Abstract
Distinuishing the species of mitis group streptococci is challenging due to ambiguous phenotypic characteristics and high degree of genetic similarity. This has been particularly true for resolving atypical Streptococcus pneumoniae and Streptococcus pseudopneumoniae. We used phylogenetic clustering to demonstrate specific and separate clades for both S. pneumoniae and S. pseudopneumoniae genomes. The genomes that clustered within these defined clades were used to extract species-specific genes from the pan-genome. The S. pneumoniae marker was detected in 8027 out of 8051 (>99.7 %) S. pneumoniae genomes. The S. pseudopneumoniae marker was specific for all genomes that clustered in the S. pseudopneumoniae clade, including unresolved species of the genus Streptococcus sequenced by the BC Centre for Disease Control Public Health Laboratory that previously could not be distinguished by other methods. Other than the presence of the S. pseudopneumoniae marker in six of 8051 (<0.08 %) S. pneumoniae genomes, both the S. pneumoniae and S. pseudopneumoniae markers showed little to no detectable cross-reactivity to the genomes of any other species of the genus Streptococcus or to a panel of over 46 000 genomes from viral, fungal, bacterial pathogens and microbiota commonly found in the respiratory tract. A real-time PCR assay was designed targeting these two markers. Genomics provides a useful technique for PCR assay design and development.
Keywords: Streptococcus pseudopneumoniae, genomics, real-time PCR
Data Summary
1. All sequencing data done by the BC Centre for Disease Control Public Health Laboratory has been deposited to NCBI’s short read archive (SRA) under BioProject: PRJNA428833
2. All supplementary data can be found in Table S1 (available in the online version of this article)
Impact Statement.
Mitis group streptococci are often difficult to distinguish with traditional biochemical assays. In particular, Streptococcus pseudopneumoniae is often hard to differentiate from atypical Streptococcus pneumoniae. Due to this, our understanding of the epidemiology and clinical significance of S. pseudopneumoniae has been limited. We sought to develop a suitable marker for distinguishing S. pseudopneumoniae from other species of the genus Streptococcus by using publicly available genomes along with phylogenetic support. This work should have broad interest for those studying species of the genus Streptococcus, in addition to being an example of how genomics can support the development of diagnostic assays.
Introduction
Streptococcus pseudopneumoniae was first reported in 2004 by Arbique et al., and was described as an acapsular, bile-insoluble and optochin-resistant bacteria when grown in CO2. It is also a member of the mitis group streptococci [1]. Members of the mitis group streptococci can be difficult to identify to the species level and often lack genetic markers for reliable discrimination. For example, Arbique et al. showed that common pneumococcal targets, such as pneumolysin (ply) and autolysin (lytA) could be detected in a few Streptococcus mitis and the majority of S. pseudopneumoniae [1]. Studies by Kawamura et al. [2] and Wessels et al. [3] further illustrate the challenges with using 16S rRNA gene sequencing [2], biochemical, MALDI–TOF MS and molecular assays [3] in discriminating between members of the mitis group streptococci.
Given the challenges in resolving mitis group streptococci, the epidemiology and clinical significance of S. pseudopneumoniae is unclear. Pathogenicity of S. pseudopneumoniae has been shown in a murine model [4], while in humans, it has been associated with chronic obstructive pulmonary disease (COPD) [5]; others did not make the same observation [6]. A common feature of S. pseudopneumoniae appears to be the prevalence of erythromycin, tetracycline and penicillin resistance [5–8]. The paucity of studies on S. pseudopneumoniae have been undoubtedly hampered by challenges in distinguishing S. pseudopneumoniae from atypical Streptococcus pneumoniae.
S. pseudopneumoniae is genetically similar to S. pneumoniae according to the results of a genomic comparison study done by Shahinas et al. [9], which documented various shared and unique features between S. pneumoniae, S. pseudopneumoniae and S. mitis. Multilocus sequence analysis (MLSA) has been successful in a number of studies in discriminating mitis group streptococci [8, 10, 11]. In the same spirit as MLSA discriminates species of the genus Streptococcus, we used phylogenetic inference to look at the population structure of mitis group streptococci, irrespective of taxonomic classification in NCBI. Ultimately, we used this clustering information to inform marker discovery that was used to develop a real-time PCR assay that discriminates between S. pneumoniae and S. pseudopneumoniae.
Methods
Streptococcus growth conditions, and isolate selection for sequencing
Members of the genus Streptococcus referred to the British Columbia Centre for Disease Control Public Health Laboratory (BCCDC PHL) were selected for study. These isolates, though identified as belonging to the mitis group streptococci by partial 16S rRNA gene sequencing, could not be classified definitively as S. pneumoniae, S. mitis or S. pseudopnuemoniae. All isolates of members of the genus Streptococcus were grown on 5 % Columbia Sheep Blood Agar (Oxoid) at 37 °C in a CO2 incubator for 18–24 h. Fifty strains of members of the genus Streptococcus isolated from various sample types were selected; three of these isolates were identified as S. pneumoniae, one as Streptococcus gordonii and one as Streptococcus australis. The remaining 44 (plus one repeated sample) isolates belong to the mitis group streptococci, but after 16S rRNA sequencing had uncertain laboratory identification beyond the viridans grouping. ATCC strains S. mitis (ATCC 49456T), Streptococcus oralis (ATCC 9811), S. pneumoniae (ATCC 49619) and S. pseudopneumoniae (ATCC BAA-960T) were included as controls for the real-time PCR.
