T, Win J, Avrova AO, Wright F, Birch PR, Kamoun S. 2005. Patterns of diversifying selection in the phytotoxin-like scr74 gene family of Phytophthora infestans. Mol Biol Evol 22:659–672. doi: 10.1093/molbev/msi049. [DOI] [PubMed] [Google Scholar]
- 30.Chen XR, Li YP, Li QY, Xing YP, Liu BB, Tong YH, Xu JY. 2016. SCR96, a small cysteine-rich secretory protein of Phytophthora cactorum, can trigger cell death in the Solanaceae and is important for pathogenicity and oxidative stress tolerance. Mol Plant Pathol 17:577–587. doi: 10.1111/mpp.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du J, Verzaux E, Chaparro-Garcia A, Bijsterbosch G, Keizer LC, Zhou J, Liebrand TW, Xie C, Govers F, Robatzek S, van der Vossen EA, Jacobsen E, Visser RG, Kamoun S, Vleeshouwers VG. 2015. Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat Plants 1:15034. doi: 10.1038/nplants.2015.34. [DOI] [PubMed] [Google Scholar]
- 32.Fellbrich G, Romanski A, Varet A, Blume B, Brunner F, Engelhardt S, Felix G, Kemmerling B, Krzymowska M, Nürnberger T. 2002. NPP1, a Phytophthora-associated trigger of plant defense in parsley and Arabidopsis. Plant J 32:375–390. doi: 10.1046/j.1365-313X.2002.01454.x. [DOI] [PubMed] [Google Scholar]
- 33.Gijzen M, Nürnberger T. 2006. Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67:1800–1807. doi: 10.1016/j.phytochem.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 34.Böhm H, Albert I, Oome S, Raaymakers TM, Van den Ackerveken G, Nürnberger T. 2014. A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog 10:e1004491. doi: 10.1371/journal.ppat.1004491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oome S, Van den Ackerveken G. 2014. Comparative and functional analysis of the widely occurring family of Nep1-like proteins. Mol Plant Microbe Interact 27:1081–1094. doi: 10.1094/MPMI-04-14-0118-R. [DOI] [PubMed] [Google Scholar]
- 36.Albert I, Böhm H, Albert M, Feiler CE, Imkampe J, Wallmeroth N, Brancato C, Raaymakers TM, Oome S, Zhang H, Krol E, Grefen C, Gust AA, Chai J, Hedrich R, Van den Ackerveken G, Nürnberger T. 2015. An RLP23-SOBIR1-BAK1 complex mediates NLP-triggered immunity. Nat Plants 1:15140. doi: 10.1038/nplants.2015.140. [DOI] [PubMed] [Google Scholar]
- 37.Chang YH, Yan HZ, Liou RF. 2015. A novel elicitor protein from Phytophthora parasitica induces plant basal immunity and systemic acquired resistance. Mol Plant Pathol 16:123–136. doi: 10.1111/mpp.12166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim YK, Wang Y, Liu ZM, Kolattukudy PE. 2002. Identification of a hard surface contact-induced gene in Colletotrichum gloeosporioidesconidia as a sterol glycosyl transferase, a novel fungal virulence factor. Plant J 30:177–187. doi: 10.1046/j.1365-313X.2002.01284.x. [DOI] [PubMed] [Google Scholar]
- 39.Förster H, Rasched I. 1985. Purification and characterization of extracellular pectinesterases from Phytophthora infestans. Plant Physiol 77:109–112. doi: 10.1104/pp.77.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Armstrong MR, Whisson SC, Pritchard L, Bos JI, Venter E, Avrova AO, Rehmany AP, Böhme U, Brooks K, Cherevach I, Hamlin N, White B, Fraser A, Lord A, Quail MA, Churcher C, Hall N, Berriman M, Huang S, Kamoun S, Beynon JL, Birch PR. 2005. An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc Natl Acad Sci U S A 102:7766–7771. doi: 10.1073/pnas.0500113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Boevink P, McLellan H, Armstrong M, Bukharova T, Qin Z, Birch PR. 2015. A host KH RNA-binding protein is a susceptibility factor targeted by an RXLR effector to promote late blight disease. Mol Plant 8:1385–1395. doi: 10.1016/j.molp.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fries M, Ihrig J, Brocklehurst K, Shevchik VE, Pickersgill RW. 2007. Molecular basis of the activity of the phytopathogen pectin methylesterase. EMBO J 26:3879–3887. doi: 10.1038/sj.emboj.7601816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mikes V, Milat ML, Ponchet M, Panabières F, Ricci P, Blein JP. 1998. Elicitins, proteinaceous elicitors of plant defense, are a new class of sterol carrier proteins. Biochem Biophys Res Commun 245:133–139. doi: 10.1006/bbrc.1998.8341. [DOI] [PubMed] [Google Scholar]
- 44.Kamoun S, van der Lee T, van den Berg-Velthuis G, de Groot KE, Govers F. 1998. Loss of production of the elicitor protein INF1 in the clonal lineage US-1 of Phytophthora infestans. Phytopathology 88:1315–1323. doi: 10.1094/PHYTO.1998.88.12.1315. [DOI] [PubMed] [Google Scholar]
- 45.Bos JI, Armstrong MR, Gilroy EM, Boevink PC, Hein I, Taylor RM, Zhendong T, Engelhardt S, Vetukuri RR, Harrower B, Dixelius C, Bryan G, Sadanandom A, Whisson SC, Kamoun S, Birch PR. 2010. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc Natl Acad Sci U S A 107:9909–9914. doi: 10.1073/pnas.0914408107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hohl HR, Stössel P. 1976. Host-parasite interfaces in a resistant and a susceptible cultivar of Solanum tuberosum inoculated with Phytophthora infestans: tuber tissue. Can J Bot 54:900–912. doi: 10.1139/b76-094. [DOI] [Google Scholar]
- 47.Shimony C, Friend J. 1975. Ultrastructure of the interaction between Phytophthora infestans and leaves of two cultivars of potato (Solanum tuberosum L.) Orion and Majestic. New Phytol 74:59–65. doi: 10.1111/j.1469-8137.1975.tb01339.x. [DOI] [Google Scholar]
