Abstract
Once mutated, a single cell must expand into a clone before becoming significant for carcinogenesis. The forces driving clonal expansion and the obstacles that must be overcome are poorly understood. In a genetic mechanism, acquiring a second mutation conferring a proliferative advantage would enable the cell to expand autonomously. If carcinogen exposure instead induced a physiological change, clonal expansion would require the carcinogen's continued presence. To determine which is the case, we studied microscopic clones of keratinocytes mutated in the p53 tumor suppressor gene. Carcinogen exposure was controlled by irradiating mice with 280–320 nm UV radiation (UVB), sunlight's principal carcinogenic component; expansion of mutant clones was observed in epidermal sheets. p53-mutant clones grew only during chronic UVB exposure. Therefore, clonal expansion was not triggered by a proliferative mutation but was instead continually driven by UVB. Unexpectedly, the clone size distribution showed periodicity with maxima at estimated intervals of 16 ± 6 cells, the size of the epidermal proliferating unit in murine dorsal skin. In the absence of UVB, rare “imprisoned clones” increased in cell number without increasing in area. We conclude that: stem cell compartments act as physical barriers to clonal expansion of a p53-mutant keratinocyte; a rate-limiting step in clonal expansion is the colonization of an adjacent compartment; and sustained UVB enables the p53-mutant keratinocyte to colonize without incurring an additional mutation.
All cells in a tumor except the first arise by clonal expansion. The initial mutant cell is innocuous, because no disease phenotype will appear at the tissue level. In diseases that require multiple mutations, clonal expansion has a second function. Because mutation frequencies are typically 10−5-10−3/gene/cell generation, even with mutagen treatment or genetic instability, clonal expansion is numerically essential to create a target large enough for one of the progeny cells to acquire the next mutation (1). Although much has been learned about genes mutated in tumors, the process of clonal expansion remains largely unexamined.
The success of the multiple genetic hit model for cancer has suggested that clonal expansion is initiated by a mutation that confers a proliferative advantage. Many genes mutated in tumors regulate the cell cycle (2). A related assumption is that mutating a stem cell is sufficient to generate an expanding mutant clone. These events would be irreversible, and the mutagen would no longer be needed for clonal expansion once the proliferative mutation was made. An alternative possibility is that clonal expansion requires physiological changes induced by the carcinogen, such as inducing growth factors, enhancing inflammation, suppressing immune surveillance, or killing flanking cells by apoptosis (3, 4). Reversible influences on skin tumor development are well known. Murine skin mutagenized with dimethyl benz(a)anthracene or 280–320 nm UV radiation (UVB) revealed the tumor promotion phenomenon, in which agents that are nonmutagenic and noncarcinogenic increase the frequency of mutagen-induced papillomas (5). Tumor promoters must be present chronically, else the precancers will regress. UVB acts as both mutagen and tumor promoter (6, 7). Promoters stimulate cell proliferation and have other effects, perhaps related, such as prooxidant activity and inhibition of gap junction intercellular communication (5, 8). Cell proliferation is also important in nonepithelial tissues (9). Yet, it is not known whether proliferation increases the likelihood of creating a new mutation when DNA replicates across a DNA lesion or instead acts to enlarge clones of existing mutant cells.
Sun-exposed human skin contains thousands of clones of p53-mutant keratinocytes, ranging in size from a few cells to several thousand (10, 11). The keratinocytes in these clones are histologically normal yet contain the same kinds of p53 mutations observed in basal and squamous cell carcinoma (12). These clones are larger in chronically sun-exposed skin than in skin exposed intermittently or sun-shielded (10). Therefore, the driver of clonal expansion for these pre-precancers is sunlight, likely via UVB, the component most carcinogenic at the earth's surface (13). Similar clones are present in mice chronically irradiated with UVB and correlate with tumor risk (14). The ability of sunlight to drive clonal expansion could have either a mutational or physiological basis. For example, frequently exposed skin would be more likely to sustain a mutation early in life, allowing a clone more time to grow by the age of examination. Here, we use such clones to directly test whether the role of UVB in clonal expansion of p53-mutant keratinocytes is to induce an irreversible mutation or reversible physiological changes.
Methods
Animals and Chronic UVB Irradiation.
