Abstract
The exposome encompasses an individual’s exposure to exogenous chemicals, as well as endogenous chemicals that are produced or altered in response to external stressors. While the exposome concept has been established for human health, its principles can be extended to include broader ecological issues. The assessment of exposure is tightly interlinked with hazard assessment. Here, we explore if mechanistic understanding of the causal links between exposure and adverse effects on human health and the environment can be improved by integrating the exposome approach with the adverse outcome pathway (AOP) concept that structures and organizes the sequence of biological events from an initial molecular interaction of a chemical with a biological target to an adverse outcome. Complementing exposome research with the AOP concept may facilitate a mechanistic understanding of stress-induced adverse effects, examine the relative contributions from various components of the exposome, determine the primary risk drivers in complex mixtures, and promote an integrative assessment of chemical risks for both human and environmental health.
Keywords: Exposome, AOP, systems toxicology, systems biology, systems chemistry, risk assessment
TOC art
Introduction
The exposome expands our perception of lifetime exposure because it integrates exogenous chemicals with genetic and external factors that generate chemicals inside the body and thereby may pose threats to human health (Miller and Jones 2014; Rappaport and Smith 2010; Wild 2012). The external contribution to the human exposome is determined by environmental exposure, also termed the eco-exposome (Lioy and Smith 2013), such as exposure via food, water, dust, air, and use of consumer products (Figure 1). Apart from environmental pollutants and their biotransformation products, the exposome includes endogenous metabolites and markers of the adaptive cellular stress responses, as well as chemicals that are generated in response to psychosocial stress and lifestyle factors. These joint exposures can be related to adverse health effects via exposome-wide association studies (EWAS; Rappaport 2012) without attempting to identify mechanistic causes (Figure 1). Importantly, these associations capture the joint effect of many stressors acting in concert, which invokes mixture effects not only in chemical space of exogenous and endogenous compounds, but also mixtures in time, including the time dependence of effects. The exposome has thus been advocated as a key to cumulative risk assessment (Smith et al. 2015).
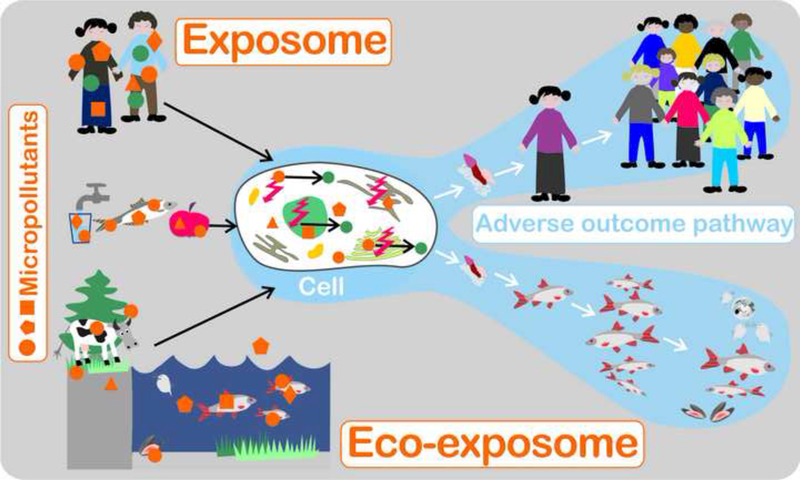
Figure 1:
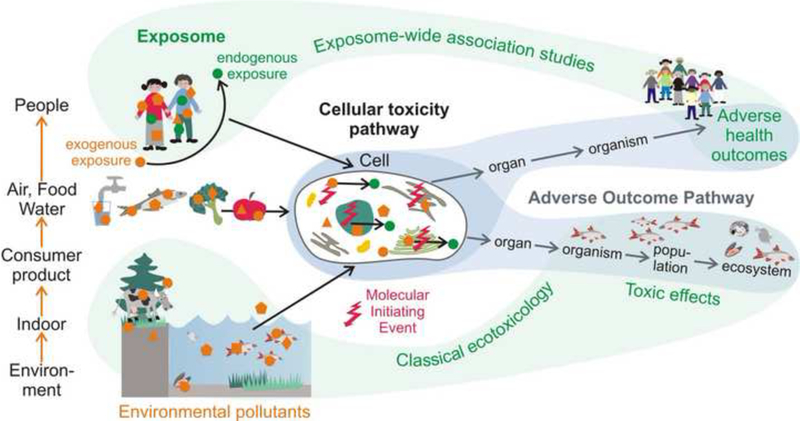
Multiple chemical exposures of the environment and their link via environmental media and the food chain to human exposure. Any type of exogenous chemical exposure will change the endogenous exposure, both of which will elicit effects on cellular toxicity pathways. The cellular level might serve as integrator to understand both, the pathways to adverse health outcomes as well as to ecosystem-level effects.
During the last decade, the exposome approach has mainly been considered in epidemiology, while the complementary concept of Adverse Outcome Pathways (AOP) has emerged in (eco)toxicology (Ankley et al. 2010). The AOP concept links the exposure of chemicals to their cellular concentrations and molecular initiating events (MIE), through network/pathway disturbances and key events (KE) to responses at the cellular, organ, organism and, finally, population and ecosystem levels (Figure 1). The AOP concept aims to enhance the utility of mechanistic data for understanding and predicting adverse effects. It also aligns in this goal with systems toxicology (Sturla et al. 2014) and the Tox21 program, a joint initiative of the US National Institute of Environmental Health Sciences (NIEHS) and the US Environmental Protection Agency (EPA) (Betts 2013; National Research Council 2007). While the concepts of AOP and cellular toxicity pathways (Patlewicz et al. 2013) account for multiple pathways triggered by a chemical or stressor and are per definition chemical agnostic, most examples that applied these concepts in risk assessment to date are limited to individual chemicals, lacking an explicit treatment of internal biotransformation and of combined effects resulting from non-chemical stressors and chemical mixtures. Moving from linear AOPs to AOP networks is one step closer towards the idea of integrating both the exposome approach and the AOP concept (Figure 1). We argue that the AOP concept can expand applications of the exposome beyond EWAS to establishing mechanistic understanding of the causal linkages between chemical exposures and adverse effects. In turn, the exposome approach can help the AOP concept grow beyond single chemicals to include the effect of jointly acting mixtures that include both chemical and non-chemical stressors.
In addition, we may profit from the analogy of the environmental and human exposure via the food chain and other uptake pathways via water, air, dust, and consumer-products (Figure 1). We consider experimental tools that can connect both research arenas to better understand well-conserved effects on the cellular level - emphasizing commonalities and differences between the anthroposphere and the ecosphere.
Here we first explore the history of the exposome and AOP concepts and then propose to use tools from systems chemistry and systems biology to integrate exposome and AOP concepts. We then conclude with recommendations for further research.
Defining relevant exposure
Exposome.
In 2005, Wild coined the term “exposome” to describe the entireness of environmental exposure that, as a complement to the genome, may provide important clues for the understanding of chronic diseases. In his concept, the exposome encompasses lifetime environmental exposures that include lifestyle factors from the prenatal period onwards (Wild 2005). From this view, an accurate assessment of a complete exposure history is required to understand the complex interplay with genetic susceptibility, since the majority of genetic alterations will contribute to population disease burden only in the presence of specific environmental exposures (Vineis et al. 2001; Wild 2005).
Rappaport and Smih refined the approach through emphasizing the role of the chemistry in the organism, defining the exposome as the totality of human exposures from all exogenous and endogenous sources in the “internal chemical environment” (Rappaport and Smith 2010). This latter definition takes into account that exposures are comprised not only of chemicals entering the body from the environment (e.g., air, water, food, dust), but also include compounds produced in the body by inflammation, (oxidative) stress, lipid peroxidation, infections, the microbiome, and other natural processes (Figure 2). The internal chemical environment is highly dynamic during lifetime due to (environmental) external and internal factors and processes such as aging, infections, lifestyle, preexisting diseases etc. (Rappaport 2011; Rappaport and Smith 2010).
Figure 2:
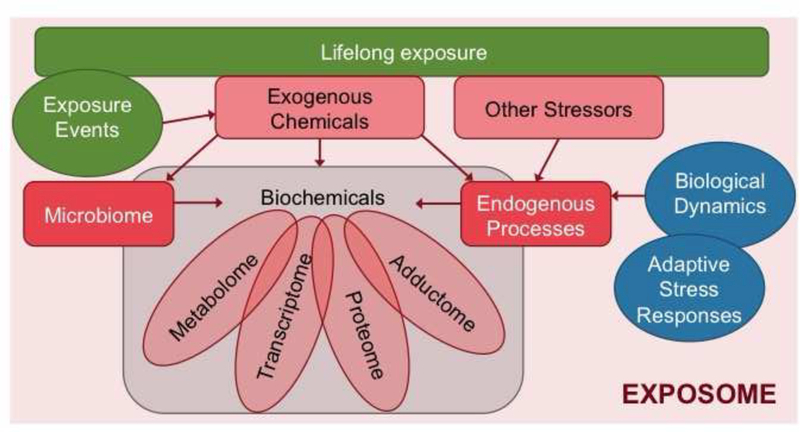
Defining the exposome.
