Abstract
Background
Most nursing facilities (NFs) lack methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE) surveillance programs due to limited resources and high costs. We investigated the utility of environmental screening of high-touch surfaces in patient rooms as a way to circumvent these challenges.
Methods
We compared MRSA and VRE culture data from high-touch surfaces in patients’ rooms (14450 samples from 6 NFs) and ranked each site’s performance in predicting patient colonization (7413 samples). The best-performing sites were included in a MRSA- and a VRE-specific panel that functioned as a proxy for patient colonization. Molecular typing was performed to confirm available concordant patient-environment pairs.
Results
We identified and validated a MRSA panel that consisted of the bed controls, nurse call button, bed rail, and TV remote control. The VRE panel included the toilet seat, bed controls, bed rail, TV remote control, and top of the side table. Panel colonization data tracked patient colonization. Negative predictive values were 89%–92% for MRSA and 82%–84% for VRE. Molecular typing confirmed a strong clonal type relationship in available concordant patient-environment pairs (98% for MRSA, 91% for VRE), pointing to common epidemiological patterns for environmental and patient isolates.
Conclusions
Environmental panels used as a proxy for patient colonization and incorporated into facility surveillance protocols can guide decolonization strategies, improve awareness of MRSA and VRE burden, and inform efforts to reduce transmission. Targeted environmental screening may be a viable surveillance strategy for MRSA and VRE detection in NFs.
Keywords: MRSA, VRE, antimicrobial resistance, surveillance, nursing facilities
Surveillance for antimicrobial-resistant organisms in nursing facilities is critically important but challenging. We developed simple panels of environmental sites that can be used as a proxy for determining nursing facility patient methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus colonization.
In the United States, nursing facilities (NFs) currently host more patients than hospitals and often suffer from similar or higher prevalence of methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE) [1–5]. Colonized patients frequently shed MRSA and VRE into the environment [6–10], which can act as a source of contamination. Therefore, understanding the role of environmental contamination is important.
Infection prevention and surveillance programs in healthcare settings traditionally rely on screening patients at key body sites to identify candidates for isolation and/or decolonization. However, such programs are seldom implemented in special settings such as NFs due to unique and pervasive challenges, including but not limited to lack of resources, high rate of patient refusal, scheduling difficulties, cost, and inconvenience of sampling some key body sites. For example, perianal or rectal swabs are considered the optimal site for VRE screening, but patients often refuse to be swabbed [2]. For MRSA, the nares are the first screening choice and usually well accepted; however, additional sites may need to be sampled in order to define MRSA prevalence and strain diversity, including sensitive sites like groin and perianal/rectal areas [11–15]. This is especially true not only in patients with gastrointestinal (GI) diseases [15] but also for specific MRSA strains that preferentially colonize the GI tract, such as a recently described ST228 variant clone [8].
Environmental microbiological screening is simpler to perform and has logistic and economic advantages over patient screening in NFs, most important being the ability to consistently sample every object of interest over time as needed, independent of patient availability. Based on our extensive data on patient and environmental MRSA and VRE colonization collected over 3 years in 6 NFs in southeast Michigan, we sought to investigate the hypothesis that environmental colonization tracks patient colonization and, consequently, that panels of selected objects can be used to shed light on the likelihood of each patient being concurrently colonized. Such panels may be used by NFs with limited resources in lieu of surveillance screening as a proxy for patient colonization, to follow facility-wide prevalence trends, and to evaluate the effectiveness of interventions.
METHODS
Study Design
Six NFs participated in our parent study. Every newly admitted patient who consented to be in the study was included, regardless of comorbidities or other patient characteristics, unless they were receiving end-of-life care. Enrolled patients had samples collected at the time of enrollment, on day 14, and monthly thereafter for a maximum of 6 months (or until death or discharge) for outcome measurements. NFs were divided in 2 cohorts: a development cohort that consisted of 4 facilities (11117 environmental and 5614 body swabs collected from 515 patients) and a validation cohort used to test our model that consisted of 2 facilities (3333 environmental and 1799 body swabs collected from 136 patients). The facilities do not cohort patients based on known colonization status. Patients were recruited at the same time in both cohorts. Ten study visits in which the patient was not cultured were excluded from our analyses.