Wild-type male or female C57BL/6 or 129 × C57BL/6 mice were used for experiments beginning at age 6–8 wks. Animals were initially in resting phase of the hair cycle, indicated by pink skin after shaving and no measurable hair regrowth the next day. Mice were shaved on the back with clippers and electric shaver under general anesthesia. Three days later, between 9:00 and 10:00 a.m., mice were exposed to UVB from three broadband FS20T12-UVB lamps filtered through a Kodacel filter (Eastman Kodak) to remove wavelengths below 290 nm. Lamp output was 250–420 nm, with peak emission at 313 nm, and after filtering contained 72.6% UVB, 27.4% 320–400 nm radiation (UVA) and 0.01% 100–280 nm radiation (UVC), as measured by an IL1700/790 spectroradiometer with double monochromator (International Light, Newburyport, MA). Dose rate was 2.2 J/m2/sec. During irradiation, animals moved freely but were prevented from standing by a 1 × 1-cm wire mesh. Mice were irradiated five times weekly; daily dose was 500–1,250 J/m2 UVB in different experiments. Mice of 129 × C57BL/6 background were irradiated on alternate days or at lower doses for the first week to prevent erythema. After 1–2 wks, the hair cycle resumed, and mice were shaved daily without anesthesia. After the indicated number of weeks, irradiation was terminated and animals kept for 3 days to allow UV-induced wild-type p53 protein to return to basal levels. Animals were killed by isofluorane anesthesia and cervical dislocation. The animal protocol was reviewed and approved by the Yale Institutional Animal Care and Use Committee.
Epidermal Sheets.
Samples (2 × 4 cm) of dorsal skin were removed and epidermal sheets prepared by a modification of previous procedures (10, 15). Tissue was scraped of excess fat and immersed in PBS/20 mM EDTA for 1–3 h at 37°C. The sample was placed epidermis side up on a glass slide, and an incision through the epidermis was made in one corner. By using curved no. 7 forceps, the epidermis was grasped from the point of the incision and gently separated from the dermis with a bent spatula. Separated epidermis was fixed in acetone at 4°C for 20 min and rinsed in PBS. Dermal fragments contained no interfollicular keratinocytes. Samples were stored up to 2 wks in 4°C PBS.
Immunohistochemistry.
Epitope was unmasked by microwaving 8 min in 10 mM citrate buffer (pH 6.0), followed by PBS wash. After quenching endogenous peroxidase activity with 0.15% H2O2/PBS, blocking nonspecific antibody binding with normal goat serum in 3% BSA/PBS, and blotting excess blocking solution, samples were incubated overnight in a humidified chamber at 4°C with rabbit polyclonal antibody CM-5 for wild-type or mutant murine p53 protein (Novocastra, Newcastle, U.K.) at 1:2,500 dilution, followed by PBS wash. Samples were incubated 45 min with 5 μl of biotinylated goat anti-rabbit antiserum (Vector Laboratories) in 1,000 μl of PBS/3% BSA and washed in PBS. Diaminobenzamide staining used ABC reagent (Vector) for 30 min and DAB reagent (Research Genetics, Huntsville, AL) for 14 min.
Clone Microdissection, DNA Amplification, and Sequencing.
After 7 or 9 wks of irradiation with 750 J/m2 UVB, epidermal sheets were isolated and 30-gauge steel needles used to microdissect samples of 15–500 cells from diamino-benzamide-stained clones ranging in size from 100 to 2,000 cells. DNA was isolated with InstaGene Matrix (Bio-Rad). Total genomic DNA was amplified by using semirandom nonamer primers of sequence 5′-NNNNNN(G/C)(G/C)(G/C)-3′ in 2.5 μl 10 × Expand High Fidelity buffer/15 mM MgCl2/7.8 μl of H2O/2.5 μl 2 mM dNTPs/1.2 μl of primer (2.5 μg)/1 μl (3.5 units) of a mix of Taq DNA polymerase and proofreading Pwo polymerase (Expand High Fidelity PCR System, Roche Diagnostics)/10 μl of template DNA (2 ng) (10). Cycling in a PTC-100 Programmable Thermal Controller (MJ Research, Cambridge, MA) was: one step-file cycle (60°C 3 min/94°C 5 min); 40 thermo-file cycles (2.9 min ramp to 94°C/94°C 10 sec/1.95 min ramp to 24°C/24°C 10 sec). Reamplification for exon 8 of the murine p53 gene used primers 5′-CTAGTTTACACACAGTCAGGATGG-3′ and 5′-AAGAGGTGACTTTGGGGTGAAGCTC-3′ and HotStartTaq DNA polymerase (Qiagen, Chatsworth, CA) for 40 cycles: 94°C 45 sec/58°C 45 sec/72°C 1 min/final extension 72°C 10 min. Negative controls for contamination were included in all reactions. DNA sequence was determined by using fluorescent forward and reverse primers and mutations confirmed with a repeat random and/or p53-specific PCR reaction.
Clone Frequency, Size, and Symmetry.
To determine cells/clone and clones/area, p53-immunopositive clones were observed at ×400, varying the focus to observe different cell layers. Clusters of ≥three adjacent cells were scored as a single clone and the number of immunopositive nuclei recorded. NIH image, ver. 1.62, analysis of digital photomicrography was used to determine biopsy sample area, clone area and perimeter, and the lengths of the two longest clone axes.
Periodicity of Clone Size.