The above-mentioned definitions of the exposome have their specific merit depending on the angle from which exposure is viewed. “Bottom-up” strategies focus on pre-selected compounds or compound adducts following targeted hypotheses. By contrast, “top- down” strategies aim at measuring all chemicals or products of their downstream processing in a subject’s biospecimen, such as blood (Rappaport 2011; Rappaport and Smith 2010), as far as technically feasible. However, the full characterization of the exposome throughout the whole lifespan remains an outstanding challenge.
Eco-exposome.
While the exposome was originally defined to characterize human exposures, the idea can certainly be adapted to consider ecosystem exposure. In 2012, the National Research Council (NRC) of the US National Academy of Sciences defined the “eco-exposome” as “the extension of exposure science from the point of contact between a stressor and receptor inward into the organism and outward to the general environment, including the ecosphere” (National Research Council (NRC) 2012). Similar to the extended definitions of the human exposome, the eco-exposome is described by both internal and external measures of exposure (Lioy and Smith 2013). Naturally, the implementation of the exposome approach is specific for each biological species. However, conservation of targets for drugs and chemicals on the cellular level (Gunnarsson et al. 2008) might allow links to be found across species and between humans and ecosystems (LaLone et al. 2014; Rand-Weaver et al. 2013). The exposome narrative could establish an important link between human and ecosystem health by examining the effect of the totality of exposure from exogenous and endogenous sources over all levels of biological organization and complexity (Figure 1). Although the eco- exposome adds another level of complexity, shared toxicity pathways and adaptive stress responses can be invoked to identify commonalities (Kramer et al. 2011), and variations in metabolic and functional traits may explain differences (Forbes and Galic 2016).
There are examples where elements of exposome research have been applied in ecotoxicology, such as monitoring contaminants in whole organisms (Houde et al. 2011; Lana et al. 2014; Lehnert et al. 2016) or correlative studies associating functional health parameters with body burdens of organic pollutants in marine wildlife (Jin et al. 2015). Similarly to exposome research in human health, there has been a focus on exposure assessment without necessarily establishing a quantitative link to adverse effects. Hence, the challenges of, both, the human exposome and eco-exposome are similar and there is mutual benefit for generating mechanistic knowledge.
Application of the AOP concept in the context of exposome research
A major motivation for developing the AOP concept was to support the risk assessment of chemicals by providing mechanistic knowledge that would enable to link in vitro (Andersen et al. 2005), in chemico (Bӧhme et al. 2009) or in silico information (Rusyn and Daston 2010), including computational biotransformation (Ji and Schüürmann 2013) and structural alerts (Schüürmann et al. 2016), to toxicity in vivo (Berggren et al. 2015; Edwards et al. 2016). The AOP concept supports the validation of predictive models based on underlying molecular mechanisms. This would allow a more efficient use of high throughput screening (HTS) to prioritize a large number of compounds for detailed testing and/or reducing the number of animal experiments (Hartung et al. 2013,b). Furthermore, within a regulatory framework of integrated assessment, AOPs can support a new, more targeted safety testing regimen that is focused on the most probable and relevant hazards (Rovida et al. 2015; Tollefsen et al. 2014). Rather than apical endpoints typically assessed for regulatory purposes, subchronic or sublethal endpoints, as well as in vitro bioassays related to acute and chronic adverse effects, may be used for establishing AOPs. The AOP concept has the inherent promise to evolve from a tool for structuring knowledge or prioritization of testing to a quantitative predictive tool to relate exposure data to adverse outcomes (AO). However, challenges remain that concern for example the incorporation of toxicokinetics as important determinant of toxicity in AOPs and extrapolations to higher levels of biological organization and across species (Groh et al. 2015). Thus, substance or species-specific differences in the toxicokinetics and genetically based differences across species can influence single events or processes within an AOP cascade, in consequence augmenting or mitigating apical adverse effects.
How could the exposome approach and the AOP concept cross-fertilize each other?
Initially, the exposome and the AOP concepts evolved separately in the fields of human toxicology and ecotoxicology, respectively. The inclusion of biological response in exposure assessment is where AOPs integrate into the exposome; the consideration of endogenous and non-chemical stresses as well as mixture effects is where the exposome can enrich the AOP concept. Per definition, the exposome is specific for individuals and integrates lifetime exposure while the AOP is conceptually focused on biological mechanisms and pathways. The temporal aspects of the exposome are at least partially implemented in the AOP concept. For instance, life-stage specificity or chronic toxicity would address effects that are related to long-term or potentially repeated exposure scenarios and biological responses.
The system-wide analysis of biological responses employing toxicogenomics represents an unbiased approach to detect chemical effects, integrating from MIEs along toxicity pathways (Berninger et al. 2014; Ellinger-Ziegelbauer et al. 2009). However, large numbers of signals compromise the response signatures (Vidal-Dorsch et al. 2016) and may lead to over-fitted associations with exposure characteristics. This calls for a reduction in data dimensionality through mechanistic reasoning and biomarker identification (Blaauboer et al. 2012) to discriminate relevant signals from epi- phenomena and random responses (Hartung et al. 2012). In this respect the combination of AOP and exposome is revisiting previous approaches to apply biomarkers in epidemiology (Toniolo et al., 1998). A biomarker may represent a KE leading to adverse outcomes but may also represent events that are without direct relevance to the adverse effect, e.g. representing a compensatory reaction. However, the AOP concept is more strict and clear with respect to the relation of events that are directly related to adverse effects and hence, its combination with the exposome could improve the relation of exposome signals to impacts on health.
AOPs could further help to anchor system-wide responses to dominant modes or mechanisms of action (Ellison et al. 2016), and thus increase the confidence in a hypothesis by providing mechanistic plausibility (Braun et al. 2016).
Aggregated exposure pathways (AEPs), AOPs, and the exposome.
The exposome may also be considered as the integration of AEPs and AOPs on (complex) mixtures if the source and pathways leading to the internal exposure are included (Figure 3). The AEP concept has been developed complementary to the AOP describing the KEs from source via external exposure (including environmental and dietary exposure) to exposure in the organism, termed target site exposure (Teeguarden et al. 2016), which is the crucial step linked to the molecular initiating events of the AOP. Similar to AOPs, the AEPs help to organize exposure information from exogenous source to internal site of action, setting the stage for inferring chemical concentrations at the internal target site and informing about expected biological effects.
Figure 3:
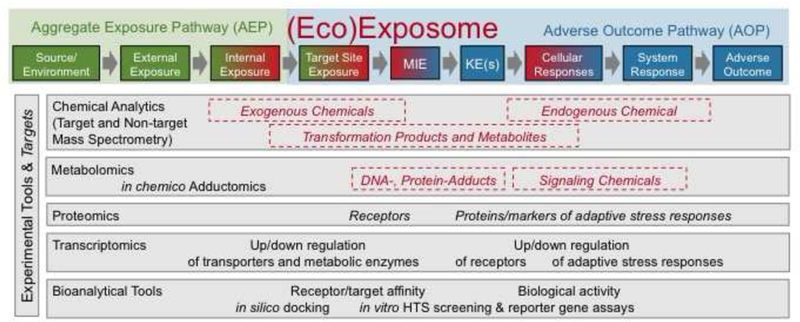
Interface between the (eco)exposome (in red), the aggregate exposure pathway (AEP, green) and adverse outcome pathway (AOP, blue). The red dashed boxes represent chemical components of the exposome. The AEP/AOP concept allows one to disentangle key events and allocate them to steps from the source of exposure to adverse effects. The grey boxes indicate experimental methods to quantify the chemical components of the exposome and the biological components of the AOP. Figure partially adapted from (Teeguarden et al. 2016).
The chemicals in the exposome constitute not only the exogenous chemicals transported into the body and their metabolites, but also adducts with cellular constituents and other endogenous compounds, as well as signaling molecules formed as part of the pathway of toxicity (Kleensang et al. 2014) or adaptive stress responses (Simmons et al. 2009; Smirnova et al. 2015) (Figure 3). The formation of chemical adducts can be considered as MIE, while endogenous chemicals (e.g. ROS, nitric oxide, ATP, glutathione) formed or changed in their levels as part of the stress response belong to the KE. Aligning the chemicals in the exposome to the various steps of the AEP/AOP (Figure 3) will help to establish a clearer a priori mechanistic link between the exposome and adverse health outcomes, with some endogenous chemicals of the exposome involved in defense against rather than in the development of the disease. However, the internal chemical response of stressed/perturbed organisms (e.g., due to an infection or an unhealthy lifestyle) may potentially lower the repair capacities, which may result in the next downstream event. These specific cases should be addressed in future studies.