Specimen Collection and Laboratory Methods
At each visit, swabs (Bacti-swabs, Remel, Lenexa, Kansas) were used to obtain samples from nares, oropharynx, groin, perianal area, and dominant hand to assess MRSA and VRE colonization. During the same visit, environmental samples were obtained from high-touch surfaces in the patient’s room by trained research personnel. Bacti-swabs were used to swab a consistent surface area (approximately 43 cm2); a separate swab was used for each item, and the same technique was used every time. Based on a review of the acute care literature for surfaces likely to be contaminated, we choose screening sites that translate well to the typical NFs room [6, 7, 9]. Among these surfaces, the bed controls, bedside table (top and bottom), nurse call button, bed curtain, toilet seat, door knob, TV remote control, bed rail, and wheelchair handles were the most consistently available and therefore were selected as candidates for our panels. More than 1 patient may have been in each room. Environmental cleaning protocols in the 6 NFs did not change during our study [16].
Patients’ nares, oral cavity, groin, and perianal site swabs were cultured on mannitol salt agar (MSA) and bile-esculin plates with 6 μg/mL vancomycin (BEV6) on the same day. Hands and environmental swabs were enriched overnight in brain heart infusion broth at 36°C before culturing on MSA and BEV6 plates. Growth suggestive of staphylococci on MSA was tested by catalase and coagulase test (Staphaurex, Remel, Lenexa, Kansas), and S. aureus isolates were screened for methicillin resistance using cefoxitin disc diffusion according to Clinical and Laboratory Standards Institute criteria. Growth suggestive of VRE on BEV6 was confirmed by pyrrolidonyl arylamidase testing (DrySlide, BD, Franklin Lakes, New Jersey), and the species was identified by growth with arginine, mannitol, and arabinose.
Molecular Typing
For pathogens with high colonization prevalence, environmental and patient colonization can be due to independent events. In this scenario, distinct strains might be found during a visit. Because the aim of the present study was to use environmental samples as a proxy for colonization in the patient occupying the room, we were especially interested in uncovering patient and environmental colonization at the same visit due to the same strain of MRSA or VRE, rather than observing coincidental but independent events. To this end, we performed molecular typing on a representative sample of patient and environmental isolates in order to estimate how often positive concordant findings are due to direct patient shedding or environmental acquisition of a specific strain.
Pulsed-field gel electrophoresis (PFGE) was performed to determine the relatedness of MRSA and VRE isolates. Genomic DNA was prepared and digested with SmaI (New England BioLabs, Beverly, Massachusetts) using a previously described method [17, 18]. SmaI fragments were separated using a CHEF DR III apparatus (BioRad, Hercules, California) and compared using BioNumerics software (Applied Maths, Belgium). All MRSA isolates in this study were also compared to MRSA strains USA100-1100 as described by the Centers for Disease Control and Prevention (CDC) [17, 19]. Isolates were placed in the same pulsotype if their SmaI restriction patterns were ≥80% similar. Polymerase chain reaction (PCR) was performed to detect the Panton-Valentine leukocidin (PVL) toxin gene using a previously described method [20]. Multiplex PCR was performed to determine staphylococcal cassette chromosome mec (SCCmec) types I, II, III, IV, and V [21–23] and accessory gene regulator (agr) type I, II, III, and IV [24, 25].
