Abstract
DNA interstrand cross-links are complex lesions that covalently bind complementary strands of DNA and whose mechanism of repair remains poorly understood. In Escherichia coli, several gene products have been proposed to be involved in cross-link repair based on the hypersensitivity of mutants to cross-linking agents. However, cross-linking agents induce several forms of DNA damage, making it challenging to attribute mutant hypersensitivity specifically to interstrand cross-links. To address this, we compared the survival of UVA-irradiated repair mutants in the presence of 8-methoxypsoralen—which forms interstrand cross-links and monoadducts—to that of angelicin—a congener forming only monoadducts. We show that incision by nucleotide excision repair is not required for resistance to interstrand cross-links. In addition, neither RecN nor DNA polymerases II, IV, or V is required for interstrand cross-link survival, arguing against models that involve critical roles for double-strand break repair or translesion synthesis in the repair process. Finally, estimates based on Southern analysis of DNA fragments in alkali agarose gels indicate that lethality occurs in wild-type cells at doses producing as few as one to two interstrand cross-links per genome. These observations suggest that E. coli may lack an efficient repair mechanism for this form of damage.
Keywords: DNA interstrand cross-link, translesion DNA synthesis, UvrD, nucleotide excision repair, RecN
DNA interstrand cross-links are highly cytotoxic lesions induced by a variety of bifunctional agents, including nitrogen mustard, cisplatin, mitomycin C, and psoralen plus UVA light (Brookes and Lawley 1960, 1961; Iyer and Szybalski 1963; Dall’Acqua et al. 1971; Dijt et al. 1988). Cytotoxicity is thought to result from the covalent linkage between both DNA strands, preventing strand separation and thus inhibiting essential processes like replication or transcription at these sites (Brookes and Lawley 1961; Lawley and Brookes 1968). Cross-linking agents are widely used as chemotherapeutics and have been effectively employed in the treatment of a range of hyperplastic or dysplastic conditions such as psoriasis or white leprosy (Pathak and Fitzpatrick 1992; Honig et al. 1994; Chakraborty et al. 1996; Huang and Li 2013). Their effectiveness in treating these disease states has led to an intense interest in understanding how these medically relevant lesions are processed by the cell, with the goal of developing novel targets or strategies for chemotherapeutics.
Although several genes have been isolated that, when mutated, render cells hypersensitive to cross-linking agents (Kohn et al. 1965; Cole 1971c; Fujiwara and Tatsumi 1977; Sinden and Cole 1978; Ishida and Buchwald 1982), many aspects of how these complex lesions are repaired and processed in cells remain unknown. Additionally, all cross-linking agents induce multiple forms of DNA damage (Cole 1971b; Eastman 1983; Metzler 1986; Povirk and Shuker 1994), making it difficult to attribute mutant hypersensitivity specifically to defective repair of the cross-link lesion. Various models for interstrand cross-link repair have been proposed, each involving multiple repair pathways that couple components of nucleotide excision repair with recombination, translesion synthesis, or other alternative nuclease complexes (Cole and Sinden 1975; Lin et al. 1977; Sinden and Cole 1978; Berardini et al. 1997, 1999; De Silva et al. 2000; Bessho 2003; Niedernhofer et al. 2004). However, after the initial incision event, all these models remain highly speculative and are hampered by the challenges of reconstituting this multi-step, multi-pathway repair process.
Based on early experiments in Escherichia coli, researchers recognized the challenge of repairing a DNA interstrand cross-link due to the covalent attachment of this adduct to both DNA strands. Researchers inferred that repair would likely require the sequential action of multiple pathways (Cole and Sinden 1975; Lin et al. 1977; Sinden and Cole 1978). The hypersensitivity of both nucleotide excision repair and recombination mutants—uvrA and recA, respectively—led initial models to propose that nucleotide excision repair may initiate incisions on one strand, and that recombination from a sister chromosome would then provide an undamaged template to replace the incised region. A second round of incisions by nucleotide excision repair could then complete the repair process (Figure 1A). In support of this model, biochemical studies found that the UvrABC nucleotide excision repair complex would recognize and incise one strand of a cross-link in vitro (Van Houten et al. 1986). Other studies demonstrated that RecA could promote strand exchange at this site in vitro if the gapped region were first expanded through exonucleolytic degradation (Sladek et al. 1989b). Although in principle this model could repair a cross-link, it should also be noted that uvrA and recA mutants are hypersensitive to almost all forms of DNA damage, not simply cross-links. And currently, no intermediates for the events following the initial incision have been characterized or observed in vivo.
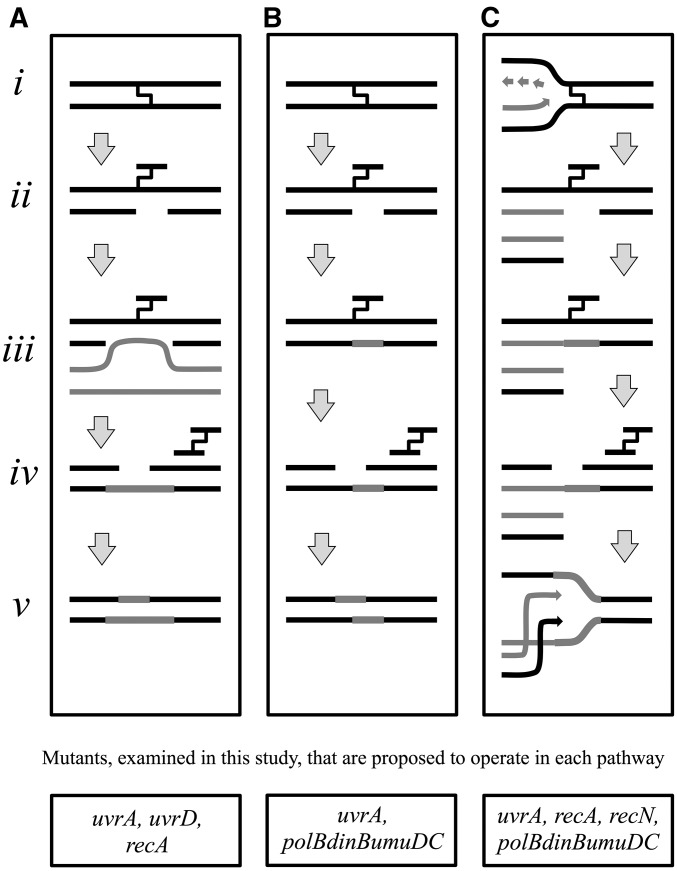
Figure 1.
Predominant models for the repair of DNA interstrand cross-links in the literature. (A) Nucleotide excision repair and homologous recombination. The interstrand cross-link (i) is initially incised by the nucleotide excision repair complex (ii), before the gap on the incised strand is filled in by recombination using a sister chromosome (iii). Nucleotide excision repair could then in theory make a second round of incisions on the opposing strand (iv), which could be filled in using the “newly formed” complementary strand as a template (v) (adapted from Cole 1973a; Van Houten et al. 1986; Sladek et al. 1989b). (B) Nucleotide excision repair and translesion synthesis. The interstrand cross-link (i) is initially incised by the nucleotide excision repair complex (ii) before translesion synthesis fills in the gap (iii). Nucleotide excision repair then makes a second round of incisions to remove the lesion (iv) before the remaining gap is resynthesized using the newly formed complementary strand as a template (v) (adapted from Berardini et al. 1997). (C) Excision-mediated double-strand break repair. Following an encounter with a replication fork (light arrows) (i), incision of the interstrand cross-link by nucleotide excision repair results in a double-strand break (ii). Translesion polymerases restore the incised template (iii) before a second round of nucleotide excision repair removes the cross-link and restores the template (iv). Recombination then repairs the double-strand break to restore the replication fork indicated by arrows (v) (adapted from De Silva et al. 2000; Bessho 2003; Niedernhofer et al. 2004). Genes examined in this work that are proposed to operate in each pathway are listed below each model.
Other models noted that DNA cross-links occurring in nonreplicating cells would not have a sister chromosome available for recombination (Berardini et al. 1997, 1999). To account for this, it was proposed that alternative DNA polymerases might replicate across the incised oligo-lesion product to provide a template for the second round of incisions (Figure 1B). In support for this idea, some early studies using plasmids containing an interstrand cross-link displayed reduced survival when transformed into a polB [polymerase (Pol) II] mutant (Berardini et al. 1999). However, several laboratories have since been unable to verify a range of phenotypes reported for this polB strain (Escarceller et al. 1994; Berardini et al. 1999; Rangarajan et al. 1999), implying the effect may have been due to secondary mutations within this particular strain (Kow et al. 1993; Courcelle et al. 2005; Kumari et al. 2008). A later biochemical study also showed that Pol IV, the dinB gene product, could synthesize through templates containing an unhooked oligo-bound cross-link in vitro, supporting the possibility that translesion synthesis could carry out this hypothetical step in cells (Kumari et al. 2008). However, to date, the potential role for translesion synthesis during cross-link repair in vivo has not been systematically examined in bacteria.
A third model for the repair of cross-links comes from studies in mammalian cells and suggests that repair is coupled to replication and proceeds through a double-strand break intermediate (Figure 1C) (De Silva et al. 2000; Bessho 2003; Niedernhofer et al. 2004). In this model, replication forks blocked at interstrand cross-links are incised by the Fanconi anemia pathway/nucleotide excision repair proteins (ERCC1/XPF endonuclease) as well as other structure-specific nucleases to create a double-strand break intermediate. Consistent with this, double-strand breaks are detected in both yeast and mammalian cells following treatment with interstrand cross-linking agents and double-strand break repair mutants are hypersensitive to these drugs (De Silva et al. 2000; Bessho 2003; Niedernhofer et al. 2004). Similarly, the bacterial UvrABC excinuclease has been shown to be capable of incising both strands of an interstrand cross-link in vitro (Sczepanski et al. 2009; Peng et al. 2010). However, whether double-strand breaks arise in vivo during the bacterial repair process has not been directly examined.