Genome sequencing
Nucleic acids were extracted from the isolates of members of the genus Streptoccocus using a DNeasy Blood and Tissue Kit (QIAgen) or a MaxMAX DNA Multi-Sample Ultra Kit (ThermoFisher). The extracted DNA was made into Illumina-compatible libraries using either a Nextera XT (Illumina), TruSeq Nano DNA Library Prep Kit for NeoPrep (Illumina) or a NxSeq AmpFREE Low DNA Library Kit (Lucigen). Libraries made with the NxSeq AmpFree DNA Library Kit were quantified using the NEBNext Library Quant Kit for Illumina (New England BioLabs). All libraries were sequenced on an Illumina MiSeq using a 500-cycle MiSeq V2 kit (Illumina). Quality of the raw sequencing reads was assessed using FastQC v0.11.5 (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and MultiQC 1.2 [12]. One isolate, BCCDCPHL-Ssp027, failed to sequence and was not further analyzed. All raw sequence data is available from the BCCDC PHL Genomic Data Bank (BioProject: PRJNA379148), specifically this study under BioProject: PRJNA428833.
Genome assembly
Raw Illumina reads were adapter and quality trimmed with Trimmomatic v0.36 [13], using the adapter sequences packaged with the A5-miseq assembly pipeline [14]. The resulting trimmed reads were assembled with the Unicycler 0.4.1 [15] assembly pipeline with the - -no_pilon option, using SPAdes v3.11.0 [16] as the assembler for the trimmed Illumina reads.
Public genome download
All available genomes of members of the genus Streptococcus from RefSeq release 84 were downloaded using ncbi-genome-download 0.2.5 (github.com/kblin/ncbi-genome-download) (n=11 455). In addition, we downloaded S. pseudopneumoiae sequence data from BioProjects PRJEB20507, PRJEB4909, PRJEB2340 and PRJNA225866, and assembled the genomes, where appropriate, as described above (n=16). In total, 52 S. pseudopneumoniae genomes (including one labelled S. mitis) were gathered (Table S1). Non-streptococci genomes that were used to assess the analytical specificity (exclusivity) were also downloaded with ncbi-genome-download (n=46 727), and included microbiota found in respiratory samples [17].
Phylogenetic inference of Streptoccocus spp
Genomes were used to reconstruct a phylogenetic tree using PhyloSift v1.0.1 [18], which places genomes phylogenetically using 37 reference markers that are found in single copies and are nearly universal. The alignment of these phylogenetic markers (21 327 nucleotide positions) were used to infer a maximum-likelihood tree using a generalized time-reversible (GTR) +GAMMA evolutionary model, and 100 bootstraps with RAxML 8.2.8 [19]. Trees were pruned and labels edited using newick_utils (github.com/tjunier/newick_utils). Phylogenetic trees were visualized using plotTree.py (github.com/katholt/plotTree), a wrapper script for Environment for Tree Exploration (ETE) [20].
Marker discovery
The pan-genome of the 34 complete RefSeq S. pneumoniae genomes and 27 S. pseudopneumoniae genomes (based on their phylogenetic placement) were generated using large-scale blast score ratio (LS-BSR) v1.011 analysis [21], predicting genes with Prodigal v2.6.3 [22] and clustering using VSEARCH v2.5.0 [23]. The LS-BSR accessory script (compare_BSR.py) was used on the resulting LS-BSR gene matrix to compare and extract genes that were unique to all 34 S. pneumoniae or 27 S. pseudopneumoniae. Candidate markers for either S. pneumoniae or S. pseudopneumoniae were selected based on having a sequence length longer than 500 nucleotides and over 99 % identity (number of identical nucleotides of query divided by subject length) when aligned back to all originating genomes using blastn v2.6.0+ [24]. Other bioinformatics software, such as bioawk (github.com/lh3/bioawk), and seqtk (github.com/lh3/seqtk) were used to filter and manage the sequence data. Candidate markers were annotated using prokka v1.12 [25].
In silico Multilocus Sequencing Typing (MLST)
Sequence types were assigned to genomes of interest using mlst 2.10-dev (github.com/tseemann/mlst) with the S. pneumoniae mlst database downloaded on October 26, 2017.
In silico pneumococcal capsule typing
Assembled genomes were used to simulate Illumina sequencing data at a sequencing depth of 150× with wgsim 0.3.2 (https://github.com/lh3/wgsim). These data were used with Pneumococcal Capsule Typing (PneumoCaT 1.0) pipeline [26] to predict pneumococcal serotypes from Illumina sequence data.
Taxonomic classification of discrepant isolates
Discrepant classifications were assessed by simulating the Illumina sequences using the assembled genome in question with wgsim 0.3.2 at a sequencing depth of 150×. The simulated reads were classified using Kraken version 1.0 [27] using the minikraken_20171019_8 GB database, and the most likely taxonomy was based on the classification with the largest number of reads assigned to it.