Statistical Methods and Selection of MRSA- and VRE-Specific Environmental Panels
Microbiological data were analyzed using Stata 13 (StataCorp, College Station, Texas), previous creation of specific variables for colonization with MRSA, and colonization with VRE at 1 or more body sites for each patient visit. Frequency tables were generated to evaluate same-visit concordance between each environmental site (excluding scarcely represented sites) with patient colonization in the 4 development cohort facilities. Environmental sites were ranked based on area under the receiver operating characteristic curve (ROC) in order to obtain a single measure for evaluating both positive concordance and negative concordance between the reference variable (patient colonization) and the tested variable (environmental site colonization). For both MRSA and VRE, initial panels consisting of the 2 highest ranked sites were created and were deemed positive if 1 or more sites were positive and negative if all sites were negative. ROC, positive and negative concordance, positive predictive value (PPV, probability that the patient is colonized when the environmental panel is positive), and negative predictive value (NPV, probability that the patient is negative when the environmental panel is negative) were calculated. After addition of the third ranked site, the above-mentioned parameters were calculated again, and the process was repeated until the addition of the next best ranked site failed to improve the predictive value of the panel. Positive and negative likelihood ratios were also calculated for the final chosen panels [26]. To estimate the influence of multiple visits of the same patient on the statistical sample size, we also calculated ROC, positive and negative concordance, and PPV and NPV on the initial visit only for every patient. Multivariable generalized linear latent and mixed models [27] analysis was used to identify patient-level risk factors for environmental panel MRSA and VRE colonization.
RESULTS
Composition of Environmental Panels
A total of 14450 environmental samples and 7413 patient body samples were collected from 651 patients during 1619 visits (515 patients, 1228 visits, 11117 environmental swabs, 5614 body swabs for model facilities and 136 patients, 391 visits, 3333 environmental swabs, 1799 body swabs for validation facilities). The average number of body sites sampled per visit was 4.59 for model facilities and 4.57 for validation facilities. The prevalence of patient colonization was 17% for MRSA and 32% for VRE.
Analysis of single sites showed that the most predictive site for patient colonization in terms of ROC was the TV remote control (ROC value of 0.71 for MRSA, 0.67 for VRE) followed by the bed rail for MRSA and the toilet seat for VRE (Table 1). Predictably, positive concordance was low for single environmental sites but improved as additional sites were added to build a panel. For both MRSA and VRE, failure to improve the panel’s predictive value was reached when sites with individual ROC values of 0.60 or less were included. Therefore, the final panels consisted of all sites with ROC 0.61 or more. For MRSA, the following 4 objects were included: bed controls, nurse call button, TV remote control, and bed rail. For VRE, the following 5 objects were selected: bed controls, side table (top surface), toilet seat, TV remote control, and bed rail.
Table 1.
Concordance of Each Environmental Site With Patient Body Colonization
| Site | Receiver Operating Characteristic Curve | (95% Confidence Interval) | Rank | Positive Concordance (%) | Positive Predictive |Value (%) | Negative Concordance (%) | Negative Predictive Value (%) |
|---|---|---|---|---|---|---|---|
| Methicillin-resistant Staphylococcus aureus | |||||||
| TV remote control | 0.71 | 0.67–0.75 | 1 | 46 | 70 | 96 | 89 |
| Bed rail | 0.66 | 0.62–0.70 | 2 | 35 | 75 | 98 | 89 |
| Nurse call button | 0.64 | 0.60–0.67 | 3 | 29 | 77 | 98 | 87 |
| Bed controls | 0.62 | 0.59–0.65 | 4 | 27 | 74 | 98 | 87 |
| Side table (top) | 0.60 | 0.58–0.63 | 5 | 23 | 68 | 98 | 86 |
| Toilet seat | 0.60 | 0.56–0.63 | 5 | 25 | 44 | 94 | 87 |
| Side table (bottom) | 0.59 | 0.56–0.61 | 7 | 19 | 71 | 98 | 85 |
| Wheelchair handles | 0.56 | 0.53–0.58 | 8 | 14 | 55 | 97 | 84 |
| Bed curtain | 0.55 | 0.53–0.58 | 9 | 13 | 55 | 98 | 85 |
| Door knob | 0.55 | 0.53–0.58 | 9 | 13 | 55 | 98 | 84 |
| Vancomycin-resistant Enterococcus | |||||||
| TV remote control | 0.67 | 0.64–0.70 | 1 | 41 | 72 | 92 | 76 |
| Toilet seat | 0.65 | 0.62–0.68 | 2 | 46 | 57 | 84 | 77 |
| Bed rail | 0.65 | 0.62–0.67 | 2 | 33 | 82 | 96 | 74 |
| Bed controls | 0.63 | 0.61–0.66 | 4 | 30 | 80 | 96 | 74 |
| Side table (top) | 0.61 | 0.59–0.63 | 5 | 26 | 74 | 95 | 73 |
| Nurse call button | 0.60 | 0.58–0.62 | 6 | 24 | 78 | 97 | 73 |
| Side table (bottom) | 0.59 | 0.57–0.62 | 7 | 21 | 80 | 97 | 72 |
| Bed curtain | 0.59 | 0.56–0.61 | 7 | 27 | 56 | 91 | 74 |
| Wheelchair handles | 0.58 | 0.56–0.60 | 9 | 19 | 75 | 97 | 70 |
| Door knob | 0.55 | 0.53–0.57 | 10 | 13 | 68 | 97 | 70 |
Sites included in our environmental panels are highlighted in bold.