Data were initially smoothed by grouping clones into bins of three cells in width; e.g., three to five cells per clone, six to eight, etc. Results were not altered by shifting bin endpoints or by choosing bin widths of two, four, or five. The periodic component of the clone size distribution was confirmed analytically by using unbinned data. Details of this statistical treatment will be described separately (D.Z. and D.E.B., unpublished results). Briefly, let xi denote the observed frequency of p53-mutant clones i cells in size, with i = 1, 2, 3, etc. We assume xi are independent, each having an underlying Poisson distribution along the frequency axis with mean λi. The model imposed is: ln λi = βi + πμi, where βi = C − αi + θ ln i represents the broad trend in clone size as the familiar Γ distribution, and πμi represents the periodic component. The periodic component is based on a series of normal density functions located at integer multiples, μ, of i. Physically, μ estimates the smallest unit of clone size. Parameters of the Γ distribution and normal density functions were estimated by using maximum Poisson likelihood in splus (MathSoft, Cambridge, MA).
Results
UVB Drives Expansion of p53-Mutant Clusters.
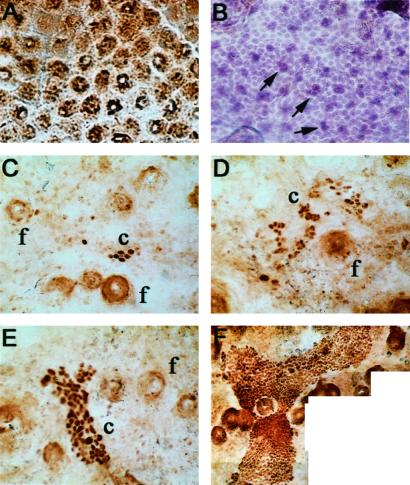
Exposing mice to daily UVB doses equivalent to a barely perceptible sunburn induced epidermal hyperproliferation (16). Despite hyperproliferation, epidermal morphology resembled the normal epidermal proliferating unit (EPU) structure of murine dorsal skin, in which 10–11 small basal keratinocytes, one of which appears to be a stem cell, underlie a stack of two to three large suprabasal keratinocytes and several nonnucleated cells (17–20). Like unirradiated skin (not shown), UVB-irradiated epidermis contained polygonal domains of nonoverlapping corneocytes (Fig. 1A). Regularly spaced groups of two to three dark overlapping nuclei, amid lightly staining nonoverlapping nuclei, are consistent with the central column of nuclei reported in a normal EPU (Fig. 1B) (18).
Figure 1.
Epidermal domains and p53-mutant clusters in epidermal sheets from UVB-irradiated mice. (A) Polygonal domains of nonoverlapping superficial corneocytes; counterstain is diaminobenzamide. (B) Keratinocyte nucleus doublets and triplets (arrows) regularly spaced amid single nuclei; hematoxylin staining. (C–F) Morphology of p53-mutant clusters. UVB dose was 1,250 J/m2 daily for 5 wks except (F) 11 wks. Mutant clusters (c) and follicles (f) are indicated. Digital photomicrography was ≈×400, except (F) ×200.
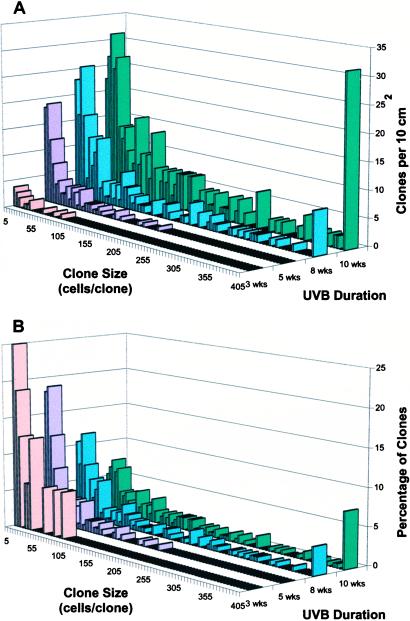
After 3–5 wks, UVB irradiation led to clusters of p53-mutant keratinocytes (Fig. 1 C–F). Clusters ranged from three (the minimum scored) to 2,000 cells. These sizes exceed that of a single normal stem cell compartment, 12–14 cells. Both the number and size of clusters increased with the duration of UVB exposure (Fig. 2A) and, less markedly, with UVB dose (500–1,250 J/m2 for 6 wks; not shown). The presence of large clusters after extended UVB reflected a shift in the size distribution of clusters, rather than resulting from the larger dataset at higher doses: chronic UVB led to a progressive increase in the percentage of large clusters and a corresponding decrease in the percentage of small clusters (Fig. 2B). Although hair follicles contain stem cells capable of populating the epidermis and participating in chemical carcinogenesis (21), follicular p53-mutant clusters were rarely seen, consistent with UVB's poor penetration into the dermis. Interfollicular clusters spared follicles (Fig. 1F), resembling actinic keratoses in human patients and suggesting that the clusters did not originate from follicles.
Figure 2.
