Abstract
A recent genome-wide association study (GWAS) of central obesity identified 27 loci, from sex-combined analysis, associated with waist-to-hip ratio adjusted for body-mass index (WHRadjBMI) in European-ancestry individuals. Nevertheless, the identified variants may not be the biological causal ones due to the presence of linkage disequilibrium (LD). To better understand the mechanisms underlying the identified loci from the GWAS meta-analysis, we first imputed summary statistics at GWAS loci to increase genetic resolution, and then we applied a Bayesian statistical fine-mapping method through PAINTOR, incorporating LD structure and functional annotations to select and prioritize the most plausible causal variants across WHRadjBMI-associated regions. Using adipose tissue- and cell-specific annotations that showed significant associations with WHRadjBMI, we identified 33 single-nucleotide polymorphisms (SNPs) from 27 sex-combined fine-mapping loci with posterior probability of causality greater than 0.9. Six of the selected 33 SNPs belong to at least one of the top five identified annotations. SNPs rs1440372 (SMAD6) and rs12608504 (JUND) are particularly important since they not only have associated functional annotations but are also GWA hits in the original study. Incorporation of functional annotations helps identify additional plausible causal variants, such as rs2213731 (DNM3-PIGC) and rs4531856 (JUND), that did not reach genome-wide significance in GWAS. Our results provide promising candidates for future functional validation experiments.
Introduction
Central obesity is known to increase the risk and mortality of metabolic and cardiovascular disease in addition to body-mass index (BMI) [1–3]. Evidence has shown that genetic effects have important roles in body fat distribution and are different from those influencing BMI and overall adiposity [4]. Waist-to-hip ratio adjusting for BMI (WHRadjBMI), a proxy for central obesity, is a heritable trait with age-adjusted heritability ranging from 36 to 61% [5]. Although recent genome-wide association studies (GWAS) have identified many genetic variants associated with WHRadjBMI [6], these variants may not be the biological causal ones due to the presence of linkage disequilibrium (LD) [7]. This circumstance is an inherent drawback of the GWAS design and leads to a challenge that needs to be solved in the post-GWAS era [8].
Identification of the true causal variants for a complex trait is fundamental to understanding the biological mechanism underlying the trait. Fine-mapping studies to select and prioritize causal variants within GWAS-associated regions is a first step toward this goal [8–11]. Statistical methods for fine-mapping studies can roughly be divided into two classes. One is selecting causal variants based on association p-values such as p < 5 × 10−8 (standard genome-wide significance). The second is a Bayesian approach that assigns posterior probabilities of causality to each variant through a Bayes Factor [8]. The Bayesian method can also directly provide a quantitative way to incorporate prior biological knowledge such as pathway analysis and functional annotations into fine-mapping studies [12]. It has been demonstrated that the Bayesian approach of fine-mapping successfully helps refine GWAS risk loci and identifies causal variants with functional interpretation in several studies [9, 11]. A recent research effort demonstrates that more dense imputation at GWAS loci with fine-mapping and genomic annotation can provide insight into the functional and regulatory mechanisms on glycemic and obesity-related traits [13]. Hence, in this study we investigate the plausible causal variants associated with WHRadjBMI through fine-mapping, using imputation and annotation.
There are numerous statistical methods for fine-mapping with a variety of publicly available software packages. For instance, SNPTEST [14] and Bim-Bam [15] assess the association strength through a Bayesian framework based on individual level genotype data. Other approaches including CAVIAR [16], PAINTOR [17], and CAVIARBF [18] only require the marginal test statistics and LD information to conduct Bayesian analysis. In addition, toolkits like fgwas [19] and PAINTOR link prior biological knowledge of these variants to the Bayesian model through selecting the most phenotype-related annotations. Moreover, PAINTOR allows more than one causal variant within each locus of interest in calculating the posterior probability of causality for each single-nucleotide polymorphism (SNP), a more biologically tenable hypothesis.
We applied PAINTOR to recent GWAS summary statistic results for WHRadjBMI [6], incorporating LD structure and various types of functional annotation information to identify the most plausible causal variants across GWAS risk loci. Our objective in this study is to better understand the results of the WHRadjBMI GWAS meta-analysis, identify the SNPs that are most likely causal with functional interpretation, and then provide a list of candidate causal SNPs for future biological validation experiments.
Materials and methods
GWAS dataset
The summary statistics used in this paper came from a 2015 GWAS meta-analysis of association for WHRadjBMI released by the Genetic Investigation of Anthropometric Traits (GIANT) Consortium [6]. This meta-analysis includes up to 210,088 European-ancestry individuals, evaluating a total of 2,542,447 autosomal SNPs from HapMap imputed data and Metabochip genotyped data. We used the sex-combined results for the European-ancestry sample since the proportion of non-European-ancestry samples is relatively small in Shungin et al. [6]. The dataset consists of p-values for SNP associations with the WHRadjBMI trait, effect sizes and their standard errors. Z-scores were computed from effect sizes divided by their standard errors.
Further imputation and defining fine-mapping loci
The original available GIANT GWAS results for this analysis were based on HapMap Phase 2 imputation or directly genotyped data from Metabochip. To increase the resolution of our data, we utilized DIST (see Web Resources) to further impute the summary statistics corresponding to the variants available in the 1000 Genomes Phase 1 EUR population reference panel [20].
