Abstract
Listeria monocytogenes is the causative agent of listeriosis, which is an uncommon but severe infection associated with high mortality rates in humans especially in high-risk groups. This bacterium survives a variety of stress conditions (e.g., high osmolality, low pH), which allows it to colonize different niches especially niches found in food processing environments. Additionally, a considerable heterogeneity in pathogenic potential has been observed in different strains. In this study, 38 isolates of L. monocytogenes collected in Chile from clinical samples (n = 22) and non-clinical samples (n = 16) were analyzed using whole genome sequencing (WGS) to determine their genomic diversity. A core genome Single Nucleotide Polymorphism (SNP) tree using 55 additional L. monocytogenes accessions classified the Chilean isolates in lineages I (n = 25) and II (n = 13). In silico, Multi-locus sequence typing (MLST) differentiated the isolates into 13 sequence types (ST) in which the most common were ST1 (15 isolates) and ST9 (6 isolates) and represented 55% of the isolates. Genomic elements associated with virulence (i.e., LIPI-1, LIPI-3, inlA, inlB, inlC, inlG, inlH, inlD, inlE, inlK, inlF, and inlJ) and stress survival (i.e., stress survival islet 1 and stress survival islet 2) were unevenly distributed among clinical and non-clinical isolates. In addition, one novel inlA premature stop codon (PMSC) was detected. Comparative analysis of L. monocytogenes circulating in Chile revealed the presence of globally distributed sequence types along with differences among the isolates analyzed at a genomic level specifically associated with virulence and stress survival.
Keywords: Listeria monocytogenes, whole genome sequencing, single nucleotide polymorphism, genomic diversity, Chile
1. Introduction
Listeria monocytogenes is a foodborne pathogen responsible for listeriosis, which is a severe disease especially in high-risk groups such as the elderly, pregnant women, and newborns [1] in which the case-fatality rate is usually up to 20–30% [2]. Furthermore, L. monocytogenes represents a major concern for the food industry due to its ubiquitously distribution in the food production environment and its ability to survive and grow in stress conditions such as acidic environments, high salt concentrations, and low temperatures [3], which are conditions usually found in food preservation barriers.
Subtyping techniques have classified L. monocytogenes into four evolutionary lineages and 13 serotypes. Most isolates from clinical cases and food belonging to lineages I and II. These two main lineages contain serotypes 1/2a, 1/2b, and 4b, which represent the most frequently reported serotypes involved in human listeriosis cases and outbreaks [4]. Multi-locus sequence typing (MLST) further subdivided L. monocytogenes into 63 phylogenetic groups known as clonal complexes (CC). Some CCs are highly prevalent [5,6] and have been associated with clinical cases worldwide [7].
Pathogenesis of L. monocytogenes is associated with their ability to invade, multiply, and survive within different non-phagocytic cells [8]. These characteristics are attributed to the presence of Listeria Pathogenicity Island-1 (LIPI-1) and the inlAB operon. Listeria Pathogenicity Island-1 contains genes that allows Listeria to escape from the phagocytic vacuole to replicate in the cytosol and to spread cell-to-cell using actin polymerization [9] and the inlAB operon encodes two internalins, which are critical for entry into non-phagocytic cells [10]. In addition, accessory internalin family members have been identified and associated with virulence [11,12,13]. Several studies have shown that subtypes of L. monocytogenes differ in their pathogenic potential [14,15,16]. For example, invasion assays in human epithelial cells have shown that some isolates have an attenuated invasion phenotype due to the presence of premature stop codon mutations (PMSC) in inlA, which leads to the production of truncated InlA. This type of mutations are mostly found in L. monocytogenes isolated from foods [17].
Multiple isolates carry genetic elements that could provide them with advantages in food processing environments such as the stress survival islet 1 (SSI-1) and the recently discovered stress survival islet 2 (SSI-2) [18]. These two islets encode genes, which allow L. monocytogenes to survive in suboptimal conditions commonly found in food processing environments (i.e., low pH, high salt concentrations, and alkaline and oxidative stress conditions) [18,19].
In Chile, two L. monocytogenes outbreaks occurred in 2008 and 2009, which were linked to the consumption of soft cheeses and sausages/meat products, respectively [20]. The pulsed field gel electrophoresis (PFGE) analysis identified two PFGE types, which are the PFGE type 9 for the 2008 outbreak and the PFGE Type 1 for the 2009 outbreak [20]. Furthermore, epidemiological surveillance has shown that sporadic listeriosis cases have slowly increased since 2010 in Chile [21]. Previous reports have described the presence of L. monocytogenes in ready-to-eat food (RTE) and raw food products [22,23,24]. In Chile, most isolates belong to serotype 4b and displayed PFGE patterns that suggested the isolates were closely related with human clinical cases [24].
To date, there is a lack of knowledge on the genomic diversity of Chilean L. monocytogenes isolates from humans and foods. This study aims to investigate the genomic diversity of L. monocytogenes from clinical cases and food in Chile and to put these isolates in a phylogenetic context with regard to isolates from other countries.
2. Materials and Methods
2.1. Listeria monocytogenes Isolates Used in This Study
A total of 38 isolates obtained from different locations and sources in Chile were selected for whole genome sequencing (WGS) and genomic analysis (Table 1 and Figure S1). These isolates were previously PFGE typed at the Chilean Institute of Public Health. A total of 22 isolates were obtained from clinical samples. In addition, 16 isolates were obtained from food and food-related environments. All isolates were selected to represent different locations in Chile, isolation years, and different PFGE types. Among them, four isolates were of PFGE types 1 and type 9, which represented the PFGE types linked to the listeriosis outbreaks occurred in 2008 and 2009 in Chile.
Table 1.