Putative AOPs in exposome assessment.
The AOP concept has been moving from the initial ideas of linear pathways to networks of pathways (Knapen et al. 2015), which accommodates the idea of multiple causes for adverse effects. Some scientists criticize that AOPs are presently often incomplete. However, partial information on KEs and their relation even in case of weak evidence can provide initial steps to prioritize areas that require further investigation, and to identify the most relevant (internal) exposure situations. The AOP definitions and development are supported and guided by the OECD on a global level (Worth et al. 2014). Putative AOPs can be validated by the same principles that apply to AOPs in general, that is, their consistency with scientific literature and evidence for mechanistic links has to be demonstrated (Bell et al. 2016; Hartung et al. 2013a). Furthermore, putative and partially incomplete AOPs with high confidence relationships between KEs can be useful in specific applications, such as predicting an adverse outcome based on an easily tested KE (Perkins et al. 2015).
Grouping of exposures with converging AOPs.
Given the large chemical variability, a full characterization of all possible MIEs and pathway-specific KEs remains one of the greatest challenges of future research. However, many AOPs converge at a higher level of biological organization and the AOP-based assessment could be conducted at more downstream KEs using cellular or organ responses. Examples include neuroactive pesticides or compounds disrupting the thyroid axis. Many pesticides act by interfering with specific steps in neural signal transduction that converge at the level of the cardiovascular system leading to a respiratory failure syndrome and finally death of the organism (Bradbury et al. 2008). Hence, characterization of behavioral responses could allow the integration of various mechanisms and to comprehensively describe exposure to neurotoxic compounds. Compounds disrupting the thyroid hormone system provide related examples in human toxicology. While various different MIEs are known, they finally converge at the intracellular or systemic thyroid hormone level (Murk et al. 2013). Hence, an integrative assessment could be based on test systems that target hormone levels in an organism, e.g., by assessment of compensatory responses to reduced thyroid hormone levels (Fetter et al. 2015).
Quantitative versus qualitative AOPs.
Most AOPs are initially developed based on qualitative, mechanistic evidence without consideration of toxicokinetics, i.e. the uptake, distribution and metabolism/elimination of a compound in an organism. However, the cellular concentration is a major driver of the magnitude of the final adverse effect. In the case of, e.g., a limited uptake or rapid metabolism, adverse effects may be mitigated even for a high affinity to the molecular target (Patlewicz et al. 2013).
Quantitative AOPs (qAOP) build on approaches for toxicokinetic-toxicodynamic (TKTD) modeling (MacKay et al. 2013) as was shown very recently for a qAOP developed for effects of synthetic glucocorticoids in fish (Margiotta-Casaluci et al. 2016). If the AOP cannot be fully described, hazard rates may be established to obtain a quantitative link to the final adverse effect (Ashauer et al. 2015).
Networks of AOPs for mixtures.
If bioassays can be mapped to KEs in AOPs of specific chemical domains, AOP networks might be used for assessing the complex mixtures of the exposome. Studies on extracts of environmental samples have demonstrated how complex chemical mixtures can be characterized using a battery of different mechanistic bioassays (Escher et al. 2014) or transcriptomic tools (Berninger et al. 2014). The levels of single chemicals will often fall below detection limits (both for chemical analysis and biological responses), but their combined exposure may nevertheless generate detectable biological responses (Altenburger et al. 2012; Silva et al. 2002). An AOP-based approach could help to interpret effects of mixtures without necessarily resolving each component, e.g., if a mixture of chemicals is extracted from biospecimens and the resulting extract is applied to cell-based bioassays (Altenburger et al. 2015). Bioanalytical assessment of these extracts with cell-based bioassays may support the identification of the most relevant chemicals and toxicity pathways interfering with human and wildlife health, provided that the bioassays are anchored in a defined step of the AOP (Busch et al. 2016). Identification of such risk drivers could define new target chemicals in (bio)monitoring programs.
The requirements for AOPs to be used for mixtures do not deviate principally from those for AOPs for single compounds (Altenburger et al. 2015). However, similar adverse outcomes may be triggered by different MIEs that could jointly affect the same KE or more complex interactions could arise as is delineated by the complex role of low-dose mixtures in carcinogenesis (Goodson et al. 2015). Hence, experimental methods should focus on those KEs that can be expected to aggregate the bioactivity of different compounds that bind to different target sites but still converge into the same adverse outcome (Figure 4A, Vogs and Altenburger 2016).
Figure 4.
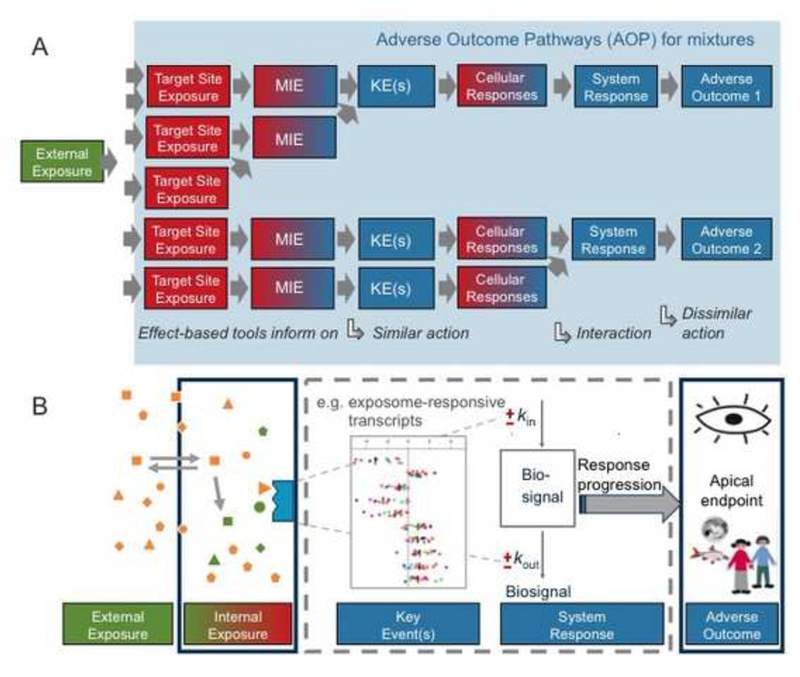
(A) AOP for mixtures and (B) qAOP mixture modelling concept for similar action (adapted with permission from Vogs, C. and Altenburger, R. (2016). Time- Dependent Effects in Algae for Chemicals with Different Adverse Outcome Pathways: A Novel Approach. Environmental Science & Technology, 50(14): 7770–7780. Copyright (2016) American Chemical Society.
Current understanding of mixture effects offers this reasoning as an explanation of why mixtures act according to concentration addition or independent action and may still show a combined effect where the individual components occur at sub-threshold concentrations (Altenburger and Greco 2009). So far, the predictive power of mixture models has been demonstrated mainly for artificially designed mixtures and defined apical endpoints (e.g., growth, survival) (Kortenkamp et al. 2009). More complex and environmentally relevant mixtures have occasionally been evaluated using the concentration addition assumption (Tang et al. 2013). In the past, mixture analysis that used endpoints anticipated to strongly relate to an MIE, such as toxicogenomic responses, were often based on poorly designed studies and therefore remained inconclusive (Altenburger et al. 2012). For the extension of the principles of mixture toxicity to the AOP concept it is thus necessary to explore how AOPs originating from different MIEs may converge at the KE and/or AO level and whether AOPs can be formulated for complex mixtures that cannot be resolved to an individual mechanism of action. However, even when a mixture effect would appear to be linked to an individual AOP, this AOP may not allow one to trace back to a certain chemical or chemical class due to the large number of chemicals and complex interactions within the overall exposome. Still, such a diagnosis would be informative because it would establish a mechanistic link between exposure and adverse effects.
Furthermore, toxicodynamic models are at present not available for mixtures. They would be an asset to quantitatively model and predict adverse outcomes or diseases resulting from mixtures for which exposure information is limited or where quantitative knowledge is available only for a limited number of KEs (Figure 4B). Such models would also open an avenue towards approaching temporal issues in adverse outcome assessment such as non-continuous or sequential exposures.
The role of systems biology and chemistry in exposome and AOP research
The methodological “glue” that links the exposome and AOPs consists, largely, of systems biological and chemical methods (Figure 3). Systems biology is characterized by (i) an initial massive parallel experimental approach, aimed at determining one or more levels of molecular signatures (e.g., transcriptome, proteome, metabolome etc.); (ii) an iterative integration of experimental approaches and computational data analysis, and modeling, and (iii) the computational generation of experimentally testable hypotheses (Garcia-Reyero and Perkins 2011; Hood et al. 2012; Ideker et al. 2001).