We identified 27 loci whose reported significances in GWAS were from sex-combined meta-analysis results in Shungin et al. [6]. We followed up each locus with fine-mapping. Each one is a 100 kb contiguous region of the genome centered on the GWA hit, the most significant SNP for the region in meta-analysis results for European ancestry. Although there is no agreed upon length to choose for each fine-mapping locus, previous studies have shown that a range of 10–30 kb for European population is relatively safe to include adequate information for LD [21]. Therefore, we conservatively chose a genome window of 100 kb with 50 kb on each side of the GWA hit to consider enough information from LD. The University of California Santa Cruz (UCSC) Genome Browser was used to locate the SNPs with missing positions. All SNPs were based on the UCSC hg19 assembly and sorted by their physical position within each locus.
Since we do not have individual level genotype data, we used PAINTOR utilities (see Web Resources) to estimate the LD structure between SNPs in each fine-mapping locus from the 1000 Genomes Phase 1 EUR population reference panel [20], consistent with the one used for imputation.
Functional annotations
It has been shown that integration of functional annotation data for SNPs can improve statistical fine-mapping performance [17, 19]. Various types of functional information were available for the purpose of fine-mapping. A total of 123 different annotations were used in this paper (Supplementary Table 1). Both general categorical annotations and tissue- and cell-specific functional annotations were included for our analysis.
The general annotations came from the Broad Institute (see Web Resources), including six primary categories: (1) coding, area overlaps exon, (2) untranslated region (UTR), a 5′ or 3′ untranslated region; (3) promoter, area within 2 kb of a transcription start site (TSS), (4) DNase I Hypersensitivity Sites (DHSs), regions that are sensitive to cleavage by DNase I enzyme and exposes the DNA for binding of transcription factors, (5) intron, area does not code proteins, and (6) intergenic, all other regions that lie between genes. In general, 88% of GWA hits lie in either intronic or intergenic regions, and only 2% of GWA hits belong to synonymous codes [22]. DHSs are important regulatory DNA regions. And many common disease and trait-associated variants identified by GWAS are more concentrated in DHSs, instead of lying in coding regions [23]. Gusev et al. reported that across 11 diseases DHS sites from 127 cell types spanned 16% of SNPs but explained an average of 79% trait heritability from imputed data [24]. Thus, recent studies tend to analyze associations between specific DHSs and their subclasses with known diseases.
We also combined 117 types of tissue- and cell-specific annotations from different resources, mainly from Roadmap Epigenomics Project [25], others from FANTOM5 project [26] and super-enhancers reported by Hnisz et al. [27]. Because the original GWA hits were shown to be significantly enriched in adipose tissue [6], we focused on adipose nuclei, mesenchymal stem cell-derived adipocyte, and adipose tissue-related regulatory and transcriptional features to human genome. Datasets of imputed and assayed histone modifications, adipose nuclei- and adipocyte-specific DHSs, and the core 15 chromatin states from Roadmap Epigenomics Project were selected. The Core 15-state model is based on the five core histone marks (H3K4me3, H3K4me1, H3K27me3, H3K9me3, and H3K36me3) across 127 epigenomes to capture the significant combinatorial interactions between different chromatin marks in their chromatin states [25]. Chromatin is a complex entity that controls gene expression and DNA replication. A cluster of histone modifications as a principal component of chromatin has a crucial role in DNA regulation [28]. Therefore, analysis of SNPs related to histone modifications helps us reveal the DNA regulation mechanism of traits. We also included datasets of adipose tissue-specific enhancers from FANTOM5 [26] and super-enhancers reported by Hnisz et al. [27]. Enhancers are DNA sequences that can bind transcription factors to increase the activity of promotor and transcription of genes [29]. Super-enhancers are large groups of transcriptional enhancers that control cell-specific gene expression [27].
Statistical analysis
We applied a Bayesian approach to prioritize causal variants in this fine-mapping study with PAINTOR software (Probabilistic Annotation Integrator version 3.0) (see Web Resources). The PAINTOR method is a probabilistic framework that integrates the association strength of genotype to phenotype with two independent sources of information, LD structure and functional annotation data to calculate the posterior probability to be a causal variant for each SNP across all fine-mapping loci [17]. In this method, the Z-scores for the SNPs at each fine-mapping locus are assumed to follow a multivariate normal distribution with LD as the covariance. Functional annotation data was introduced into the model through a logistic function. Let ϒ = (ϒ1, ϒ2, …, ϒk) be a vector of the effects of every functional annotation on the probability of causality. The Expectation Maximization (EM) algorithm is used to yield the maximum likelihood estimation over the effect size parameter ϒ for each annotation across all fine-mapping loci. Therefore, the effect of each annotation on the causal probability is determined by the data itself. A likelihood ratio test is applied to determine which annotation has a significant effect with p-value threshold of 0.05 on the probability of causality and then the algorithm chooses those significant annotations to estimate the probability of causality. Bayes theorem is implemented to compute posterior probabilities of causality for SNPs belonging to each causal configuration over the set of all possible causal configurations. Individual posterior probabilities for each SNP to be causal are obtained by marginalizing across all causal variants at each locus.