Real-time PCR assay
A TaqMan assay was developed for the S. pneumoniae marker (SPN0001) and S. pseudopneumoniae marker (SPS0002). Primers and probes were designed using Geneious 9.0.4 (www.geneious.com, [28]) (Table 1), IDT OligoAnalyzer 3.1 was used to assess primer interactions, and Thermofisher Primer Express 3.0.1 to predict primer and probe melting temperatures. Real-time PCR reactions were performed on an ABI 7500 with recommended Fast thermal-cycling conditions in a 20 µl final volume using TaqMan Fast Advanced Master Mix (Life Technologies), PCR-grade water and primers and probes in a 20× mix. The 20× multiplex mix consists of each primer and probe resuspended in IDTE (pH 8.0) at a final concentration of 200 nM each for SPN-F and SPN-R, 100 nM for SPN-P, 500 nM each for SPS-F and SPS-R and 250 nM for SPS-P oligonucleotides.
Table 1. Oligonucleotides and probes for amplification of SPN0001 and SPS0002.
| Target | Organism | Name | Sequence | Probe |
|---|---|---|---|---|
| SPN0001 | S. pneumoniae | SPN-F | AATATCTGAAGATGCTCATTCTACAATT | |
| SPN-R | ATAAGGTTTACCGTCAATAATACGCAG | |||
| SPN-P | AACTACAGGTCGCTTTGCAGAGTCCAGTTT | 6FAM/ZEN | ||
| SPS0002 | S. pseudopneumoniae | SPS2-F | GTTCGGACTGGAGAGGAAGC | |
| SPS2-R | AAGCTACGAATCTTGTCAATAATGTCTT | |||
| SPS2-P | ACAGATCATTTCGCAATTT | VIC MGB |
Streptococcus lysates were prepared by either re-suspending a half loop (0.01 ml) of bacterial growth from isolated colonies into 1 ml of PCR-grade water in a micro-centrifuge tube and heating in a dry bath at 100 °C for 8 min, or using Instagene following the manufacture’s protocol. A 2 µl aliquot of sample lysate was used in each real-time PCR reaction.
Results and discussion
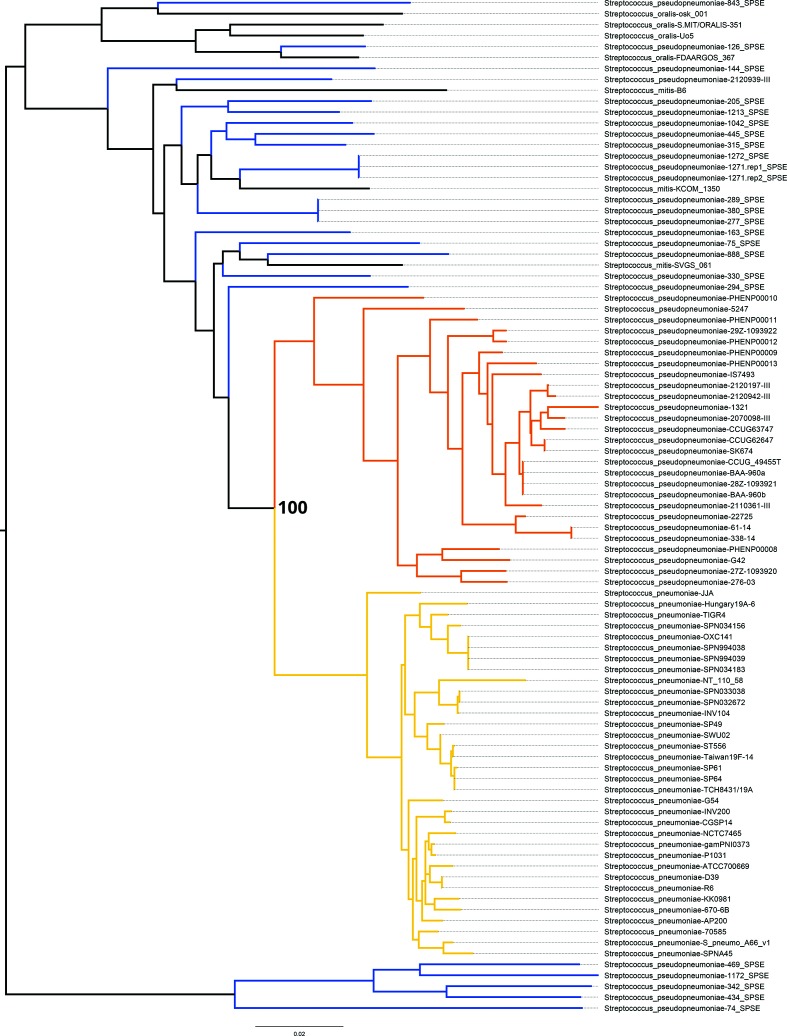
We set out to look for species-specific markers that would unambiguously distinguish S. pseudopneumoniae from S. pneumoniae and other members of the mitis group streptococci. Given the challenges related to accurately identifying S. pseudopneumoniae, we elected to first generate a phylogenetic tree of species of the genus Streptoccocus downloaded from RefSeq complete, including 34 S. pneumoniae, four S. oralis and three S. mitis, as well as 36 S. pseudopneumoniae and 16 more from various BioProjects (Table S1). Fig. 1 illustrates that 25 S. pseudopneumoniae cluster (blue branches) among S. mitis, and S. oralis. Of the 52 S. pseudopneumoniae isolates 27 clustered together on the tree in a clade (orange branches) near the S. pneumoniae clade (gold branches), including two S. pseudopneumoniae ATCC BAA-960T genomes that had been sequenced by different groups. The majority (24 out of 25) of the S. pseudopneumoniae that were not part of the large (orange) S. pseudopnuemoniae clade were isolated and sequenced during a study in one intensive care unit population [29]. In that study, the original laboratory identification of these S. pseudopneumoniae were mostly ‘Strep Viridans’, Neisseria, Enterococcus fecaelis or Staphylococcus aureus, while the authors used the average nucleotide identity (ANI) to find the best match of these 24 genomes to S. pseudopneumoniae IS7493 in the NCBI database. The taxonomy that was applied to these genomes and uploaded to NCBI was based on the ANI, which ranged from 0.82 and 0.93. It has been suggested that an ANI of at least 0.95 is needed for classification of isolates as members of the same species [30–32]. Given that the S. pseudopneumoniae classification from the Roach et al. study [29] is not strongly supported, we decided to use the S. pseudopneumoniae genomes that clustered within that defined clade (Fig. 1; orange clade). S. pseudopneumoniae 2120939-III (BioProject: PRJEB4909), also did not cluster within the defined clade and was excluded from the marker discovery process (Fig. 1, and Table S1).