The complementary view on systems involving molecules with their compartmental partitioning and reaction networks is called systems chemistry. It addresses the organization of molecular feedback, amplification and information gain, and how these translate into the emergence of system-level properties (Ludlow and Otto 2008; Nitschke 2009; Whitesides 2015). In the context of the exposome, systems chemistry may describe the interplay between exogenous and endogenous compounds including their dependence on the spatially varying dose as well as on reactivity, time, and further system properties.
Since chemistry underpins biological processes both in organisms and in the environment, a way forward is to combine omics and analytics with computational tools. Interdisciplinary approaches and bioinformatics tools may serve to identify perturbations in organisms and in the environment, to define biomarkers of exposure and disease, and to integrate information from all relevant levels of organization (Smith et al. 2015). AOPs can be regarded as biological roadmaps along which chemistry mediates the development of toxicological effects. Identification and quantification of AOPs involve chemicals as both triggers and modulators of toxicity, and employ systems biology methods to reconstruct regulatory pathways, and to assess perturbations and rewiring in biological networks (Figure 3).
What to measure?
The first step is to use advanced analytics for identifying and quantifying exogenous and endogenous chemicals (Figure 3). EWAS are limited by the inherent definition of ‘exposure-wide’ that calls for untargeted analytical approaches (Patel 2016). Despite enormous advancement in analytical methodologies in recent years, problems persist and only a very small number of the thousands of compounds detectable in a sample can actually be identified, leaving the largest fraction of chemicals at the level of a known accurate mass (or molecular formula) and retention time. Any improvements here rely strongly on a better assignment of likely structures for these peaks based on a prediction of fragmentation, ionization, or chromatographic retention times supported by more comprehensive mass spectra databases. In contrast to non- targeted analysis, targeted approaches require initial hypotheses regarding classes of analytes and thus cannot carry through on the promise of ‘exposure-wide’ detection. Currently, efforts are under way to develop automated workflows to analyze large analytical datasets, including multivariate statistical approaches dealing with patterns of chemical signals in relationship to adverse outcomes without attempting identification and quantification of individual chemicals (Patel 2016).
The characterization of the impact of external factors on the internal chemical environment calls for the use of omics and analytical techniques (Figure 3). In recent years, substantial progress has been made in measuring small molecules and metabolites (metabolomics) (Athersuch and Keun 2015), DNA adducts (adductomics) (Phillips et al. 2014; Rappaport et al. 2012) or large molecules, such as proteins and peptides (proteomics) (Stallman Brown 2012).
Transcriptomics - except emerging techniques to detect chemically modified RNA - does not identify products of external stress directly, but has proven valuable for characterizing responses to exposure at the pathway level. The transcriptome is easily assessed - also in cohort studies - since it can be applied even if only little material down to single cells is available. By combining transcriptomics with classifier analysis, more targeted endpoints can be developed (de Boer et al. 2015).
Also of interest is the assessment of environment-induced epigenetic changes, some of which have been shown to persist for prolonged periods of time (Bauer et al. 2016; Guida et al. 2015). Epigenomics may thus provide stable biomarkers for past exposures that are not detectable due to a limited sampling scheme by other omics layers. However, interpretation of epigenomic data remains challenging and functional assessment of epigenomic changes frequently requires integration with other omics data sets.
To assist in interpreting large-scale exposure data, the AOP concept may help to establish mechanistic links to external perturbation, for example by high-throughput screening of chemical interference with specific cellular toxicity pathways. Reporter gene assays based on nuclear receptors and transcription factors of cellular toxicity pathways have become popular tools not only to quantify exposure, but also to identify relevant steps of the AOP triggered by chemicals and their complex environmental mixtures (Figure 3).
Where to measure?
In human exposomics, several biospecimens are appropriate for assessing the internal chemical environment. Urine can be sampled non-invasively, but favors the detection of water-soluble chemicals, including metabolites and conjugates. Measuring the blood exposome is a sensible approach compared to organ-specific samples (Rappaport et al. 2014). Blood transports chemicals to and from tissues and represents a reservoir of many endogenous and exogenous chemicals in the body at a given time (Nicholson et al. 2012). Blood samples are collected in most cohort studies, are therefore easily accessible, and sometimes a drop of blood is sufficient for biomonitoring studies (Mao and Wang 2015).
In eco-exposomics, integrating external with internal exposure has practical but also theoretical implications: In small aquatic invertebrates or fish embryos, the whole organisms need to be extracted. Analysis of whole organisms is based on extracts of a poorly defined mixture of lipids, proteins, carbohydrates, and bodily fluids. For larger organisms with more complex exposure pathways, such as mammals, larger fish or birds, the exposome may be investigated in body fluids similar to the human exposome approach.
The AOP components involving MIEs and pathway responses in key processes, such as development, are often evolutionarily conserved between human and model organism. Cross-species comparison focused on conserved KEs allows for AOP anchoring, assuming that evolutionary conserved proteins may have conserved functions. Therefore identification of protein orthologs through sequence similarity or other methods might be helpful to infer susceptibility, particularly if an AOP has already been identified (Perkins et al. 2013). Difference between species that cannot be captured by this approach would be toxicokinetic differences, especially with respect to metabolism.
When to measure?
The human exposome is highly variable and dynamic throughout the human lifespan. The time-dependence of the exposome poses a challenge for the integration with the AOP concept. AOPs so far do not adequately reflect when KEs or KERs are only valid during a certain life stage or developmental phase.
Routine measurements, in particular during critical life stages, such as fetal development (cord blood analysis), early childhood, and puberty, are key to establishing a personalized picture of specific individuals’ exposures (Rappaport and Smith 2010). Time-sequenced information on individual exposome at different time points prior to disease onset would be of immense relevance for identifying environmental causes of disease.
Human life-long exposure may leave its traces in the exposome, while a retrospective analysis of external exposure along all uptake pathways is impossible. For small and short-lived aquatic organisms, however, intelligent environmental sampling regimes yield time-integrated and peak exposures, bringing us close to the ideal of an assessment of life-long exposure to external chemicals.
From exposome to adverse outcomes.
Systems biology and chemistry tools may be linked to prioritize compounds for chemical analytics by a tiered approach: Starting from a population cohort with a nested case-control study - “meet in the middle” approach that combines bottom-up and top-down approaches (Vineis et al. 2013) - omics data from epigenomics, proteomics or transcriptomics are generated and used to identify pathways/networks potentially affected by exposures and subsequently driving disease risk. For these pathways, in silico exploitation of (toxicological) databases and chemical bioactivity from HTS, reporter gene assays and docking studies will yield pathway- associated exogenous and endogenous chemicals (Figure 3).
Tox 21 and ToxCast have demonstrated the practicality of combining (hundreds of) HTS screens with in vitro bioassays for more than 8000 single compounds (Tice et al. 2013). While the current focus is in vitro to in vivo extrapolation using toxicokinetic models (Phillips et al. 2016; Wetmore et al. 2013), these in vitro bioassays can also be applied to monitor unknown mixtures in environmental samples from water (Escher et al. 2014) to biota (Jin et al. 2015) and human specimens. In this regard, in vitro assays may be useful to capture endogenous exposures and changes in internal of stress. Methods to link cause with effect, such as effect-directed analysis, are well established for the analysis of the external exposure in water (Brack et al. 2016) or sediment (Brack et al. 2005) and are increasingly applied to mammals and fish bio-fluids and tissues. Mode-of-action-specific bioassays have been used together with fractionation approaches and untargeted analysis to identify drivers of potential adverse effects in wildlife (Houtman et al. 2007; Simon et al. 2013).
Certainly, not all prerequisites for such an approach are currently available. The link between pathway perturbation and MIE may not be traceable for a particular omics data set, specifically when lacking data for the appropriate time points. However, the increasing capability to acquire multi-omic data sets at several time-points in population cohorts and environmental populations, combined with the use of defined in vitro HTS tools (Tufi et al. 2016), may alleviate this issue.
Systems biology challenges in exposome research.
Depending on the kinetics between initial exposure, MIEs and subsequent adaptations, different levels of molecular responses may require analysis (Yugi et al. 2016). Regarding prenatal exposure, longterm health effects might be accessible through epigenetics, whereas the metabolome level appears best for immediate effects induced by environmental chemicals.
Cross-omics data integration is particularly relevant for AOP construction: the multiscale nature of AOPs calls for integrating more than one omics layer. Single-cell pathway level effects will most likely be studied using respective proteomics or transcriptomics, whereas the organismal and population effects might be more easily captured using serum metabolomics. Currently available approaches reach their limits when the need arises to capture complex non-linear relationships between different omics data sets such as feedback loops.
Omics data used for network inference and pathway analysis should ideally be assessed in a time-resolved manner (Bar-Joseph et al. 2012). Most methods used for network reconstruction do not model time explicitly (Hempel et al. 2011) and more recent methods that do may demand further analysis to fully assess their potential (Le Novere 2015).
