Abstract
Fluoroquinolone (FQ)-resistant Salmonella spp. were listed by the WHO in 2017 as priority pathogens for which new antibiotics were urgently needed. The overall global burden of Salmonella infections is high, but differs per region. Whereas typhoid fever is most prevalent in South and South-East Asia, non-typhoidal salmonellosis is prevalent across the globe and associated with a mild gastroenteritis. By contrast, invasive non-typhoidal Salmonella cause bloodstream infections associated with high mortality, particularly in sub-Saharan Africa. Most Salmonella strains from clinical sources are resistant to first-line antibiotics, with FQs now being the antibiotic of choice for treatment of invasive Salmonella infections. However, FQ resistance is increasingly being reported in Salmonella, and multiple molecular mechanisms are already described. Whole-genome sequencing (WGS) is becoming more frequently used to analyse bacterial genomes for antibiotic-resistance markers, and to understand the phylogeny of bacteria in relation to their antibiotic-resistance profiles. This mini-review provides an overview of FQ resistance in Salmonella, guided by WGS studies that demonstrate that WGS is a valuable tool for global surveillance.
Keywords: Salmonella, fluoroquinolones, ciprofloxacin, antibiotic resistance, whole-genome sequencing
Data Summary
Supplementary material is available with the online version of this article.
Impact Statement.
In 2016, the United Nations General Assembly underlined the threat of antibiotic resistance and committed to join forces to combat this threat. Antibiotic resistance could cause a predicted 10 million deaths and have detrimental economic effects by 2050 if no actions are taken (https://amr-review.org/sites/default/files/160518_Final paper_with cover.pdf). In 2017, the WHO published a priority list of antibiotic-resistant bacteria, to support research and development of new antibiotics. Fluoroquinolone (FQ)-resistant Salmonella spp. were listed as a high priority. FQs have broad-spectrum activity and good pharmacokinetics for clinical use, and are important antibiotics for treatment of invasive bacterial infections, such as typhoid fever and invasive non-typhoidal Salmonella (iNTS) infections. Enteric fever (caused by the Salmonella enterica subspecies enterica serotypes Typhi and Paratyphi A, B and C) and iNTS (mainly caused by the serotypes Typhimurium and Enteritidis) have the highest impact and mortality in low- and middle-income countries. However, the resistance of Salmonella against FQs has been increasingly reported. The understanding of the FQ-resistance mechanisms and spread in Salmonella has significantly advanced through the implementation of whole-genome sequencing (WGS) during the past 5 years. Here, we review the genetic mechanisms of FQ resistance reported by WGS studies on Salmonella.
Introduction
Salmonellae are Gram-negative bacteria, and strains that are pathogenic to humans are traditionally subdivided into two major groups based on their clinical presentation: typhoidal Salmonella and non-typhoidal Salmonella (NTS). Typhoidal Salmonella, comprising the Salmonella enterica subspecies enterica (hereafter Salmonella) serovars Typhi and Paratyphi A, B and C, cause a systemic disease, also known as enteric fever [1, 2]. Human-restricted Salmonella Typhi is the dominant cause of typhoid fever, with an estimated number of cases between 21.7 million [3] and 26.9 million per year [4], and an estimated 217 000 deaths per year [3]. The diverse group of NTS strains consists of more than 2500 serovars, which generally have different animals as hosts, and cause milder gastro-intestinal infections in humans, resulting in an estimated 93.8 million cases and 155 000 deaths each year [5]. However, some NTS strains, referred to as invasive NTS (iNTS), cause bloodstream infections with invasion of other organs. The global yearly burden of iNTS is estimated at 3.4 million infections and 681 316 deaths [6], and iNTS is highly prevalent in sub-Saharan Africa, where malnutrition, malaria and human immunodeficiency virus infections form major risk factors [7–9]. In sub-Saharan Africa, specific lineages of Salmonella serovars Typhimurium and Enteritidis have undergone genomic evolution associated with niche adaptation towards invasive disease in humans [10–13].
Multidrug resistance (MDR) in Salmonella is defined as co-resistance to the first-line antibiotics ampicillin, chloramphenicol and trimethoprim/sulfamethoxazole. The high prevalence of MDR in typhoidal Salmonella and iNTS necessitates the use of second-line antibiotics [14]. The fluoroquinolone (FQ) ciprofloxacin and the third-generation cephalosporin ceftriaxone are now the recommended drugs to treat invasive Salmonella infections or patients at risk of developing an invasive infection [15]. The macrolide antibiotic azithromycin can be used as an alternative [14]. Resistance to these recommended antibiotics is, however, increasingly described in Salmonella [9, 14, 16, 17]. The U.S. National Antimicrobial Resistance Monitoring System (NARMS) reported an increase in the percentage of Salmonella isolates that are non-susceptible [i.e. with minimum inhibitory concentration (MIC) values above the susceptibility breakpoint, see Supplementary Data S1, available with the online version of this article] to ciprofloxacin from <0.5 % up to 3.5 % since 1996 [18, 19]. Moreover, 6 % of Salmonella isolates were non-susceptible to ciprofloxacin in the EUCAST (European Committee on Antimicrobial Susceptibility Testing) database in 2015 [19].
In 2017, the WHO specifically ranked FQ resistant Salmonella as a high priority pathogen for the research and development of new antibiotics [20]. This ranking was based on ten criteria, of which FQ-resistant Salmonellae rank high for: (1) prevalence in the community, (2) transmissibility and zoonotic potential, (3) length of hospitalization after infection, and (4) unlikeliness of development of alternative antibiotics in the nearby future. Additional important criteria are the 10 year prevalence of FQ resistance among Salmonella Typhi and Paratyphi strains in the Americas, South Asia and South-East Asia, and the high mortality rates (up to 20 % associated with iNTS in sub-Saharan Africa [7, 20]). In this mini-review, we present and discuss the current situation of FQ resistance in Salmonella, guided by WGS studies, with a focus on molecular mechanisms.