Fig. 1.
Phylogenetic tree of selected species of the genus Streptococcus. Phylosift was used to place 34 S. pneumoniae (gold branches), 52 S. pseudopneumoniae, three S. mitis and four S. oralis based on NCBI taxonomy (RefSeq release 84). Note that one S. mitis strain (1042_SPSE) was included as a S. pseudopneumoniae due to information in supplemental data from [29]. The S. pneumonaie cluster is shown with gold branches, while the major S. pseudopneumoniae cluster (including two S. pseudopneumoniae ATCC BAA-960T genomes) is shown with orange branches. S. pseudopneumoniae that fall outside of the orange S. pseudopneumoniae clade are denoted by blue branches and were ultimately excluded from the marker discovery process. Bootstrap support values are indicated in black bold type at the node that separates the S. pneumoniae and S. pseudopneumoniae clades.
S. pneumoniae and S. pseudopneumoniae have specific markers
We next wanted to look for S. pneumoniae and S. pseudopneumoniae species-specific markers. To accomplish this, we took all genomes complete RefSeq S. pneumoniae (n=34) and publicly available S. pseudopneumoniae genomes that clustered within the S. pseudopneumoniae phylogenetic clade (Fig. 1; n=27) and identified the pan-genome using LS-BSR [21]. After filtering, 13 candidate S. pneumoniae and four S. pseudopneumoniae genes that were greater than 500 nucleotides in length were identified. These had at least a 99 % identical match across their intended targets (Table S1). We decided to further investigate a single candidate marker from both S. pneumoniae (centroid_2470; 729 nt; GtnR-family transcriptional regulator; SPN0001) and S. pseudopneumoniae (centroid_2440; 735 nt; kdpDE—an osmosensitive potassium channel histidine kinase/response regulator; SPS0002). Analytical specificity (inclusivity) was assessed by looking for blast hits for each marker against all species of the genus Streptococcus (n=11 455) and the non-RefSeq S. pseudopneumoniae (n=16), to look for any potential-cross reactivity within the genus.
The S. pneumoniae marker was found in 8019 out of 8066 (99.41 %) of the S. pneumoniae genomes in RefSeq, it was not found in any non-pneumococcal genomes. The criteria for a genome containing a marker required at least 99 % nucleotide identity across the length of the SPN0001 target. If we looked at the raw blastn output before applying the strict 99 % identity across the entire gene, there were 11 out of 47 S. pneumoniae genomes that had blastn matches with fewer identical nucleotides. When we queried these 11 S. pneumoniae genomes with the 154 base pair (bp) real-time PCR marker sequence (see below), eight of them matched the SPN0001 target with at least 99 % nucleotide coverage (Table S1; spn0001_discrepant). The remaining 3 S. pneumoniae isolates had short blastn matches, which may be reflective of misassemblies or inadequate genome sequencing coverage prior to assembly. On the basis of the results of the the in silico analysis using the SPN0001 PCR target sequence, the adjusted specificity of SPN0001 would improve to 8027 out of 8066 (99.52 %).
The S. pseudopneumoniae marker was found in all 27 S. pseudopneumoniae used to discern the marker, as expected, and did not have any matches in the 25 so-called S. pseudopneumoniae that did not cluster within the major S. pseudopneumoniae clade (Fig. 1). There were, however, 20 non-pseudopneumoniae matches: two Streptococcus canis and four Streptococcus pseudoporcinus that shared approximately 80 % identical nucleotide sequence to SPS0002, and 14 S. pneumoniae genomes (14 out of 8066; 0.173 %). We looked at the MLST of the 14 S. pneumoniae and five of them represent ST5107 (non-typeable according to PneumoCaT [26]), isolated from Thailand during a study by Chewapreecha et al. [33], and one was a ST2971 from China (also non-typeable). The remaining eight genomes belong to various unknown sequence types and were all non-typeable except for one serotype 37 (Table S1; sps0002_discrepant), a serotype that has been described in non-pneumococcal streptococci [34].
We also looked at the exclusivity of these two markers by assessing any blast hits to known viral, fungal and bacterial (microbiota and pathogens) genomes associated with sputum and nasopharyngeal samples (Table S1; exclusivity). Of the 46 727 genomes queried, only the S. pseudopneumoniae marker, SPS0002, matched 38 identical nucleotides (38 out of 735 nt; 5 %) in four species of the genus Enterococcus (Table S1; sps0002_discrepants). On the basis of analytical specificity inclusivity and exclusivity results, both the S. pneumoniae SPN0001 and S. pseudopneumoniae SPS0002 markers have high specificity to their respective species and were considered useful targets for a real-time PCR assay.