In our study, we first ran PAINTOR independently on different types of functional annotations and obtained effect size estimates ϒ and the sum of log-Bayes Factors (BFs). Let ϒ0 be the baseline estimate when there is no annotation at all. The prior causal probability for any SNP belonging to the kth annotation was estimated through ϒk in a logistic model. The log2 relative causal probability for kth annotation, a measure for annotation effect, is equal to the log2 of the ratio of prior causal probability between the kth annotation and baseline. We used log2 relative causal probability to compare the effects between different annotations on causal probability for SNPs. Since BFs are proportional to the full likelihood, the sum of log-BFs is used to construct a LRT to assess significance of annotation effect sizes. We then selected the top significant annotations (usually no more than five) to calculate the posterior probability for each SNP within our fine-mapping loci by PAINTOR.
Results
Fine-mapping loci for WHRadjBMI
The GWAS-identified 27 sex-combined loci located on 15 different chromosomes [6]. Each fine-mapping locus was defined as a 100 kb region surrounding the GWA hit in the center. There are 3374 SNPs from the original summary data within 27 fine-mapping loci. After using Direct Imputation of Summary Statistics (DIST) [30] to perform further imputation with the 1000 Genome as a reference panel, we obtained 10,725 SNPs for the sex-combined fine-mapping loci with an average (SD) of 397 (247) per locus (Table 1). Among all SNPs, 801 reach genome-wide significance (i.e., p < 5 × 10−8). There is great variability in the number of significant SNPs within each locus. For example, region RSPO3 has as many as 150 significant SNPs out of total 321 SNPs in contrast to region SPATA5-FGF2 which has only one SNP reaching the genome-wide significance.
Table 1.
Sex-combined fine-mapping loci for WHRadjBMI with different counts of SNPs related to imputation, original GWAS p-values, and final results calculated by PAINTOR
| Locus | Nearest gene | Chr | Index GWA hits rs ID (hg19 HGVS identifier) | Number of SNPs before imputation | Number of SNPs after DIST imputation | Number of SNPs achieving genome-wide significance | Number of SNPs Identified by PAINTOR |
|---|---|---|---|---|---|---|---|
| 1 | TBX15-WARS2 | 1 | rs2645294 chr1:g.119574587C>T | 161 | 365 | 55 | 2 |
| 2 | DNM3-PIGC | 1 | rs714515 chr1:g.172352990G>A | 93 | 236 | 51 | 1 |
| 3 | MEIS1 | 2 | rs1385167 chr2:g.66200648A>G | 162 | 431 | 14 | 1 |
| 4 | CALCRL | 2 | rs1569135 chr2:g.188115398A>G | 88 | 317 | 3 | 2 |
| 5 | PBRM1 | 3 | rs2276824 chr3:g.52637486C>G | 93 | 295 | 5 | 1 |
| 6 | LEKR1 | 3 | rs17451107 chr3:g.156797609T>C | 83 | 276 | 15 | 0 |
| 7 | SPATA5-FGF2 | 4 | rs303084 chr4:g.124066948G>A | 115 | 427 | 1 | 0 |
| 8 | CPEB4 | 5 | rs7705502 chr5:g.173320815G>A | 189 | 391 | 72 | 2 |
| 9 | FGFR4 | 5 | rs6556301 chr5:g.176527577G>T | 51 | 362 | 1 | 0 |
| 10 | LY86 | 6 | rs1294410 chr6:g.6738752T>C | 155 | 402 | 50 | 0 |
| 11 | BTNL2 | 6 | rs7759742 chr6:g.32381736T>A | 352 | 1586 | 19 | 2 |
| 12 | RSPO3 | 6 | rs1936805 chr6:g.127452116C>T | 182 | 321 | 150 | 1 |
| 13 | HOXA11 | 7 | rs10245353 chr7:g.25858614C>A | 131 | 276 | 84 | 2 |
| 14 | NFE2L3 | 7 | rs7801581 chr7:g.27223771C>T | 121 | 247 | 2 | 2 |
| 15 | MSC | 8 | rs12679556 chr8:g.72514228T>G | 157 | 379 | 5 | 2 |
| 16 | ABCA1 | 9 | rs10991437 chr9:g.107735920C>A | 131 | 569 | 7 | 0 |
| 17 | ITPR2-SSPN | 12 | rs10842707 chr12:g.26471364C>T | 44 | 371 | 52 | 1 |
| 18 | CCDC92 | 12 | rs4765219 chr12:g.124440110C>A | 68 | 284 | 116 | 2 |
| 19 | KLF13 | 15 | rs8042543 chr15:g.31708263C>T | 83 | 350 | 1 | 1 |
| 20 | RFX7 | 15 | rs8030605 chr15:g.56504598G>A | 134 | 385 | 2 | 1 |
| 21 | SMAD6 | 15 | rs1440372 chr15:g.67033151T>C | 121 | 329 | 2 | 1 |
| 22 | PEMT | 17 | rs4646404 chr17:g.17420199G>A | 82 | 304 | 10 | 1 |
| 23 | JUND | 19 | rs12608504 chr19:g.18389135A>G | 84 | 385 | 2 | 2 |
| 24 | CEBPA | 19 | rs4081724 chr19:g.33824946G>A | 64 | 392 | 33 | 2 |
| 25 | BMP2 | 20 | rs979012 chr20:g.6623374T>C | 142 | 303 | 13 | 2 |
| 26 | EYA2 | 20 | rs6090583 chr20:g.45558831A>G | 170 | 378 | 27 | 2 |
| 27 | ZNRF3 | 22 | rs2294239 chr22:g.29449477A>G | 118 | 364 | 9 | 0 |
| Total | 3374 | 10,725 | 801 | 33 |
We used PAINTOR utilities to calculate the LD correlation matrix for each locus using the 1000 Genome Phase 1 European (EUR) population [20] as the reference panel. Ambiguous SNPs that are not bi-allelic or do not match the reference panel were discarded. We also dropped SNPs whose positions cannot be identified. We finally had 7693 SNPs with 27 LD matrices corresponding to the sex-combined fine-mapping loci.