FQ: activity and resistance
Quinolones, such as nalidixic acid, are antibiotics that target the bacterial type II topoisomerases, and more specifically the DNA gyrase and the DNA topoisomerase IV [21]. Both proteins are encoded by the gyrA, gyrB and parC, parE genes, respectively, and modulate DNA supercoiling. Quinolones inhibit these enzymes, resulting in disrupted chromosome replication and rapid bacterial death [22–24]. FQs are quinolones with a single fluorine substituent, which increases DNA gyrase inhibitory activity and facilitates penetration into the bacterial cell [25–27]. While levofloxacin, gatifloxacin, moxifloxacin and gemifloxacin show the highest efficacy against Gram-positive bacteria, ciprofloxacin is most effective against Gram-negative bacteria, such as Salmonella [25].
Multiple resistance mechanisms against quinolones have been described in bacteria. First, mutations in the quinolone-resistance-determining regions (QRDRs) of the chromosomal gyr and par genes result in a lower quinolone-binding affinity of the topoisomerase enzymes [21, 28, 29]. Secondly, plasmid-mediated quinolone resistance (PMQR) involves acquisition of (i) qnr genes (qnrA, qnrB, qnrS, qnrC, qnrD), encoding topoisomerase-binding proteins that provide physical protection from quinolones [22, 30–32], (ii) the aac(6′)-lb-cr gene, encoding a modifying enzyme that decreases FQ activity [21, 23], and (iii) oqxAB and qepA, encoding quinolone efflux pumps [21, 25]. Finally, downregulation and upregulation of chromosome-encoded porins or multidrug efflux pumps (e.g. AcrAB-TolC), respectively, lower the cellular FQ concentrations [21, 22, 25].
Resistance against FQs is determined phenotypically, and the reference method uses measurement of the MIC for ciprofloxacin. Standardized cut-off values are provided by the Clinical and Laboratory Standards Institute (CLSI) and EUCAST. Resistance is defined as ciprofloxacin MIC values ≥1 µg ml−1, while MIC values ≤0.06 µg ml−1 indicate susceptibility [33]. Intermediate values are associated with treatment failure in Salmonella [34, 35], and are referred to as decreased ciprofloxacin susceptibility (DCS). A practical introduction to in vitro FQ susceptibility testing in Salmonella is provided in Supplementary Data S1. Detailed information on the definitions, molecular mechanisms and clinical impact of FQ susceptibility, DCS and FQ resistance is presented in Table S1. In this mini-review, we use the term ‘FQ resistance markers’ to group all molecular mechanisms that cause resistance to quinolones and non-susceptibility to FQs.
FQ resistance in typhoidal Salmonella
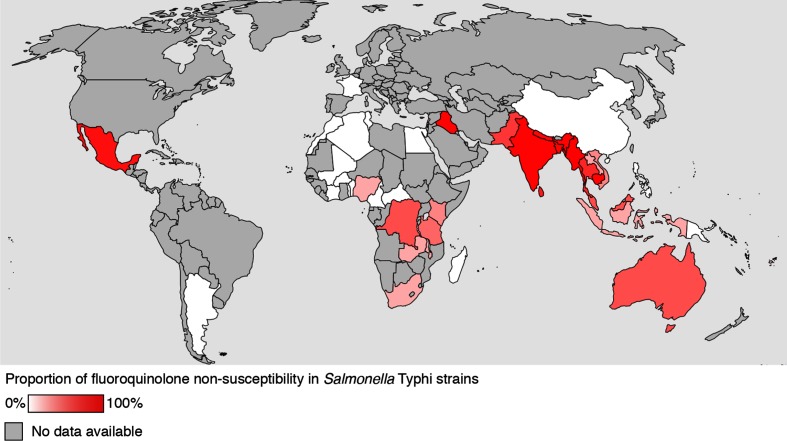
The implementation of WGS opened out our understanding of the prevalence and spread of FQ resistance in Salmonella Typhi. FQ resistance mechanisms in Salmonella Typhi as reported by WGS are summarized in Table 1 and the global distribution of FQ resistance in Salmonella Typhi is shown on the map in Fig. 1. In 2015, a large collaborative effort using WGS on 1832 isolates from 63 countries unravelled the global population structure of Salmonella Typhi [36]. The authors reported the spread of the dominant multidrug-resistant Salmonella Typhi clade H58 from Asia to East Africa and Oceania, which is more significantly associated with QRDR mutations (predominantly Ser83Phe, i.e. a point mutation in codon 83, resulting in a serine to phenylalanine amino acid change) compared to other Salmonella Typhi [36]. Multiple subsequent studies using WGS have reported FQ resistance markers in additional Salmonella Typhi isolates [36–40] (Table 1). Interestingly, accumulating mutations in the QRDR caused Salmonella Typhi to incrementally evolve towards increasing MIC values. Ciprofloxacin-susceptible strains (MIC≤0.06 µg ml−1) acquired a gyrA Ser83Phe single mutation causing DCS (MIC=0.12–0.5 µg ml−1) and additional gyrA and parC mutations, encoding Asp87Asn and Ser80Ile, respectively, caused high-level FQ resistance (MIC≥4 µg ml−1) [40]. Strains with multiple gyr and par mutations were reported from Cambodia, India and Nepal [36–39] (Table 1). Additionally, the in vitro evidence that QRDR mutations increase the fitness of Salmonella Typhi [41] is indicative that FQ resistance is irreversible and likely to remain.