Number and size of p53-mutant clones increase with UVB irradiation. Clone number is a visual assay for initial mutational events; clone size is an assay of subsequent clonal expansion. (A) Time course. Mice were irradiated daily with 1,250 J/m2 UVB for the indicated number of weeks. Clone frequency and size were scored in epidermal sheets immunostained with CM5 antibody for normal and mutant p53 protein. (B) Data normalized to percentage of total clone number, showing the increased proportion of large p53-mutant clones with irradiation.
UVB-Induced Expansion Is Clonal.
p53-mutant clusters might grow by clonal expansion or by independently mutating adjacent EPUs. Independent mutation of 10–100 adjacent EPUs (≈140–1,400 cells) is not expected, because only 1.2% of EPUs were mutant at 11 wks (mutant clusters occupied 1.2% of the epidermis). Clusters have been observed to merge at sizes and densities exceeding those reached here (14).
The clonality of expansion was tested directly by determining the DNA sequence of p53 mutations. If a cluster arose by clonal expansion of one p53-mutant cell, it would have that same mutation throughout. But if a 140-cell cluster grew by independent mutation of 10 adjacent EPUs, the joint cluster would have 10 independent mutations. Seventy-four percent of p53-immunopositive clusters carried a detectable mutation in exon 8 (Table 1); no mutations were seen in p53-immunonegative skin. One-fifth of mutant clusters showed loss of the normal allele. Point mutations were UVB-like (1), with a predominance of C→T transitions at sites of adjacent pyrimidines (Table 1). The mutation spectrum was dominated by a known murine UVB hotspot at codon 270 (22). To test clonality, we first excised samples 100–500 cells in size from six clusters (Table 1: 1, 2, 4, 8, 10 L, 11 L). In every case, the mutation (or lack of one) was present throughout each sample because the sequencing band intensity approximated that of the other allele. The discriminatory power of this test depends on the frequency of the particular mutation involved and on the cluster size. For codon 270 (mutated in 63% of clusters), independently mutated EPUs would fortuitously all carry the same mutation in 4% of 100-cell samples (Table 1: 4, 11 L) and 10−7 of 500-cell samples (Table 1: 1, 10 L). For codon 271 (mutated in 5% of clusters, 250-cell sample), the probability is 10−24. Second, we microdissected biopsies of ≥40 cells from opposite ends of six clusters (Table 1, L and R). Each pair carried at least one identical mutation. The probability that two biopsies, each 3 EPU in size, would fortuitously have the same mutation, ranged from 0.06 for clusters 3 and 10 carrying the hotspot mutation down to 10−5-10−9 for clusters 11, 15, and 16 carrying two mutations. These results imply clonal growth.
Table 1.
Clonality of p53 mutations in exon 8 of p53-immunopositive clones
| Clone number | Codon | Normal sequence | Base change | Amino acid change |
|---|---|---|---|---|
| Normal | wt | |||
| Normal | wt | |||
| Normal | wt | |||
| 1 | 270 | gttCgt | C→T/wt | Arg→Cys |
| 2 | wt | |||
| 3 L | 270 | gttCgt | C→T/wt | Arg→Cys |
| 3 R | 270 | gttCgt | C→T/wt | Arg→Cys |
| 4 | 270 | gttCgt | C→T/wt | Arg→Cys |
| 5 | 270 | gttCgt | C→T/wt | Arg→Cys |
| 6 | wt | |||
| 7 L | wt | |||
| 7 R | wt | |||
| 8 | 271 | cgtGTTtgt | GTT→TGG/wt | Val→Trp |
| 9 | wt | |||
| 10 L | 270 | gttCgt | C→T/0 | Arg→Cys |
| 10 R | 270 | gttCgt | C→T/wt | Arg→Cys |
| 11 L | 270 | gttCgt | C→T/wt | Arg→Cys |
| 275 | tgccCt | C→T/wt | Pro→Leu | |
| 11 R | 270 | gttCgt | C→T/wt | Arg→Cys |
| 275 | tgccCt | C→T/wt | Pro→Leu | |
| 12 | wt | |||
| 13 | 270 | gttCgt | C→T/wt | Arg→Cys |
| 14 | 270 | gttCgt | C→T/0 | Arg→Cys |
| 15 L | 270 | gttCgt | C→T/0 | Arg→Cys |
| 15 R | 270 | gttCgt | C→T/0 | Arg→Cys |
| 16 L | 270 | gttCgt | C→T/0 | Arg→Cys |
| 16 R | 270 | gttCgt | C→T/0 | Arg→Cys |
| 17 | 270 | gttCgt | C→T/wt | Arg→Cys |
| 284 | gaaGaa | G→A/wt | Glu→Lys | |
| 18 | 270 | gttCgt | C→T/wt | Arg→Cys |
| 19 | 278 | agaGac | G→A/wt | Asp→Asn |
wt, wild type.
Clonal Expansion Is Not Cell-Autonomous.