Abbreviation: TV, television.
Predictive Value of MRSA and VRE Panels
Our panels were tested on 4 facilities to determine overall performance. Two facilities were used to validate our model.
Analysis of 2 × 2 tables for the 4 facilities used to build our model showed a marked trend toward concordance between body and environmental panel colonization at the same visit. Overall values for positive concordance and likelihood ratio, negative concordance and likelihood ratio, and PPV and NPV of our MRSA and VRE panels are shown in Table 2. Results for the validation facilities are shown in Table 3. Concordance was more marked for MRSA, and NPV values were especially high.
Table 2.
Performance of Methicillin-Resistant Staphylococcus aureus and Vancomycin-Resistant Enterococcus Environmental Panels in Predicting Same-Visit Colonization of the Patient in Model Development Facilities
| Performance measurement | Methicillin-Resistant Staphylococcus aureus Panel | 95% CI | Vancomycin-Resistant Enterococcus Panel | 95% CI |
|---|---|---|---|---|
| Receiver operating characteristic curve | 0.78 | 0.74–0.81 | 0.70 | 0.68–0.73 |
| Positive concordance, % | 62 | 55–69 | 66 | 61–70 |
| Negative concordance, % | 93 | 92–95 | 75 | 72–78 |
| Positive predictive value, % | 66 | 60–72 | 56 | 53–59 |
| Negative predictive value, % | 92 | 91–93 | 82 | 80–84 |
| Positive likelihood ratio | 9.6 | 7.4–12.4 | 2.7 | 2.4–3.2 |
| Negative likelihood ratio | 0.40 | 0.34–0.48 | 0.45 | 0.39–0.52 |
Abbreviation: CI, confidence interval.
Table 3.
Performance of Methicillin-Resistant Staphylococcus aureus and Vancomycin-Resistant Enterococcus Environmental Panels in Predicting Same-Visit Colonization of the Patient in Validation Facilities
| Performance measurement | Methicillin-Resistant Staphylococcus aureus Panel | 95% CI | Vancomycin-Resistant Enterococcus Panel | 95% CI |
|---|---|---|---|---|
| Receiver operating characteristic curve | 0.71 | 0.65–0.77 | 0.72 | 0.68–0.77 |
| Positive concordance, % | 48 | 36–60 | 68 | 59–76 |
| Negative concordance, % | 93 | 90–96 | 77 | 71–82 |
| Positive predictive value, % | 61 | 49–72 | 58 | 52–64 |
| Negative predictive value, % | 89 | 87–91 | 84 | 80–87 |
| Positive likelihood ratio | 7.4 | 4.5–11.9 | 2.9 | 2.3–3.8 |
| Negative likelihood ratio | 0.56 | 0.44–0.70 | 0.42 | 0.32–0.54 |
Abbreviation: CI, confidence interval.