Metadata and molecular characterization of L. monocytogenes isolates used in this study.
| Isolates | Origin 1 | Source | Isolation Date | Geographic Location 2 | Pulse Type 3 | Serogroup 4 | Sequence Type 4 | Clonal Complex 4 |
|---|---|---|---|---|---|---|---|---|
| T1-001 | Clinical | Amniotic fluid | 2009 | Santiago | 167 | IIc | ST9 | CC9 |
| T1-002 | Clinical | Blood | 2010 | Santiago | 3 | IIa | ST7 | CC7 |
| T1-003 | Clinical | Cerebrospinal fluid | 2011 | Los Lagos | 114 | IIa | ST8 | CC8 |
| T1-004 | Clinical | Cerebrospinal fluid | 2011 | Santiago | 260 | IIb | ST392 | - |
| T1-005 | Clinical | Cerebrospinal fluid | 2010 | Valparaiso | 19 | IIb | ST5 | CC5 |
| T1-006 | Clinical | Blood | 2010 | Aysén | 48 | IVb | ST1 | CC1 |
| T1-007 | Clinical | Blood | 2010 | Santiago | 197 | IVb | ST1 | CC1 |
| T1-008 | Clinical | Cerebrospinal fluid | 2011 | Araucanía | 235 | IVb | ST1 | CC1 |
| T1-009 | Clinical | Amniotic fluid | 2011 | Santiago | 252 | IVb | ST1 | CC1 |
| T1-010 | Clinical | Blood | 2011 | O’Higgins | 264 | IVb | ST1 | CC1 |
| T1-011 | Food | Ham | 2010 | Santiago | 167 | IIa | ST8 | CC8 |
| T1-012 | Food | Sausage | 2010 | Los Lagos | 147 | IIb | ST3 | CC3 |
| T1-013 | Food | Ice cream | 2010 | Santiago | 212 | IIb | ST5 | CC5 |
| T1-014 | Food | Sausage | 2010 | Los Lagos | 210 | IIc | ST9 | CC9 |
| T1-016 | Food | Ham | 2010 | Santiago | 211 | IVb | ST1207 | CC6 |
| T1-017 | Food | Ham | 2010 | Santiago | 99 | IIc | ST9 | CC9 |
| T1-018 | Food | Ice cream | 2010 | Santiago | 156 | IVb | ST1395 7 | CC6 |
| T1-019 | Environment | Food plant | N/A | Bío-Bío | N/A | IVb | ST6 | CC6 |
| T1-020 | Environment | Food plant | N/A | Bío-Bío | N/A | IIa | ST121 | CC121 |
| T1-022 | Clinical | Blood | 2011 | Valparaiso | 256 | IVb | ST1 | CC1 |
| T1-023 | Clinical | Blood | 2008 | Santiago | 9 5 | IVb | ST1 | CC1 |
| T1-024 | Clinical | Blood | 2008 | O’Higgins | 9 5 | IVb | ST1 | CC1 |
| T1-025 | Clinical | Blood | 2010 | Santiago | 99 | IVb | ST1 | CC1 |
| T1-026 | Food | Sausage | 2010 | Los Lagos | 2 | IIc | ST9 | CC9 |
| T1-027 | Clinical | Blood | 2010 | Santiago | 46 | IIa | ST7 | CC7 |
| T1-028 | Clinical | Blood | 2008 | Bío-Bío | 9 5 | IVb | ST1 | CC1 |
| T1-029 | Food | Pork pate | 2010 | Araucanía | 126 | IVb | ST2 | CC2 |
| T1-030 | Food | Sausage | 2009 | Bío-Bío | 1 6 | IIa | ST9 | CC9 |
| T1-031 | Clinical | Human | 2010 | O’Higgins | 20 | IVb | ST1 | CC1 |
| T1-033 | Clinical | Blood | 2010 | Araucanía | 133 | IVb | ST1 | CC1 |
| T1-034 | Food | Ice cream | 2010 | Santiago | 64 | IIb | ST5 | CC5 |
| T1-037 | Clinical | Peritoneal fluid | 2011 | Bío-Bío | 137 | IIa | ST37 | CC37 |
| T1-038 | Food | Ham | 2010 | Santiago | 53 | IIa | ST121 | CC121 |
| T1-039 | Clinical | Cerebrospinal fluid | 2010 | Santiago | 209 | IVb | ST2 | CC2 |
| T1-040 | Food | Beef | 2009 | Bío-Bío | 1 6 | IIc | ST9 | CC9 |
| T1-041 | Clinical | Blood | 2011 | Bío-Bío | 245 | IVb | ST1 | CC1 |
| T1-042 | Food | Cheese | 2009 | Santiago | 9 5 | IVb | ST1 | CC1 |
| T1-043 | Clinical | Blood | 2010 | Santiago | 58 | IVb | ST1 | CC1 |
N/A: Not available. 1 All clinical cases were obtained from humans. 2 For details on geographic origin within Chile, see Figure S1. 3 Pulsed Field Gel Electrophoresis (PFGE) were typed at the Chilean Institute of Public Health. 4 Identified in this study. 5 PFGE type was involved in outbreak 2008. 6 PFGE type was involved in outbreak 2009. 7 Novel sequence type.
2.2. Genome Sequencing and Annotation
For DNA purification, the DNeasy Blood and Tissue kit (Qiagen, Valencia, CA, USA) was used. The QUBIT fluorimeter (Life Technologies, Carlsbad, CA, USA) was used to quantify the DNA. The Nextera XT DNA library Preparation kit (Illumina, San Diego, CA, USA) was used for library preparation and DNA sequencing was performed on the NextSeq500 (Illumina Inc., San Diego, CA, USA). Sequencing was conducted at the Food and Drug Administration (FDA) Center for Food Safety and Applied Nutrition. Listeria genomes were sequenced with a 2 × 150-bp paired-end run. Adapters of the obtained reads were removed and quality trimmed with Trimmomatic (v.0.35) [25]. Reads were analyzed and checked for quality using FastQC (v0.11.4) [26]. The reads were de novo assembled using SPAdes (V3.7.1) [27]. Assemblies were obtained by setting k-mer lengths of 21, 33, 55, and 77 for read lengths between 150 and 300 bp (default settings). Contigs were annotated using a combination of annotation with RAST [28] and automatic annotation with the National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP) [29]. All genomes were deposited at the DDBJ/ENA/GenBank (See Table S1 for accession numbers).