Integrating functional annotations
We evaluated general annotations and adipose tissue- and cell-specific annotations. For general annotations, we used data from the Broad Institute (https://data.broadinstitute.org/alkesgroup/). The six general categories of annotation cover all SNPs in our fine-mapping loci. About 80% of the SNPs lie in intronic and intergenic regions (Table 2). In order to know the effects of functional annotations on the causal probability for each SNP in the fine-mapping loci and to test whether these effects of annotations are statistically significant for our trait, we ran PAINTOR individually on each of the six annotations. None of these annotations has a great effect on the prior probability of causality for the SNPs residing in the fine-napping regions. The log2 relative causal probability, a measure for the annotation effect, ranges from −0.41 to 0.93. SNPs that belong to promoters are most likely to be causal. Through testing each functional dataset by the likelihood ratio test (LRT), we found that none of these annotations has a significant enrichment to improve information on the causal probability for the WHRadjBMI trait, indicating that more specific annotation information may be required to increase the accuracy of selecting the potential true causal variants by PAINTOR.
Table 2.
General functional annotations with frequencies, effect size estimates, and related p-values from likelihood ratio tests
| Annotation class | Frequency | Estimate of annotation effect (ϒk) | Prior probability | Log2 relative causal probability | LRT p-value |
|---|---|---|---|---|---|
| Coding | 0.95% | 0.28 | 5.34 × 10−3 | −0.41 | 0.84 |
| UTR | 1.07% | −0.23 | 9.45 × 10−3 | 0.43 | 0.81 |
| Promoter | 5.43% | −0.65 | 1.28 × 10−2 | 0.93 | 0.11 |
| DHSs | 25.50% | −0.36 | 9.08 × 10−3 | 0.51 | 0.17 |
| Intron | 38.70% | −0.03 | 7.19 × 10−3 | 0.05 | 0.89 |
| Intergenic | 43.10% | 0.27 | 5.41 × 10−3 | −0.38 | 0.24 |
We then extended the range of annotations to be tissue- and cell-specific. It has been reported that DNA regulatory regions are highly cell-specific and using more phenotype-related tissue-specific annotation can further improve the performance of signal prioritization in fine-mapping studies [23, 24]. We focused on annotation data related to adipocyte and adipose tissue for our phenotype of WHRadjBMI, evaluating each type separately. Among the 117 types of adipose nuclei, mesenchymal stem cell-derived adipocyte, and adipose tissue-specific annotations, six showed significant effects on the probability of causality as tested by the LRT individually, including histone modification H2AK9ac of adipose nuclei, histone modifications H2BK20ac and H2AK5ac, Genic Enhancers, DHSs, and strong transcription factors of mesenchymal stem cell-derived adipocyte (Table 3). We chose the top five annotations for our final model as recommended by PAINTOR, covering 16.2% of the SNPs in our fine-mapping loci. The effects of the top five annotations on the prior probability of causality are higher on average than those from general categorical annotations, ranging from 0.83 to 2.76 as evaluated by log2 relative causal probability and all these effects are positive, indicating that the SNPs lying in these selected five annotations are more likely to be causal variants. The histone modification H2AK9ac of adipose nuclei from imputed narrow peak data has the most significant improvement on causal probability with p-value of 7.85 × 10−4.
Table 3.
Significant annotations among 117 types of adipose related tissue- and cell-specific functional annotations with frequencies, effect size estimates, and related p-values from likelihood ratio tests
| Annotation class | Frequency | Estimate of annotation effect (ϒk) | Prior probability | Log2 relative causal probability | LRT p-value |
|---|---|---|---|---|---|
| Adipose Nuclei-H2AK9ac (imputed)a | 1.11% | −1.95 | 4.48 × 10−2 | 2.76 | 7.85 × 10−4 |
| Adipocyte-H2BK20ac (imputed)a | 1.56% | −1.79 | 3.84 × 10−2 | 2.53 | 3.99 × 10−3 |
| Adipocyte-EnhG (core 15-state model)a | 1.44% | −1.51 | 2.98 × 10−2 | 2.15 | 1.29 × 10−2 |
| Adipocyte-H2AK5ac (imputed)a | 3.88% | −0.95 | 1.71 × 10−2 | 1.36 | 3.37 × 10−2 |
| Adipocyte-DNase (imputed)a | 8.18% | −0.58 | 1.17 × 10−2 | 0.83 | 4.09 × 10−2 |
| Adipocyte-Tx (core 15-state model) | 4.51% | 4.29 | 1.01 × 10−4 | −6.19 | 4.27 × 10−2 |
a Top five annotations used in the final PAINTOR model
Identified SNPs with highest posterior probabilities
For the 27 fine-mapping loci, there are 33 SNPs with posterior probability of causality greater than 0.9 computed by PAINTOR (Supplementary Table 2). Nine of the fine-mapping loci have one selected SNP per locus, and another twelve loci have more than one selected variant likely to be causal per locus. The remaining six loci have no selected SNPs based on the posterior probability of causality >0.9. At four loci, the original reported GWA hits remained the lead SNP for that region as ranked by posterior probability: rs8042543 at KLF13; rs8030605 at RFX7; rs1440372 at SMAD6; and rs12608504 at JUND. Among the remaining selected SNPs, their original GWAS p-values are not always significant. Sixteen of the original p-values did not reach genome-wide significance and would likely be ignored in further analyses based on GWAS association p-values alone.