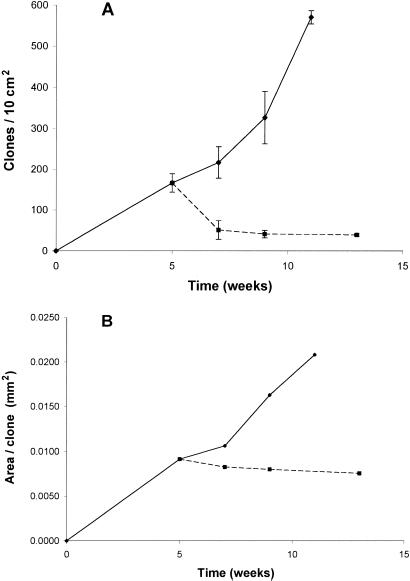
To directly test the mutational and physiological models for UVB-induced clonal expansion, we generated p53-mutant clones, terminated UVB, and observed the subsequent evolution of clone size. After UVB ended, the number of clones decreased rapidly (Fig. 3A). About 20% of the p53-mutant clones resisted regression and so are probably the clones of interest for cancer. Clone area, in contrast, remained constant after UVB ended (Fig. 3B), which was true for both stable and transient clones. The difference between the two branches of the curve in Fig. 2B is statistically significant (Wilcoxon rank sum test, two-sided P values are P = 0.003 at 9 wks and P = 0.0002 for 11 vs. 13 wks). The fact that expansion of a clone's territory required sustained UVB rules out the possibility that clonal expansion was cell-autonomous, initiated irreversibly by UVB-induced mutations.
Figure 3.
Sustained UVB is required for clonal expansion. (A) Clone number declines rapidly after cessation of daily 1,250 J/m2 irradiation (dashed line); ≈20% of p53-mutant clones stably resist regression for up to 13 wks. Mean clone number ± SEM. (B) Clone area does not grow after UVB ends. Each point on the graph has a broad distribution of clone sizes, as in Fig. 1; mean clone size is plotted. The difference between the two branches of the curve is statistically significant (P = 0.003 at 9 wks and P = 0.0002 for 11 vs. 13 wks, Wilcoxon rank sum test two-sided). One of two similar experiments is presented.
The mutational and physiological models of clonal expansion make weak predictions about the geometry of the clones. In cell-autonomous expansion, a cell mutated by UVB clonally expands without the aid of any additional effect of UVB on its neighbors. Clonal expansion would thus tend to be radially symmetric, even if regularly spaced epidermal features such as hair follicles forced it to deviate from circularity. In contrast, p53-mutant clones were asymmetric (Fig. 1 C–F) with most clones fitting a 2:1 length/width regression line (not shown). The axes of nearby clones were not aligned.
Quantized Clone Size.
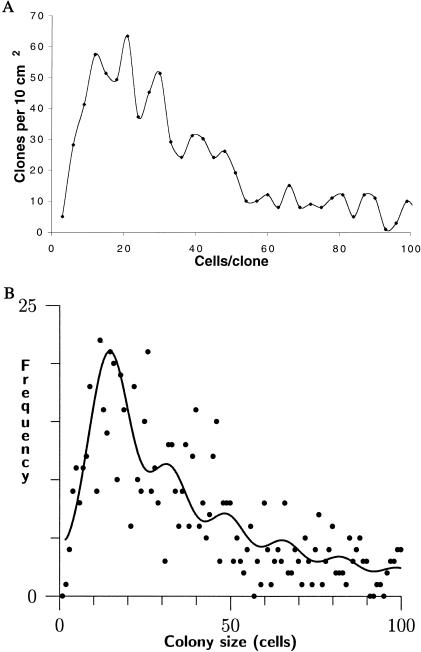
The morphology of p53-mutant clones also sheds light on the obstacles that must be overcome during clonal expansion and the reason sustained UVB exposure is required. Close examination of clone sizes revealed a fine structure to the clone size distribution. A purely invasive mode of clone expansion would be expected to generate a smooth size distribution, as clones of any size would be possible. But to our surprise, it was possible to observe a quantization in clone size (Fig. 4A). Moreover, the quantization corresponded to multiples of ≈12–16 cells (depending on the treatment of doublets), similar to the number of nucleated cells in a murine EPU. This qualitative observation motivated a search for periodicity in the data by using analytical methods. In the raw data, the periodicity is less visually apparent but is readily detectable statistically (Fig. 4B). The data fit the product of a Γ distribution and a periodic series of evenly spaced normal distributions (see Methods), leading to a spacing parameter μ of 16 ± 6 cells with a P value for the contribution of the periodic component of <10−16. The quantization of clone size implies that stepwise increases in the number of stem cell compartments involved in a p53-mutant clone occurred relatively slowly, whereas filling up of a stem cell compartment with mutant cells occurred rapidly once entered. That is, clonal expansion proceeded by successively colonizing adjacent stem cell compartments rather than by continuous invasion. It is not possible to determine whether mutant colonies are composed of multiple EPUs, each containing a single stem cell, because p53-mutant cells and stem cell compartments cannot be compared directly; available epidermal stem cell markers are incompatible with p53 immunostaining or rely on immunonegativity (23, 24).