- 48.Kanneganti TD, Huitema E, Cakir C, Kamoun S. 2006. Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nepl-like protein PiNPP1.1 and INF1 elicitin. Mol Plant Microbe Interact 19:854–863. doi: 10.1094/MPMI-19-0854. [DOI] [PubMed] [Google Scholar]
- 49.Wawra S, Trusch F, Matena A, Apostolakis K, Linne U, Zhukov I, Stanek J, Koźmiński W, Davidson I, Secombes CJ, Bayer P, van West P. 2017. The RxLR motif of the host targeting effector AVR3a of Phytophthora infestans is cleaved before secretion. Plant Cell 29:1184–1195. doi: 10.1105/tpc.16.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grenville-Briggs LJ, Avrova AO, Bruce CR, Williams A, Whisson SC, Birch PR, van West P. 2005. Elevated amino acid biosynthesis in Phytophthora infestans during appressorium formation and potato infection. Fungal Genet Biol 42:244–256. doi: 10.1016/j.fgb.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 51.Judelson HS, Tyler BM, Michelmore RW. 1991. Transformation of the oomycete pathogen, Phytophthora infestans. Mol Plant Microbe Interact 4:602–607. doi: 10.1094/MPMI-4-602. [DOI] [PubMed] [Google Scholar]
- 52.Karimi M, Depicker A, Hilson P. 2007. Recombinational cloning with plant gateway vectors. Plant Physiol 145:1144–1154. doi: 10.1104/pp.107.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Avrova AO, Venter E, Birch PRJ, Whisson SC. 2003. Profiling and quantifying differential gene transcription in Phytophthora infestans prior to and during the early stages of potato infection. Fungal Genet Biol 40:4–14. doi: 10.1016/S1087-1845(03)00063-X. [DOI] [PubMed] [Google Scholar]
- 54.Bruck RI, Fry WE, Apple AE, Mundt CC. 1981. Effect of protectant fungicides on the developmental stages of Phytophthora infestans in potato foliage. Phytopathology 71:164–166. doi: 10.1094/Phyto-71-164. [DOI] [Google Scholar]
- 55.Leal JA, Gallegly ME, Lilly VG. 1971. The value of 21 amino acids as nitrogen sources for Phytophthora cactorum and P. heveae. Can J Microbiol 17:1319–1325. doi: 10.1139/m71-211. [DOI] [PubMed] [Google Scholar]
- 56.Boevink PC, Wang X, McLellan H, He Q, Naqvi S, Armstrong MR, Zhang W, Hein I, Gilroy EM, Tian Z, Birch PR. 2016. A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat Commun 7:10311. doi: 10.1038/ncomms10311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLellan H, Boevink PC, Armstrong MR, Pritchard L, Gomez S, Morales J, Whisson SC, Beynon JL, Birch PR. 2013. An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog 9:e1003670. doi: 10.1371/journal.ppat.1003670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 59.Johnson LS, Eddy SR, Portugaly E. 2010. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinformatics 11:431. doi: 10.1186/1471-2105-11-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vizcaíno JA, Csordas A, del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, Xu QW, Wang R, Hermjakob H. 2016. 2016 update of the PRIDE database and related tools. Nucleic Acids Res 44(D1):D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of the efficacy of BFA treatments of the samples of the transformant expressing EPIC1-mRFP (9) used for the LC-MS/MS assay. Download FIG S1, PDF file, 0.2 MB (216.9KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
(A) List of proteins identified from culture filtrate of Phytophthora infestans transformant expressing EPIC1-mRFP with or without brefeldin A treatment. (B) List of proteins with signal peptide or transmembrane domain. (C) List of proteins enriched in the absence of brefeldin A treatment fractions. Download TABLE S1, XLSX file, 0.3 MB (291.8KB, xlsx) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
INF4 and pectinesterase (PE) are highly upregulated during infection. Download FIG S2, PDF file, 0.3 MB (266.8KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
INF4 and PE enhance P. infestans colonization. Download FIG S3, PDF file, 0.2 MB (221KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
An independent biological replicate of in vitro BFA treatment. Download FIG S4, PDF file, 0.2 MB (169.8KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phytophthora infestans RXLR effector Pi22926 localizes to the nucleus and accumulates in the nucleolus. Download FIG S5, PDF file, 0.2 MB (166.9KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Confocal projection of an additional transformant expressing SP-Pi22926-mRFP. Download FIG S6, PDF file, 0.3 MB (288.4KB, pdf) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotide primers for qRT-PCR and constructs for P. infestans transformation. Download TABLE S2, DOCX file, 0.01 MB (13.1KB, docx) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
INF4-mRFP fluorescence during infection following treatment with BFA. Download MOVIE S1, AVI file, 14 MB (14.4MB, avi) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
PE1-mRFP fluorescence during infection following treatment with BFA. Download MOVIE S2, AVI file, 14 MB (14.4MB, avi) .
Copyright © 2018 Wang (王姝梅) et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE (60) partner repository with the data set identifier PXD009802.