To further characterize the pathway and gene products involved in the repair of DNA interstrand cross-links, we compared the survival of E. coli cultures irradiated with UVA in the presence of 8-methoxypsoralen or angelicin. The comparison between 8-methoxypsoralen and angelicin allowed us to differentiate between genes that are involved in the general repair of monoadducts from those that have a specific role in interstrand cross-link repair (Kaye et al. 1980). DNA monoadducts and interstrand cross-links are formed by 8-methoxypsoralen in a ratio of ∼4:1 (Kanne et al. 1982; Tessman et al. 1985). Angelicin is a congener of psoralen that, due to its angular structure, is unable to form cross-links and produces almost entirely monoadducts (Dall’Acqua et al. 1971; Bordin et al. 1976; Ashwood-Smith and Grant 1977; Perera et al. 2016). Using this approach, mutants lacking gene products that specifically contribute to the repair of interstrand cross-links would be expected to exhibit a greater sensitivity to 8-methoxypsoralen relative to angelicin when compared to sensitivities of wild-type cultures. Here, we examined several candidate mutants of genes that have been proposed to be involved in cross-link repair based on current models in bacteria or mammals.
Materials and Methods
Bacterial strains
The parent strain used in this study is SR108, a thyA36 deoC2 derivative of W3110 (Courcelle et al. 1997). Isogenic strains lacking uvrA (HL952), uvrD (CL1302), and polB dinB umuDC (CL646) were constructed using standard P1 transduction methods and have been described previously (Courcelle et al. 2005; Newton et al. 2012). CL912 (DY329 recN::cat) was constructed using the recombineering strain DY329 (Yu et al. 2000). The chloramphenicol resistance gene was amplified from TP507 (Murphy et al. 2000) using the following PCR primers: recNF-catF 5′ GTAATGGTTTTTCATACAGGAAAACGACTATGTTGGCACAATGAGACGTTGATCGGCAC 3′, and recNR-catR 5′ GCAGGAAAAAAGTTTACGCTGCAAGCAGTTCTTTCGCATTCTTTCGAATTTCTGCCATT 3′. PCR product was then transformed into DY329 to generate CL912, selecting for chloramphenicol resistance. The gene replacement was transferred into SR108 using standard P1 transduction to generate CL915 (SR108 recN::cat). pBR322 is a medium copy number, ColE1-based, 4.4-kb plasmid (Promega, Madison, WI).
Psoralen-UVA and angelicin-UVA survival assays
Fresh overnight cultures were diluted 1:100 in Davis medium (Davis 1949) supplemented with 0.4% glucose, 0.2% Casamino Acids, and 10 μg/ml thymine (DGCthy) and grown at 37° to an optical density of 0.4 at 600 nm (OD600). At this time, 10 μg/ml of 8-methoxypsoralen (item 298-81-7; Acros Organics) or 40 μg/ml of angelicin (item A0956; Sigma-Aldrich) was added to the cultures and incubation continued for 10 min. Cells were then irradiated using two 32-W UVA bulbs (Sylvania) with a peak emittance of 320 nm at an incident dose of 6.3 J/m2/sec. At the doses indicated, 100-µl aliquots were removed from each culture and serially diluted in 10-fold increments. Triplicate 10-μl aliquots of each dilution were spotted onto Luria-Bertani agar plates supplemented with 10 μg/ml thymine (LBthy) and incubated at 37°. Viable colonies were counted the next day to determine the surviving fraction.
Survival assays using nonreplicating cells were done as described above, except that subcultures were grown for 24 hr prior to treatment and irradiation.
The lethal dose was determined, based on the Poisson expression, P(i) = xie−x/i!, as the fraction of surviving cells (i.e., those having zero lethal lesions) at a dose where cells in the population have an average of one lethal lesion, i = 0, x = 1.
In vivo detection of DNA interstrand cross-links
Cultures containing the plasmid pBR322 were grown overnight at 37° in DGCthy medium supplemented with 50 μg/ml ampicillin. A 0.1-ml aliquot from this culture was pelleted and resuspended in 10-ml DGCthy medium without ampicillin and grown in a 37° shaking water bath to an OD600 of 0.4.
Cultures used for the incision/recovery assay were exposed to 20 μg/ml 8-methoxypsoralen for 10 min at 37° and subsequently irradiated with a UVA dose of 5.7 kJ/m2. Irradiated cultures were then incubated for a recovery period of 120 min where 0.75-ml aliquots were collected at the indicated times and then transferred to an equal volume of ice-cold 4× NET buffer (10 mM Tris, pH 8.0, 40 mM EDTA, 400 mM NaCl).
Cultures used for the dose-dependent induction of cross-links were exposed to 10 μg/ml 8-methoxypsoralen for 10 min at 37° and UVA irradiated. Aliquots (0.75 ml) were collected during the course of the UVA treatment at the doses indicated and transferred to an equal volume of ice-cold 4× NET buffer.
Cells were pelleted, resuspended in 130-µl lysis buffer (1 mg/ml lysozyme, 0.5 mg/ml RNaseA in 10 mM Tris, 1 mM EDTA, pH 8.0), and incubated at 37° for 30 min. Then, 10 µl of 10 mg/ml proteinase K and 10 µl of 20% Sarkosyl were added to the samples, and incubation continued for 1 hr at 37°. Samples were extracted with four volumes of phenol:chloroform, followed by two volumes of chloroform, and then dialyzed against 200 ml of 1 mM Tris (pH 8.0), 1 mM EDTA for 45 min using 47-mm Millipore 0.025-µm pore disks. The DNA was digested with HindIII (Thermo Fisher) overnight at 37°. Samples were electrophoresed on a 0.75% alkaline agarose gel in 30 mM NaOH, 1 mM EDTA at 1 V/cm for 16 hr. DNA in the gels was then transferred to Hybond N+ nylon membranes (GE Healthcare) using standard Southern blotting techniques. The plasmid DNA was detected by probing with 32P-labeled pBR322 which was prepared by nick translation (Roche) using α-32P-dCTP >6000Ci/mmol (Perkin Elmer-Cetus, Norwalk, CT). Southern blots were visualized using a Storm 840 PhosphorImager (GE Biosciences) and its associated ImageQuant analysis software.
The fraction of 8-methoxypsoralen cross-links formed at each time point was calculated as the ratio of the DNA band running at about twice the molecular weight of the linear band to the total DNA per lane and normalized to the fraction of cross-links in untreated samples.
where XD represents cross-linked DNA and TD represents total DNA.
The best fit line for the fraction of lesion-free plasmid was calculated from the Poisson distribution, based on the assumption that two photons must be absorbed to form a cross-link (Cassuto et al. 1977; Lin et al. 1977; Kanne et al. 1982), where y = P(0) + P(1) = e−(m)x + (m)xe−(m)x.
Data availability
Strains and plasmids are available upon request. Supplemental material (comprising the following files: SupplementalFiguresAndTable.pdf contains Supplemental Material, Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, and Table S1; Exp1WTleftUvrAright.tif; Exp2WTtopUvrAbottom.tif; and Exp3WTleftUvrAright.tif) available at Figshare: https://doi.org/10.25386/genetics.6847547.
Results
uvrD mutants are hypersensitive to monoadducts, but are almost as resistant as wild-type cells when both monoadducts and cross-links are present
A feature common to most models of DNA interstrand cross-link repair is that the process is initiated by the nucleotide excision repair pathway. Nucleotide excision repair is the primary pathway by which a diverse range of base damage and monoadducts are repaired (Sancar and Tang 1993; Sancar 1996). In E. coli, UvrA, UvrB, and UvrC form an excinuclease that makes dual incisions surrounding the adduct on the damage-containing strand (Sancar and Rupp 1983). Cells deficient in any one of these gene products fail to make incisions and exhibit elevated levels of recombination, genomic rearrangements, and cell lethality (Setlow et al. 1963; Howard-Flanders et al. 1969; Courcelle et al. 1999, 2003). UvrD is a helicase that promotes removal of the incised strand and releases the excinuclease complex from the DNA before DNA Pol I fills in the gap and DNA ligase seals the nick to complete repair (Caron et al. 1985; Husain et al. 1985; Orren et al. 1992). In the absence of UvrD, the excinuclease complex remains bound to the initial adduct (Rothman and Clark 1977; Kuemmerle and Masker 1980; Van Sluis et al. 1983; Crowley and Hanawalt 2001). As a result, mutants lacking UvrD appear defective at making incisions and are almost as hypersensitive and defective at removing lesions from the genome as those lacking UvrA, -B, or -C.
Several studies, both in vitro and in vivo, suggest that nucleotide excision repair acts to initiate the processing of interstrand cross-links as uvrA, uvrB, or uvrC mutants are hypersensitive to DNA interstrand cross-linking agents and fail to incise cross-links in vivo and in vitro (Cole 1971a,c; Cole et al. 1976; Cassuto et al. 1977; Lin et al. 1977; Van Houten et al. 1986; Sladek et al. 1989b; Perera et al. 2016). Cho, an alternative nuclease with homology to UvrC, has also recently been shown to contribute to survival in the presence of 8-methoxypsoralen, perhaps by enhancing the rate of the initial incision (Moolenaar et al. 2002; Perera et al. 2016). However, the role of UvrD in interstrand cross-link repair has not yet been examined. In theory, following the initial incision, the interstrand cross-link remains bound to the second DNA strand, preventing displacement by helicases. However, at monoadducts, UvrD is still required for excinuclease turnover, and turnover may still be required to allow further cross-link repair events to occur. UvrD may also be required for strand displacement following the second round of incisions if repair proceeds through mechanisms similar to those depicted in Figure 1A or Figure 1B.