Conclusions
How the AOP concept enhances understanding the exposome and its impact on adverse outcome.
An inspection of publications on AOP and exposome from the past decade indicates that the concepts are widely accepted and intensively discussed but that examples of practical applications remain scarce. It can be anticipated that this will change in the near future provided that application of the concepts in research are intensified. While both concepts are in the midst of development, their integration might lead to synergy, which will be promoted by early harmonization of data collection and terminology.
The exposome approach and the AOP concept are essentially orthogonal with the AOPs covering single pathways over the entire effect chain while the exposome covers a multitude of pathways, with the exposure in principle integrated over the lifetime but in practice as cross sectional studies at a fixed time over a population. Thus the crossing points of AOP and exposome will need to be expanded in both dimensions.
To further explore the utility and benefit of future application of AOPs in exposome assessment the following research topics need to be strengthened:
-
(1)
More AOPs should be developed and deposited in central databases (e.g., https://aopwiki.org/). With every new AOP developed the capacity of exposome assessment based on biological responses will increase significantly.
-
(2)
Evolutionarily conserved cellular toxicity pathways may serve as common denominators for integrated effect assessments. We advocate the use of KEs across species for a systems biology-assisted approach to exposome assessment.
-
(3)
Chronic exposure (both in terms of exposure duration and delay of effects) represents the typical environmental situation and is most relevant for human health. AOPs for chronic toxicity are at present not well-described (Groh et al. 2015). However, one example for a chronic AOP is described in the AOP wiki, i.e. “Chronic binding of antagonist to N-methyl-D-aspartate receptors (NMDARs) during brain development induces impairment of learning and memory abilities” (https://aopwiki.org). Furthermore, many other endpoints, such as reproductive dysfunction originating from endocrine disruption are of relevance only in case of a chronic exposure since short-term exposures may not lead to significant population decline. Hence, an AOP-based exposome assessment should target chronic endpoints and disease outcomes.
-
(4)
The research question is an important driver of the type of exposome/AOP research, such as the identification of the main chemical risk drivers in relation to mode of action, the complex multifactorial influences of mixtures or identification of a threshold level that can explain adverse effects.
Mixtures.
Existing mixture models have to be adapted and tested with regard to the AOP and exposome concepts. How established mixture toxicity models can be applied to different KEs that lead to the same adverse outcome remains a key question for future research.
Exposome characterization as a driver for risk assessment.
Characterizing the exposome via untargeted measurement of an internal chemical environment promotes data-driven discoveries of causal factors for human diseases or effects on the ecosystem. To identify the sources and to develop prevention strategies, the main exposures have to be characterized and validated by targeted techniques. Moreover, the discrimination of different exposures also provides the basis for hypothesis-driven research to promote mechanistic understanding. A promising approach to further develop such a mechanistic understanding will be the use of the AOP concept as long as it is evolving further from a linear pathway analysis to a tool to organize the complex networks of toxicity pathways.
Acknowledgements
This review was informed by a Workshop organized by the Integrated Project EXPOSOME at the UFZ Helmholtz Centre for Environmental Research in Leipzig, Germany, in December 2015. The workshop participants comprised all coauthors plus Stephen M. Rappaport (University of California, Berkeley), Kai-Uwe Goss (UFZ) and Urs Berger (UFZ). We thank the Integrated Project “Exposome” at UFZ for funding of the workshop. We thank Stephen M. Rappaport for helpful discussions and review of the manuscript. This document has been subjected to review by the U.S. EPA and approved for publication. The views expressed in this paper are those of the authors and do not necessarily reflect the views and policies of the European Commission, the German Environmental Agency or the U.S. EPA. The authors declare they have no actual or potential competing financial interests.
Abbreviations:
- AEP
aggregated exposure pathway
- AO
adverse outcome
- AOP
adverse outcome pathway
- EWAS
exposome-wide association studies
- KE
key event
- HTS
high throughput screening
- ΜIΕ
molecular initiating event
- TD
toxicodynamic
- TK
toxicokinetic
References
- Altenburger R; Ait-Aissa S; Antczak P; Backhaus T; Barcelo D; Seiler T-B; Brion F; Busch W; Chipman K; de Alda ML; de Aragao Umbuzeiro G; Escher BI; Falciani F; Faust M; Focks A; Hilscherova K; Hollender J; Hollert H; Jager F; Jahnke A; Kortenkamp A; Krauss M; Lemkine GF; Munthe J; Neumann S; Schymanski EL; Scrimshaw M; Segner H; Slobodnik J; Smedes F; Kughathas S; Teodorovic I; Tindall AJ; Tollefsen KE; Walz K-H; Williams TD; Van den Brink PJ; van Gils J; Vrana B; Zhang X; Brack W Future water quality monitoring - Adapting tools to deal with mixtures of pollutants in water resource management. Sci Tot Environ 2015;512–513:540–551. [DOI] [PubMed] [Google Scholar]
- Altenburger R; Greco WR. Extrapolation Concepts for Dealing with Multiple Contamination in Environmental Risk Assessment. Integr Environ Assess Manag 2009;5:62–68. [DOI] [PubMed] [Google Scholar]
- Altenburger R; Scholz S; Schmitt-Jansen M; Busch W; Escher BI. Mixture toxicity revisited from a toxicogenomic perspective. Environ Sci Technol 2012;46:2508–2522. [DOI] [PubMed] [Google Scholar]
- Andersen ME; Dennison JE; Thomas RS; Conolly RB. New directions in incidence- dose modeling. Trends Biotechnol 2005;23:122–127. [DOI] [PubMed] [Google Scholar]
- Ankley GT; Bennett RS; Erickson RJ; Hoff DJ; Hornung MW; Johnson RD; Mount DR; Nichols JW; Russom CL; Schmieder PK; Serrrano JA; Tietge JE; Villeneuve DL. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ Tox Chem 2010;29:730–741. [DOI] [PubMed] [Google Scholar]
- Ashauer R; O’Connor I; Hintermeister A; Escher BI. Death Dilemma and Organism Recovery in Ecotoxicology. Environ Sci Technol 2015;49:10136–10146. [DOI] [PubMed] [Google Scholar]
- Athersuch TJ; Keun HC. Metabolic profiling in human exposome studies. Mutagenesis 2015;30:755–762. [DOI] [PubMed] [Google Scholar]
- Bar-Joseph Z; Gitter A; Simon I STUDY DESIGNS Studying and modelling dynamic biological processes using time-series gene expression data. Nature Rev Genet 2012;13:552–564. [DOI] [PubMed] [Google Scholar]
- Bauer T; Trump S; Ishaque N; Thurmann L; Gu L; Bauer M; Bieg M; Gu Z; Weichenhan D; Mallm J-P; Roder S; Herberth G; Takada E; Mucke O; Winter M; Junge KM; Grutzmann K; Rolle-Kampczyk U; Wang Q; Lawerenz C; Borte M; Polte T; Schlesner M; Schanne M; Wiemann S; Georg C; Stunnenberg HG; Plass C; Rippe K; Mizuguchi J; Herrmann C; Eils R; Lehmann I Environment-induced epigenetic reprogramming in genomic regulatory elements in smoking mothers and their children. Mol Sys Biol 2016;12:861–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SM; Angrish MM; Wood CE; Edwards SW. Integrating Publicly Available Data to Generate Computationally Predicted Adverse Outcome Pathways for Fatty Liver. Tox Sci 2016;150:510–520. [DOI] [PubMed] [Google Scholar]