Table 1. FQ-resistance markers in typhoidal salmonellae, reported by WGS.
n is the number of isolates sequenced, with superscript letters indicating whether the isolates were serotype Typhi (T) or Paratyphi A (P). The ‘% H58’ column indicates the percentage of Typhi isolates that were identified as part of the H58 clade for each region. The percentage of sequenced isolates containing FQ-resistance markers is reported under ‘% FQR markers’. The right panel of the table provides an overview of the identified FQ-resistance mechanisms per study. Each line represents a combination of FQ markers that was observed in the respective study. Mutations in gyrase (gyr) and topoisomerase IV (par) encoding genes are provided as resulting changes in residue, and presented per gene and per identified combination. na, Not available.
| Reference | Region or country | nT or P | % H58 | % FQR markers | FQ–resistance marker | ||||
|---|---|---|---|---|---|---|---|---|---|
| PMQR | Mutations in gyr and par | ||||||||
| gyrA | gyrB | parC | parE | ||||||
| [36]* | 63 countries (Africa; Asia) | 1832 (371; 1061) |
47 (46; 62) |
34 (10; 49) |
– | Ser83Phe | – | – | – |
| – | Ser83Phe | – | – | Asp420Asn | |||||
| – | Asp87Tyr | – | – | – | |||||
| – | – | Ser464Phe | – | – | |||||
| – | Ser83Tyr+Ser83Phe | – | Ser80Ile | – | |||||
| qnrS | – | – | – | – | |||||
| [39] | Cambodia | 64T | 98 | 97 | – | Ser83Phe | – | – | – |
| – | – | Ser464Phe | – | – | |||||
| – | Ser83Phe+Asp87Asn | – | – | – | |||||
| 21P | 0 | 100 | – | Ser83Phe | – | – | – | ||
| – | Asp87Gly | – | – | – | |||||
| [40]† | South Asia and South-East Asia | 107T | 73 | na | – | Ser83Phe | – | – | – |
| – | Ser83Tyr | – | – | – | |||||
| – | Asp87Asn | – | – | – | |||||
| – | Ser83Phe | – | Glu84Gly | – | |||||
| – | Ser83Phe | – | – | Asp420Asn | |||||
| – | Ser83Phe | Asp87Asn | Ser80Ile | – | |||||
| [37] | Nepal | 78T | 83 | 81 | – | Ser83Phe | – | – | – |
| – | Ser83Phe+Asp87Asn | – | Ser80Ile | – | |||||
| – | Ser83Phe+Asp87Val | – | Ser80Ile | Ala364Val | |||||
| [38] | Cambodia | 209T | 97 | 95 | – | Ser83Phe | – | – | – |
| [43] | Nigeria | 128T | 0 | 5 | qnrS | – | – | – | – |
| – | Ser83Phe | – | – | – | |||||
| – | Ser83Tyr | – | – | – | |||||
| [42]† | Zambia | 32T | 100 | 4 | – | Ser83Tyr | – | – | – |
| – | Asp87Asn | – | – | – | |||||
| [46]† | DR Congo | 1T | 0 | 100 | – | Ser83Phe | – | – | – |
| [47]† | India, New Delhi | 4T | na | 75 | qnrB | – | – | – | – |
| [49]† | Pakistan | 87T | 100 | 100 | qnrS | Ser83Phe | – | – | – |
*Detailed information is provided at: www.stoptyphoid.org.
†Isolates were selected for their resistance properties prior to sequencing, i.e. implicates biased sampling.
Fig. 1.
Percentage of FQ-resistance markers identified in whole-genome sequenced Salmonella Typhi isolates per country. The percentage of isolates carrying resistance markers are indicated with a colour gradient from 0 % (white) to 100 % (dark red). Countries for which no sequencing data is available are marked in grey. Data originates from the following studies: Wong et al. [36]; International Typhoid Consortium 2016 [43]; Hendriksen et al. [42]; Pham Thanh et al. [37]; Kuijpers et al. [39].
In Africa, FQ-resistance markers were present in Salmonella Typhi H58 from Kenya, Tanzania, Malawi South Africa and Zambia [36, 42]. Interestingly, QRDR mutations were also reported in non-H58 Typhi in the Democratic Republic of the Congo (DR Congo) [36] and Nigeria [43] (Table 1). These studies suggest a lower prevalence and spread of FQ-resistance markers in Africa compared to Asia (Table 1, Fig. 1). Also in Africa, the gyrA Ser83Phe mutation was most frequently observed [44]. This may in part reflect the adaptability of Salmonella Typhi to changing antibiotic pressures with less FQ being used in Africa compared to Asia. However, given the varying incidence of typhoid fever between African regions [45] and the unavailability of bloodstream infection surveillance in large parts of Africa, the exact proportion of FQ-resistant strains in Africa remains elusive. For example, recently a single Salmonella Typhi isolate showing a Ser83Phe mutation in gyrA causing DCS, in combination with extended-spectrum β-lactamase (ESBL) production, was identified in the DR Congo [46]; in remote areas (such as in this report), it remains unclear whether such an isolate is part of a larger undetected outbreak with increased resistance.
Overall, PQMR in Salmonella Typhi is more rare than QRDR mutations and has been identified using WGS in isolates from Bangladesh [qnrS1 on IncFIB(K) plasmid, n=5], South Africa [qnrS2 on IncFIB(K) plasmid, n=1], India (qnrB7 on IncX3 plasmid, n=4) and Nigeria (qnrS on Kpn3 plasmid, n=1) [36, 43, 47] (Table 1). This low prevalence of PMQR is in line with a recent meta-analysis of FQ-resistant Salmonella in Africa [44], and reports from Asia [48]. However, an ongoing outbreak of extensively drug-resistant and ESBL-producing Salmonella Typhi H58 from Pakistan was associated with QRDR mutations and the qnrS gene [49]. The presence of PMQR can provide a favourable environment for the selection of chromosomal QRDR mutations in Salmonella [19], which was also observed for other Enterobacteriaceae [50, 51].
Less WGS data are available for Salmonella Paratyphi A. In Cambodia, a recent increase of DCS in Salmonella Paratyphi A was predominantly associated with a Ser83Phe mutation in gyrA [39]. This is of significant interest, since Salmonella Paratyphi A infection is advancing in Asia [16, 17], while increasing DCS has been observed using conventional microbiological methods [52–55].
FQ resistance in NTS
Foodborne infections with NTS are especially well documented in Europe and the USA, where frequencies of DCS and FQ resistance vary per serovar and country or region [56, 57]. Resistance at the human–animal interface is especially important for NTS, which have both animals and humans as potential hosts. Potential transmission of resistance is exemplified by recent findings that the resistance of Salmonella Typhimurium against ampicillin in the 1960s was related to the use of penicillin in animal feed in the late 1950s [58, 59]. Nowadays, FQs are extensively used in agriculture, and they additionally show a relatively low biodegradability [60]. FQs are still extensively used for animal production in several countries, e.g. for disease prevention and treatment in poultry [61]. Moreover, banning the use of FQs in food animals in Australia correlated with reduced FQ resistance in bacteria isolated from food, food animals and patients [62, 63].