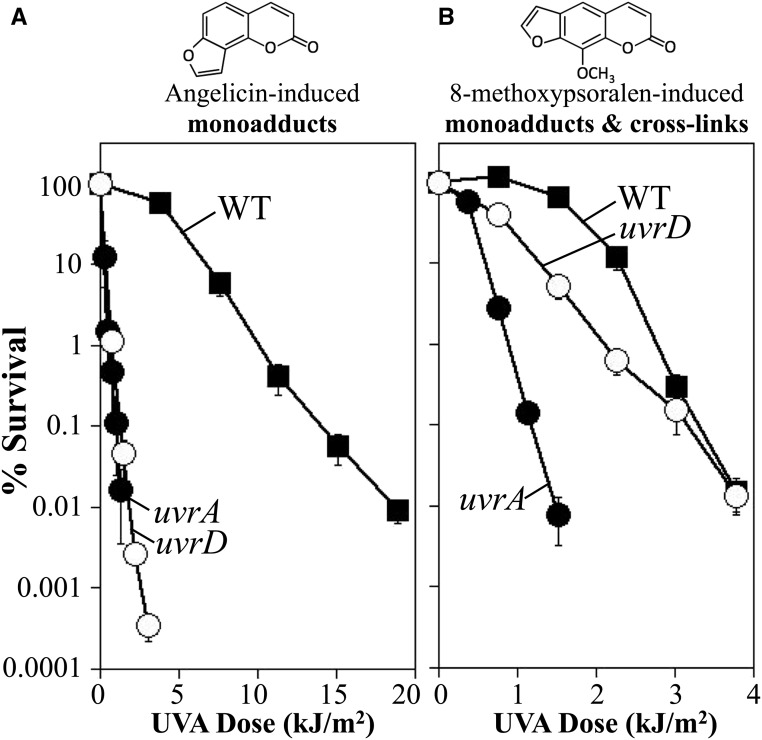
To investigate the role of UvrD in cross-link repair, we examined the survival of uvrD mutants following UVA irradiation in the presence of either 10 μg/ml 8-methoxypsoralen or 40 μg/ml angelicin. We elected to use a concentration of angelicin fourfold higher than that of 8-methoxypsoralen. These concentrations were chosen so that more similar amounts of adducts would be formed per given dose of UVA, guided by studies by Bordin et al. (1976) where psoralen, which is similar but not identical to 8-methoxypsoralen, was found to form adducts approximately fivefold more efficiently than angelicin (Bordin et al. 1976). Under these conditions, the survival of wild-type cultures in the presence of angelicin was reduced by more than two orders of magnitude within 12 kJ/m2 of UVA irradiation (Figure 2A). Whereas in the presence of 8-methoxypsoralen a similar loss of viability was observed within 3 kJ/m2, consistent with the high lethality associated with the formation of DNA interstrand cross-links (Figure 2B). By comparison, uvrA mutants were hypersensitive to both angelicin and 8-methoxypsoralen, reducing survival by more than two orders of magnitude after 1 kJ/m2 in each case.
Figure 2.
UvrD contributes to survival in the presence of monoadducts, but not DNA interstrand cross-links. The survival of wild type (WT) (▪), uvrA (●), and uvrD (○) mutants following UVA irradiation in the presence of (A) 40-μg/ml angelicin or (B) 10-μg/ml 8-methoxypsoralen is plotted. Plots represent the average of at least four experiments. Error bars represent SEM. The structure of angelicin and 8-methoxypsoralen are shown above each graph.
When we examined uvrD cultures, we observed that these mutants were nearly as hypersensitive to angelicin-induced monoadducts as uvrA mutants (Figure 2A), consistent with what has been observed for other forms of base damage and monoadducts, such as pyrimidine dimers and N-acetoxy-N-2-acetylaminofluorene (Granger-Schnarr et al. 1986; Washburn and Kushner 1991; Crowley and Hanawalt 2001). Surprisingly, however, uvrD mutants were nearly as resistant as wild-type cells when both monoadducts and interstrand cross-links were induced with 8-methoxypsoralen (Figure 2B). We interpret the modest hypersensitivity of uvrD cells at UVA doses ≤2 kJ/m2 to result from the preferential formation of monoadducts at these low UVA doses (Cassuto et al. 1977; Lin et al. 1977). At higher doses of UVA, where DNA interstrand cross-links are formed, no significant difference is observed between wild-type and uvrD cultures. Two inferences can be made from this observation. First, the comparable resistance of wild type and uvrD mutants implies that UvrD is not required for interstrand cross-link repair. If nucleotide excision repair initiates repair at cross-links, one can infer that UvrD is not required for either oligo removal or excinuclease turnover during the first incision, nor would it be required if a second round of incisions occurs as is postulated in many models such as those shown in Figure 1.
Second, it is notable that uvrD mutants are hypersensitive in the presence of monoadducts (Figure 2A), but as resistant as wild-type cells when both monoadducts and interstrand cross-links are present. This allows us to infer that the lethality observed in wild-type cells is caused almost exclusively by the presence of DNA interstrand cross-links. Thus, although 8-methoxypsoralen monoadducts are formed at higher frequencies than interstrand cross-links, wild-type cultures are able to efficiently repair and survive in the presence of these lesions and lethality results from the presence of interstrand cross-links.
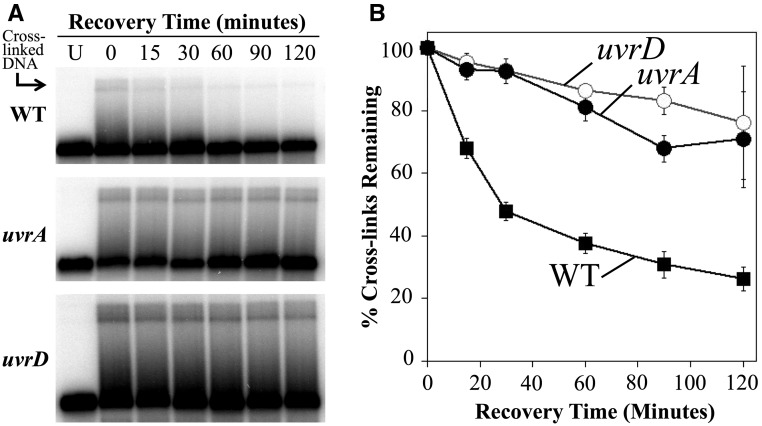
The nearly normal resistance of uvrD mutants to interstrand cross-links might suggest that incision occurs normally in these mutants. To examine this question, we compared the rate that interstrand cross-links were incised in wild type, uvrD, and uvrA mutants. To this end, cultures containing the plasmid pBR322 were treated with 8-methoxypsoralen and UVA light and then allowed to recover. At various times during the recovery period, total genomic DNA was purified from aliquots of the culture. The purified DNA was then restricted with HindIII to linearize the plasmid and examined by Southern analysis following alkali denaturing agarose gel electrophoresis to determine the rate that the DNA interstrand cross-links were incised over time (Figure 3A). In wild-type cultures immediately following UVA irradiation, 3.6% of the plasmids contained a DNA interstrand cross-link. The fraction of DNA migrating in the cross-link region of the gel decreased over the recovery period, with more than three-quarters of the cross-links being incised by the end of the 120-min time course (Figure 3B). In uvrA mutants, 7.9% of the molecules initially contained DNA interstrand cross-links following UVA irradiation. In these cultures, the incisions of cross-links were severely impaired as most cross-links persisted in the DNA throughout the 120-min recovery period, consistent with previous studies showing defective incisions in these mutants. However, the hypersensitivity of uvrA mutants to monoadducts makes it impossible to determine whether these incisions are necessary for surviving cross-link repair. Surprisingly, although uvrD cultures are almost as resistant to cross-links as wild-type cultures, the mutants were impaired in their ability to make incisions, similar to uvrA mutants. The elevated resistance of uvrD mutants to cross-links, despite their inability to incise or unhook these lesions, raises the possibility that although nucleotide excision repair recognizes and incises interstrand DNA cross-links, the incisions may not be productive and do not necessarily contribute to the survival of the cell.
Figure 3.
uvrD mutants are resistant to 8-methoxypsoralen despite an inability to incise interstrand cross-links. (A) Cultures containing the plasmid pBR322 were UVA irradiated with 5.7 kJ/m2 in the presence of 20-μg/ml 8-methoxypsoarlen and then allowed to recover. To observe the cross-links remaining in the DNA at each time point, total genomic and plasmid DNA was purified from cells, restricted with HindIII to linearize the plasmid, and examined by Southern analysis following alkali gel electrophoresis using 32P-labeled pBR322 as a probe. Lane U, untreated cells. (B) The fraction of cross-links remaining in the DNA during the recovery period is plotted for wild type (WT) (▪), uvrA (●), and uvrD (○). The initial percentage of cross-linked plasmids immediately following UVA irradiation were 3.6 ± 1.0% for wild type, 7.9 ± 2.3% for uvrA, and 6.6 ± 0.7% for uvrD. Plots represent the average of seven experiments for wild type and four experiments for uvrA and uvrD. Error bars represent SEM.
Translesion DNA polymerases do not contribute to survival in the presence of interstrand cross-links
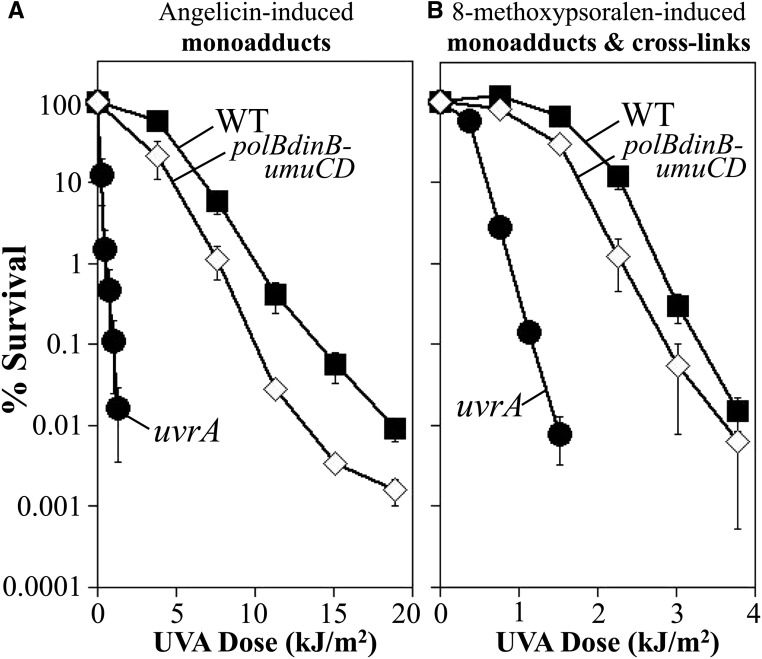
Following the initial incisions by nucleotide excision repair, models in both bacteria and mammals have proposed that translesion synthesis by alternative polymerases function to replicate across an incised oligo-cross-link intermediate to provide a template for the second round of incisions (Berardini et al. 1997, 1999; Sarkar et al. 2006; Kumari et al. 2008; Mogi et al. 2008; Ho and Scharer 2010; Sharma and Canman 2012; Tomicic et al. 2014). Biochemically, Pol IV—the dinB gene product—is capable of synthesizing through templates containing an unhooked oligo-bound cross-link in vitro, supporting the possibility that translesion synthesis could carry out this hypothetical step (Kumari et al. 2008). In mammals, several polymerases have been speculated to participate in cross-link repair based on mutant hypersensitivity to chemicals that form cross-links in addition to in vitro evidence for the ability of certain polymerases to synthesize past partially incised cross-links (Sarkar et al. 2006; Mogi et al. 2008; Ho and Scharer 2010; Sharma and Canman 2012; Tomicic et al. 2014). However, the potential role for translesion polymerases during cross-link repair in vivo has not been examined in bacteria. To this end, the comparative survival of a mutant lacking all three translesion polymerases—Pol II, Pol IV, and Pol V—was examined following UVA irradiation in the presence of both angelicin and 8-methoxypsoralen. Relative to wild-type cells, the absence of translesion polymerases rendered cells modestly sensitive to monoadducts formed by angelicin (Figure 4A). Comparatively, in the presence of both monoadducts and DNA interstrand cross-links, the hypersensitivity remained nearly identical (Figure 4B). Thus, the presence of DNA interstrand cross-links does not further sensitize the polymerase mutants beyond that seen in the presence of monoadducts alone.