2.3. Lineage Determination and Phylogenetic Analysis
Lineage determination was performed using Parsnp from software Harvest suite tools [30]. A core genome alignment of the 38 Chilean isolates along with 55 publicly available sequences (See Table S2) was performed. These isolates of L. monocytogenes were used as references for three major lineages (I, II, and III). The phylogenetic relationship of the 38 Chilean isolates was inferred using single nucleotide polymorphisms (SNPs) with Call SNPSs & Infer Phylogeny (CSI) Phylogeny v.1.4, which creates a maximum likelihood tree [31] using L. monocytogenes strain EGD-e (NCBI: NC_003210.1) as a reference genome by using default settings (10× or at least 10% of the average depth). A separate phylogenetic analysis was performed for a group of 15 isolates using CSI phylogeny v.1.4 [31], but F2365 (NCBI: AE017262.2) was used as a phylogenetically closer reference.
2.4. Subtyping
When silico serotyping was performed with the LisSero v.0.1, [32] a script predicting serogroups for L. monocytogenes simulating a PCR of five regions of DNA (lmo118, lmo0737, ORF2110, ORF2829, and prs as an internal amplification control) [32]. This scheme classified isolates on four molecular serogroups known as IIa:1/2a, 3a; IIb:1/2b, 3b, 7; IIc:1/2c, 3c and IVb:4b, 4d, 4e. The sequence type was inferred from WGS data using the program MLST 1.8 from the Center for Genomic Epidemiology [33] and was revised for updated assignments and Clonal complexes by using the Institut Pasteur whole genome MLST database [16]. One novel ST was identified, which was submitted to the Pasteur Institute database to confirm the new assignment. Prophage analysis was performed using PHASTER [34]. The diversity of plasmids was conducted with PlasmidFinder [35] and the presence of antimicrobial resistance genes was screened with ResFinder [36].
2.5. Screening of Virulence Genes and Stress-Related Elements
To analyze genes related to virulence and stress survival islets, the BLAST algorithm from NCBI was used [37]. The strain of Listeria monocytogenes EGD-e (NCBI: NC_003210.1) was used as a reference for the analysis of SSI-1, LIPI-1, inlAB operon, and other internalins (inlC, inlG, inlH, inlE, inlF, inlK, inlJ, and inlD). For inlA characterization, sequences were aligned and screened for non-sense mutations causing premature stop codons or amino acid deletion using the software ClustalO 2.1 [38]. Listeria monocytogenes strain F2365 4b was used as a reference for LIPI-3 (NCBI: NC_002973.6) and L. monocytogenes strain CDL64 (NCBI: HQ179545.1) was used as a reference for SSI-2. Additionally, VirulenceFinder 1.5 of Listeria was used to screen for 81 distinct virulence genes in their database [39].
3. Results and Discussion
The present study characterized the genomic diversity of 38 isolates of L. monocytogenes from clinical (human) and non-clinical (food and food related environment) samples obtained in different regions of Chile between 2008 and 2011. Genome sizes ranged between 2.89 Mb and 3.11 Mb. The average G + C content was 37.9%. De novo assembly ranged from 12 contigs to 61 contigs with an average mean of length of the contigs or N50 of 454,984 bp (Table S1).
Major findings of this study include: (i) L. monocytogenes isolates are mostly represented by CCs distributed worldwide and involved in human infections and outbreaks, (ii) isolates of the PFGE type causing the 2008 to 2009 outbreaks showed genetic relatedness to other worldwide clinical isolates, (iii) clinical and non-clinical L. monocytogenes isolates showed distinct virulence and stress survival genetic elements, and (iv) the presence of one novel PMSC mutation in the inlA gene along with additional PMSC already reported in other countries in isolates from non-clinical samples.
3.1. Listeria monocytogenes Isolates Are Mostly Represented by Clonal Complexes Distributed Worldwide and Involved in Human Infections and Outbreaks
A rapid core genome alignment classified the 38 isolates in two lineages (I and II). The majority of clinical isolates grouped in Lineage I (n = 25) while the majority of non-clinical isolates grouped in Lineage II (n = 13) (Figure S2). Between these two lineages, isolates were classified in four serogroups: serogroup IVb (52.6%), serogroup IIa (21.1%), and serogroups IIb and IIc (13.1% each). Serogroup IVb and IIb belong to Lineage I and serogroup IIa and IIc belong to Lineage II (Table 1 and Figure 1). Strains of serotype 4b (belonging to serogroup IVb) have been responsible for the majority of human listeriosis outbreaks worldwide [40] even though serotype 1/2a and 1/2b have been also involved in outbreaks especially in Europe [41]. In this study, most of the isolates sequenced represented serogroup IVb. Most of the clinical isolates (65%) were of this serogroup while serogroups IIa and IIb were represented by 17% and 13% of the isolates, respectively. One of the clinical isolates was classified as serogroup IIc, which is an uncommon serotype in human clinical cases [42]. Among isolates obtained from non-clinical samples, serogroup IVb were also the most common (33%). This distribution is similar to results previously obtained by Montero et al. (2015) in Chile where serotype 4b was the most prevalent in isolates from food (46%) [24]. Similar findings were reported in China in 2010 [43]. Previous studies have shown that serotypes 1/2b and 4b are the most prevalent in food in Uruguay [44] and serotypes 1/2c and 4b are frequent in food samples from Brazil [45]. Within isolates classified as serogroup IVb, a search for an atypical IVb variant 1 (IVb-1) was conducted [46]. This variant has recently been linked to several outbreaks in the United States [47]. However, none of the isolates were found to represent this variant. While the IVb-1 is considered rare, a recent study identified this variant in isolates of L. monocytogenes isolated from frozen prawns in Chile [48].