PMQR can play an important role in spreading FQ resistance among strains at the human–animal interface. This is reflected by the higher numbers of the PMQR genes qnr and oqx detected by WGS studies in NTS (Table 2) compared to Salmonella Typhi (Table 1). In 2017, an integrated surveillance by several European reference laboratories allowed the linkage of an outbreak of Salmonella Chester to a food chain in Morocco [64]. One epidemic clone contained almost exclusively (87 %, n=96) isolates with PMQR markers [64] (Table 2). Toro et al. reported two Salmonella Enteritidis isolates from poultry in Chile carrying the qnrB gene [65]. One of the top five Salmonella serovars detected in humans in the USA is monophasic Salmonella Typhimurium, serotype 4,[4],12:i:- [56]. A recent WGS study (n=659) identified PMQR determinants in isolates from one multidrug-resistant clade of Salmonella serotype 4,[4],12:i:- originating from swine (Table 2), and the authors highlighted the risk as a potential reservoir for human infections [66].
Table 2. FQ-resistance markers in NTS, reported by WGS.
The number of isolates sequenced is indicated by ‘n’. ‘Source’ indicates whether samples were of human (H) or animal (A) origin. The percentage of sequenced isolates containing FQ-resistance markers is reported under ‘% FQR markers’. The right panel of the table provides an overview of the identified (combinations of) FQ-resistance mechanisms. Each line represents a combination of FQ markers that was observed in the respective study. Mutations in gyrase (gyr) and topoisomerase IV (par) encoding genes are provided as resulting changes in residue, and presented per gene and per identified combination. na, Not available.
| Reference | Region or country | n | NTS serovar | Source | % FQR markers | FQ-resistance marker | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| PMQR | Mutations in gyr and par | |||||||||
| gyrA | gyrB | parC | parE | |||||||
| [67] | Scotland | 290 | Typhimurium DT104 | H, A | 13 | – | Ser83Phe | – | – | – |
| – | Asp87G | – | – | – | ||||||
| – | Asp87Asn | – | – | – | ||||||
| [10] | Africa, Asia, Europe, Americas | 675 | Enteritidis | H, A | 0.15 | – | – | – | – | – |
| Africa | 496 | qnrS | – | – | – | – | ||||
| [69] | USA | 640 | 12 NTS serotypes | H | 3 | – | Asp87Tyr | – | – | – |
| – | Ser83Phe | – | – | – | ||||||
| – | Asp87Tyr+Ser83Phe | – | Ser80Ile | – | ||||||
| qnrS | – | – | – | – | ||||||
| qnrB | – | – | – | – | ||||||
| qnrB+oqxA+oqXB | – | – | – | – | ||||||
| [68] | USA (New York and Washington) | 90 | Typhimurium | H | 7 | qnrS | – | – | – | – |
| oqxA, oqxB | Asp87Tyr | – | – | – | ||||||
| oqxA, oqxB | Ser83Tyr | – | – | – | ||||||
| qnrB | – | – | – | – | ||||||
| oqxA, oqxB | Asp87Asn | – | – | – | ||||||
| – | Asp87Asn | – | – | – | ||||||
| [66] | USA, Europe | 659 | 4,[4],12:i:- | A | 5 | qnrB, qnrS | – | – | – | – |
| [64] | Morocco, unknown | 153 | Chester | H | 54 | qnrS, qnrB | – | – | – | – |
| [81] | South Asia, South-East Asia and Oceania | 115 | Weltevreden | H, A | na | qnrD, qnrS | – | – | – | – |
| oqxA, oqxB | – | – | – | – | ||||||
| [80] | Southern China | 44 | Weltevreden | H | 5 | qnrD | – | – | – | – |
| qnrS | – | – | – | – | ||||||
| [65] | Chile | 30 | Enteritidis | A | 7 | qnrB | – | – | – | – |
| 2018* | Vietnam | na | Typhimurium | H | na | qnrS | Asp87Asn | – | – | – |
*S. Baker, personal communication (2018).
In contrast, a retrospective study from Scotland stated little evidence of Salmonella Typhimurium DT104 transmission between human and animal reservoirs; some strains also contained FQ-resistance markers [67] (Table 2). Similar results were reported for Salmonella Typhimurium in the USA, in which strains isolated from humans contained a more diverse repertoire of resistance markers, including QRDR mutations (Table 2), compared to bovine isolates [68]. In a WGS study from the USA on NTS isolated from retail meat and human patients, only strains isolated from humans contained FQ-resistance markers [69]. WGS allows the study of transmission events with an unprecedented resolution, but interdisciplinary and inter-sectorial research will be required to fully elucidate and monitor the drivers of resistance in NTS.
In lower-income and middle-income countries, iNTS infection is highly prevalent and associated with high mortality [8]. For invasive Salmonella Enteritidis in Africa, the prevalence of FQ-resistance markers is low (Table 2). Among 496 Salmonella Enteritidis isolates originating from African countries, only 1 isolate had a qnrS gene [10] (Table 2). Large studies focussing on Salmonella Typhimurium and other NTS serotypes are limited, and only a few have applied WGS. Although FQ-resistance levels are low in most studies in Africa [70, 71], several small-scale studies report FQ-resistance markers in specific areas, ranging from mutations conferring DCS [44, 70, 72–77], up to high-level FQ resistance conferred by two gyrA mutations (Ser83Phe and Asp87Gly), a parC (Ser80Ile) mutation and an additional PMQR gene [aac(6′)-Ib-cr] [78]. In Asia, the burden of iNTS is much lower than the burden of typhoid fever [79]. PQMR genes have been reported in isolates of Salmonella Weltevreden from Asia (Table 2), a serotype that can potentially cause invasive infections [80, 81]. In Vietnam, WGS revealed a new clone of invasive Salmonella Typhimurium, which is associated with human immunodeficiency virus patients, and some isolates showing QRDR mutations and PMQR (S. Baker, personal communication) (Table 2).