Figure 4.
The translesion DNA polymerases do not contribute to survival in the presence of 8-methoxypsoralen-induced DNA interstrand cross-links. The survival of wild type (WT) (▪), uvrA (●), and polB dinB umuDC (⋄) mutants following UVA irradiation in the presence of (A) 40-μg/ml angelicin or (B) 10-μg/ml 8-methoxypsoralen is plotted. Plots represent the average of at least three experiments. Error bars represent SEM. Wild type and uvrA from Figure 2 shown for comparison.
Some models, of both bacteria and humans, propose that translesion synthesis may operate primarily in the absence of replication (Berardini et al. 1997, 1999; Sarkar et al. 2006; Kumari et al. 2008; Mogi et al. 2008; Ho and Scharer 2010; Sharma and Canman 2012; Tomicic et al. 2014). If true, and if a prominent replication-coupled repair pathway exists, then it is possible that the contribution from translesion polymerases would be missed in exponentially growing cultures. To address this, we repeated this analysis using stationary phase cultures. Similar to that seen in replicating cultures, the presence of interstrand cross-links did not affect the survival of the translesion polymerase mutants beyond that seen in the presence of monoadducts alone (Figure S1). If the predominant mechanism of cross-link repair proceeded through translesion synthesis as depicted in the model of Figure 1B, one would predict that these mutants should exhibit a greater hypersensitivity to 8-methoxypsoralen. The observation that they do not show this hypersensitivity does not support the idea that these enzymes are required for the repair. Taken together, these observations imply that the translesion polymerases are not contributing to the survival of interstrand cross-links in vivo. However, it also does not exclude the possibility that alternative (or secondary) mechanisms exist that can compensate in their absence.
Double-strand breaks are not a prominent intermediate in the repair of interstrand cross-links
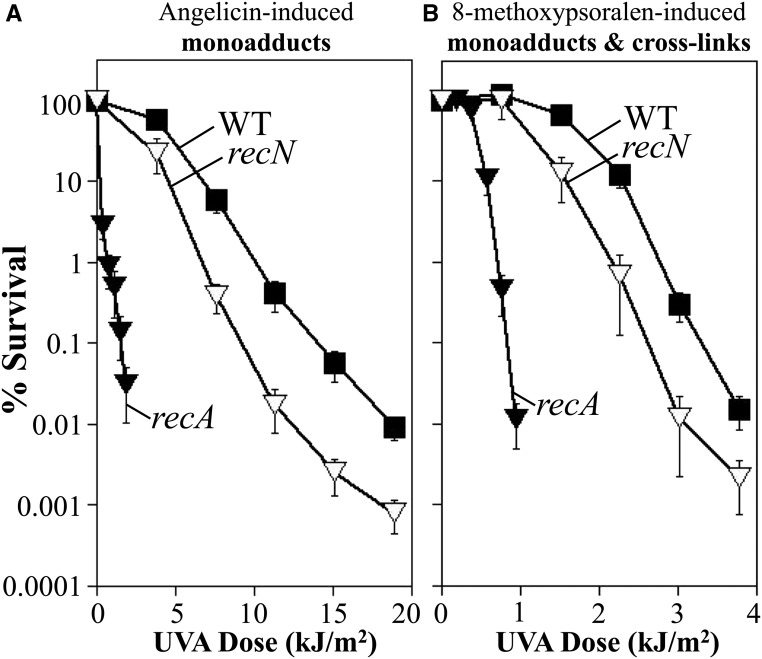
Some interstrand cross-link repair models propose that double-strand breaks arise as an intermediate and that recombination is required for repair to occur. However, other evidence suggests that the breaks observed may arise as pathological aberrancies and occur primarily after repair fails or is impeded from occurring normally (De Silva et al. 2000; McHugh et al. 2000; Bessho 2003; Niedernhofer et al. 2004; Rothfuss and Grompe 2004; Mogi and Oh 2006; Sczepanski et al. 2009; Peng et al. 2010; Vare et al. 2012). In E. coli, several genes, when mutated, render cells hypersensitive to double-strand breaks. However, most of these mutants, including recA, are hypersensitive to multiple forms of DNA damage. recN mutants are unique in that they are hypersensitive to agents that generate double-strand breaks—such as gamma irradiation, mitomycin C, nalidixic acid, or enzymatic restriction—but are resistant to other forms of DNA damage that form single-strand lesions or monoadducts such as UV (Picksley et al. 1984). Consistent with this role in repairing double-strand breaks, purified RecN binds and protects double-strand DNA ends and interacts with RecA to stimulate its ATPase activity and facilitate its loading at these sites (Grove et al. 2009; Keyamura et al. 2013; Uranga et al. 2017). If double-strand break intermediates form during the processing of interstrand cross-links, then one would expect recN mutants to be sensitive to 8-methoxypsoralen, but not to angelicin. To examine this directly, we compared the survival of recN mutants after UVA irradiation in the presence of angelicin and 8-methoxypsoralen. As shown in Figure 5A, recN mutants were modestly hypersensitive to monoadducts formed in the presence of angelicin when compared to wild-type cultures. Cells lacking RecN were also modestly hypersensitive to 8-methoxypsoralen treatment, however the degree of hypersensitivity of this strain, relative to wild-type cells, was similar irrespective of the agent used (Figure 5B). The similar sensitivity of recN mutants relative to wild-type cells, in the presence of monoadducts alone or both monoadducts and cross-links, suggests that recN is not contributing to survival in the presence of interstrand cross-links. Although RecN is required for resistance in all known cases where double-strand breaks are generated, we cannot rule out the possibility that breaks associated with processing cross-links would prevent access or not require RecN. By comparison, recA mutants were hypersensitive to treatments that produced cross-links and monoadducts as well as monoadducts alone (Figure 5).
Figure 5.
recN, which is required for resistance to double-strand breaks, does not contribute to survival in the presence of DNA interstrand cross-links. The survival of wild type (WT) (▪), recA (▾), and recN (▿) mutants following UVA irradiation in the presence of (A) 40-μg/ml angelicin or (B) 10-μg/ml 8-methoxypsoralen is plotted. Plots represent the average of at least three experiments. Error bars represent SEM. Wild type from Figure 2 shown for comparison.
In some cases, double-strand breaks have been postulated to arise in the absence of replication, if recognition and incision were to occur on both DNA strands (Van Houten et al. 1986; Sladek et al. 1989b). To test this possibility, we also examined whether RecN affected cell survival in nonreplicating cells. However, the hypersensitivity of recN mutants, relative to wild-type cells, was not altered by the presence or absence of replication (Figure S2). Taken together, these observations would argue against models such as those depicted in Figure 1C, in which double-strand breaks arise as an intermediate in the repair of interstrand cross-links in E. coli.
Repair of interstrand cross-links is inefficient
The data presented above suggests that neither incisions by nucleotide excision repair, nor double-strand break repair, nor translesion synthesis significantly contribute to the survival of E. coli in the presence of interstrand cross-links. We next quantified the cross-links formed in vivo under the conditions used in our assays to determine how efficiently cells are able to repair these lesions. To quantify interstrand cross-links in vivo, we examined their formation as a function of dose on an endogenous plasmid in uvrA cultures. The higher copy number of the plasmid, relative to the chromosome, increases our ability to detect cross-links in a specific sequence. Since uvrA mutants fail to incise cross-links (Figure 2), these lesions accumulate and persist on the plasmid and can be directly quantified. To this end, cultures containing the plasmid pBR322 were grown in media containing 10 μg/ml 8-methoxypsoralen and UVA irradiated with increasing doses. Total (genomic and plasmid) DNA was then purified and digested with the HindIII restriction endonuclease to linearize the plasmid. The samples were then electrophoresed in an alkaline denaturing agarose gel and Southern analysis was used to identify and quantify the cross-links formed at each dose (Islas et al. 1991; Perera et al. 2016). The percent of cross-link-free plasmid was plotted at each UVA dose. Interstrand cross-links require adsorption of two photons and, as demonstrated by a number of studies, form with second-order kinetics as a function of dose (Cassuto et al. 1977; Lin et al. 1977; Sinden and Cole 1978). Absorption of photons during UVA irradiation should follow a normal distribution. We therefore used the Poisson expression to approximate the normal distribution and determined the best fit line for cross-link-free plasmids as those containing fewer than two photon hits (the sum of the 0 and 1 classes). The number of cross-links per base pair could then be determined directly from the proportion of plasmids observed to contain a single cross-link. By example, a dose which generates a cross-link on 10% of the endogenous 4.4-kb plasmids would represent a cross-link frequency of 1 cross-link/10 plasmids, or 1 cross-link/44 kb.