Six of the selected 33 SNPs belong to at least one of the five annotation categories, accounting for ~18% of the total selected SNPs (Table 4). SNPs rs1440372 at locus SMAD6 (Fig. 1) and rs12608504 at locus JUND are particularly important since they are not only located in several functional annotation groups but are also GWA hits in the original study. In contrast, SNP rs1884897 at locus BMP2 residing in an adipocyte DHS region has more than 99% probability to be causal even though it is not the index SNP in the original GWAS study. SNPs rs2213731 at DNM3-PIGC (Fig. 2) and rs4531856 at JUND are selected with posterior probability >0.9, although their original GWAS p-values did not reach genome-wide significance. In this circumstance, functional information has a major role in prioritizing them as the most likely causal variants. The locus JUND has more than one selected variant likely to be causal with high posterior probability and annotations.
Table 4.
Six selected SNPs lying in the top five annotations with posterior probabilities for causality >0.9
| rs ID | hg19 HGVS identifier | Nearest gene | Chr | Original p-value | Posterior probability | Annotations |
|---|---|---|---|---|---|---|
| rs1884897 | chr20:g.6612832A>G | BMP2 | 20 | 4.57 × 10−13 | >0.99 | DNase |
| rs2213731 | chr1:g.172359815C>A | DNM3-PIGC | 1 | 2.67 × 10−01 | >0.99 | H2AK5ac, DNase |
| rs13083798 | chr3:g.52649748A>G | PBRM1 | 3 | 6.68 × 10−12 | >0.99 | EnhG |
| rs1440372a | chr15:g.67033151T>C | SMAD6 | 15 | 1.34 × 10−10 | 0.99 | H2BK20ac, EnhG, H2AK5ac, DNase |
| rs12608504a | chr19:g.18389135A>G | JUND | 19 | 4.95 × 10−10 | 0.98 | H2AK9ac, H2AK5ac, DNase |
| rs4531856 | chr19:g.18388383C>T | JUND | 19 | 2.49 × 10−06 | 0.97 | DNase |
a Denotes the SNPs that are also original GWA hits
Fig. 1.
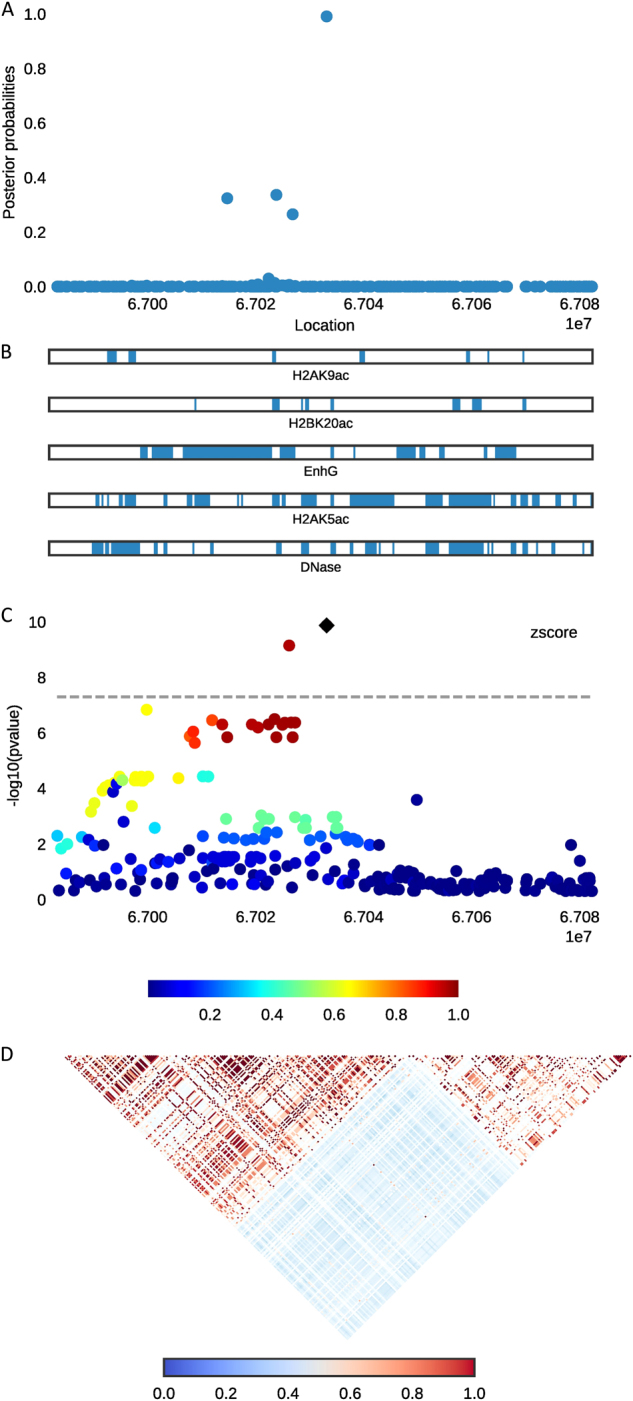
Combining regional plots of locus SMAD6 with variants’ scatterplot of posterior probabilities, annotation bars, scatterplot of original signals, and correlation heat-map of LD matrix. Top, a The posterior probability for each variant calculated by PAINTOR after integrating functional annotations and LD correlation. b The relative coverage and location of the top five selected annotations corresponding to the variants above. c A scatterplot that shows the original GWAS signals of each variant in −log10 (p-value) unit. Bottom, d The heat-map of LD structure estimated from 1000 Genome Phase 1 European population
Fig. 2.