Figure 4.
Quantized distribution of sizes of p53-mutant clones. (A) In a qualitative analysis of 868 clones grouped into size bins three cells in width, maxima occurred in the clone size distribution at intervals of ≈12–16 cells/clone. Clones larger than 100 cells are not shown. (B) Statistical analysis of clone size frequencies as the product of the aperiodic Γ distribution and a periodic series of evenly spaced normal distributions (see Methods) leads to an estimated spacing parameter μ of 16 ± 6 cells (P < 10−16). Because an epidermal stem cell compartment in murine dorsal skin contains ≈12–14 nucleated keratinocytes, these peaks correspond to individual mutant stem cell compartments. The height of successive peaks decreased in ratios of 0.5–0.8, implying that a clone of size n compartments has an ≈2/3 probability of expanding to size n + 1 under these irradiation conditions. All clones were included in the analysis; not shown because of space are clones >100 cells and three points corresponding to high-frequency small clones.
In the clone size distribution, the height of successive peaks decreased in a ratio of 0.5–0.8, as determined from Γ distribution parameters. This result implies that a clone of size n compartments has an ≈2/3 probability of expanding to size n + 1 under these irradiation conditions. This frequency is too high to be caused by point mutation, again implicating the physiological effect of UVB.
“Imprisoned Clones.”
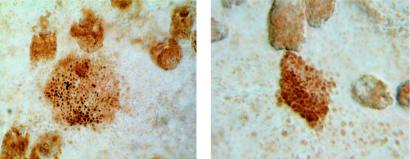
In the UVB withdrawal experiment, clone size measured as area was found to remain constant in the absence of UVB (Fig. 3B). When clone size was instead measured as the number of cells per clone, there was a slight increase after UVB ended that was statistically significant (two-sided P = 0.003, Wilcoxon rank sum test; not shown). This increase came from rare clones that continued to proliferate while not expanding in area, containing densely packed cells with very little cytoplasm (Fig. 5). These “imprisoned clones” were observed only in the absence of UVB, suggesting that epidermal stem cell compartments constitute physical barriers to the expansion of a p53-mutant keratinocyte.
Figure 5.
Imprisoned clones. In the absence of UVB, rare p53-mutant clones achieved high cell density without a corresponding increase in clone area. Clonal expansion was thus spatially restricted in the absence of UVB exposure. UVB (1,250 J/m2) daily for 5 wks, followed by 0 J/m2 for 8 wks (×400).
Discussion
UVB Exposure Allows p53-Mutant Keratinocytes to Clonally Expand Beyond Their Stem Cell Compartments.
Several lines of evidence indicate that the dorsal skin of the mouse is organized into hexagonal domains termed “EPUs” (17, 18). Morphologically, the domains are marked by silver's tendency to stain the edges of large squaming keratinized cells. These hexagons overlie smaller and more numerous proliferative keratinocytes. Functionally, the basal layer of each hexagon contains a central cell that replicates infrequently, loses radiolabeled DNA slowly, and is presumed to be a stem cell. Thus, EPUs are considered stem cell compartments. Surrounding the central cell are more rapidly proliferating keratinocytes that give rise to the vertically migrating differentiating keratinocytes and are termed “transit amplifying cells.” These two proliferation classes correspond to the observation that 10% of keratinocytes can form colonies after x-irradiation (17). The entire EPU contains ≈12–14 nucleated cells and six to eight overlying differentiating cells. Recent experiments with keratinocytes transduced with retroviruses containing the β-galactosidase gene show vertical columns of labeled cells arising from the basal layer (19, 20). Hair follicles also contain stem cells (21). Human epidermis contains EPU-like clusters of integrin-expressing nonproliferating basal keratinocytes that have high proliferative potential in vitro. These putative stem cells seem to emanate from a single central cell and are surrounded by basal cells that do not express integrin as highly, are often proliferating, and have lower long-term proliferative potential in vitro (putative amplifying cells) (23). These domains, centered at particular morphological sites such as the tips of dermal papillae in scalp or foreskin (23, 25), contain ≈150 cells, larger than murine EPUs.
The p53-mutant clones generated by several weeks of UVB irradiation ranged in size from 3 to 2,000 cells (Figs. 1 and 2). Approximately half were greater than 20 cells in size (Figs. 2 and 5) and so must have expanded beyond their initial stem cell compartment. After a mutation, a mutant cell's clonal expansion would have two steps: (i) proliferation of the mutant stem cell to fill its own EPU, and (ii) subsequent escape from the mutant EPU to colonize or displace an adjacent one. In vitro, a single stem cell can recreate over 150 cell domains resembling stem cell compartments (26). Mutant transit amplifying or suprabasal cells may also be capable of founding clones (23, 26, 27). The observation of quantized clone sizes indicates that it is the second step, escape from the stem cell compartment, that is rate limiting. The observed size of 16 ± 6 nucleated cells per mutant compartment is consistent with the 12–14 range in normal skin, particularly because irradiated epidermis is hyperproliferative. Although the EPU becomes subdivided in hyperproliferative epidermis induced by abrasion or tape stripping, because of the increased number of smaller cells (28), after UVB the normal morphology and stem-amplifying cell mechanism appear to be retained (Fig. 1 A and B and ref. 29).