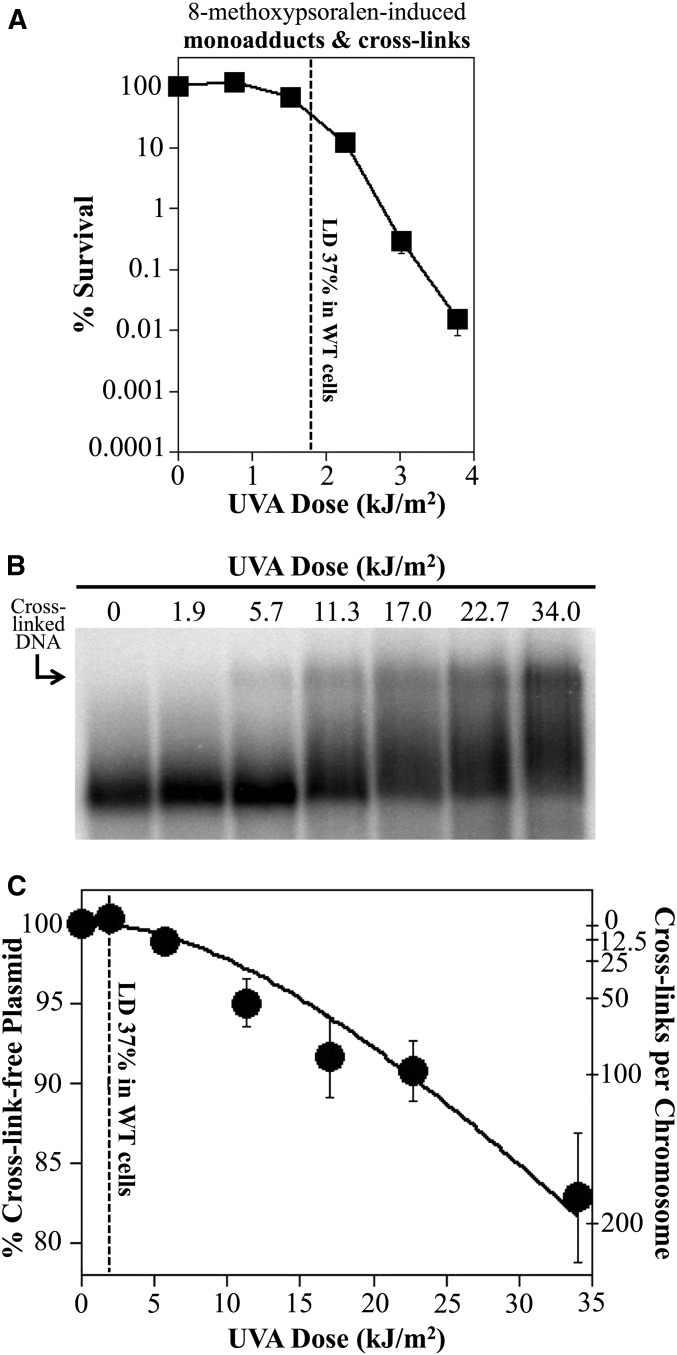
In our analysis of wild-type cultures, ∼1.9 kJ/m2 UVA radiation was required to reduce survival to 37%, where cells in the culture incur one lethal lesion on average (Figure 6A). Based on the dose curve shown in Figure 6B, this dose generated 0.23 cross-links/Mb which would correspond to ∼1.1 cross-links per 4.6-Mb chromosome in E. coli. Thus, by this estimate wild-type cells would lack or have an extremely inefficient system for repairing this form of damage. In uvrA mutants, lethality occurred at ∼0.4 kJ/m2, which would correspond to ∼0.04 cross-links per genome. This likely argues that uvrA mutants are killed by monoadducts, which form at these low doses, before any cross-links arise in the cell. It is possible that nucleotide excision repair processes cross-links on plasmid substrates with different efficiencies than it does on the chromosome. Although such a comparison has not been made for interstrand cross-links, we have measured the incision and repair rate of cyclobutane pyrimidine dimers on both chromosome and plasmid substrates, and found them to be similar (Koehler et al. 1996; Courcelle et al. 1999, 2003). It is important to note that this estimate is based on an extrapolation of our dose response curve which was generated from a set of three separate trials (Figure S3). Using Southern analysis of the alkali agarose gels, we can easily detect and quantify cross-links when ∼1% of the plasmids have a cross-link, which corresponds to ∼11 cross-links/genome and exceeds the lethal dose in wild-type cultures. Irrespective of this limitation, it is clear that the cell’s repair capacity for interstrand cross-links is extremely limited. We infer from these results that E. coli may lack an effective, error-free mechanism for repairing interstrand cross-links.
Figure 6.
As few as one to two cross-links are sufficient to inactivate our parental strain of E. coli. (A) The survival of wild-type (WT) cells in the presence of 10 μg/ml 8-methoxypsoralen and increasing doses of UVA light is plotted. Dotted line represents the UVA dose at which survival is reduced to 37% and occurs at ∼1.9 kJ/m2. Data replotted from Figure 2. (B) Dose response of interstrand cross-link formation in vivo. uvrA mutants containing plasmid pBR322 were UVA irradiated at the indicated dose in the presence of 10 μg/ml 8-methoxypsoarlen. DNA from cultures were then immediately purified and analyzed as in Figure 3 to observe the interstrand cross-links formed at each dose. (C) The percent of cross-link-free 4.4-kb plasmids remaining at each dose is plotted. The corresponding number of cross-links this represents per 4.6-Mb genome is also shown. Plots represent the average of three experiments. Error bars represent SEM. The best fit line was calculated from the Poisson expression based on the assumption that absorption of two-photon events is required to form an interstrand cross-link (Cassuto et al. 1977; Lin et al. 1977; Kanne et al. 1982). The fraction of cross-link-free plasmids decreased at a rate of y = e−(m)x + (m)xe−(m)x, where under our irradiation conditions m = 0.023. LD, lethal dose.
Discussion
We show that although uvrD mutants are hypersensitive to monoadducts, they are nearly as resistant as wild-type cells in the presence of cross-links. uvrD mutants are resistant to cross-links despite an impaired ability to incise these lesions, similar to uvrA mutants. In vitro, it is well established that nucleotide excision repair is able to recognize and incise psoralen interstrand cross-links (Van Houten et al. 1986; Mu et al. 2000), and mutants lacking these enzymes fail to incise cross-links and are hypersensitive to psoralen in vivo (Cole 1971a,c; Cole et al. 1976; Cassuto et al. 1977; Lin et al. 1977; Van Houten et al. 1986; Sladek et al. 1989b; Perera et al. 2016). This has led to the proposal in most current models that nucleotide excision repair initiates repair of these lesions. However, nucleotide excision repair mutants are also hypersensitive to the presence of monoadducts. Psoralen, as with all cross-linking agents, induces monoadducts at higher frequencies than interstrand cross-links, which hampers the ability to determine whether nucleotide excision repair mutants are specifically sensitive to cross-links. The near wild-type resistance of uvrD mutants occurs despite an inability to make incisions, demonstrating that UvrD does not necessarily contribute to the repair process, and that incisions by nucleotide excision repair enzymes are not essential for resistance to psoralen-induced cross-links. These results imply that the incisions by nucleotide excision repair are not significantly contributing to survival in the presence of interstrand cross-links.
We also tested other aspects commonly proposed to operate in many interstrand cross-link repair models, including the presence of a double-strand break intermediate and the participation of translesion DNA polymerases. In both cases, we observed that mutants impaired for double-strand break repair, or lacking the translesion DNA polymerases, did not exhibit any specific hypersensitivity to cross-links beyond that observed when exposed to monoadducts alone. Furthermore, the hypersensitivity of these mutants was relatively modest when either monoadducts alone or monoadducts and cross-links were present. Taken together, this comparative method would argue that many of the proteins and pathways frequently proposed to participate in the repair process—including nucleotide excision repair, double-strand break repair, or translesion synthesis—do not significantly improve cell survival in the presence of these lesions.
The lack of contribution by these pathways led us to examine the cell’s overall repair capacity for interstrand cross-links. Our estimates, based on the differential mobility of cross-linked fragments in alkali agarose gels, suggest that UVA doses which generate only one to two cross-links per genome are sufficient to inactivate our wild-type strain. If these estimates are accurate, this would be consistent with our observations that neither nucleotide excision repair, nor double-strand break repair, nor translesion synthesis contribute to survival. Further, they would argue that E. coli lacks an efficient or effective mechanism for repairing this form of damage.
Early studies for the repair of cross-links in E. coli began with the assumption that an effective repair pathway is likely to exist (Cole 1971c, 1973a,b; Cole and Sinden 1975; Bordin et al. 1976; Cole et al. 1976; Ashwood-Smith and Grant 1977; Cassuto et al. 1977; Lin et al. 1977; Seki et al. 1978; Sinden and Cole 1978; Grover et al. 1981). This assumption has persisted in more recent work, including from our laboratory (Perera et al. 2016). Several of these studies have characterized the survival and recovery of cultures that had been treated with psoralen plus UVA. Many of these studies reported that repair mutants such as recA and uvrA were hypersensitive to psoralen and that incision intermediates appeared and were joined over time. Although these observations were attributed to the formation and removal of interstrand cross-links, it seems likely that most of these phenotypes could also be attributed to a compromised ability to process monoadducts, which are generated at higher frequencies and require these same enzymes for repair. These studies were generally unable to differentiate between cellular effects caused by monoadducts or cross-links, and this issue remains a challenge in present studies.
Although our results would suggest repair of psoralen interstrand cross-links is inefficient in E. coli, other cross-linking agents may yield different results and be processed more efficiently than the psoralen compound characterized here. A recent study demonstrated that the bacteria, Streptomyces sahachiroi, produces azinomycin, a cross-linking agent, as an antimicrobial. The bacteria additionally encodes a protein that confers self-resistance against this toxin (Wang et al. 2016). Initial characterizations suggest the protein has activities that protect cells from forming cross-links, but could also potentially be involved in processing either monoadducts or cross-links when they form (Wang et al. 2016; Mullins et al. 2017). Further characterization of how this organism processes these lesions, as well as other compounds, will be important to determine how the lesions affect cell viability and genome integrity in humans and other organisms.
Previous studies have used angelicin and psoralen in a comparative manner to study interstrand cross-link repair or recovery, as we have here, but these analyses were limited to comparing the overall toxicity and mutagenicity of monoadducts to cross-links (Bordin et al. 1976; Ashwood-Smith and Grant 1977; Seki et al. 1978; Venturini et al. 1980). Interestingly, in all cases there was consensus that lethality could be attributed to the presence of cross-links, consistent with the general idea that the repair capacity of these lesions is limited. In one of the few studies that measured cross-links directly, Lin et al. (1977) observed that the surviving fraction of psoralen–UVA-treated phage was always equivalent to the fraction of phage that remained cross-link-free, consistent with the idea that cells lack an efficient repair mechanism for cross-links.