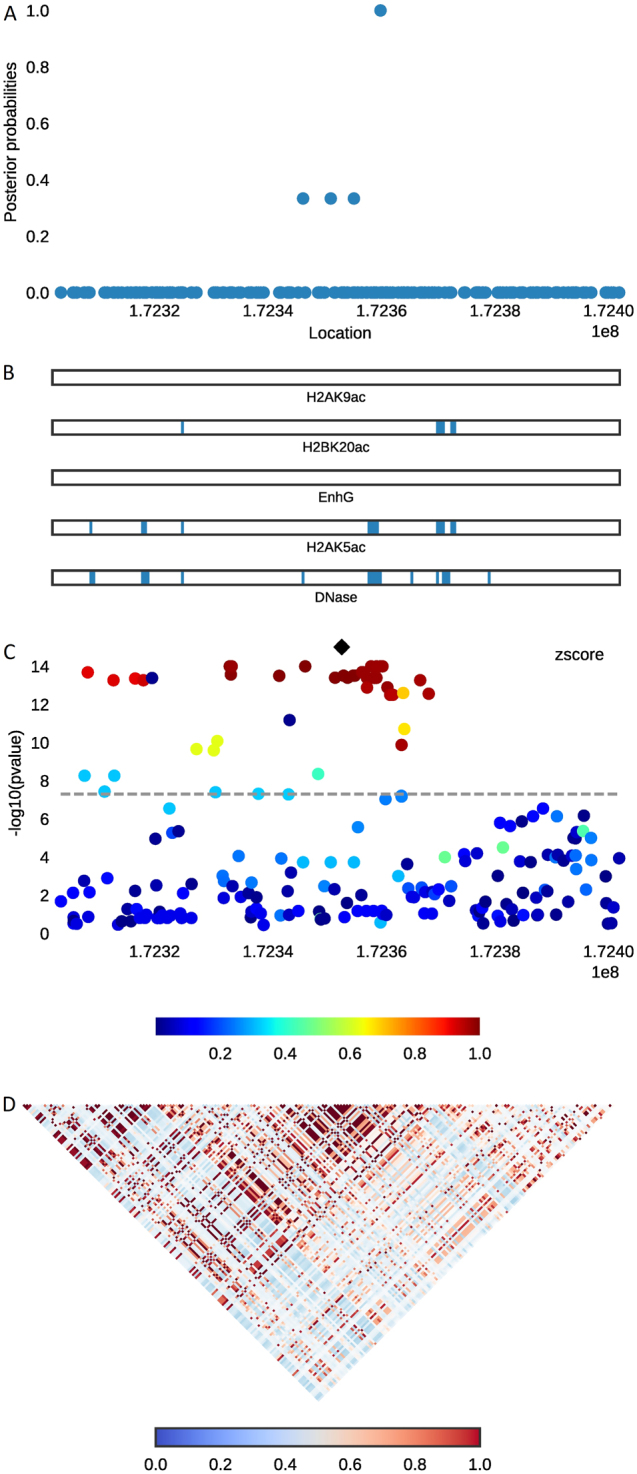
Combining regional plots of locus DNM3-PIGC with variants’ scatterplot of posterior probabilities, annotation bars, scatterplot of original signals, and correlation heat-map of LD matrix. Top, a The posterior probability for each variant calculated by PAINTOR after integrating functional annotations and LD correlation. b The relative coverage and location of top five selected annotations corresponding to the variants above. c A scatterplot that shows the original GWAS signals of each variant in −log10 (p-value) unit. Bottom, d The heat-map of LD structure estimated from 1000 Genome Phase 1 European population
Discussion
In this statistical fine-mapping study of central obesity loci, we performed a Bayesian approach using PAINTOR and incorporated functional annotations. We further refined the results from the original GWAS analysis and provided a more biologically meaningful set of SNPs with high probability to be causally associated with WHRadjBMI. Meanwhile, we also identified some variants whose GWAS association p-values did not reach genome-wide significance and some variants not included in the original HapMap or Metabochip meta-analysis data (Supplementary Table 2). Our identified variants are valuable for future functional validation experiments to assess the true trait-associated variants.
The Bayesian approach in PAINTOR only requires association signals from GWAS summary statistics and LD structure. It also can incorporate functional annotations directly to improve the accuracy of prioritization plausible causal variants. As shown in Table 2, we observed that general categorical functional annotations did not show significant effects on the probability of causality for our trait. A similar result was reported by Greenbaum et al. [31] in a fine-mapping study related to bone mineral density. We then extended the annotations to adipose tissue- and cell-specific since many studies revealed that DNA regulation regions are highly cell-specific [23, 24]. Several adipose tissue- and cell-specific annotations were identified by PAINTOR with significant enrichment and the top five were selected to calculate the posterior probabilities (Table 3). Therefore, we recommend choosing trait-associated tissue- and cell-specific functional annotation data when conducting a fine-mapping study.