Presence/absence of S. pneumoniae (SPN0001) and S. pseudopneumoniae (SPS0002) PCR marker is concordant with phylogenetic placement of clinical isolates of species of the genus Streptococcus
The initial impetus for this study was to develop molecular markers that would distinguish S. pneumoniae from S. pseudopneumoniae. Since the SPN0001 and SPS0002 markers were used to look for presence and absence of all RefSeq streptococci genomes, we added discrepant results and more genomes to the reference phylogenetic tree (Fig. 1) to further understand the relationship of these markers to the clustering of the genomes. We included all S. pneumoniae isolates that lacked the SPN0001 PCR target (154 nucleotide sequence; n=39; originally 47, but excluding the eight S. pneumoniae that had truncated SPN0001, but that contained the PCR marker as described above), as well as all S. pneumoniae isolates that contained the SPS0002 PCR target (119 nucleotide sequence; n=14). Two S. mitis, one S. oralis and five S. infantis genomes randomly picked from RefSeq were added to populate the tree such that each species was represented by five members of these mitis group streptococci. Finally, BCCDC PHL sequenced streptococci isolates were also included on the tree, but are described below.
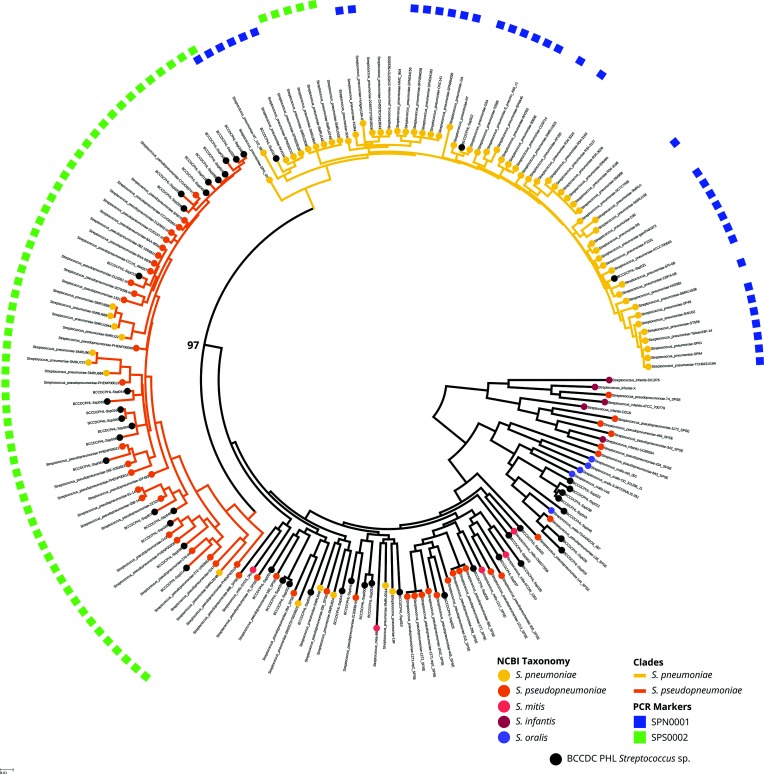
The first tree that was generated had two isolates that were distantly related to the other isolates on the tree. We were suspicious of the classification of these organisms, which were labelled S. pneumoniae in the NCBI records. We simulated raw reads from both of these two genomes and used Kraken to classify the simulated reads. One isolate was classified as Streptococcus salivarius, while the other was classified as a Staphylococcus species. Further evidence that these were not S. pneumoniae genomes came from the in silico MLST results; the S. salivarius genome had no matches to any S. pneumoniae MLST markers, while the Staphylococcus sp. genome best matched MLST markers from Stapyhlococcus hominis (Table S1). We pruned these two genomes from the phylogenetic tree and regenerated the tree, decorated with blast results for both the SPN0001 and SPS0002 real-time PCR sequence target (Fig. 2).
Fig. 2.
Phylogenetic tree of members of the genus Streptococcus and the presence/absence of the SPN0001 and SPS0002 PCR marker. A phylogenetic tree was reconstructed to show the relationship of the SPN0001 S. pneumoniae (blue squares) and SPS0002 S. pseudopneumoniae (green squares) PCR markers and the placement of the genomes on the tree. Note the S. gordonii, and S. australis isolates were excluded from this tree. S. pneumoniae (gold nodes); S. pseudopneumoniae (orange nodes); S. mitis (coral nodes); S. oralis (mauve nodes); S. infantis (magenta nodes); and BC Streptococcus sp. (black nodes). Bootstrap support value is indicated in black bold type at the node that separates the S. pneumoniae and S. pseudopneumoniae clades. All SPN0001-positive genomes clustered in the S. pneumoniae clade (yellow branches) while almost all SPS0002-positive genomes clustered in the S. pseudopneumoniae clade (orange branches). The three BC S. pneumoniae clinical isolates clustered with the other S. pneumoniae, while 22 BC SPS0002-positive Streptoccocus sp. clustered with the S. pseudopneumoniae clade. This tree also shows all discrepant results (excluding non-Streptococcus organisms and S. salivarius), such as SPN0001-negative S. pneumoniae and SPS0002-positive S. pneumoniae. Note that only RefSeq complete S. pneumoniae and discrepant S. pneumoniae genomes are shown as a representation of all 8051 S. pneumoniae genomes in RefSeq. Overall, 8027 out of 8051 S. pneumoniae were SPN0001-positive.