Finally, based on our ability to detect cross-links using alkali agarose gels and Southern analysis, we estimated that only one or two lesions were sufficient to inactivate our wild-type cells. Our limit of detection for cross-links in our assay was ∼11 cross-links per chromosome, which occurred at a dose well beyond the lethal dose for our wild-type culture. Thus, if our method of quantifying cross-links is accurate, the ability of E. coli to repair these lesions is <11 per chromosome. Based on the extrapolation of our dose curve, lethality is predicted to occur after one or two lesions have formed. The only previous estimate of cross-link repair capacity in E. coli was from an early study by Cole (1971c), who reported that E. coli strain AB1157 was capable of surviving treatments generating 67 cross-links. Cole stated that these estimates were derived from the elution pattern of single- and double-strand DNA from hydroxyapatite columns, but the data and methodology were not shown. Additionally, Cole inferred that cross-links formed at similar rates in UVA-irradiated cells as when DNA was purified in solution, and that cross-link formation increased linearly with UVA dose (Cole 1970, 1971b,c). Neither of these assumptions turned out to be correct and both would significantly overestimate the number of cross-links in samples. Subsequent work from the group of Howard-Flanders questioned the accuracy of the hydroxyapatite method and used alkali sucrose gradient sedimentation to demonstrate that psoralen cross-links form with second-order kinetics, consistent with their required absorption of two photons (Cassuto et al. 1977; Lin et al. 1977; Vare et al. 2014). These observations are important because many studies using high UVA doses often infer that cross-link induction forms linearly with dose (Cole 1970, 1971c; Cao et al. 2008; Lai et al. 2008; Liu and Wang 2013), leading to significant overestimations of cross-links, particularly at low doses where lethality is observed. Although the Howard-Flanders studies quantified psoralen-induced cross-links and demonstrated a single cross-link was sufficient to inactivate λ-phage, they did not determine the lethal number of cross-links for the bacteria.
Although few studies have quantified the induction rate of cross-links in E. coli, a number of studies have examined this question in human cells. Using a liquid chromatography–mass spectrometry-based approach, separate studies from the Wang laboratory reported that cells treated with 1 μg/ml 8-methoxypsoralen and 5 kJ/m2 UVA in PBS formed between ∼4 and ∼40 cross-links/Mb (Cao et al. 2008; Lai et al. 2008). A subsequent study from this group reported ∼7 cross-links/Mb when treating cells with 2.5 μg/ml 8-methoxypsoralen and 20 kJ/m2 UVA in media (Liu and Wang 2013). While the different cell types and media do not allow one to compare results directly, we observed ∼3 and ∼22 cross-links/Mb in 10 μg/ml 8-methoxypsoralen when irradiating with 5.7 and 22.7 kJ/m2 UVA, respectively (Figure S4). Thus, our induction frequencies are in a range similar to that obtained with other methodologies.
Surprisingly, few studies have looked at the repair capacity for cross-links in human cells. One study from the Jenssen laboratory quantified cross-links using the hydroxyapatite method reported that 3500 psoralen cross-links were lethal in the human VH10/HAEB cell line (Vare et al. 2014). Notably, they found that cells were able to unhook >20,000 cross-links/hr and cell survival did not correlate with incision or unhooking. This observation is consistent with what we report in E. coli. Wild-type E. coli are able to unhook cross-links at doses far beyond where lethality occurs, and uvrD mutants are noticeably more resistant to cross-linking agents than uvrA, despite the inability to unhook cross-links. Thus, similar to E. coli, the ability of nucleotide excision repair to incise cross-links is not the factor limiting survival in the presence of cross-links. The lethal dose reported by Vare et al. would also suggest that human cells are able to survive ∼0.5 cross-links/Mb DNA, a value that is not dissimilar from that which we observe in E. coli. Other studies in mammalian cells have suggested that cross-links formed by other agents, such as cisplatin or nitrogen mustard, may be even more lethal than those formed by psoralen, with lethality occurring between 200 and 900 lesions per cell (Crathorn and Roberts 1966; Ball and Roberts 1970; Roberts et al. 1986; Knox et al. 1991).
A limitation in all previous studies, as well as the study presented here, is that current methods depend on extrapolation to determine the repair capacity of cells in culture, since lethality occurs at a UVA dose that is slightly below the limit one can detect cross-links directly. Thus, the lethal dose we report here, like previous studies, depends on the sensitivity and accuracy of the quantitative methods used (Figure S3). While the precise number of cross-links that can be repaired varies modestly between these methods of estimation, it is clear that the repair capacity for these interstrand cross-links is very limited. The comparable resistance between uvrD and wild-type strains and the low estimations of psoralen cross-link repair capacity presented here would suggest that psoralen plus UVA may be more comparable to treatments such as nalidixic acid or rifampicin. Unlike irradiation with UVC, when these drugs effectively target their respective essential cellular functions, cells lack true repair pathways for clearing this form of damage. Earlier work noted that while highly lethal overall, cells surviving cross-links contain high frequencies of deletions and point mutations that only appeared in nucleotide excision-proficient strains (Cassuto et al. 1977; Sladek et al. 1989a), consistent with the idea that a significant portion of these repair attempts are not accurate. In mammalian cells, double-strand breaks are proposed to form prominently following treatment with cross-linking agents (Bessho 2003; Mogi and Oh 2006; Vare et al. 2012). It may be that the presence of double-strand end-joining activity and a large proportion of noncoding DNA allow mammalian cells to survive inaccurate or deletion-prone repair events better than E. coli. This will be an addressable and important question to focus on in future work.
Acknowledgments
This work was supported by the National Institute of Environmental Health Sciences (grant R15 ES-025953) and the National Science Foundation (grant MCB0130486).
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25386/genetics.6847547.
Communicating editor: A. Britt
Literature Cited
- Ashwood-Smith M. J., Grant E., 1977. Conversion of psoralen DNA monoadducts in E. coli to interstrand DNA cross links by near UV light (320–360 nm): inability of angelicin to form cross links, in vivo. Experientia 33: 384–386. 10.1007/BF02002841 [DOI] [PubMed] [Google Scholar]
- Ball C. R., Roberts J. J., 1970. DNA repair after mustard gas alkylation by sensitive and resistant Yoshida sarcoma cells in vitro. Chem. Biol. Interact. 2: 321–329. 10.1016/0009-2797(70)90054-2 [DOI] [PubMed] [Google Scholar]
- Berardini M., Mackay W., Loechler E. L., 1997. Evidence for a recombination-independent pathway for the repair of DNA interstrand cross-links based on a site-specific study with nitrogen mustard. Biochemistry 36: 3506–3513. 10.1021/bi962778w [DOI] [PubMed] [Google Scholar]
- Berardini M., Foster P. L., Loechler E. L., 1999. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J. Bacteriol. 181: 2878–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho T., 2003. Induction of DNA replication-mediated double strand breaks by psoralen DNA interstrand cross-links. J. Biol. Chem. 278: 5250–5254. 10.1074/jbc.M212323200 [DOI] [PubMed] [Google Scholar]
- Bordin F., Carlassare F., Baccichetti F., Anselmo L., 1976. DNA repair and recovery in Escherichia coli after psoralen and angelicin photosensitization. Biochim. Biophys. Acta 447: 249–259. 10.1016/0005-2787(76)90048-4 [DOI] [PubMed] [Google Scholar]
- Brookes P., Lawley P. D., 1960. The reaction of mustard gas with nucleic acids in vitro and in vivo. Biochem. J. 77: 478–484. 10.1042/bj0770478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brookes P., Lawley P. D., 1961. The reaction of mono- and di-functional alkylating agents with nucleic acids. Biochem. J. 80: 496–503. 10.1042/bj0800496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H., Hearst J. E., Corash L., Wang Y., 2008. LC-MS/MS for the detection of DNA interstrand cross-links formed by 8-methoxypsoralen and UVA irradiation in human cells. Anal. Chem. 80: 2932–2938. 10.1021/ac7023969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron P. R., Kushner S. R., Grossman L., 1985. Involvement of helicase II (uvrD gene product) and DNA polymerase I in excision mediated by the uvrABC protein complex. Proc. Natl. Acad. Sci. USA 82: 4925–4929. 10.1073/pnas.82.15.4925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassuto E., Gross N., Bardwell E., Howard-Flanders P., 1977. Genetic effects of photoadducts and photocross-links in the DNA of phage lambda exposed to 360 nm light and tri-methylpsoralen or khellin. Biochim. Biophys. Acta 475: 589–600. 10.1016/0005-2787(77)90319-7 [DOI] [PubMed] [Google Scholar]
- Chakraborty D. P., Roy S., Chakraborty A. K., 1996. Vitiligo, psoralen, and melanogenesis: some observations and understanding. Pigment Cell Res. 9: 107–116. 10.1111/j.1600-0749.1996.tb00098.x [DOI] [PubMed] [Google Scholar]
- Cole R. S., 1970. Light-induced cross-linking of DNA in the presence of a furocoumarin (psoralen). Studies with phage lambda, Escherichia coli, and mouse leukemia cells. Biochim. Biophys. Acta 217: 30–39. 10.1016/0005-2787(70)90119-X [DOI] [PubMed] [Google Scholar]
- Cole R. S., 1971a Properties of F′ factor deoxyribonucleic acid transferred from ultraviolet-irradiated donors: photoreactivation in the recipient and the influence of recA, recB, recC, and uvr genes. J. Bacteriol. 106: 143–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R. S., 1971b Psoralen monoadducts and interstrand cross-links in DNA. Biochim. Biophys. Acta 254: 30–39. 10.1016/0005-2787(71)90111-0 [DOI] [PubMed] [Google Scholar]