Conclusions
FQ resistance in Salmonella seriously compromises treatment options, especially for invasive salmonellosis. The dominant presence of the Salmonella Typhi H58 clade associated with QRDR mutations jeopardizes effective FQ treatment of typhoid fever in Asia. Recent reports from Nepal indicated that even the fourth-generation FQ gatifloxacin has lost its effectiveness due to high-level FQ resistance [52, 82]. WGS data on FQ-resistant iNTS are rare and this can be due to the low resistance levels reported in most studies in Africa, while the burden of iNTS is the highest in this region. Because FQ resistance may be emerging [70], large multi-country studies are required to monitor the presence and spread of FQ resistance in iNTS in Africa. For NTS, both animals and humans are potential hosts, and from the existing literature, it is clear that there is a higher diversity of PMQR mechanisms in NTS compared to typhoidal Salmonella. This might be linked to a diverse host niche, including several animal reservoirs, indicative of the need for a ‘one health’ approach to efficiently monitor the spread and source of FQ resistance.
The increasing use of WGS provides new molecular surveillance approaches to monitor and understand the spread of FQ resistance in Salmonella. Whereas originally predominantly used for research, WGS is becoming more available in diagnostic laboratories across the world and tools are being developed to facilitate the data analyses (such as www.WGSA.net).
In summary, FQ resistance in Salmonella spp. is rising towards critical levels and there is need for alternatives, such as last resort antibiotics and the development of new antibiotics, as stated by the WHO in 2017 [20]. Further monitoring will be critical in the coming years to analyse the evolution of Salmonella strains and their resistance patterns. Hereto, the implementation of WGS provides new opportunities for surveillance.
Funding information
This work was financially supported by the Baillet Latour Fund [The Bacterial Infections in the Tropics (BIT) research cluster at ITM Antwerp, Belgium], the Research Foundation – Flanders (FWO SB PhD fellowship 1S40018N to W. L. C.), the Wellcome Trust (E. J. K. and V. K. W), and the Department of Economy, Science and Innovation in Flanders, Belgium (EWI funding to S. V. P).
Acknowledgements
We thank Kris Laukens and Pieter Meysman for their critical reading of the manuscript.
Conflicts of interest
The authors declare no conflicts of interest.
Supplementary Data
Footnotes
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
Abbreviations: DCS, decreased ciprofloxacin susceptibility; DR Congo, Democratic Republic of the Congo; ESBL, extended-spectrum β-lactamase; EUCAST, European Committee on Antimicrobial Susceptibility Testing; FQ, fluoroquinolone; iNTS, invasive non-typhoidal Salmonella; MDR, multidrug resistance; MIC, minimum inhibitory concentration; NTS, non-typhoidal Salmonella; PMQR, plasmid-mediated quinolone resistance; QRDR, quinolone-resistance-determining region; WGS, whole-genome sequencing.
References
- 1.Parry CM, Hien TT, Dougan G, White NJ, Farrar JJ, et al. Typhoid fever. N Engl J Med. 2002;347:1770–1782. doi: 10.1056/NEJMra020201. [DOI] [PubMed] [Google Scholar]
- 2.Parry CM, Basnyat B, Crump JA. The management of antimicrobial-resistant enteric fever. Expert Rev Anti Infect Ther. 2013;11:1259–1261. doi: 10.1586/14787210.2013.858019. [DOI] [PubMed] [Google Scholar]
- 3.Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–353. [PMC free article] [PubMed] [Google Scholar]
- 4.Buckle GC, Walker CL, Black RE. Typhoid fever and paratyphoid fever: systematic review to estimate global morbidity and mortality for 2010. J Glob Health. 2012;2:10401. doi: 10.7189/jogh.01.010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majowicz SE, Musto J, Scallan E, Angulo FJ, Kirk M, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882–889. doi: 10.1086/650733. [DOI] [PubMed] [Google Scholar]
- 6.Ao TT, Feasey NA, Gordon MA, Keddy KH, Angulo FJ, et al. Global burden of invasive nontyphoidal Salmonella disease, 2010. Emerg Infect Dis. 2015;21:941–949. doi: 10.3201/eid2106.140999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gordon MA. Salmonella infections in immunocompromised adults. J Infect. 2008;56:413–422. doi: 10.1016/j.jinf.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 8.Feasey NA, Dougan G, Kingsley RA, Heyderman RS, Gordon MA. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kariuki S, Gordon MA, Feasey N, Parry CM. Antimicrobial resistance and management of invasive Salmonella disease. Vaccine. 2015;33:C21–C29. doi: 10.1016/j.vaccine.2015.03.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Feasey NA, Hadfield J, Keddy KH, Dallman TJ, Jacobs J, et al. Distinct Salmonella Enteritidis lineages associated with enterocolitis in high-income settings and invasive disease in low-income settings. Nat Genet. 2016;48:1211–1217. doi: 10.1038/ng.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carden SE, Walker GT, Honeycutt J, Lugo K, Pham T, et al. Pseudogenization of the secreted effector gene ssei confers rapid systemic dissemination of S. Typhimurium ST313 within migratory dendritic cells. Cell Host Microbe. 2017;21:182–194. doi: 10.1016/j.chom.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44:1215–1221. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crump JA, Sjölund-Karlsson M, Gordon MA, Parry CM. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin Microbiol Rev. 2015;28:901–937. doi: 10.1128/CMR.00002-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shane AL, Mody RK, Crump JA, Tarr PI, Steiner TS, et al. Infectious Diseases Society of America clinical practice guidelines for the diagnosis and management of infectious diarrhea. Clin Infect Dis. 2017;65:e45. doi: 10.1093/cid/cix669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wain J, Hendriksen RS, Mikoleit ML, Keddy KH, Ochiai RL. Typhoid fever. Lancet. 2015;385:1136–1145. doi: 10.1016/S0140-6736(13)62708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crump JA, Mintz ED. Global trends in typhoid and paratyphoid fever. Clin Infect Dis. 2010;50:241–246. doi: 10.1086/649541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.NARMS . National Antimicrobial Resistance Monitoring System: Enteric Bacteria – Human Isolates Final Report. Atlanta, GA: Centers for Disease Control and Prevention; 2013. [Google Scholar]