In particular, integration of functional data into PAINTOR helps to identify additional plausible causal variants even when original GWAS p-values did not meet the genome-wide significant threshold. SNPs rs2213731 at DNM3-PIGC and rs4531856 at JUND, lying in at least one of the five selected annotations, were selected in this study with posterior probability >0.9, although their original GWAS p-values were greater than 5 × 10−8. They could be easily neglected by traditional GWAS analysis, especially when further studies are based on p-value alone.
Aside from functional data, selection of plausible causal variants by PAINTOR is also influenced by LD structure in each fine-mapping locus. Since we do not have access to individual level of genome data, all LD matrices were estimated based on the 1000 Genome Phase 1 EUR population reference panel [20]. We observed that some of our fine-mapping loci had strong regional association statistics and LD, consistent with results from a previous study [32]. This circumstance makes it difficult for PAINTOR to distinguish the true causal variants from a batch of similar tagging SNPs (i.e., similar z-scores and LD). It becomes even worse when no extra information such as functional annotations can be introduced into the model. For instance, the locus MEIS1 in our study has many tagging SNPs with similar Z-scores and LD structures, but with no annotations. Hence, the selected SNPs within this locus may not be very informative.
The six selected SNPs in Table 4 have posterior probability greater than 0.9 for causality, lying in at least one of the five selected annotations. Five candidate genes were identified. The top selected SNP rs1884897 is located at the DHS site for BMP2 of mesenchymal stem cell-derived adipocytes. BMP2 is a member of the transforming growth factor beta (TGF-β) superfamily of genes. High levels of bone morphogenetic protein 2 (BMP2) inhibit adipogenesis and induce chondrogenesis or osteogenesis on mesenchymal stem cells [33]. Another candidate gene SMAD6 is closely related to BMP2 [34]. The proteins encoded by SMAD6 negatively regulate BMPs and TGF-β-signaling, which influence the formation of adipose tissue [33]. Meanwhile, the identified SNP rs1440372 at locus SMAD6 is the index SNP in the original GWAS analysis, belonging to four functional classes of annotations, including histone modifications H2BK20ac and H2AK5ac, generic enhancer, and DHSs of adipocyte. Therefore, further study on this variant may be crucial to figuring out the biological mechanism of central obesity. Also, SMAD6 and JUND encode transcriptional regulators at WHRadjBMI loci, affecting differentiation and proliferation of adipocyte [6, 35]. SNP rs12608504 at locus JUND is also a GWA hit in the original study. In addition, the locus JUND has two selected variants likely to be causal with high posterior probability and annotations. Thus, further investigation of the gene JUND would be worthwhile to uncover the functional mechanisms of central obesity.
Since the objective of this study was to further understand the GWAS results, we identified our fine-mapping loci based on the reported GWA hits with 50 kb windows on both sides. Even though the length of our fine-mapping loci is relatively conservative in order to consider all plausible causal variants [21], there still may exist other causal variants outside our fine-mapping loci. Also, some LD in our fine-mapping loci may span further than 100 kb region, such as DNM3-PIGC, CALCRL, and CCDC92. As whole genome sequence (WGS) is more and more popular these days, future studies could take advantage of such data and enlarge the length of fine-mapping loci in order to include all possible true causal variants. We chose GWA hits based on the original sex-combined meta-analysis results for European ancestry reported by Shungin et al. [6]. We suggest further fine-mapping studies could focus on loci showing sex-specific or sex-interaction effects.
It is difficult for this statistical fine-mapping method to distinguish real causal variants among a set of SNPs with similar association signals and LD structure. Hence integration of extra information is critical. However, PAINTOR recommends no more than five annotations in the final model to help prioritize plausible causal variants. We believe that expanding the available number of annotations may improve the accuracy of selecting potential causal variants in the future studies. We did not adjust for multiple testing when choosing the top annotations as we regarded this work as leading to further research. Multiple-testing adjustment may be needed in future studies. Our study is also limited by using European data only. Due to a relatively small number of non-European samples included in Shungin et al. [6], we failed to impute the multi-ethic summary statistics. Future fine-mapping studies that incorporate all samples across different populations may gain more power.
In summary, we performed a Bayesian statistical fine-mapping study for WHRadjBMI with existing results from meta-analysis GWAS through PAINTOR. We further refined the results from original GWAS analysis and provided a list of biologically meaningful variants and genes with high probability of causality. Through integration of functional information, we also identified some additional plausible causal variants that may be neglected by traditional GWAS analysis. Overall, our results are valuable for future studies to determine the true causal variants and to further understand the mechanisms of central obesity.
Web resources
Genetic Investigation of Anthropometric Traits (GIANT) consortium data: http://portals.broadinstitute.org/collaboration/giant/index.php/GIANT_consortium_data_files
DIST: https://dleelab.github.io/dist/
PAINTOR: https://github.com/gkichaev/PAINTOR_V3.0
Annotation from Broad Institute: https://data.broadinstitute.org/alkesgroup/ANNOTATIONS/
Functional Annotations: https://github.com/gkichaev/PAINTOR_V3.0/wiki/2b.-Overlapping-annotations
Electronic supplementary material
Acknowledgements
We would like to thank Gleb Kichaev and Donghyung Lee for responses to questions related to PAINTOR and DIST software packages. And we also thank Virginia A. Fisher for technical assistance.