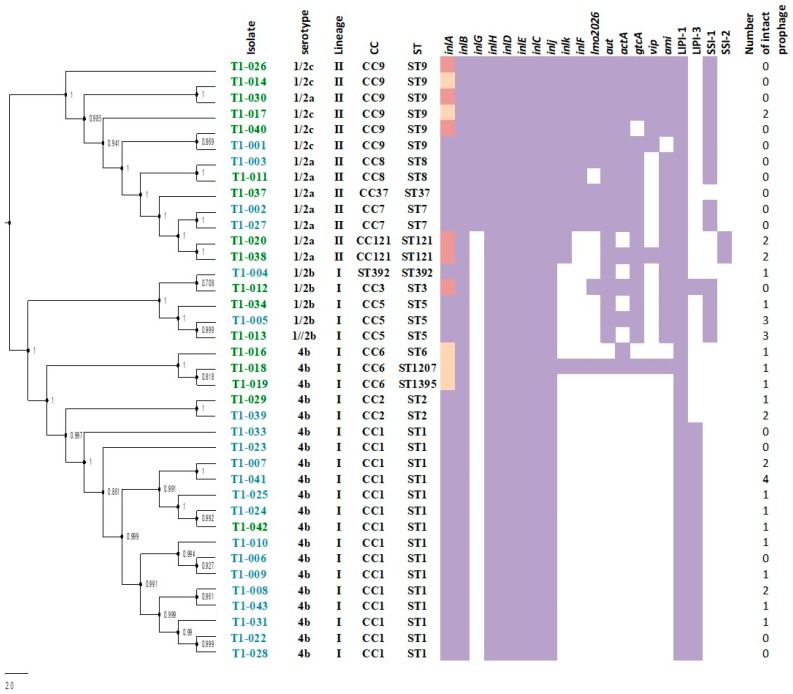
Figure 1.
Phylogenetic tree of 38 L. monocytogenes isolates from Chile inferred from single nucleotide polymorphisms (SNPs) of whole genome sequencing data and the distribution of their genetic elements associated with virulence and stress survival. Isolates from clinical samples are colored in blue and isolates from non-clinical samples are colored in green. Columns on the right of the tree indicate the presence (purple) or absence (white) of genetic elements. Isolates with premature stop codon mutations (PMSCs) in inlA are in pink and isolates with 3 aa deletions are yellow. The number of intact prophages predicted with PHAST is added.
The MLST analysis differentiated the 38 Chilean isolates into 13 sequence types (STs). A total of 12 STs had been previously reported and were present in the Institut Pasteur whole genome MLST database and one novel ST was assigned (ST1395) (Table 1). Additionally, the 13 STs were grouped in 11 CCs and 1 ST represented a singleton (Figure 1 and Table 1). In Lineage I, isolates were grouped in CC1 (n = 15), CC2 (n = 2), CC3 (n = 1), CC5 (n = 3), CC6 (n = 3), and one singleton (ST392). In Lineage II, isolates were grouped in CC7 (n = 2), CC8 (n = 2), CC9 (n = 6), CC37 (n = 1), and CC121 (n = 2). The most common CCs in Lineage I and Lineage II were CC1 (60%) and CC9 (46%), respectively. Most of the CCs identified among Chilean isolates represent the most common CCs worldwide with CC1 and CC9 as the most commonly and widely reported CCs in Europe and South/Central America [5,6]. Importantly, CC1 has also been associated with hyper virulence [16]. In addition, CCs were not equally distributed among isolates from different origins. CC1 was more common on isolates from clinical cases and CC9 in non-clinical isolates. This result is in agreement with a recent retrospective study that analyzed over 6000 strains of L. monocytogenes in France [16].
3.2. Isolates of the Pulsed Field Gel Electrophoresis Type Causing the 2008–2009 Outbreaks Showed Genetic Relatedness to Other Worldwide Clinical Isolates
The phylogenetic analysis of the core genome alignment identified a group within Lineage I of clinical isolates (T1-006, T1-007, T1-008, T1-009, T1-010, T1-022, T1-025, T1-031, T1-033, T1-041, and T1-043) and one non-clinical isolate (T1-042) that were clustered together (Figures S2 and S3, Table S3). All these isolates belong to the same CC1 and were obtained in different years (2008–2011) and displayed different PFGE types. To gain insights into relatedness between these isolates, an analysis of the whole genome to determine SNP differences among them was conducted using the L. monocytogenes strain F2365 as the most closely related reference. SNP differences of these isolates ranged from 17 SNPs to 198 SNPs (Table S3). Within this subgroup, T1-023, T1-024, and T1-028 presented the same PFGE type as the isolates that caused the 2008 outbreak (type 9). However, these three isolates were found to differ between 66 SNPs to 122 SNPs (Table S3) from each other. A previous study using WGS showed a lower diversity among epidemiologically linked isolates (same PFGE type) with SNP differences less than 10 SNPs [49]. A cluster of three non-clinical isolates (T1-016, T1-018, and T1-019), which clustered in CC6 and showed SNP differences that ranged from three SNPs to 12 SNPs was found, which suggests these isolates are highly related even though these isolates were not epidemiologically linked in this study. Prophage analysis on these isolates showed one intact prophage identified in T1-016, T1-018, and T1-019. These two isolates (T1-018 and T1-019) were different by only three SNPs.