- 19.Fang FC. Fluoroquinolone resistance in Salmonella and the utility of pefloxacin disk diffusion. J Clin Microbiol. 2015;53:3401–3404. doi: 10.1128/JCM.02270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 21.Correia S, Poeta P, Hébraud M, Capelo JL, Igrejas G. Mechanisms of quinolone action and resistance: where do we stand? J Med Microbiol. 2017;66:551–559. doi: 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 22.Redgrave LS, Sutton SB, Webber MA, Piddock LJ. Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol. 2014;22:438–445. doi: 10.1016/j.tim.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Aldred KJ, Kerns RJ, Osheroff N. Mechanism of quinolone action and resistance. Biochemistry. 2014;53:1565–1574. doi: 10.1021/bi5000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Drlica K, Malik M, Kerns RJ, Zhao X. Quinolone-mediated bacterial death. Antimicrob Agents Chemother. 2008;52:385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooper DC, Jacoby GA. Topoisomerase inhibitors: fluoroquinolone mechanisms of action and resistance. Cold Spring Harb Perspect Med. 2016;6:a025320. doi: 10.1101/cshperspect.a025320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfson JS, Hooper DC. The fluoroquinolones: structures, mechanisms of action and resistance, and spectra of activity in vitro. Antimicrob Agents Chemother. 1985;28:581–586. doi: 10.1128/AAC.28.4.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andriole VT. The quinolones: past, present, and future. Clin Infect Dis. 2005;41:S113–S119. doi: 10.1086/428051. [DOI] [PubMed] [Google Scholar]
- 28.Hopkins KL, Davies RH, Threlfall EJ. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int J Antimicrob Agents. 2005;25:358–373. doi: 10.1016/j.ijantimicag.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 29.Madurga S, Sánchez-Céspedes J, Belda I, Vila J, Giralt E. Mechanism of binding of fluoroquinolones to the quinolone resistance-determining region of DNA gyrase: towards an understanding of the molecular basis of quinolone resistance. Chembiochem. 2008;9:2081–2086. doi: 10.1002/cbic.200800041. [DOI] [PubMed] [Google Scholar]
- 30.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein Qnr with Escherichia coli DNA gyrase. Antimicrob Agents Chemother. 2005;49:118–125. doi: 10.1128/AAC.49.1.118-125.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran JH, Jacoby GA, Hooper DC. Interaction of the plasmid-encoded quinolone resistance protein QnrA with Escherichia coli topoisomerase IV. Antimicrob Agents Chemother. 2005;49:3050–3052. doi: 10.1128/AAC.49.7.3050-3052.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robicsek A, Jacoby GA, Hooper DC. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect Dis. 2006;6:629–640. doi: 10.1016/S1473-3099(06)70599-0. [DOI] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2018. CLSI supplement M100. [Google Scholar]
- 34.Parry CM, Threlfall EJ. Antimicrobial resistance in typhoidal and nontyphoidal Salmonellae. Curr Opin Infect Dis. 2008;21:531–538. doi: 10.1097/QCO.0b013e32830f453a. [DOI] [PubMed] [Google Scholar]
- 35.Humphries RM, Fang FC, Aarestrup FM, Hindler JA. In vitro susceptibility testing of fluoroquinolone activity against Salmonella: recent changes to CLSI standards. Clin Infect Dis. 2012;55:1107–1113. doi: 10.1093/cid/cis600. [DOI] [PubMed] [Google Scholar]
- 36.Wong VK, Baker S, Pickard DJ, Parkhill J, Page AJ, et al. Phylogeographical analysis of the dominant multidrug-resistant H58 clade of Salmonella Typhi identifies inter- and intracontinental transmission events. Nat Genet. 2015;47:632–639. doi: 10.1038/ng.3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pham Thanh D, Karkey A, Dongol S, Ho Thi N, Thompson CN, et al. A novel ciprofloxacin-resistant subclade of H58 Salmonella Typhi is associated with fluoroquinolone treatment failure. Elife. 2016;5:1–13. doi: 10.7554/eLife.14003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pham Thanh D, Thompson CN, Rabaa MA, Sona S, Sopheary S, et al. The molecular and spatial epidemiology of typhoid fever in rural Cambodia. PLoS Negl Trop Dis. 2016;10:e0004785. doi: 10.1371/journal.pntd.0004785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuijpers LMF, Phe T, Veng CH, Lim K, Ieng S, et al. The clinical and microbiological characteristics of enteric fever in Cambodia, 2008-2015. PLoS Negl Trop Dis. 2017;11:e0005964. doi: 10.1371/journal.pntd.0005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matono T, Morita M, Yahara K, Lee KI, Izumiya H, et al. Emergence of resistance mutations in Salmonella enterica serovar Typhi against fluoroquinolones. Open Forum Infect Dis. 2017;4:1–17. doi: 10.1093/ofid/ofx230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baker S, Duy PT, Nga TV, Dung TT, Phat VV, et al. Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. Elife. 2013;2:e01229. doi: 10.7554/eLife.01229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hendriksen RS, Leekitcharoenphon P, Lukjancenko O, Lukwesa-Musyani C, Tambatamba B, et al. Genomic signature of multidrug-resistant Salmonella enterica serovar Typhi isolates related to a massive outbreak in Zambia between 2010 and 2012. J Clin Microbiol. 