Funding
This research was in part support by the grant NIH R01 DK089256.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
The online version of this article (10.1038/s41431-018-0168-5) contains supplementary material, which is available to authorized users.
References
- 1.Haslam DW, James WPT. Obesity. Lancet. 2005;366:1197–209. doi: 10.1016/S0140-6736(05)67483-1. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Rimm EB, Stampfer MJ, Willett WC, Hu FB. Comparison of abdominal adiposity and overall obesity in predicting risk of type 2 diabetes among men. Am J Clin Nutr. 2005;81:555–63. doi: 10.1093/ajcn/81.3.555. [DOI] [PubMed] [Google Scholar]
- 3.Pischon T, Boeing H, Hoffmann K, et al. General and abdominal adiposity and risk of death in Europe. N Engl J Med. 2008;359:2105–20. doi: 10.1056/NEJMoa0801891. [DOI] [PubMed] [Google Scholar]
- 4.Heid IM, Jackson AU, Randall JC, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat Genet. 2010;42:949–60. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rose KM, Newman B, Mayer-Davis EJ, Selby JV. Genetic and behavioral determinants of waist-hip ratio and waist circumference in women twins. Obes Res. 1998;6:383–92. doi: 10.1002/j.1550-8528.1998.tb00369.x. [DOI] [PubMed] [Google Scholar]
- 6.Shungin D, Winkler TW, Croteau-Chonka DC, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–96. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Hum Mol Genet. 2015;24:R111–9. doi: 10.1093/hmg/ddv260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Onengut-Gumuscu S, Chen WM, Burren O, et al. Fine mapping of type 1 diabetes susceptibility loci and evidence for colocalization of causal variants with lymphoid gene enhancers. Nat Genet. 2015;47:381–6. doi: 10.1038/ng.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kote-Jarai Z, Saunders EJ, Leongamornlert DA, et al. Fine-mapping identifies multiple prostate cancer risk loci at 5p15, one of which associates with TERT expression. Hum Mol Genet. 2013;22:2520–8. doi: 10.1093/hmg/ddt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wellcome Trust Case Control C, Maller JB, McVean G, et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stephens M, Balding DJ. Bayesian statistical methods for genetic association studies. Nat Rev Genet. 2009;10:681–90. doi: 10.1038/nrg2615. [DOI] [PubMed] [Google Scholar]
- 13.Horikoshi M, Mgi R, van de Bunt M, et al. Discovery and fine-mapping of glycaemic and obesity-related trait loci using high-density imputation. PLoS Genet. 2015;11:e1005230. doi: 10.1371/journal.pgen.1005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 15.Servin B, Stephens M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 2007;3:e114. doi: 10.1371/journal.pgen.0030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hormozdiari F, Kostem E, Kang EY, Pasaniuc B, Eskin E. Identifying causal variants at loci with multiple signals of association. Genetics. 2014;198:497–508. doi: 10.1534/genetics.114.167908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kichaev G, Yang WY, Lindstrom S, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen W, Larrabee BR, Ovsyannikova IG, et al. Fine mapping causal variants with an approximate Bayesian method using marginal test statistics. Genetics. 2015;200:719–36. doi: 10.1534/genetics.115.176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am J Hum Genet. 2014;94:559–73. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Genomes Project C, Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- 22.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–7. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–5. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gusev A, Lee SH, Trynka G, et al. Partitioning heritability of regulatory and cell-type-specific variants across 11 common diseases. Am J Hum Genet. 2014;95:535–52. doi: 10.1016/j.ajhg.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH roadmap epigenomics mapping consortium. Nat Biotechnol. 2010;28:1045–8. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lizio M, Harshbarger J, Shimoji H, et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hnisz D, Abraham BJ, Lee TI, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pennacchio LA, Bickmore W, Dean A, Nobrega MA, Bejerano G. Enhancers: five essential questions. Nat Rev Genet. 2013;14:288–95. doi: 10.1038/nrg3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee D, Bigdeli TB, Riley BP, Fanous AH, Bacanu SA. DIST: direct imputation of summary statistics for unmeasured SNPs. Bioinformatics. 2013;29:2925–7. doi: 10.1093/bioinformatics/btt500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greenbaum J, Deng HW. A statistical approach to fine mapping for the identification of potential causal variants related to bone mineral density. J Bone Miner Res. 2017;32:1651–8. doi: 10.1002/jbmr.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu CT, Buchkovich ML, Winkler TW, et al. Multi-ethnic fine-mapping of 14 central adiposity loci. Hum Mol Genet. 2014;23:4738–44. doi: 10.1093/hmg/ddu183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skillington J, Choy L, Derynck R. Bone morphogenetic protein and retinoic acid signaling cooperate to induce osteoblast differentiation of preadipocytes. J Cell Biol. 2002;159:135–46. doi: 10.1083/jcb.200204060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takase M, Imamura T, Sampath TK, et al. Induction of Smad6 mRNA by bone morphogenetic proteins. Biochem Biophys Res Commun. 1998;244:26–9. doi: 10.1006/bbrc.1998.8200. [DOI] [PubMed] [Google Scholar]
- 35.Wang H, Scott RE. Adipocyte differentiation selectively represses the serum inducibility of c-jun and junB by reversible transcription-dependent mechanisms. Proc Natl Acad Sci USA. 1994;91:4649–53. doi: 10.1073/pnas.91.11.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