3.3. Clinical and Non-Clinical L. monocytogenes Isolates Showed Distinct Virulence and Stress Survival Genetic Elements
In this study, the distribution of selected virulence genes and genetic elements related with stress survival were surveyed. Genes encoded in LIPI-1 (prfA, plcA, plcB, hly, and mpl) and the inlAB operon, which encodes internalin A and B, were present in all 38 isolates (Figure 1). A previous study conducted in Chile of L. monocytogenes isolated from foods (e.g., raw meat, cheese, and frozen seafood) reported a different distribution of these genomic elements. In the previous study, the LIPI-1 cluster and the inlAB operon were found to be associated with a given serotype and food group [24]. However, methodologies and isolates between our study and this previous are different, which may explain the difference in the results. Reports in France and China found these genes in all isolates and both studies used WGS [16,50]. Other internalin family members (inlC, inlJ, inlH, inlD, inlE, and inlJ) were detected in all isolates (Figure 1). However, inlG, inlF, and inlK were found not evenly distributed among isolates. The genes inlG and inlK were found in 14 isolates and inlF in 12 isolates. Most of these isolates were obtained from non-clinical sources, which are commonly represented by Lineage II. The presence of inlG seems to be associated with Lineage II and our result agree with previous studies using PCR that associated the presence of these internalins exclusively with Lineage II [51,52]. Additionally, an analysis looking at 81 distinct genes in the database of VirulenceFinder for Listeria identified seven genes distinct to internalins (i.e., lmo2026, aut, actA, gtcA, vip, and ami) that showed diversity in their content (Figure 1). Other virulence markers possibly associated with lineage is LIPI-3, which was exclusively found in 15 isolates of the Lineage I of serotype 4b and one from serotype 1/2b (Figure 1). LIPI-3 has been previously associated with Lineage I, serotype 4b [53,54], and with Lineage III and Listeria innocua [55,56]. Conversely, the stress survival associated gene cluster known as SSI-1 was found in both lineages. SSI-1 was found in 37% of isolates and most of these isolates belonged to CC9, CC8, and CC7 of Lineage II and to CC5 and CC3 (both Lineage I). However, SSI-2 was found only in two isolates from CC121 (Lineage II). This is consistent with previous reports that indicate SSI-2 to be only associated with CC121 isolates [18]. The analysis of the plasmids identified that none of the isolates contained a known plasmid. In addition, the only antimicrobial resistance gene identified was the gene fosX, which confers resistance to fosfomycin identified in all 38 isolates. The number of prophages detected with PHAST was very diverse and ranged from 0 to 4 intact prophages detected (Figure 1).
3.4. Presence of One Novel PMSC Mutation in the inlA Gene Along with Additional PMSC Reported in Other Countries in Isolates from Non-Clinical Samples
The WGS analysis showed that most of the isolates of this study (68%) contained a complete InlA. All clinical isolates have a full-length inlA gene while 11 isolates from non-clinical samples carried a PMSC mutation (Figure 1). Five isolates harbored a previously reported 9 nucleotide deletion, which was predicted to encode a shorter (797 amino acids) version of InlA. This variant of inlA is predicted to be fully functional. In vitro invasion assays have shown that these shorter variants have an invasion ability comparable with that of full-length inlA isolates [57,58] and also have been reported in isolates from clinical cases [14]. This type of deletion was found in isolates of serotypes 1/2c and 4b and have been found in isolates of serotypes 1/2b in USA [59] as well as serotypes 4c and 1/2a in Canada and Switzerland [60]. Additionally, six PMSCs were detected exclusively in isolates from non-clinical origin, which belongs to serotypes 1/2c (3), 1/2a (2), and 1/2b (1) (Table 2). These mutations were classified into four PMSC types that were previously described, which include one type 19 (resulting in 325 aa protein product), one type 13 (resulting in a 527 aa protein product), two type 6 (resulting in 491 aa protein product), and one type 11 (resulting in a 684 aa protein product) [61,62,63,64]. In addition, the presence of a novel PMSC type was found in one isolate (T1-012), which carried a non-sense mutation at position 821 where one adenine base was deleted. This resulted in a frameshift mutation, which codes for a 277-protein product (Table 2). This truncated protein might result in low virulence in in vitro invasion assays due to the lack of the LPXTG motif, which is involved in anchoring the protein to peptidoglycan in the cell wall [65]. Further studies are essential to confirm this.
Table 2.
Length of InlA among Chilean L. monocytogenes.
| Number of Isolates | InlA Length (aa) | Mutation Type (PMSC) | Nucleotide Mutation Position | Functional | Reference |
|---|---|---|---|---|---|
| 27 | 800 | - | - | yes | Glaser et al., 2001 |
| 5 | 797 | NA | NA | yes | Chen et al., 2011 |
| 1 | 684 | 11 | 2054 (G to A) | no | Rousseaux et al., 2002 |
| 1 | 527 | 13 | 1579 (Ato T) | no | Handa-Miya et al., 2007 |
| 2 | 491 | 6 | 1474 (Cto T) | no | Olier et al., 2002 |
| 1 | 325 | 19 | 976 (G to T) | no | Gelbicova et al., 2015 |
| 1 | 277 | Novel | 821 (deletion A) | no | In this study |
4. Conclusions
Whole genome sequencing has proven to be a powerful subtyping tool for L. monocytogenes especially in reference centers in North America and Europe. This study provides the first characterization at a genomic level using WGS of clinical and non-clinical isolates of L. monocytogenes isolated from Chile. Our results show the presence of isolates from Chile that represent clonal groups associated with listeriosis worldwide, which supports the global distribution of key human diseases associated with L. monocytogenes clonal groups. This study is the first of this type in South America, so further efforts are necessary in order to implement WGS in routine surveillance in South America.