- Cole R. S., 1971c Inactivation of Escherichia coli, F′ episomes at transfer, and bacteriophage lambda by psoralen plus 360-nm light: significance of deoxyribonucleic acid cross-links. J. Bacteriol. 107: 846–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R. S., 1973a Repair of DNA containing interstrand crosslinks in Escherichia coli: sequential excision and recombination. Proc. Natl. Acad. Sci. USA 70: 1064–1068. 10.1073/pnas.70.4.1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole R. S., 1973b Repair of interstrand cross-links in DNA induced by psoralen plus light. Yale J. Biol. Med. 46: 492. [PMC free article] [PubMed] [Google Scholar]
- Cole R. S., Sinden R. R., 1975. Repair of cross-linked DNA in Escherichia coli. Basic Life Sci. 5B: 487–495. [DOI] [PubMed] [Google Scholar]
- Cole R. S., Levitan D., Sinden R. R., 1976. Removal of psoralen interstrand cross-links from DNA of Escherichia coli: mechanism and genetic control. J. Mol. Biol. 103: 39–59. 10.1016/0022-2836(76)90051-6 [DOI] [PubMed] [Google Scholar]
- Courcelle C. T., Belle J. J., Courcelle J., 2005. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J. Bacteriol. 187: 6953–6961. 10.1128/JB.187.20.6953-6961.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Carswell-Crumpton C., Hanawalt P. C., 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc. Natl. Acad. Sci. USA 94: 3714–3719. 10.1073/pnas.94.8.3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Crowley D. J., Hanawalt P. C., 1999. Recovery of DNA replication in UV-irradiated Escherichia coli requires both excision repair and recF protein function. J. Bacteriol. 181: 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J., Donaldson J. R., Chow K. H., Courcelle C. T., 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299: 1064–1067. 10.1126/science.1081328 [DOI] [PubMed] [Google Scholar]
- Crathorn A. R., Roberts J. J., 1966. Mechanism of the cytotoxic action of alkylating agents in mammalian cells and evidence for the removal of alkylated groups from deoxyribonucleic acid. Nature 211: 150–153. 10.1038/211150a0 [DOI] [PubMed] [Google Scholar]
- Crowley D. J., Hanawalt P. C., 2001. The SOS-dependent upregulation of uvrD is not required for efficient nucleotide excision repair of ultraviolet light induced DNA photoproducts in Escherichia coli. Mutat. Res. 485: 319–329. 10.1016/S0921-8777(01)00068-4 [DOI] [PubMed] [Google Scholar]
- Dall’Acqua F., Marciani S., Ciavatta L., Rodighiero G., 1971. Formation of inter-strand cross-linkings in the photoreactions between furocoumarins and DNA. Z. Naturforsch. B 26: 561–569. [DOI] [PubMed] [Google Scholar]
- Davis B. D., 1949. The isolation of biochemically deficient mutants of bacteria by means of penicillin. Proc. Natl. Acad. Sci. USA 35: 1–10. 10.1073/pnas.35.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva I. U., Mchugh P. J., Clingen P. H., Hartley J. A., 2000. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol. Cell. Biol. 20: 7980–7990. 10.1128/MCB.20.21.7980-7990.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dijt F. J., Fichtinger-Schepman A. M., Berends F., Reedijk J., 1988. Formation and repair of cisplatin-induced adducts to DNA in cultured normal and repair-deficient human fibroblasts. Cancer Res. 48: 6058–6062. [PubMed] [Google Scholar]
- Eastman A., 1983. Characterization of the adducts produced in DNA by cis-diamminedichloroplatinum(II) and cis-dichloro(ethylenediamine)platinum(II). Biochemistry 22: 3927–3933. 10.1021/bi00285a031 [DOI] [PubMed] [Google Scholar]
- Escarceller M., Hicks J., Gudmundsson G., Trump G., Touati D., et al. , 1994. Involvement of Escherichia coli DNA polymerase II in response to oxidative damage and adaptive mutation. J. Bacteriol. 176: 6221–6228. 10.1128/jb.176.20.6221-6228.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., Tatsumi M., 1977. Cross-link repair in human cells and its possible defect in Fanconi’s anemia cells. J. Mol. Biol. 113: 635–649. 10.1016/0022-2836(77)90227-3 [DOI] [PubMed] [Google Scholar]
- Granger-Schnarr M., Daune M. P., Fuchs R. P., 1986. Specificity of N-acetoxy-N-2-acetylaminofluorene-induced frameshift mutation spectrum in mismatch repair deficient Escherichia coli strains mutH, L, S and U. J. Mol. Biol. 190: 499–507. 10.1016/0022-2836(86)90018-5 [DOI] [PubMed] [Google Scholar]
- Grove J. I., Wood S. R., Briggs G. S., Oldham N. J., Lloyd R. G., 2009. A soluble RecN homologue provides means for biochemical and genetic analysis of DNA double-strand break repair in Escherichia coli. DNA Repair (Amst.) 8: 1434–1443. 10.1016/j.dnarep.2009.09.015 [DOI] [PubMed] [Google Scholar]
- Grover N. B., Margalit A., Zaritsky A., Ben-Hur E., Hansen M. T., 1981. Sensitivity of exponentially growing populations of Escherichia coli to photo-induced psoralen-DNA interstrand crosslinks. Biophys. J. 33: 93–106. 10.1016/S0006-3495(81)84874-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho T. V., Scharer O. D., 2010. Translesion DNA synthesis polymerases in DNA interstrand crosslink repair. Environ. Mol. Mutagen. 51: 552–566. 10.1002/em.20573 [DOI] [PubMed] [Google Scholar]
- Honig B., Morison W. L., Karp D., 1994. Photochemotherapy beyond psoriasis. J. Am. Acad. Dermatol. 31: 775–790. 10.1016/S0190-9622(94)70240-3 [DOI] [PubMed] [Google Scholar]
- Howard-Flanders P., Theriot L., Stedeford J. B., 1969. Some properties of excision-defective recombination-deficient mutants of Escherichia coli K-12. J. Bacteriol. 97: 1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Li L., 2013. DNA crosslinking damage and cancer - a tale of friend and foe. Transl. Cancer Res. 2: 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I., Van Houten B., Thomas D. C., Abdel-Monem M., Sancar A., 1985. Effect of DNA polymerase I and DNA helicase II on the turnover rate of UvrABC excision nuclease. Proc. Natl. Acad. Sci. USA 82: 6774–6778. 10.1073/pnas.82.20.6774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida R., Buchwald M., 1982. Susceptibility of Fanconi’s anemia lymphoblasts to DNA-cross-linking and alkylating agents. Cancer Res. 42: 4000–4006. [PubMed] [Google Scholar]
- Islas A. L., Vos J. M., Hanawalt P. C., 1991. Differential introduction and repair of psoralen photoadducts to DNA in specific human genes. Cancer Res. 51: 2867–2873. [PubMed] [Google Scholar]
- Iyer V. N., Szybalski W., 1963. A molecular mechanism of mitomycin action: linking of complementary DNA strands. Proc. Natl. Acad. Sci. USA 50: 355–362. 10.1073/pnas.50.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanne D., Straub K., Rapoport H., Hearst J. E., 1982. Psoralen-deoxyribonucleic acid photoreaction. Characterization of the monoaddition products from 8-methoxypsoralen and 4,5′8-trimethylpsoralen. Biochemistry 21: 861–871. 10.1021/bi00534a008 [DOI] [PubMed] [Google Scholar]
- Kaye J., Smith C. A., Hanawalt P. C., 1980. DNA repair in human cells containing photoadducts of 8-methoxypsoralen or angelicin. Cancer Res. 40: 696–702. [PubMed] [Google Scholar]
- Keyamura K., Sakaguchi C., Kubota Y., Niki H., Hishida T., 2013. RecA protein recruits structural maintenance of chromosomes (SMC)-like RecN protein to DNA double-strand breaks. J. Biol. Chem. 288: 29229–29237. 10.1074/jbc.M113.485474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox R. J., Lydall D. A., Friedlos F., Basham C., Rawlings C. J., et al. , 1991. The Walker 256 carcinoma: a cell type inherently sensitive only to those difunctional agents that can form DNA interstrand crosslinks. Mutat. Res. 255: 227–240. 10.1016/0921-8777(91)90026-L [DOI] [PubMed] [Google Scholar]
- Koehler D. R., Courcelle J., Hanawalt P. C., 1996. Kinetics of pyrimidine(6–4)pyrimidone photoproduct repair in Escherichia coli. J. Bacteriol. 178: 1347–1350. 10.1128/jb.178.5.1347-1350.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn K. W., Steigbigel N. H., Spears C. L., 1965. Cross-linking and repair of DNA in sensitive and resistant strains of E. coli treated with nitrogen mustard. Proc. Natl. Acad. Sci. USA 53: 1154–1161. 10.1073/pnas.53.5.1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kow Y. W., Faundez G., Hays S., Bonner C. A., Goodman M. F., et al. , 1993. Absence of a role for DNA polymerase II in SOS-induced translesion bypass of phi X174. J. Bacteriol. 175: 561–564. 10.1128/jb.175.2.561-564.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmerle N. B., Masker W. E., 1980. Effect of the uvrD mutation on excision repair. J. Bacteriol. 142: 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A., Minko I. G., Harbut M. B., Finkel S. E., Goodman M. F., et al. , 2008. Replication bypass of interstrand cross-link intermediates by Escherichia coli DNA polymerase IV. J. Biol. Chem. 283: 27433–27437. 10.1074/jbc.M801237200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C., Cao H., Hearst J. E., Corash L., Luo H., et al. , 2008. Quantitative analysis of DNA interstrand cross-links and monoadducts formed in human cells induced by psoralens and UVA irradiation. Anal. Chem. 80: 8790–8798. 10.1021/ac801520m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley P. D., Brookes P., 1968. Cytotoxicity of alkylating agents towards sensitive and resistant strains of Escherichia coli in relation to extent and mode of alkylation of cellular macromolecules and repair of alkylation lesions in deoxyribonucleic acids. Biochem. J. 109: 433–447. 10.1042/bj1090433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin P. F., Bardwell E., Howard-Flanders P., 1977. Initiation of genetic exchanges in lambda phage–prophage crosses. Proc. Natl. Acad. Sci. USA 74: 291–295. 10.1073/pnas.74.1.291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Wang Y., 2013. A quantitative mass spectrometry-based approach for assessing the repair of 8-methoxypsoralen-induced DNA interstrand cross-links and monoadducts in mammalian cells. Anal. Chem. 85: 6732–6739. 10.1021/ac4012232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHugh P. J., Sones W. R., Hartley J. A., 2000. Repair of intermediate structures produced at DNA interstrand cross-links in Saccharomyces cerevisiae. Mol. Cell. Biol. 20: 3425–3433. 10.1128/MCB.20.10.3425-3433.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler M., 1986. DNA adducts of medicinal drugs: some selected examples. J. Cancer Res. Clin. Oncol. 112: 210–215. 10.1007/BF00395914 [DOI] [PubMed] [Google Scholar]