- Berggren E; Amcoff P; Benigni R; Blackburn K; Carney E; Cronin M; Deluyker H; Gautier F; Judson RS; Kass GEN; Keller D; Knight D; Lilienblum W; Mahony C; Rusyn I; Schultz T; Schwarz M; Schüürmann G; White A; Burton J; Lostia AM; Munn S; Worth A Chemical Safety Assessment Using Read- Across: Assessing the Use of Novel Testing Methods to Strengthen the Evidence Base for Decision Making. Environ Health Persp 2015;123:1232–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berninger JP; Martinovic-Weigelt D; Garcia-Reyero N; Escalon L; Perkins EJ; Ankley GT; Villeneuve DL. Using Transcriptomic Tools to Evaluate Biological Effects Across Effluent Gradients at a Diverse Set of Study Sites in Minnesota, USA. Environ Sci Technol 2014;48:2404–2412. [DOI] [PubMed] [Google Scholar]
- Betts KS. Tox21 to Date Steps toward Modernizing Human Hazard Characterization. Environ Health Persp 2013;121:A228–A228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaauboer BJ; Boekelheide K; Clewell HJ; Daneshian M; Dingemans MM; Goldberg AM; Heneweer M; Jaworska J; Kramer NI; Leist M The use of biomarkers of toxicity for integrating in vitro hazard estimates into risk assessment for humans. Altex 2012;29:411–425. [DOI] [PubMed] [Google Scholar]
- Bӧhme A; Thӓns D; Paschke A; Schüürmann G Kinetic Glutathione Chemoassay To Quantify Thiol Reactivity of Organic Electrophiles-Application to alpha,beta- Unsaturated Ketones, Acrylates, and Propiolates. Chem Res Toxicol 2009;22:742–750. [DOI] [PubMed] [Google Scholar]
- Brack W; Ait-Aissa S; Burgess RM; Busch W; Creusot N; Di Paolo C; Escher BI; Hewitt LM; Hilscherova K; Hollender J; Hollert H; Jonker W; Kool J; Lamoree M; Muschket M; Neumann S; Rostkowski P; Ruttkies C; Schollee J; Schymanski EL; Schulze T; Seiler T-B; Tindall AJ; Umbuzeiro GDA; Vrana B; Krauss M Effect-directed analysis supporting monitoring of aquatic environments - An in-depth overview. Sci Tot Environ 2016;544:1073–1118. [DOI] [PubMed] [Google Scholar]
- Brack W; Schirmer K; Erdinger L; Hollert H Effect-directed analysis of mutagens and ethoxyresorufm-O-deethylase inducers in aquatic sediments. Environ Tox Chem 2005;24:2445–2458. [DOI] [PubMed] [Google Scholar]
- Bradbury SP; Carlson RW; Henry TR; Padilla S; Cowden J Toxic responses of the fish nervous system. The toxicology of fishes 2008:417–455. [Google Scholar]
- Braun JM; Gennings C; Hauser R; Webster TF. What Can Epidemiological Studies Tell Us about the Impact of Chemical Mixtures on Human Health? Environ Health Persp 2016;124:A6–A9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch W; Schmidt S; Kühne R; Schulze T; Krauss M; Altenburger R Micropollutants in European rivers: A mode of action survey to support the development of effect-based tools for water monitoring. Environ Tox Chem 2016;35:1887–1899. [DOI] [PubMed] [Google Scholar]
- de Boer TE; Janssens TKS; Legler J; van Straalen NM; Roelofs D Combined Transcriptomics Analysis for Classification of Adverse Effects As a Potential End Point in Effect Based Screening. Environ Sci Technol 2015;49:14274–14281. [DOI] [PubMed] [Google Scholar]
- Edwards SW; Tan Y-M; Villeneuve DL; Meek M; McQueen CA Adverse Outcome Pathways—Organizing Toxicological Information to Improve Decision Making. J Pharmacol Exp Therapeut 2016;356:170–181. [DOI] [PubMed] [Google Scholar]
- Ellinger-Ziegelbauer H; Aubrecht J; Kleinjans JC; Ahr H-J Application of toxicogenomics to study mechanisms of genotoxicity and carcinogenicity. Toxicol Lett 2009;186:36–44. [DOI] [PubMed] [Google Scholar]
- Ellison CM; Piechota P; Madden JC; Enoch SJ; Cronin MTD. Adverse Outcome Pathway (AOP) Informed Modeling of Aquatic Toxicology: QSARs, Read- Across, and Interspecies Verification of Modes of Action. Environ Sci Technol 2016;50:3995–4007. [DOI] [PubMed] [Google Scholar]
- Escher B; Allinson M; Altenburger R; Bain P; Balaguer B; Busch W; Crago J; Humpage A; Denslow N; Dopp E; Hilscherova K; Kumar A; Grimaldi M; Jayasinghe B; Jarosova B; Jia A; Makarov S; Maruya K; Medvedev A; Mehinto A; Mendez J; Poulsen A; Prochazka E; Richard J; Schifferli A; Schlenk D; Scholz S; Shiraishi F; Snyder S; Su G; Tang J; van der Burg B; van der Linden S; Werner I; Westerheide S; Wong C; Yang M; Yeung B; Zhang X; Leusch F Benchmarking organic micropollutants in wastewater, recycled water and drinking water with in vitro bioassays. Environ Sci Technol 2014;48:1940–1956. [DOI] [PubMed] [Google Scholar]
- Fetter E; Baldauf L; Da Fonte DF; Ortmann J; Scholz S Comparative analysis of goitrogenic effects of phenylthiourea and methimazole in zebrafish embryos. Reprod Toxicol 2015;57:10–20. [DOI] [PubMed] [Google Scholar]
- Forbes VE; Galic N Next-generation ecological risk assessment: Predicting risk from molecular initiation to ecosystem service delivery. Env Int 2016;91:215–219. [DOI] [PubMed] [Google Scholar]
- Garcia-Reyero N; Perkins EJ. Systems biology: leading the revolution in ecotoxicology. Environ Tox Chem 2011;30:265–273. [DOI] [PubMed] [Google Scholar]
- Goodson WH III; Lowe L; Carpenter DO; Gilbertson M; Ali AM; de Cerain Salsamendi AL; Lasfar A; Carnero A; Azqueta A; Amedei A; Charles AK; Collins AR; Ward A; Salzberg AC; Colacci A; Olsen A-K; Berg A; Barclay BJ; Zhou BP; Blanco-Aparicio C; Baglole CJ; Dong C; Mondello C; Hsu C-W; Naus CC; Yedjou C; Curran CS; Laird DW; Koch DC; Carlin DJ; Felsher DW; Roy D; Brown DG; Ratovitski E; Ryan EP; Corsini E; Rojas E; Moon E-Y; Laconi E; Marongiu F; Al-Mulla F; Chiaradonna F; Darroudi F; Martin FL; Van Schooten FJ; Goldberg GS; Wagemaker G; Nangami G; Calaf GM; Williams G; Wolf GT; Koppen G; Brunborg G; Lyerly HK; Krishnan H; Ab Hamid H; Yasaei H; Sone H; Kondoh H; Salem HK; Hsu H-Y; Park HH; Koturbash I; Miousse IR; Scovassi AI; Klaunig JE; Vondracek J; Raju J; Roman J; Wise JP Sr.; Whitfield JR; Woodrick J; Christopher JA; Ochieng J; Fernando Martinez-Leal J; Weisz J; Kravchenko J; Sun J; Prudhomme KR; Narayanan KB; Cohen-Solal KA; Moorwood K; Gonzalez L; Soucek L; Jian L; D’Abronzo LS; Lin L-T; Li L; Gulliver L; McCawley LJ; Memeo L; Vermeulen L; Leyns L; Zhang L; Valverde M; Khatami M; Romano MF; Chapellier M; Williams MA; Wade M; Manjili MH; Lleonart M; Xia M; Gonzalez MJ; Karamouzis MV; Kirsch-Volders M; Vaccari M; Kuemmerle NB; Singh N; Cruickshanks N; Kleinstreuer N; van Larebeke N; Ahmed N; Ogunkua O; Krishnakumar PK; Vadgama P; Marignani PA; Ghosh PM; Ostrosky-Wegman P; Thompson P; Dent P; Heneberg P; Darbre P; Leung PS; Nangia-Makker P; Cheng Q; Robey RB; Al-Temaimi R; Roy R; Andrade- Vieira R; Sinha RK; Mehta R; Vento R; Di Fiore R; Ponce-Cusi R; Dometshuber-Fleiss R; Nahta R; Castellino RC; Palorini R; Abd Hamid R; Langie SAS; Eltom S; Brooks SA; Ryeom S; Wise SS; Bay SN; Harris SA; Papagerakis S; Romano S; Pavanello S; Eriksson S; Forte S; Casey SC; Luanpitpong S; Lee T-J; Otsuki T; Chen T; Massfelder T; Sanderson T; Guamieri T; Hultman T; Dormoy V; Odero-Marah V; Sabbisetti V; Maguer- Satta V; Rathmell WK; Engstrom W; Decker WK; Bisson WH; Rojanasakul Y; Luqmani Y; Chen Z; Hu Z Assessing the carcinogenic potential of low-dose exposures to chemical mixtures in the environment: the challenge ahead. Carcinogenesis 2015;36:S254–S296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groh KJ; Carvalho RN; Chipman JK; Denslow ND; Haider M; Murphy CA; Roelofs D; Rolaki A; Schirmer K; Watanabe KH. Development and application of the adverse outcome pathway framework for understanding and predicting chronic toxicity: F Challenges and research needs in ecotoxicology. Chemosphere 2015;120:764–777. [DOI] [PubMed] [Google Scholar]