Acknowledgments
We thank Marc Allard from FDA for kindly sequencing the isolates analyzed in this study.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/9/8/396/s1. Figure S1. Location and distribution of isolates used in this study. (a) Map of Chile divided by regions and red dots represent the locations where the isolates were obtained. (b) Circular representation of the percent of isolates sequenced that were obtained from the different regions within Chile. Figure S2. Maximum likelihood phylogenetic tree based on SNPs of core genome of Chilean L. monocytogenes using 55 L. monocytogenes sequences as references. Phylogenetic tree representing the two lineages identified in this study, ID of the isolates from clinical samples are colored in blue and ID of the isolates from non-clinical samples are colored in green. Figure S3. L. monocytogenes phylogeny based on single nucleotide polymorphism (SNP). Phylogenetic tree of the 15 isolates that clustered together that were obtained from clinical samples, mostly of the CC1. Isolates with their ID in blue were obtained from human clinical samples and the one in green from non-clinical samples. The bootstrap was added to the clades. Table S1. Sequencing statistics of 38 isolates of L. monocytogenes obtained from clinical and non-clinical samples sequenced in this study. Table S2. L. monocytogenes used as references in the core genome alignment for lineage determination. Table S3. Pairwise distance matrix of SNP number differences for 15 Chilean isolates of L. monocytogenes clustered in CC1.
Author Contributions
V.T. and A.I.M.-S. conceived this project and designed the analysis. V.T. and H.C.d.B. performed the bioinformatics analysis. J.C.H., G.G.-R., H.B.-T. contributed with material. V.T., G.G.-R., H.B.-T., M.T., and A.I.M.-S. contributed to the writing of this manuscript.
Funding
The study was funded by the Universidad Andres Bello Grant no. DI-638-15/I to V.T.
Conflicts of Interest
The authors declared no conflict of interest.
References
- 1.Swaminathan B., Gerner-Smidt P. The epidemiology of human listeriosis. Microbes Infect. 2007;9:1236–1243. doi: 10.1016/j.micinf.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 2.Mead P.S., Slutsker L., Dietz V., McCaig L.F., Bresee J.S., Shapiro C., Griffin P.M., Tauxe R.V. Food-related illness and death in the United States. Emerg. Infect. Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Farber J.M., Peterkin P.I. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 1991;55:476–511. doi: 10.1104/pp.110.161547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLauchlin J., Mitchell R.T., Smerdon W.J., Jewell K. Listeria monocytogenes and listeriosis: A review of hazard characterisation for use in microbiological risk assessment of foods. Int. J. Food Microbiol. 2004;92:15–33. doi: 10.1016/S0168-1605(03)00326-X. [DOI] [PubMed] [Google Scholar]
- 5.Ragon M., Wirth T., Hollandt F., Lavenir R., Lecuit M., Le Monnier A., Brisse S. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chenal-Francisque V., Lopez J., Cantinelli T., Caro V., Tran C., Leclercq A., Lecuit M., Brisse S. Worldwide distribution of major clones of Listeria monocytogenes. Emerg. Infect. Dis. 2011;17:1110–1112. doi: 10.3201/eid/1706.101778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantinelli T., Chenal-Francisque V., Diancourt L., Frezal L., Leclercq A., Wirth T., Lecuit M., Brisse S. Epidemic clones of Listeria monocytogenes are widespread and ancient clonal groups. J. Clin. Microbiol. 2013;51:3770–3779. doi: 10.1128/JCM.01874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vázquez-Boland J.A., Kuhn M., Berche P., Chakraborty T., Domi G., González-Zorn B., Wehland J. Listeria pathogenesis and molecular virulence determinants Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez-Boland J.A., Dominguez-Bernal G., Gonzalez-Zorn B., Kreft J., Goebel W. Pathogenicity islands and virulence evolution in Listeria. Microbes Infect. 2001;3:571–584. doi: 10.1016/S1286-4579(01)01413-7. [DOI] [PubMed] [Google Scholar]
- 10.Gaillard J.L., Jaubert F., Berche P. The inlAB locus mediates the entry of Listeria monocytogenes into hepatocytes in vivo. J. Exp. Med. 1996;183:359–369. doi: 10.1084/jem.183.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffelsbauer D., Bubert A., Engelbrecht F., Scheinpflug J., Simm A., Hess J., Kaufmann S.H.E., Goebel W. The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol. Genet. Genom. 1998;260:144–158. doi: 10.1007/s004380050880. [DOI] [PubMed] [Google Scholar]
- 12.Sabet C., Lecuit M., Cabanes D., Cossart P. LPXTG protein InlJ, a newly identified internalin involved in Listeria monocytogenes virulence. Infect. Immun. 2005;73:6912–6922. doi: 10.1128/IAI.73.10.6912-6922.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neves D., Job V., Dortet L., Cossart P., Dessen A. Structure of internalin InlK from the human pathogen Listeria monocytogenes. J. Mol. Biol. 2013;425:4520–4529. doi: 10.1016/j.jmb.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 14.Chen Y., Strain E.A., Allard M., Brown E.W. Genome sequence of L. monocytogenes Strains J1816 and J1-220, associated with human outbreaks. J. Bacteriol. 2011;193:3424–3425. doi: 10.1128/JB.05048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roche S.M., Grépinet O., Kerouanton A., Ragon M., Leclercq A., Témoin S., Schaeffer B., Skorski G., Mereghetti L., Le Monnier A., et al. Polyphasic characterization and genetic relatedness of low-virulence and virulent Listeria monocytogenes isolates. BMC Microbiol. 2012;12 doi: 10.1186/1471-2180-12-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maury M., Tsai Y.H., Charlier C., Touchon M., Chenal-Francisque V., Leclercq A., Criscuolo A., Gaultier C., Roussel S., Brisabois A., et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016;48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nightingale K.K., Windham K., Martin K.E., Yeung M., Wiedmann M. Select Listeria monocytogenes subtypes commonly found in foods carry disctinct nonsense mutations in inlA. Appl. Environ. Microbiol. 2005;71:8764–8772. doi: 10.1128/AEM.71.12.8764-8772.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harter E., Wagner E.M., Zaiser A., Halecker S., Wagner M., Rychli K. The novel stress survival islet 2 (SSI-2), predominantly present in Listeria monocytogenes strains of ST121, is involved in alkaline and oxidative stress response. Appl. Environ. Microbiol. 2017 doi: 10.1128/AEM.00827-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryan S., Begley M., Hill C., Gahan C.G.M. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010;109:984–995. doi: 10.1111/j.1365-2672.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- 20.MINSAL, Departamento de Epidemiología Informe Listeriosis Actualizado el 15 de Septiembre 2011. [(accessed on 31 July 2018)]; Available online: http://www.ispch.cl/sites/default/files/documento/2011/09/listeria2011.pdf.