- Mogi S., Oh D. H., 2006. gamma-H2AX formation in response to interstrand crosslinks requires XPF in human cells. DNA Repair (Amst.) 5: 731–740. 10.1016/j.dnarep.2006.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogi S., Butcher C. E., Oh D. H., 2008. DNA polymerase eta reduces the gamma-H2AX response to psoralen interstrand crosslinks in human cells. Exp. Cell Res. 314: 887–895. 10.1016/j.yexcr.2007.10.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moolenaar G. F., Van Rossum-Fikkert S., Van Kesteren M., Goosen N., 2002. Cho, a second endonuclease involved in Escherichia coli nucleotide excision repair. Proc. Natl. Acad. Sci. USA 99: 1467–1472. 10.1073/pnas.032584099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D., Bessho T., Nechev L. V., Chen D. J., Harris T. M., et al. , 2000. DNA interstrand cross-links induce futile repair synthesis in mammalian cell extracts. Mol. Cell. Biol. 20: 2446–2454. 10.1128/MCB.20.7.2446-2454.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullins E. A., Warren G. M., Bradley N. P., Eichman B. F., 2017. Structure of a DNA glycosylase that unhooks interstrand cross-links. Proc. Natl. Acad. Sci. USA 114: 4400–4405. 10.1073/pnas.1703066114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K. C., Campellone K. G., Poteete A. R., 2000. PCR-mediated gene replacement in Escherichia coli. Gene 246: 321–330. 10.1016/S0378-1119(00)00071-8 [DOI] [PubMed] [Google Scholar]
- Newton K. N., Courcelle C. T., Courcelle J., 2012. UvrD participation in nucleotide excision repair is required for the recovery of DNA synthesis following UV-induced damage in Escherichia coli. J. Nucleic Acids 2012: 271453 10.1155/2012/271453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niedernhofer L. J., Odijk H., Budzowska M., Van Drunen E., Maas A., et al. , 2004. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol. Cell. Biol. 24: 5776–5787. 10.1128/MCB.24.13.5776-5787.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orren D. K., Selby C. P., Hearst J. E., Sancar A., 1992. Post-incision steps of nucleotide excision repair in Escherichia coli. Disassembly of the UvrBC-DNA complex by helicase II and DNA polymerase I. J. Biol. Chem. 267: 780–788. [PubMed] [Google Scholar]
- Pathak M. A., Fitzpatrick T. B., 1992. The evolution of photochemotherapy with psoralens and UVA (PUVA): 2000 BC to 1992 AD. J. Photochem. Photobiol. B 14: 3–22. 10.1016/1011-1344(92)85080-E [DOI] [PubMed] [Google Scholar]
- Peng X., Ghosh A. K., Van Houten B., Greenberg M. M., 2010. Nucleotide excision repair of a DNA interstrand cross-link produces single- and double-strand breaks. Biochemistry 49: 11–19. 10.1021/bi901603h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera A. V., Mendenhall J. B., Courcelle C. T., Courcelle J., 2016. Cho endonuclease functions during DNA interstrand cross-link repair in Escherichia coli. J. Bacteriol. 198: 3099–3108. 10.1128/JB.00509-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley S. M., Attfield P. V., Lloyd R. G., 1984. Repair of DNA double-strand breaks in Escherichia coli K12 requires a functional recN product. Mol. Gen. Genet. 195: 267–274. 10.1007/BF00332758 [DOI] [PubMed] [Google Scholar]
- Povirk L. F., Shuker D. E., 1994. DNA damage and mutagenesis induced by nitrogen mustards. Mutat. Res. 318: 205–226. 10.1016/0165-1110(94)90015-9 [DOI] [PubMed] [Google Scholar]
- Rangarajan S., Woodgate R., Goodman M. F., 1999. A phenotype for enigmatic DNA polymerase II: a pivotal role for pol II in replication restart in UV-irradiated Escherichia coli. Proc. Natl. Acad. Sci. USA 96: 9224–9229. 10.1073/pnas.96.16.9224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. J., Friedlos F., Scott D., Ormerod M. G., Rawlings C. J., 1986. The unique sensitivity of Walker rat tumour cells to difunctional agents is associated with a failure to recover from inhibition of DNA synthesis and increased chromosome damage. Mutat. Res. 166: 169–181. [DOI] [PubMed] [Google Scholar]
- Rothfuss A., Grompe M., 2004. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol. Cell. Biol. 24: 123–134. 10.1128/MCB.24.1.123-134.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman R. H., Clark A. J., 1977. The dependence of postreplication repair on uvrB in a recF mutant of Escherichia coli K-12. Mol. Gen. Genet. 155: 279–286. 10.1007/BF00272806 [DOI] [PubMed] [Google Scholar]
- Sancar A., 1996. DNA excision repair. Annu. Rev. Biochem. 65: 43–81. 10.1146/annurev.bi.65.070196.000355 [DOI] [PubMed] [Google Scholar]
- Sancar A., Rupp W. D., 1983. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell 33: 249–260. 10.1016/0092-8674(83)90354-9 [DOI] [PubMed] [Google Scholar]
- Sancar A., Tang M. S., 1993. Nucleotide excision repair. Photochem. Photobiol. 57: 905–921. 10.1111/j.1751-1097.1993.tb09233.x [DOI] [PubMed] [Google Scholar]
- Sarkar S., Davies A. A., Ulrich H. D., Mchugh P. J., 2006. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 25: 1285–1294. 10.1038/sj.emboj.7600993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sczepanski J. T., Jacobs A. C., Van Houten B., Greenberg M. M., 2009. Double-strand break formation during nucleotide excision repair of a DNA interstrand cross-link. Biochemistry 48: 7565–7567. 10.1021/bi901006b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki T., Nozu K., Kondo S., 1978. Differential causes of mutation and killing in Escherichia coli after psoralen plus light treatment: monoadducts and cross-links. Photochem. Photobiol. 27: 19–24. 10.1111/j.1751-1097.1978.tb07559.x [DOI] [PubMed] [Google Scholar]
- Setlow R. B., Swenson P. A., Carrier W. L., 1963. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science 142: 1464–1466. 10.1126/science.142.3598.1464 [DOI] [PubMed] [Google Scholar]
- Sharma S., Canman C. E., 2012. REV1 and DNA polymerase zeta in DNA interstrand crosslink repair. Environ. Mol. Mutagen. 53: 725–740. 10.1002/em.21736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Cole R. S., 1978. Repair of cross-linked DNA and survival of Escherichia coli treated with psoralen and light: effects of mutations influencing genetic recombination and DNA metabolism. J. Bacteriol. 136: 538–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek F. M., Melian A., Howard-Flanders P., 1989a Incision by UvrABC excinuclease is a step in the path to mutagenesis by psoralen crosslinks in Escherichia coli. Proc. Natl. Acad. Sci. USA 86: 3982–3986. 10.1073/pnas.86.11.3982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sladek F. M., Munn M. M., Rupp W. D., Howard-Flanders P., 1989b In vitro repair of psoralen-DNA cross-links by RecA, UvrABC, and the 5′-exonuclease of DNA polymerase I. J. Biol. Chem. 264: 6755–6765. [PubMed] [Google Scholar]
- Tessman J. W., Isaacs S. T., Hearst J. E., 1985. Photochemistry of the furan-side 8-methoxypsoralen-thymidine monoadduct inside the DNA helix. Conversion to diadduct and to pyrone-side monoadduct. Biochemistry 24: 1669–1676. 10.1021/bi00328a015 [DOI] [PubMed] [Google Scholar]
- Tomicic M. T., Aasland D., Naumann S. C., Meise R., Barckhausen C., et al. , 2014. Translesion polymerase eta is upregulated by cancer therapeutics and confers anticancer drug resistance. Cancer Res. 74: 5585–5596. 10.1158/0008-5472.CAN-14-0953 [DOI] [PubMed] [Google Scholar]
- Uranga L. A., Reyes E. D., Patidar P. L., Redman L. N., Lusetti S. L., 2017. The cohesin-like RecN protein stimulates RecA-mediated recombinational repair of DNA double-strand breaks. Nat Comms 8: 15282 10.1038/ncomms15282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Houten B., Gamper H., Holbrook S. R., Hearst J. E., Sancar A., 1986. Action mechanism of ABC excision nuclease on a DNA substrate containing a psoralen crosslink at a defined position. Proc. Natl. Acad. Sci. USA 83: 8077–8081. 10.1073/pnas.83.21.8077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sluis C. A., Moolenaar G. F., Backendorf C., 1983. Regulation of the uvrC gene of Escherichia coli K12: localization and characterization of a damage-inducible promoter. EMBO J. 2: 2313–2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vare D., Groth P., Carlsson R., Johansson F., Erixon K., et al. , 2012. DNA interstrand crosslinks induce a potent replication block followed by formation and repair of double strand breaks in intact mammalian cells. DNA Repair (Amst.) 11: 976–985. 10.1016/j.dnarep.2012.09.010 [DOI] [PubMed] [Google Scholar]
- Vare D., Johansson F., Persson J. O., Erixon K., Jenssen D., 2014. Quantification and repair of psoralen-induced interstrand crosslinks in human cells. Toxicol. Lett. 226: 343–350. 10.1016/j.toxlet.2014.01.019 [DOI] [PubMed] [Google Scholar]
- Venturini S., Tamaro M., Monti-Bragadin C., Bordin F., Baccicchetti F., et al. , 1980. Comparative mutagenicity of linear and angular furocoumarins in Escherichia coli strains deficient in known repair functions. Chem. Biol. Interact. 30: 203–207. 10.1016/0009-2797(80)90126-X [DOI] [PubMed] [Google Scholar]
- Wang S., Liu K., Xiao L., Yang L., Li H., et al. , 2016. Characterization of a novel DNA glycosylase from S. sahachiroi involved in the reduction and repair of azinomycin B induced DNA damage. Nucleic Acids Res. 44: 187–197. 10.1093/nar/gkv949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washburn B. K., Kushner S. R., 1991. Construction and analysis of deletions in the structural gene (uvrD) for DNA helicase II of Escherichia coli. J. Bacteriol. 173: 2569–2575. 10.1128/jb.173.8.2569-2575.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Ellis H. M., Lee E. C., Jenkins N. A., Copeland N. G., et al. , 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97: 5978–5983. 10.1073/pnas.100127597 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Strains and plasmids are available upon request. Supplemental material (comprising the following files: SupplementalFiguresAndTable.pdf contains Supplemental Material, Figure S1, Figure S2, Figure S3, Figure S4, Figure S5, and Table S1; Exp1WTleftUvrAright.tif; Exp2WTtopUvrAbottom.tif; and Exp3WTleftUvrAright.tif) available at Figshare: https://doi.org/10.25386/genetics.6847547.