Clonal Expansion Is Not Driven by a UVB-Induced Proliferative Mutation.
In these experiments, sustained UVB was required for a clone's territory to expand (Fig. 3B). This requirement rules out the possibility that clonal expansion was cell-autonomous, i.e., that it was initiated by irreversible UVB-induced proliferative mutations in the p53-mutant keratinocyte. The p53 mutation itself is one of those ruled out, a significant result because p53 can induce asymmetric stem-cell-like division (30). Constant clone area cannot result from shrinkage being offset by UVB-independent expansion; this could at best be transient, because clones expand indefinitely but regressing clones can only shrink to zero size. The nonautonomous nature of clonal expansion is supported by the asymmetric morphology of the individual clones. Murine precancers also require UVB for growth (31). There is precedent for physiological events driving clonal expansion. For tumors, which have already escaped regulation by adjacent cells, hypoxia-induced apoptosis acts as a selection pressure favoring clonal expansion of apoptosis-resistant p53-mutant cells (32). A similar model has been proposed for mutant keratinocytes in normal skin exposed to UVB (3).‡‡
Epidermal Stem Cell Compartments Act as Barriers to Proliferation of Aberrant Cells, Breachable by UVB.
What does sustained UVB exposure achieve, if not a mutation conferring cell-autonomous proliferation? Cairns noted that stem cells pose a threat to multicellular organisms and suggested they might be constrained by stem cell compartments (33). Two present results support this concept. First, the quantization of clone sizes indicates that escape of a p53-mutant keratinocyte from a stem cell compartment is rate-limiting (Fig. 4). Second, in the absence of UVB, several clones were constrained in clone area despite increasing in cell number (Fig. 5). Thus, after mutating the p53 gene, chronic carcinogen exposure evidently allows cells to escape a barrier presented by the stem cell compartment arrangement. This barrier may simply be the flanking stem cell compartments, rather than a physical or physiological structure within each compartment, because in chimeric embryos stem cell progeny can migrate across EPU borders (34). Clonal expansion of p53-mutant keratinocytes emerges as an interplay between the obstacle presented by stem cell compartments and the driving force of physiological changes induced by sustained UVB exposure, which allow repeated breaching of this barrier.
Acknowledgments
We thank M. Zhu and C. Adrada for technical assistance, Drs. R. E. Tigelaar, A. R. Oseroff, P. D. Forbes, D. B. Yarosh, S. Takeuchi, P. Havre, and F. R. de Gruijl for scientific advice, and Dr. R. E. Tarone for statistical analysis. This work was supported by National Institutes of Health Grants CA78735 and CA55737, by a Robert Leet and Clara Guthrie Patterson Trust award to D.E.B., and by National Institutes of Health Grant P30-CA16359 to the Yale Cancer Center.
Abbreviations
- UVB
280–320 nm UV radiation
- UVA
320–400 nm radiation
- UVC
100–280 nm radiation
- EPU
epidermal proliferating unit
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
The 2/3 frequency of escaping from one stem cell compartment to the next might be achievable by mutation if UVB induces allelic loss in a succession of flanking stem cell compartments, each loss conferring susceptibility to colonization. The known cycle times for transit amplifying and stem cells dictate that the allelic loss frequency be ≥10−3/gene/cell generation and keratinocytes have ≥100 active genes capable of conferring a proliferative advantage on losing only one of the two alleles.