2015;53:262–272. doi: 10.1128/JCM.02026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.International Typhoid Consortium. Wong VK, Holt KE, Okoro C, Baker S, et al. Molecular surveillance identifies multiple transmissions of typhoid in West Africa. PLoS Negl Trop Dis. 2016;10:e0004781. doi: 10.1371/journal.pntd.0004781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tadesse G, Tessema TS, Beyene G, Aseffa A. Molecular epidemiology of fluoroquinolone resistant Salmonella in Africa: a systematic review and meta-analysis. PLoS One. 2018;13:e0192575. doi: 10.1371/journal.pone.0192575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dougan G. Typhoid in Africa and vaccine deployment. Lancet Glob Health. 2017;5:e236. doi: 10.1016/S2214-109X(17)30045-1. [DOI] [PubMed] [Google Scholar]
- 46.Phoba MF, Barbé B, Lunguya O, Masendu L, Lulengwa D, et al. Salmonella enterica serovar Typhi producing CTX-M-15 extended spectrum β-Lactamase in the Democratic Republic of the Congo. Clin Infect Dis. 2017;65:1229–1231. doi: 10.1093/cid/cix342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodrigues C, Kapil A, Sharma A, Devanga Ragupathi NK, Inbanathan FY, et al. Whole-genome shotgun sequencing of cephalosporin-resistant Salmonella enterica serovar Typhi. Genome Announc. 2017;5:e01639-16. doi: 10.1128/genomeA.01639-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chiou CS, Lauderdale TL, Phung DC, Watanabe H, Kuo JC, et al. Antimicrobial resistance in Salmonella enterica serovar Typhi isolates from Bangladesh, Indonesia, Taiwan, and Vietnam. Antimicrob Agents Chemother. 2014;58:6501–6507. doi: 10.1128/AAC.03608-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klemm EJ, Shakoor S, Page AJ, Qamar FN, Judge K, et al. Emergence of an extensively drug-resistant Salmonella enterica serovar Typhi clone harboring a promiscuous plasmid encoding resistance to fluoroquinolones and third-generation cephalosporins. MBio. 2018;9:e00105-18. doi: 10.1128/mBio.00105-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robicsek A, Sahm DF, Strahilevitz J, Jacoby GA, Hooper DC, et al. Broader distribution of plasmid-mediated quinolone resistance in the United States. Antimicrob Agents Chemother. 2005;49:3001–3003. doi: 10.1128/AAC.49.7.3001-3003.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Poirel L, Cattoir V, Nordmann P. Is plasmid-mediated quinolone resistance a clinically significant problem? Clin Microbiol Infect. 2008;14:295–297. doi: 10.1111/j.1469-0691.2007.01930.x. [DOI] [PubMed] [Google Scholar]
- 52.Thompson CN, Karkey A, Dongol S, Arjyal A, Wolbers M, et al. Treatment response in enteric fever in an era of increasing antimicrobial resistance: an individual patient data analysis of 2092 participants enrolled into 4 randomized, controlled trials in Nepal. Clin Infect Dis. 2017;64:1522–1531. doi: 10.1093/cid/cix185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Joshi S, Amarnath SK. Fluoroquinolone resistance in Salmonella typhi and S. paratyphi A in Bangalore, India. Trans R Soc Trop Med Hyg. 2007;101:308–310. doi: 10.1016/j.trstmh.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Dutta S, Das S, Mitra U, Jain P, Roy I, et al. Antimicrobial resistance, virulence profiles and molecular subtypes of Salmonella enterica serovars Typhi and Paratyphi A blood isolates from Kolkata, India during 2009–2013. PLoS One. 2014;9:e101347. doi: 10.1371/journal.pone.0101347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zellweger RM, Basnyat B, Shrestha P, Prajapati KG, Dongol S, et al. A 23-year retrospective investigation of Salmonella Typhi and Salmonella Paratyphi isolated in a tertiary Kathmandu hospital. PLoS Negl Trop Dis. 2017;11:e0006051. doi: 10.1371/journal.pntd.0006051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Michael GB, Schwarz S. Antimicrobial resistance in zoonotic nontyphoidal Salmonella: an alarming trend? Clin Microbiol Infect. 2016;22:968–974. doi: 10.1016/j.cmi.2016.07.033. [DOI] [PubMed] [Google Scholar]
- 57.Veldman K, Cavaco LM, Mevius D, Battisti A, Franco A, et al. International collaborative study on the occurrence of plasmid-mediated quinolone resistance in Salmonella enterica and Escherichia coli isolated from animals, humans, food and the environment in 13 European countries. J Antimicrob Chemother. 2011;66:1278–1286. doi: 10.1093/jac/dkr084. [DOI] [PubMed] [Google Scholar]
- 58.Tran-Dien A, Le Hello S, Bouchier C, Weill FX. Early transmissible ampicillin resistance in zoonotic Salmonella enterica serotype Typhimurium in the late 1950s: a retrospective, whole-genome sequencing study. Lancet Infect Dis. 2018;18:207–214. doi: 10.1016/S1473-3099(17)30705-3. [DOI] [PubMed] [Google Scholar]
- 59.van Puyvelde S, Deborggraeve S, Jacobs J. Why the antibiotic resistance crisis requires a one health approach. Lancet Infect Dis. 2018;18:1–2. doi: 10.1016/S1473-3099(17)30704-1. [DOI] [PubMed] [Google Scholar]
- 60.Pikkemaat M, Yassin H, van der Fels-Klerx H, Berendsen BJA. Antibiotic Residues and Resistance in the Environment. Wageningen: RIKILT Wageningen UR; 2016. [Google Scholar]
- 61.Roth N, Mayrhofer S, Gierus M, Weingut C, Schwarz C, et al. Effect of an organic acids based feed additive and enrofloxacin on the prevalence of antibiotic-resistant E. coli in cecum of broilers. Poult Sci. 2017;96:4053–4060. doi: 10.3382/ps/pex232. [DOI] [PubMed] [Google Scholar]
- 62.Collignon PC, Conly JM, Andremont A, McEwen SA, Aidara-Kane A, et al. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies to control antimicrobial resistance from food animal production. Clin Infect Dis. 2016;63:1087–1093. doi: 10.1093/cid/ciw475. [DOI] [PubMed] [Google Scholar]