- Guida F; Sandanger TM; Castagne R; Campanella G; Polidoro S; Palli D; Krogh V; Tumino R; Sacerdote C; Panico S; Severi G; Kyrtopoulos SA; Georgiadis P; Vermeulen RCH; Lund E; Vineis P; Chadeau-Hyam M Dynamics of smoking- induced genome-wide methylation changes with time since smoking cessation. Hum Mol Genet 2015;24:2349–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunnarsson L; Jauhiainen A; Kristiansson E; Nerman O; Larsson DGJ. Evolutionary conservation of human drug targets in organisms used for environmental risk assessments. Environ Sci Technol 2008;42:5807–5813. [DOI] [PubMed] [Google Scholar]
- Hartung T; Hoffmann S; Stephens M Food for Thought …Mechanistic Validation. ALTEX 2013a;30:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T; Luechtefeld T; Maertens A; Kleensang A Food for Thought … Integrated Testing Strategies for Safety Assessments. ALTEX 2013b;30:3–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung T; van Vliet E; Jaworska J; Bonilla L; Skinner N; Thomas R Systems toxicology. ALTEX 2012;29:119–128. [DOI] [PubMed] [Google Scholar]
- Hempel S; Koseska A; Nikoloski Z; Kurths J Unraveling gene regulatory networks from time-resolved gene expression data - a measures comparison study. BMC Bioinformatics 2011;12:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood L; Balling R; Auffray C Revolutionizing medicine in the 21st century through systems approaches. Biotechnol J 2012;7:992–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houde M; De Silva AO; Muir DCG; Letcher RJ. Monitoring of Perfluorinated Compounds in Aquatic Biota: An Updated Review PFCs in Aquatic Biota. Environ Sci Technol 2011;45:7962–7973. [DOI] [PubMed] [Google Scholar]
- Houtman CJ; Booij P; van der Valk KM; van Bodegom PM; van den Ende F; Gerritsen AAM; Lamoree MH; Legler J; Brouwer A Biomonitoring of estrogenic exposure and identification of responsible compounds in bream from Dutch surface waters. Environ Tox Chem 2007;26:898–907. [DOI] [PubMed] [Google Scholar]
- Ideker T; Galitski T; Hood L A new aproach to decoding life: Systems Biology. Annu Rev Genomics Hum Genet 2001;2:343–372. [DOI] [PubMed] [Google Scholar]
- Ji L; Schüürmann G Model and Mechanism: N-Hydroxylation of Primary Aromatic Amines by Cytochrome P450. Angew Chem Int Ed 2013;52:744–748. [DOI] [PubMed] [Google Scholar]
- Jin L; Gaus C; Escher B Bioanalytical Approaches to Understanding Toxicological Implications of Mixtures of Persistent Organic Pollutants in Marine Wildlife in: Zheng E, ed. Comprehensive Analytical Chemistry Persistent Organic Pollutants (POPs): Analytical Techniques, Environmental Fate and Biological Effects: Elsevier; 2015. [Google Scholar]
- Kleensang A; Maertens A; Rosenberg M; Fitzpatrick S; Lamb J; Auerbach S; Brennan R; Crofton KM; Gordon B; Fornace AJ Jr.; Gaido K; Gerhold D; Haw R; Henney A; Ma’ayan A; McBride M; Monti S; Ochs MF; Pandey A; Sharan R; Stierum R; Tugendreich S; Willett C; Wittwehr C; Xia J; Patton GW; Arvidson K; Bouhifd M; Hogberg HT; Luechtefeld T; Smirnova L; Zhao L; Adeleye Y; Kanehisa M; Carmichael P; Andersen ME; Hartung T Pathways of Toxicity. ALTEX 2014;31:53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapen D; Vergauwen L; Villeneuve DL; Ankley GT. The potential of AOP networks for reproductive and developmental toxicity assay development. Reprod Toxicol 2015;56:52–55. [DOI] [PubMed] [Google Scholar]
- Kortenkamp, A; Backhaus, T; Faust, M. State of the art report on mixture toxicity. European Commission 070307/2007/485103/ETU/D.1; 2009.
- Kramer VJ; Etterson MA; Hecker M; Murphy CA; Roesijadi G; Spade DJ; Spromberg JA; Wang M; Ankley GT. Adverse outcome pathways and ecological risk assessment bridging to population-level effects. Environ Tox Chem 2011;30:64–76. [DOI] [PubMed] [Google Scholar]
- LaLone CA; Berninger JP; Villeneuve DL; Ankley GT. Leveraging existing data for prioritization of the ecological risks of human and veterinary pharmaceuticals to aquatic organisms. Phil Trans Royal Soc B-Biol Sci 2014;369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lana NB; Berton P; Covaci A; Ciocco NF; Barrera-Oro E; Atencio A; Altamirano JC. Fingerprint of persistent organic pollutants in tissues of Antarctic notothenioid fish. Sci Tot Environ 2014;499:89–98. [DOI] [PubMed] [Google Scholar]
- Le Novere N Quantitative and logic modelling of molecular and gene networks. Nature Rev Genet 2015;16:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert K; Ronnenberg K; Weijs L; Covaci A; Das K; Hellwig V; Siebert U Xenobiotic and Immune-Relevant Molecular Biomarkers in Harbor Seals as Proxies for Pollutant Burden and Effects. Arch Environ Contam Toxicol 2016;70:106–120. [DOI] [PubMed] [Google Scholar]
- Lioy PJ; Smith KR. A Discussion of Exposure Science in the 21st Century: A Vision and a Strategy. Environ Health Persp 2013;121:405–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludlow RF; Otto S Systems chemistry. Chem Soc Rev 2008;37:101–108. [DOI] [PubMed] [Google Scholar]
- MacKay C; Davies M; Summerfield V; Maxwell G From Pathways to People: Applying the Adverse Outcome Pathway (AOP) for Skin Sensitization to Risk Assessment. ALTEX 2013;30:473–486. [DOI] [PubMed] [Google Scholar]
- Mao P; Wang D Biomonitoring of Perfluorinated Compounds in a Drop of Blood. Environ Sci Technol 2015;49:6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margiotta-Casaluci L; Owen SF; Huerta B; Rodriguez-Mozaz S; Kugathas S; Barcelo D; Rand-Weaver M; Sumpter JP. Internal exposure dynamics drive the Adverse Outcome Pathways of synthetic glucocorticoids in fish. Scientific Reports 2016:DOI: 10.1038/srep21978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller GW; Jones DP. The Nature of Nurture: Refining the Definition of the Exposome. Toxicol Sci 2014;137:1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murk AJ; Rijntjes E; Blaauboer BJ; Clewell R; Crofton KM; Dingemans MM; Furlow JD; Kavlock R; Kohrle J; Opitz R; Traas T; Visser TJ; Xia M; Gutleb AC. Mechanism-based testing strategy using in vitro approaches for identification of thyroid hormone disrupting chemicals. Toxicol In Vitro 2013;27:1320–1346. [DOI] [PubMed] [Google Scholar]
- National Research Council. Toxicity testing in the 21st century: A vision and a strategy ed^eds. Washington, DC: National Academies Press; 2007. [Google Scholar]
- National Research Council (NRC). Exposure science in the 21st century: a vision and a strategy. Washington, DC: National Academic Press 2012; [PubMed] [Google Scholar]
- Nicholson JK; Holmes E; Kinross JM; Darzi AW; Takats Z; Lindon JC. Metabolic phenotyping in clinical and surgical environments. Nature 2012;491:384–392. [DOI] [PubMed] [Google Scholar]
- Nitschke JR. Systems chemistry: Molecular networks come of age. Nature 2009;462:736–738. [DOI] [PubMed] [Google Scholar]
- Patel CJ. Analytical Complexity in Detection of Gene Variant-by-Environment Exposure Interactions in High-Throughput Genomic and Exposomic Research. Curr Env Health Rep 2016;3:64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patlewicz G; Simon T; Goyak K; Phillips RD; Rowlands JC; Seidel SD; Becker RA. Use and validation of HT/HC assays to support 21st century toxicity evaluations. Regul Toxicol Pharmacol 2013;65:259–268. [DOI] [PubMed] [Google Scholar]
- Perkins EJ; Ankley GT; Crofton KM; Garcia-Reyero N; LaLone CA; Johnson MS; Tietge JE; Villeneuve DL. Current Perspectives on the Use of Alternative Species in Human Health and Ecological Hazard Assessments. Environ Health Persp 2013;121:1002–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins EJ; Antczak P; Burgoon L; Falciani F; Garcia-Reyero N; Gutsell S; Hodges G; Kienzler A; Knapen D; McBride M; Willett C Adverse Outcome Pathways for Regulatory Applications: Examination of Four Case Studies With Different Degrees of Completeness and Scientific Confidence. Toxicol Sci 2015;148:14–25. [DOI] [PubMed] [Google Scholar]
- Phillips DH; Preston G; Sozeri O Adductomics - measurement of DNA and protein adducts in human tissues as an approach to defining the exposome. Mutagenesis 2014;29:502–502. [Google Scholar]