References
- 1.Brash D E. Trends Genet. 1997;13:410–414. doi: 10.1016/s0168-9525(97)01246-8. [DOI] [PubMed] [Google Scholar]
- 2.Sherr C J. Science. 1996;274:1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 3.Ziegler A, Jonason A S, Leffell D J, Simon J A, Sharma H W, Kimmelman J, Remington L, Jacks T, Brash D E. Nature (London) 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 4.Ullrich S E. In: Biochemical Modulation of Skin Reactions. Kydonieus A F, Wille J J, editors. Boca Raton, FL: CRC; 2000. pp. 281–300. [Google Scholar]
- 5.DiGiovanni J. Pharmacol Ther. 1992;54:63–128. doi: 10.1016/0163-7258(92)90051-z. [DOI] [PubMed] [Google Scholar]
- 6.Epstein J H, Epstein W L. J Invest Dermatol. 1962;39:455–460. [Google Scholar]
- 7.Blum H F. In: The Biologic Effects of Ultraviolet Radiation. Urbach F, editor. Oxford, U.K.: Pergamon; 1969. pp. 543–549. [Google Scholar]
- 8.Ruch R J. In: Gap Junctions–Molecular Basis of Cell Communication in Health and Disease. Peracchia C, editor. San Diego: Academic; 2000. pp. 535–554. [Google Scholar]
- 9.Cohen S M, Ellwein L B. Science. 1990;249:1007–1011. doi: 10.1126/science.2204108. [DOI] [PubMed] [Google Scholar]
- 10.Jonason A S, Kunala S, Price G J, Restifo R J, Spinelli H M, Persing J A, Leffell D J, Tarone R E, Brash D E. Proc Natl Acad Sci USA. 1996;93:14025–14029. doi: 10.1073/pnas.93.24.14025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ren Z P, Hedrum A, Ponten F, Nister M, Ahmadian A, Lundeberg J, Uhlen M, Ponten J. Oncogene. 1996;12:765–773. [PubMed] [Google Scholar]
- 12.Brash D E, Ziegler A, Jonason A, Simon J A, Kunala S, Leffell D J. J Invest Dermatol Symp Proc. 1996;1:136–142. [PubMed] [Google Scholar]
- 13.de Gruijl F R, Forbes P D. BioEssays. 1995;17:651–660. doi: 10.1002/bies.950170711. [DOI] [PubMed] [Google Scholar]
- 14.Rebel H, Mosnier L O, Berg R J, Westerman-de Vries A, van Steeg H, van Kranen H J, de Gruijl F R. Cancer Res. 2001;61:977–983. [PubMed] [Google Scholar]
- 15.Bergstresser P R, Juarez D V. Methods Enzymol. 1984;108:683–691. doi: 10.1016/s0076-6879(84)08127-1. [DOI] [PubMed] [Google Scholar]
- 16.Ouhtit A, Muller H K, Davis D W, Ullrich S E, McConkey D, Ananthaswamy H N. Am J Pathol. 2000;156:201–207. doi: 10.1016/S0002-9440(10)64720-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potten C S. In: Stem Cells: Their Identification and Characterization. Potten C S, editor. New York: Churchill–Livingstone; 1983. pp. 200–232. [Google Scholar]
- 18.Morris R J, Fischer S M, Slaga T J. J Invest Dermatol. 1985;84:277–281. doi: 10.1111/1523-1747.ep12265358. [DOI] [PubMed] [Google Scholar]
- 19.Mackenzie I C. J Invest Dermatol. 1997;109:377–383. doi: 10.1111/1523-1747.ep12336255. [DOI] [PubMed] [Google Scholar]
- 20.Kolodka T M, Garlick J A, Taichman L B. Proc Natl Acad Sci USA. 1998;95:4356–4361. doi: 10.1073/pnas.95.8.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor G, Lehrer M S, Jensen P J, Sun T T, Lavker R M. Cell. 2000;102:451–461. doi: 10.1016/s0092-8674(00)00050-7. [DOI] [PubMed] [Google Scholar]
- 22.You Y H, Szabo P E, Pfeifer G P. Carcinogenesis. 2000;21:2113–2117. doi: 10.1093/carcin/21.11.2113. [DOI] [PubMed] [Google Scholar]
- 23.Jensen U B, Lowell S, Watt F M. Development (Cambridge, UK) 1999;126:2409–2418. doi: 10.1242/dev.126.11.2409. [DOI] [PubMed] [Google Scholar]
- 24.Tani H, Morris R J, Kaur P. Proc Natl Acad Sci USA. 2000;97:10960–10965. doi: 10.1073/pnas.97.20.10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller S J, Lavker R M, Sun T T. Semin Dev Biol. 1993;4:217–240. [Google Scholar]
- 26.Jones P H. BioEssays. 1997;19:683–690. doi: 10.1002/bies.950190808. [DOI] [PubMed] [Google Scholar]
- 27.Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- 28.Potten C S, Allen T D. J Cell Sci. 1975;17:413–447. doi: 10.1242/jcs.17.3.413. [DOI] [PubMed] [Google Scholar]
- 29.Al-Barwari S E, Potten C S. Cell Tissue Kinet. 1979;12:281–289. doi: 10.1111/j.1365-2184.1979.tb00150.x. [DOI] [PubMed] [Google Scholar]
- 30.Sherley J L, Stadler P B, Johnson D R. Proc Natl Acad Sci USA. 1995;92:136–140. doi: 10.1073/pnas.92.1.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Gruijl F R, van der Leun J C. Cancer Res. 1991;51:979–984. [PubMed] [Google Scholar]
- 32.Graeber T G, Peterson J F, Tsai M, Monica K, Fornace A F, Giaccia A J. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cairns J. Nature (London) 1975;255:197–200. doi: 10.1038/255197a0. [DOI] [PubMed] [Google Scholar]
- 34.Schmidt G H, Blount M A, Ponder B A. Development (Cambridge, UK) 1987;100:535–541. doi: 10.1242/dev.100.3.535. [DOI] [PubMed] [Google Scholar]