- 21.MINSAL, Departamento de Epidemiología. Informe año 2017 Situación Epidemiológica de Listeriosis en Chile. [(accessed on 31 July 2018)];2017 :1–11. Available online: http://epi.minsal.cl/wp-content/uploads/2018/04/INFORME-ANUAL-LISTERIOSIS-2017_2018-03-09-RevIRO-SAF.pdf.
- 22.Saludes M., Troncoso M., Figueroa G. Presence of Listeria monocytogenes in Chilean food matrices. Food Control. 2015;50:331–335. doi: 10.1016/j.foodcont.2014.08.008. [DOI] [Google Scholar]
- 23.Cordano A.M., Jacquet C. Listeria monocytogenes isolated from vegetable salads sold at supermarkets in Santiago, Chile: Prevalence and strain characterization. Int. J. Food Microbiol. 2009;132:176–179. doi: 10.1016/j.ijfoodmicro.2009.04.008. [DOI] [PubMed] [Google Scholar]
- 24.Montero D., Bodero M., Riveros G., Lapierre L., Gaggero A., Vidal R.M., Vidal M. Molecular epidemiology and genetic diversity of Listeria monocytogenes isolates from a wide variety of ready-to-eat foods and their relationship to clinical strains from listeriosis outbreaks in Chile. Front. Microbiol. 2015;6:1–8. doi: 10.3389/fmicb.2015.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolger A.M., Lohse M., Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andrews S. FastQC: A Quality Control Tool for High Throughput Sequence Data. [(accessed on 31 July 2018)]; Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 27.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D., et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatusova T., Dicuccio M., Badretdin A., Chetvernin V., Nawrocki E.P., Zaslavsky L., Lomsadze A., Pruitt K.D., Borodovsky M., Ostell J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44:6614–6624. doi: 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Treangen T.J., Ondov B.D., Koren S., Phillippy A.M. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaas R.S., Leekitcharoenphon P., Aarestrup F.M., Lund O. Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9:1–8. doi: 10.1371/journal.pone.0104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doumith M., Buchrieser C., Glaser P., Jacquet C., Martin P. Differentiation of the major Listeria monocytogenes serovars by multiplex PCR. J. Clin. Microbiol. 2004;42:3819–3822. doi: 10.1128/JCM.42.8.3819-3822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Larsen M.V., Cosentino S., Rasmussen S., Friis C., Hasman H., Marvig R.L., Jelsbak L., Sicheritz-Pontén T., Ussery D.W., Aarestrup F.M., et al. Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 2012;50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arndt D., Grant J.R., Marcu A., Sajed T., Pon A., Liang Y., Wishart D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016;44:W16–W21. doi: 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carattoli A., Zankari E., García-Fernández A., Larsen M.V., Lund O., Villa L., Aarestrup F.M., Hasman H. In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 2014;58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madden T. The BLAST Sequence Analysis Tool. In: McEntyre J., editor. The NCBI Handbook National Library of Medicine (US) National Center for Biotechnology Information; Bethesda, MD, USA: 2013. [Google Scholar]
- 38.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joensen K.G., Scheutz F., Lund O., Hasman H., Kaas R.S., Nielsen E.M., Aarestrup F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014;52:1501–1510. doi: 10.1128/JCM.03617-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cartwright E.J., Jackson K.A., Johnson S.D., Graves L.M., Silk B.J., Mahon B.E. Listeriosis outbreaks and associated food vehicles, United States, 1998–2008. Emerg. Infect. Dis. 2013;19:1–9. doi: 10.3201/eid1901.120393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orsi R.H., den Bakker H.C., Wiedmann M. Listeria monocytogenes lineages: Genomics, evolution, ecology, and phenotypic characteristics. Int. J. Med. Microbiol. 2011;301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 42.Graves L.M., Swaminathan B. PulseNet standardized protocol for subtyping Listeria monocytogenes by macrorestriction and pulsed-field gel electrophoresis. Int. J. Food Microbiol. 2001;65:55–62. doi: 10.1016/S0168-1605(00)00501-8. [DOI] [PubMed] [Google Scholar]
- 43.Chen J., Chen Q., Jiang J., Hu H., Ye J., Fang W. Serovar 4b complex predominates among Listeria monocytogenes isolates from imported aquatic products in China. Foodborne Pathog. Dis. 2010;7:31–41. doi: 10.1089/fpd.2009.0353. [DOI] [PubMed] [Google Scholar]
- 44.Braga V., Vázquez S., Vico V., Pastorino V., Mota M.I., Legnani M., Schelotto F., Lancibidad G., Varela G. Prevalence and serotype distribution of Listeria monocytogenes isolated from foods in Montevideo-Uruguay. Braz. J. Microbiol. 2017;48:689–694. doi: 10.1016/j.bjm.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vallim D.C., Barroso Hofer C., Lisbôa R.D.C., Victor Barbosa A., Alves Rusak L., Dos Reis C.M.F., Hofer E. Twenty years of Listeria in Brazil: Occurrence of Listeria species and Listeria monocytogenes serovars in food samples in Brazil between 1990 and 2012. Biomed. Res. Int. 2015 doi: 10.1155/2015/540204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Leclercq A., Chenal-Francisque V., Dieye H., Cantinelli T., Drali R., Brisse S., Lecuit M. Characterization of the novel Listeria monocytogenes PCR serogrouping profile IVb-v1. Int. J. Food Microbiol. 2011;147:74–77. doi: 10.1016/j.ijfoodmicro.