The presence and absence of the S. pneumoniae (SPN0001) and S. pseudopneumoniae (SPS0002) PCR marker sequences correlated almost exclusively with the genomes that clustered in the S. pneumoniae (Fig. 2; gold clade) and S. pseudopneumoniae (Fig. 2; orange clade) clades. One exception was a S. pneumoniae genome (ST2971) that was both SPN0001- and SPS0002-positive, but clustered with the S. pneumoniae clade.
We noted that many of the discrepant genomes identified above may be due to incorrect classification in the NCBI RefSeq record, as they clustered according to the presence of either the SPN0001 or SPS0001 PCR markers. Many of the taxonomic classification associated with genomes in NCBI RefSeq originate from the submitting laboratory, and the two genomes (S. salivarius and the species of the genus Staphylococcus) that were pruned from the final tree (Fig. 2) made us suspicious of some of the other discrepant genomes. To provide a reference method to classify these genomes, we used the top match from Kraken classification to support or refute the classification provided by NCBI. For S. pneumoniae that were SPN0001-negative, 15 genomes clustered outside of the S. pneumoniae clade (Fig. 2; gold clade): two were pruned from Fig. 2 (S. salivarius and the member of the genus Staphylococcus), five were classified as S. mitis and eight were classified as S. pseudopneumoniae (Table S1). The eight genomes that were classified as S. pseudopneumoniae clustered with the S. pseudopneumoniae clade (Fig. 2; orange clade) and were SPS0002-positive. The remaining 24 discrepant S. pneumoniae genomes clustered with the S. pneumoniae clade, but were SPN0001-negative. Those 24 S. pneumoniae consisted of 11 different MLST sequences types, and one unknown sequence type among these 24 S. pneumoniae genomes (Table S1). Notably, ST425 (n=6), ST5107 (n=5), and ST2705 (n=3) are present multiple times. In terms of serotype, 9 out of 24 genomes were predicted to be non-typeable whereas the rest were assigned a predicted serotype of 19F (n=7), 33F (n=3), 3, 06E, 14, 23F and 32F (Table S1; spn0001_discrepants). The five S. pneumoniae ST5107 isolates were SPN0002-positive, and along with the S. pneumoniae ST2971 (SPN0001- and SPS0002-positive) are the only instances of the SPS0002 PCR marker having a match outside of genomes in the S. pseudopneumoniae clade. This is possibly due to recombination, common in S. pneumoniae and particularly in acapsular lineages [33]. With the support of the Kraken classification and the tree placement for 15 discrepant S. pneumoniae genomes, we readjusted the SPN0001 specificity to 8027 out of 8051 (99.70 %). Likewise, the S. pseudopneumoniae SPS0002 marker specificity to S. pneumoniae was adjusted based on 8 out of 14 S. pneumoniae genomes probably being S. pseudopneumoniae (they cluster within the S. pseudopneumoniae clade and were SPS0002-positive), and SPS0002 marker was detected in 6 out of 8051 (0.074 %) of S. pneumoniae genomes.
Over a three-year period, the BCCDC PHL collected isolates of members of the genus Streptococcus that could not be classified to species by 16S rRNA gene sequencing. We took this collection of unknown isolates of members of the genus Streptococcus and sequenced their genomes to see where they clustered phylogenetically, and if that clustering was supported by the expected matches to the SPN0001 and SPS0002 PCR markers. We grew 44 ambiguous isolates of members of the genus Streptococcus from April 10 2014 to June 1, 2017 for genome sequencing, as well three laboratory-confirmed S. pneumoniae isolates, one S. gordonii isolate and one S. australis isolate. All isolates, with the omission of BCCDCPHL-Ssp027 (failed sequencing), S. gordonii, and S. australis, were added to the reference phylogenetic tree seen in Fig. 1 to generate the final tree (Fig. 2). The three clinically identified S. pneumoniae clustered with the other S. pneumoniae genomes and were positive for the SPN0001 PCR marker. The remaining 45 (44 plus one repeat) genomes of members of the genus Streptococcus clustered throughout the phylogenetic tree, however, all SPS0002-positive genomes (n=22) clustered with the S. pseudopneumoniae clade, indicating that they are probably isolates of S. pseudopneumoniae. Together, the SPS0002 PCR marker was detected in 57 out of 57 (27 RefSeq genomes, eight Kraken genomes classified as S. pseudopneumoniae and 22 BC sequenced genomes) Streptococcus genomes when they clustered within the S. pseudopneumoniae clade. Finally, we looked at the presence/absence, of the S. pneumoniae R6 lytA gene (Accession number: NC_003098.1; locus_tag:spr1754) among the isolates included on the tree in Fig. 2. This lytA gene was detected in all genomes from both the S. pneumoniae and S. pseudopneumoniae clades at over 98 % and approximately 82 % nucleotide identity, respectively (Table S1). However, lytA was also detected in 20 SPN0001- and SPS0002-negative genomes, such as S. mitis B6, at approximately 82 % nucleotide identity. These data further support the usefulness of SPS0002 for distinguishing S. pseudopneumoniae from S. pneumoniae.