- Phillips MB; Leonard JA; Grulke CM; Chang DT; Edwards SW; Brooks R; Goldsmith M-R; El-Masri H; Tan Y-M. A Workflow to Investigate Exposure and Pharmacokinetic Influences on High-Throughput in Vitro Chemical Screening Based on Adverse Outcome Pathways. Environ Health Persp 2016;124:53–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand-Weaver M; Margiotta-Casaluci L; Patel A; Panter GH; Owen SF; Sumpter JP. The Read-Across Hypothesis and Environmental Risk Assessment of Pharmaceuticals. Environ Sci Technol 2013;47:11384–11395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM. Implications of the exposome for exposure science. J Expo Sci Environ Epidemiol 2011;21:5–9. [DOI] [PubMed] [Google Scholar]
- Rappaport SM. Biomarkers intersect with the exposome. Biomarkers 2012;17:483–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM; Barupal DK; Wishart D; Vineis P; Scalbert A The Blood Exposome and Its Role in Discovering Causes of Disease. Environ Health Persp 2014;122:769–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM; Li H; Grigoryan H; Funk WE; Williams ER. Adductomics: Characterizing exposures to reactive electrophiles. Toxicol Lett 2012;213:83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappaport SM; Smith MT. Environment and Disease Risks. Science 2010;330:460–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovida C; Alepee N; Api AM; Basketter DA; Bois FY; Caloni F; Corsini E; Daneshian M; Eskes C; Ezendam J; Fuchs H; Hayden P; Hegele-Hartung C; Hoffmann S; Hubesch B; Jacobs MN; Jaworska J; Kleensang A; Kleinstreuer N; Lalko J; Landsiedel R; Lebreux F; Luechtefeld T; Locatelli M; Mehling A; Natsch A; Pitchford JW; Prater D; Prieto P; Schepky A; Schuurmann G; Smirnova L; Toole C; van Vliet E; Weisensee D; Hartung T Integrated Testing Strategies (ITS) for Safety Assessment. ALTEX 2015;32:25–40. [DOI] [PubMed] [Google Scholar]
- Rusyn I; Daston GP. Computational Toxicology: Realizing the Promise of the Toxicity Testing in the 21st Century. Environ Health Persp 2010;118:1047–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüürmann G; Ebert R-U; Tluczkiewicz I; Escher SE; Kühne R Inhalation threshold of toxicological concern (TTC) - Structural alerts discriminate high from low repeated-dose inhalation toxicity. Env Int 2016;88:123–132. [DOI] [PubMed] [Google Scholar]
- Silva E; Rajapakse N; Kortenkamp A Something from “nothing”- eight weak estrogenic chemicals combined at concentrations below NOECs produce significant mixture effects. Environ Sci Technol 2002;36:1751–1756. [DOI] [PubMed] [Google Scholar]
- Simmons SO; Fan CY; Ramabhadran R Cellular stress response pathway system as a sentinel ensemble in toxicological screening. Toxicol Sci 2009;111:202–225. [DOI] [PubMed] [Google Scholar]
- Simon E; van Velzen M; Brandsma SH; Lie E; Loken K; de Boer J; Bytingsvik J; Jenssen BM; Aars J; Hamers T; Lamoree MH. Effect-Directed Analysis To Explore the Polar Bear Exposome: Identification of Thyroid Hormone Disrupting Compounds in Plasma. Environ Sci Technol 2013;47:8902–8912. [DOI] [PubMed] [Google Scholar]
- Smirnova L; Harris G; Leist M; Hartung T Cellular Resilience. ALTEX 2015;32:247–260. [DOI] [PubMed] [Google Scholar]
- Smith MT; de la Rosa R; Daniels SI. Using exposomics to assess cumulative risks and promote health. Environ Mol Mutagen 2015;56:715–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallman Brown E Measuring individual exposomes. Emerg Sci Environ Health Decis 2012:1–13. [Google Scholar]
- Sturla SJ; Boobis AR; FitzGerald RE; Hoeng J; Kavlock RJ; Schirmer K; Whelan M; Wilks MF; Peitsch MC. Systems Toxicology: From Basic Research to Risk Assessment. Chem Res Toxicol 2014;27:314–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang JYM; McCarty S; Glenn E; Neale PA; Warne MS; Escher BI. Mixture effects of organic micropollutants present in water: towards the development of effect-based water quality trigger values for baseline toxicity. Water Research 2013;47:3300–3314. [DOI] [PubMed] [Google Scholar]
- Teeguarden JG; Tan C; Edwards S; Leonard JA; Anderson KA; Corley RA; Harding A; Kile M; LMS S; Stone D; Waters KM; Harper S; Williams DE; Tanguay RL. Completing the Link between Exposure Science and Toxicology for Improved Environmental Health Decision Making: The Aggregate Exposure Pathway Framework. Environ Sci Technol 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tice RR; Austin CP; Kavlock RJ; Bucher JR. Improving the Human Hazard Characterization of Chemicals: A Tox21 Update. Environ Health Persp 2013;121:756–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollefsen KE; Scholz S; Cronin MT; Edwards SW; de Knecht J; Crofton K; Garcia- Reyero N; Hartung T; Worth A; Patlewicz G Applying Adverse Outcome Pathways (AOPs) to support Integrated Approaches to Testing and Assessment (IATA). Regul Toxicol Pharmacol 2014;70:629–640. [DOI] [PubMed] [Google Scholar]
- Toniolo P; Boffetta P; Shuker D; Rothman N; Hulka B; Pearce N (eds.) Application of Biomarkers in Cancer Epidemiology IARC Scientific Publications No. 142 International Agency for Research on Cancer, Lyon, France; 1997. [Google Scholar]
- Tufi S; Wassenaar PNH; Osorio V; De Boer J; Leonards PEG; Lamoree MH. Pesticide Mixture Toxicity in Surface Water Extracts in Snails (Lymnaea stagnalis) by an in Vitro Acetylcholinesterase Inhibition Assay and Metabolomics. Environ Sci Technol 2016;50:3937–3944. [DOI] [PubMed] [Google Scholar]
- Vidal-Dorsch DE; Bay SM; Moore S; Layton B; Mehinto AC; Vulpe CD; Brown- Augustine M; Loguinov A; Poynton H; Garcia-Reyero N; Perkins EJ; Escalon L; Denslow ND; Cristina C-DR; Doan T; Shukradas S; Bruno J; Brown L; Van Agglen G; Jackman P; Bauer M Ecotoxicogenomics: Microarray interlaboratory comparability. Chemosphere 2016;144:193–200. [DOI] [PubMed] [Google Scholar]
- Vineis P; Schulte P; McMichael AJ. Misconceptions about the use of genetic tests in populations. Lancet 2001;357:709–712. [DOI] [PubMed] [Google Scholar]
- Vineis P; van Veldhoven K; Chadeau-Hyam M; Athersuch TJ. Advancing the application of omics-based biomarkers in environmental epidemiology. Environ Mol Mutagen 2013;54:461–467. [DOI] [PubMed] [Google Scholar]
- Vogs C; Altenburger R Time-Dependent Effects in Algae for Chemicals with Different Adverse Outcome Pathways: A Novel Approach. Environ Sci Technol 2016;50:7770–7780. [DOI] [PubMed] [Google Scholar]
- Wetmore BA; Wambaugh JF; Ferguson SS; Li L; Clewell HJ III; Judson RS; Freeman K; Bao W; Sochaski MA; Chu T-M; Black MB; Healy E; Allen B; Andersen ME; Wolfinger RD; Thomas RS Relative Impact of Incorporating Pharmacokinetics on Predicting In Vivo Hazard and Mode of Action from High- Throughput In Vitro Toxicity Assays. Toxicol Sci 2013;132:327–346. [DOI] [PubMed] [Google Scholar]
- Whitesides GM. Reinventing Chemistry. Angew Chem Int Ed 2015;54:3196–3209. [DOI] [PubMed] [Google Scholar]
- Wild CP. Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol Biomarkers Prev 2005;14:1847–1850. [DOI] [PubMed] [Google Scholar]
- Wild CP. The exposome: from concept to utility. Intl J Epidem 2012;41:24–32. [DOI] [PubMed] [Google Scholar]
- Worth, A; Barroso, J; Bremer, S; Burton, J; Casati, S; Coecke, S; Corvi, R; Desprez, B; Dumont, C; Gouliarmou, V; Goumenou, M; Grӓpel, R; Griesinger, C; Haider, M; Roi, AJ; Kienzler, A; Madia, F; Munn, S; Nepelska, M; Paini, A; Price, A; Prieto, P; Rolaki, A; Schӓffer, M; Triebe, J; Whelan, M; Wittwehr, C; Zuang, V. Alternative methods for regulatory toxicology - a state-of-the-art review. Available at echa.europe.eu. 2014.
- Yugi K; Kubota H; Hatano A; Kuroda S Trans-Omics: How To Reconstruct Biochemical Networks Across Multiple ‘Omic’ Layers. Trends Biotechnol 2016:10.1016/j.tibtech.2015.1012.1013. [DOI] [PubMed] [Google Scholar]