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 47.Burall L.S., Grim C.J., Datta A.R. A clade of Listeria monocytogenes serotype 4b variant strains linked to recent listeriosis outbreaks associated with produce from a defined geographic region in the US. PLoS ONE. 2017;12 doi: 10.1371/journal.pone.0176912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Burall L.S., Grim C.J., Mammel M.K., Datta A.R. A Comprehensive evaluation of the genetic relatedness of Listeria monocytogenes serotype 4b variant strains. Front. Public Health. 2017;5:1–10. doi: 10.3389/fpubh.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kwong J.C., Mercoulia K., Tomita T., Easton M., Li H.Y., Bulach D.M., Stinear T.P., Seemann T., Howden B.P. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J. Clin. Microbiol. 2016;54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J., Cao G., Xu X., Allard M., Li P., Brown E., Yang X., Pan H., Meng J. Evolution and diversity of Listeria monocytogenes from clinical and food samples in Shanghai, China. Front. Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jia Y., Nightingale K.K., Boor K.J., Ho A., Wiedmann M., McGann P. Distribution of internalin gene profiles of Listeria monocytogenes isolates from different sources associated with phylogenetic lineages. Foodborne Pathog. Dis. 2007;4:222–232. doi: 10.1089/fpd.2006.0081. [DOI] [PubMed] [Google Scholar]
- 52.Su X., Zhang J., Shi W., Yang X., Li Y., Pan H., Kuang D., Xu X., Shi X., Meng J. Molecular characterization and antimicrobial susceptibility of Listeria monocytogenes isolated from foods and humans. Food Control. 2016;70:96–102. doi: 10.1016/j.foodcont.2016.04.020. [DOI] [Google Scholar]
- 53.Clayton E.M., Hill C., Cotter P.D., Ross R.P. Real-time PCR assay to differentiate listeriolysin S-positive and -negative strains of Listeria monocytogenes. Appl. Environ. Microbiol. 2011;77:163–171. doi: 10.1128/AEM.01673-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cotter P.D., Draper L.A., Lawton E.M., Daly K.M., Groeger D.S., Casey P.G., Ross R.P., Hill C. Listeriolysin S, a novel peptide haemolysin associated with a subset of lineage I Listeria monocytogenes. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen J., Rump L., Zhang Y., Chen Y., Wang X., Meng J. Molecular subtyping and virulence gene analysis of Listeria monocytogenes isolates from food. Food Microbiol. 2013;35:58–64. doi: 10.1016/j.fm.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 56.Clayton E.M., Daly K.M., Guinane C.M., Hill C., Cotter P.D., Ross P.R. Atypical Listeria innocua strains possess an intact LIPI-3. BMC Microbiol. 2014;14 doi: 10.1186/1471-2180-14-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kanki M., Naruse H., Taguchi M., Kumeda Y. Characterization of specific alleles in inlA and prfA of Listeria monocytogenes isolated from foods in Osaka, Japan and their ability to invade Caco-2 cells. Int. J. Food Microbiol. 2015;211:18–22. doi: 10.1016/j.ijfoodmicro.2015.06.023. [DOI] [PubMed] [Google Scholar]
- 58.Kovacevic J., Arguedas-Villa C., Wozniak A., Tasara T., Allen K.J. Examination of food chain-derived Listeria monocytogenes strains of different serotypes reveals considerable diversity in inlA genotypes, mutability, and adaptation to cold temperatures. Appl. Environ. Microbiol. 2013;79:1915–1922. doi: 10.1128/AEM.03341-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gorski L., Parker C.T., Liang A.S., Walker S., Romanolo K.F. The majority of genotypes of the virulence gene inlA are intact among natural watershed isolates of Listeria monocytogenes from the central California Coast. PLoS ONE. 2016;11:e0167566. doi: 10.1371/journal.pone.0167566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hingston P., Chen J., Dhillon B.K., Laing C., Bertelli C., Gannon V., Tasara T., Allen K., Brinkman F.S.L., Hansen L.T., et al. Genotypes associated with Listeria monocytogenes isolates displaying impaired or enhanced tolerances to cold, salt, acid, or desiccation stress. Front. Microbiol. 2017;8:369. doi: 10.3389/fmicb.2017.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olier M., Pierre F., Lemaître J.P., Divies C., Rousset A., Guzzo J. Assessment of the pathogenic potential of two Listeria monocytogenes human faecal carriage isolates. Microbiology. 2002;148:1855–1862. doi: 10.1099/00221287-148-6-1855. [DOI] [PubMed] [Google Scholar]
- 62.Rousseaux S., Olier M., Piveteau P., Guzzo J. Use of PCR-restriction fragment length polymorphism of inlA for rapid screening of Listeria monocytogenes strains deficient in the ability to invade Caco-2 cells. Appl. Environ. Microbiol. 2004;70:2180–2185. doi: 10.1128/AEM.70.4.2180-2185.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Handa-Miya S., Kimura B., Takahashi H., Sato M., Ishikawa T., Igarashi K., Fujii T. Nonsense-mutated inlA and prfA not widely distributed in Listeria monocytogenes isolates from ready-to-eat seafood products in Japan. Int. J. Food Microbiol. 2007;117:312–318. doi: 10.1016/j.ijfoodmicro.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Gelbíčová T., Koláčková I., Pantůček R., Karpíšková R. A novel mutation leading to a premature stop codon in inlA of Listeria monocytogenes isolated from neonatal listeriosis. New Microbiol. 2015;38:293–296. [PubMed] [Google Scholar]
- 65.Dramsi S., Trieu-Cuot P., Bierne H. Sorting sortases: A nomenclature proposal for the various sortases of Gram-positive bacteria. Res. Microbiol. 2005;156:289–297. doi: 10.1016/j.resmic.2004.10.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.