Real-time PCR assay results agree with phylogenetic placement of S. pneumoniae and S. pseudopneumoniae isolates
Given the in silico specificity of the SPN0001 PCR marker to S. pneumoniae and specificity of the SPS0002 PCR marker to S. pseudopneumoniae, these sequences were designed as a real-time PCR assay (Table 1), which can be run as a singleplex or a duplex. We had three different panels: (1) A well-characterized panel (n=36), consisting of S. pneumoniae serotyped at the National Microbiology Laboratory (Winnipeg, Manitoba, Canada), ATCC and DSMZ isolates; (2) a clinical panel made up of 103 clinical isolates of members of the genus Streptococcus identified by the BCCDC PHL and; (3) the 49 (plus one repeat) isolates of members of the genus Streptococcus that had been sequenced on the Illumina MiSeq (Table S1).
SPN0001 was detected in all known isolates of S. pneumoniae in all three panels (29 out of 29) with 100 % accuracy and analytical specificity; no cross-reactivity was observed in the remaining 159 isolates of members of the genus Streptococcus (Table S1). SPS0002 could be detected in only S. pseudopneumoniae ATCC BAA-960T, as expected, from the well-characterized panel. In the clinical panel, ten isolates of the S. mitis group were positive for SPS0002, indicating that they were probably S. pseudopneumoniae. However, because of the lack of a reference assay that could reliably confirm the species classification of mitis group streptococci, the detection of the SPS0002 marker in the genome sequencing panel and where the SPS0002-positive isolates clustered on the tree was important. The real-time PCR results confirmed the clustering of the sequenced isolates of members of the genus Streptococcus on the phylogenetic tree in Fig. 2; all genomes of members of the genus Streptococcus that clustered within the S. pseudopneumoniae clade (Fig. 2; orange clade) were SPS0002-positive, while any members of the genus Streptococcus not clustering within the S. pseudopneumoniae cluster were PCR negative for SPS0002. These data support the hypothesis that the SPN0001 and SPS0002 markers identified using comparative genomics are suitable markers for distinguishing S. pneumoniae and S. pseudopneumoniae from other mitis group streptococci.
In this study we used the power of genomics to identify molecular specific markers capable of reliably differentiating S. pneumoniae and S. pseudopneumoniae. These markers were used as the basis for development of a real-time PCR assay, providing the clinical, microbiology and epidemiological communities a robust tool for reliable differentiation of S. pneumoniae and S. pseudopneumoniae. Given the abundance of misidentification of mitis group streptococci in NCBI RefSeq, phylogenetic inference was helpful in separating species to give us confidence in the dataset that we used to capture specific markers from the pan-genome. The phylogenetic inference was also helpful for looking at discrepant results as we were testing our markers in silico. For example, during our in silico analysis of the S. pseudopneumoniae marker, 14 S. pneumoniae matches (out of 8051) were found. Clustering of these 14 discrepant S. pneumoniae genomes placed eight of them in the S. pseudopneumoniae clade, and these eight genomes were also negative for the S. pneumoniae marker. This highlights the importance of alternate methods to investigate discrepant genomes from public databases, such as NCBI RefSeq. Database issues aside, this approach has broad applications for other diagnostics, including targeted assay design for outbreaks or surveillance, similar to that described by Bowers et al. [35].
Data bibliography
Croxen MA, Lee TD, Azana R, Hoang LM. BioProject:PRJNA428833 (2018).
Croxen MA, Lee TD, Azana R, Hoang LM. Supplemental Table 1 (2018).
NCBI RefSeq release 84. www.ncbi.nlm.nih.gov/refseq/ (2017).
University of Oxford PubMLST. www.pubmlst.org (October 26, 2017).
Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM et al. https://doi.org/10.1371/journal.pgen.1005413.s012 (2015).
Public Health England BioProject: PRJEB20507 (2017).
Wellcome Trust Sanger Institute BioProject:PRJEB4909 (2014), used with permission.
Ikryannikova LN, Ischenko DS, Lominadze GG, Kanygina AV, Karpova IY et al. BioProject: PRJNA225866 (2013).
Wellcome Trust Sanger Institute Bioproject: PRJEB2340 (2013), used with permission.
Funding information
This work was supported by in-kind mandate-dedicated funding from the BCCDC Public Health Laboratory.
Acknowledgements
We would like to thank the technical staff at the British Columbia Centre for Disease Control Public Health Laboratory - Public Health Advanced Bacteriology and Mycology Program. Additionally, we would like to thank Irene Martin’s group at the National Microbiology Laboratory, Winnipeg, Manitoba, Canada for special identification testing of members of the genus Streptococcus.
Conflicts of interest
The authors declare that there are no conflicts of interests.
Ethical statement
No human or animal work was done as part of this study.
Supplementary Data
Footnotes
Abbreviations: BCCDC PHL, British Columbia Centre for Disease Control Public Health Laboratory; MALDI–TOF MS, matrix-assisted laser desorption/ionization time-of-flight mass spectrometer; MLST, multilocus sequence typing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Supplementary material is available with the online version of this article.