- 63.Cheng AC, Turnidge J, Collignon P, Looke D, Barton M, et al. Control of fluoroquinolone resistance through successful regulation, Australia. Emerg Infect Dis. 2012;18:1453–1460. doi: 10.3201/eid1809.111515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fonteneau L, Jourdan da Silva N, Fabre L, Ashton P, Torpdahl M, et al. Multinational outbreak of travel-related Salmonella Chester infections in Europe, summers 2014 and 2015. Euro Surveill. 2017;22:1–11. doi: 10.2807/1560-7917.ES.2017.22.7.30463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toro M, Retamal P, Ayers S, Barreto M, Allard M, et al. Whole-genome sequencing analysis of Salmonella enterica serovar enteritidis isolates in Chile provides insights into possible transmission between gulls, poultry, and humans. Appl Environ Microbiol. 2016;82:6223–6232. doi: 10.1128/AEM.01760-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Elnekave E, Hong S, Mather AE, Boxrud D, Taylor AJ, et al. Salmonella enterica serotype 4,[5],12:i:- in swine in the United States Midwest: an emerging multidrug resistant clone. Clin Infect Dis. 2018;66:877–885. doi: 10.1093/cid/cix909. [DOI] [PubMed] [Google Scholar]
- 67.Mather AE, Reid SWJ, Maskell DJ, Parkhill J, Fookes MC, et al. Distinguishable epidemics within different hosts of the multidrug resistant zoonotic pathogen Salmonella Typhimurium. Science. 2013;341:1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carroll LM, Wiedmann M, den Bakker H, Siler J, Warchocki S, et al. Whole-genome sequencing of drug-resistant Salmonella enterica isolates from dairy cattle and humans in New York and Washington States reveals source and geographic associations. Appl Environ Microbiol. 2017;83:e00140-17. doi: 10.1128/AEM.00140-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, et al. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother. 2016;60:5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lunguya O, Lejon V, Phoba MF, Bertrand S, Vanhoof R, et al. Antimicrobial resistance in invasive non-typhoid Salmonella from the Democratic Republic of the Congo: emergence of decreased fluoroquinolone susceptibility and extended-spectrum beta lactamases. PLoS Negl Trop Dis. 2013;7:e2103. doi: 10.1371/journal.pntd.0002103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marks F, von Kalckreuth V, Aaby P, Adu-Sarkodie Y, El Tayeb MA, et al. Incidence of invasive salmonella disease in sub-Saharan Africa: a multicentre population-based surveillance study. Lancet Glob Health. 2017;5:e310. doi: 10.1016/S2214-109X(17)30022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kariuki S, Onsare RS. Epidemiology and genomics of invasive nontyphoidal Salmonella infections in Kenya. Clin Infect Dis. 2015;61:S317–S324. doi: 10.1093/cid/civ711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Harrois D, Breurec S, Seck A, Delauné A, Le Hello S, et al. Prevalence and characterization of extended-spectrum β-lactamase-producing clinical Salmonella enterica isolates in Dakar, Senegal, from 1999 to 2009. Clin Microbiol Infect. 2014;20:O109–O116. doi: 10.1111/1469-0691.12339. [DOI] [PubMed] [Google Scholar]
- 74.Fashae K, Ogunsola F, Aarestrup FM, Hendriksen RS. Antimicrobial susceptibility and serovars of Salmonella from chickens and humans in Ibadan, Nigeria. J Infect Dev Ctries. 2010;4:484–494. doi: 10.3855/jidc.909. [DOI] [PubMed] [Google Scholar]
- 75.Eguale T, Birungi J, Asrat D, Njahira MN, Njuguna J, et al. Genetic markers associated with resistance to beta-lactam and quinolone antimicrobials in non-typhoidal Salmonella isolates from humans and animals in central Ethiopia. Antimicrob Resist Infect Control. 2017;6:1–10. doi: 10.1186/s13756-017-0171-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Beyene G, Nair S, Asrat D, Mengistu Y, Engers H, et al. Multidrug resistant Salmonella Concord is a major cause of salmonellosis in children in Ethiopia. J Infect Dev Ctries. 2011;5:23–33. doi: 10.3855/jidc.906. [DOI] [PubMed] [Google Scholar]
- 77.Eibach D, Al-Emran HM, Dekker DM, Krumkamp R, Adu-Sarkodie Y, et al. The emergence of reduced ciprofloxacin susceptibility in Salmonella enterica causing bloodstream infections in rural Ghana. Clin Infect Dis. 2016;62:S32–S36. doi: 10.1093/cid/civ757. [DOI] [PubMed] [Google Scholar]
- 78.Hendriksen RS, Joensen KG, Lukwesa-Musyani C, Kalondaa A, Leekitcharoenphon P, et al. Extremely drug-resistant Salmonella enterica serovar Senftenberg infections in patients in Zambia. J Clin Microbiol. 2013;51:284–286. doi: 10.1128/JCM.02227-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gordon MA. Invasive non-typhoidal Salmonella disease – epidemiology, pathogenesis and diagnosis. Curr Opin Infect Dis. 2012;24:484–489. doi: 10.1097/QCO.0b013e32834a9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li B, Yang X, Tan H, Ke B, He D, et al. Whole genome sequencing analysis of Salmonella enterica serovar Weltevreden isolated from human stool and contaminated food samples collected from the southern coastal area of China. Int J Food Microbiol. 2018;266:317–323. doi: 10.1016/j.ijfoodmicro.2017.10.032. [DOI] [PubMed] [Google Scholar]
- 81.Makendi C, Page AJ, Wren BW, Le Thi Phuong T, Clare S, et al. A phylogenetic and phenotypic analysis of Salmonella enterica serovar Weltevreden, an emerging agent of diarrheal disease in tropical regions. PLoS Negl Trop Dis. 2016;10:e0004446. doi: 10.1371/journal.pntd.0004446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Arjyal A, Basnyat B, Nhan HT, Koirala S, Giri A, et al. Gatifloxacin versus ceftriaxone for uncomplicated enteric fever in Nepal: an open-label, two-centre, randomised controlled trial. Lancet Infect Dis. 2016;16:535–545. doi: 10.1016/S1473-3099(15)00530-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.