References
- 1.Arbique JC, Poyart C, Trieu-Cuot P, Quesne G, Carvalho MG, et al. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J Clin Microbiol. 2004;42:4686–4696. doi: 10.1128/JCM.42.10.4686-4696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawamura Y, Hou XG, Sultana F, Miura H, Ezaki T. Determination of 16S rRNA sequences of Streptococcus mitis and Streptococcus gordonii and phylogenetic relationships among members of the genus Streptococcus. Int J Syst Bacteriol. 1995;45:406–408. doi: 10.1099/00207713-45-2-406. [DOI] [PubMed] [Google Scholar]
- 3.Wessels E, Schelfaut JJ, Bernards AT, Claas EC. Evaluation of several biochemical and molecular techniques for identification of Streptococcus pneumoniae and Streptococcus pseudopneumoniae and their detection in respiratory samples. J Clin Microbiol. 2012;50:1171–1177. doi: 10.1128/JCM.06609-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harf-Monteil C, Granello C, Le Brun C, Monteil H, Riegel P. Incidence and pathogenic effect of Streptococcus pseudopneumoniae. J Clin Microbiol. 2006;44:2240–2241. doi: 10.1128/JCM.02643-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keith ER, Podmore RG, Anderson TP, Murdoch DR. Characteristics of Streptococcus pseudopneumoniae isolated from purulent sputum samples. J Clin Microbiol. 2006;44:923–927. doi: 10.1128/JCM.44.3.923-927.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laurens C, Michon AL, Marchandin H, Bayette J, Didelot MN, et al. Clinical and antimicrobial susceptibility data of 140 Streptococcus pseudopneumoniae isolates in France. Antimicrob Agents Chemother. 2012;56:4504–4507. doi: 10.1128/AAC.06374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keith ER, Murdoch DR. Antimicrobial susceptibility profile of Streptococcus pseudopneumoniae isolated from sputum. Antimicrob Agents Chemother. 2008;52:2998. doi: 10.1128/AAC.01526-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rolo D, S Simões A, Domenech A, Fenoll A, Liñares J, et al. Disease isolates of Streptococcus pseudopneumoniae and non-typeable S. pneumoniae presumptively identified as atypical S. pneumoniae in Spain. PLoS One. 2013;8:e57047. doi: 10.1371/journal.pone.0057047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shahinas D, Thornton CS, Tamber GS, Arya G, Wong A, et al. Comparative genomic analyses of Streptococcus pseudopneumoniae provide insight into virulence and commensalism dynamics. PLoS One. 2013;8:e65670. doi: 10.1371/journal.pone.0065670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leegaard TM, Bootsma HJ, Caugant DA, Eleveld MJ, Mannsåker T, et al. Phenotypic and genomic characterization of pneumococcus-like streptococci isolated from HIV-seropositive patients. Microbiology. 2010;156:838–848. doi: 10.1099/mic.0.035345-0. [DOI] [PubMed] [Google Scholar]
- 11.van der Linden M, Otten J, Bergmann C, Latorre C, Liñares J, et al. Insight into the diversity of penicillin-binding protein 2x alleles and mutations in viridans streptococci. Antimicrob Agents Chemother. 2017;61:e02646-16. doi: 10.1128/AAC.02646-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coil D, Jospin G, Darling AE. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 2015;31:587–589. doi: 10.1093/bioinformatics/btu661. [DOI] [PubMed] [Google Scholar]
- 15.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabrera-Rubio R, Garcia-Núñez M, Setó L, Antó JM, Moya A, et al. Microbiome diversity in the bronchial tracts of patients with chronic obstructive pulmonary disease. J Clin Microbiol. 2012;50:3562–3568. doi: 10.1128/JCM.00767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darling AE, Jospin G, Lowe E, Matsen FA, Bik HM, et al. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ. 2014;2:e243. doi: 10.7717/peerj.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huerta-Cepas J, Dopazo J, Gabaldón T. ETE: a python environment for tree exploration. BMC Bioinformatics. 2010;11:24. doi: 10.1186/1471-2105-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sahl JW, Caporaso JG, Rasko DA, Keim P. The large-scale blast score ratio (LS-BSR) pipeline: a method to rapidly compare genetic content between bacterial genomes. PeerJ. 2014;2:e332. doi: 10.7717/peerj.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 25.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 26.Kapatai G, Sheppard CL, Al-Shahib A, Litt DJ, Underwood AP, et al. Whole genome sequencing of Streptococcus pneumoniae: development, evaluation and verification of targets for serogroup and serotype prediction using an automated pipeline. PeerJ. 2016;4:e2477. doi: 10.7717/peerj.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15:R46. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roach DJ, Burton JN, Lee C, Stackhouse B, Butler-Wu SM, et al. A year of infection in the intensive care unit: Prospective whole genome sequencing of bacterial clinical isolates reveals cryptic transmissions and novel microbiota. PLoS Genet. 2015;11:e1005413. doi: 10.1371/journal.pgen.1005413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richter M, Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High-throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. bioRxiv. 2017;27:225342. doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci USA. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, et al. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet. 2014;46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheppard CL, Kapatai G, Broughton K, Schaefer U, Hannah M, et al. Clinical streptococcal isolates, distinct fromStreptococcus pneumoniae, but containing the β-glucosyl transferase tts gene and expressing serotype 37 capsular polysaccharide. PeerJ. 2017;5:e3571. doi: 10.7717/peerj.3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bowers JR, Lemmer D, Sahl JW, Pearson T, Driebe EM, et al. KlebSeq, a diagnostic tool for surveillance, detection, and monitoring of Klebsiella pneumoniae. J Clin Microbiol. 2016;54:2582–2596. doi: 10.1128/JCM.00927-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.