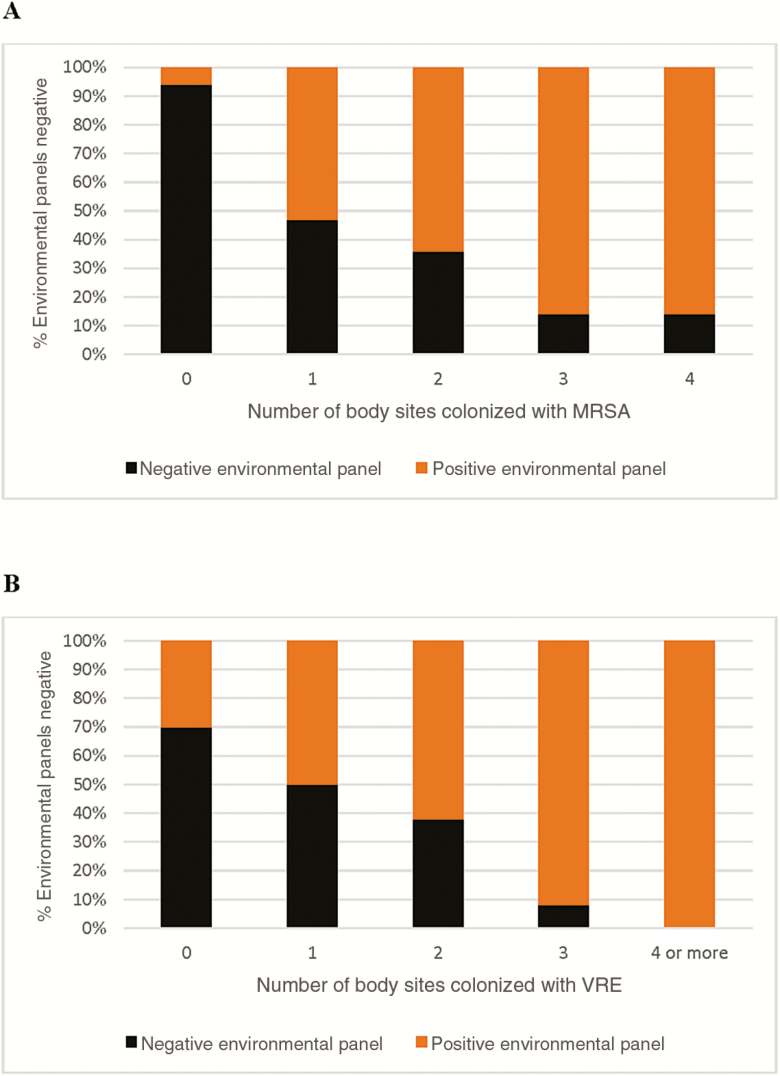
To estimate the impact of patient colonization burden on the likelihood of false-negative results, we also calculated the distribution of negative and positive environmental panel results according to the number of patient body sites found to be colonized (Figure 1). The percentage of false negatives decreased with the increase of patient colonization burden.
Figure 1.
Cumulative percent distribution of negative and positive environmental panel contamination results based on the number of patient body sites colonized at the same visit (logistic regression P < .001 for both methicillin-resistant Staphylococcus aureus (A) and vancomycin-resistant Enterococcus (B). Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; VRE, vancomycin-resistant Enterococcus.
To estimate the influence of multiple visits of the same patient on the statistical sample size, we also calculated ROC, positive and negative concordance, and PPV and NPV on the initial visit only for every patient (Supplementary Table 1). The results were similar, indicating that influence of repeated visits on the statistical sample size is minimal. We also report the performance of a small panel that included only the 3 sites that performed best for both MRSA and VRE as an example that may be of interest for facilities interested in screening both MRSA and VRE using the minimum amount of resources (Supplementary Table 2). For MRSA, the performance of such a panel was worse for model facilities and similar for validation facilities, while for VRE the PPV and PLR were better but NPV and NLR were worse in both model and validation facilities.
Molecular Typing, Same-Strain Association
A total of 354 MRSA strains from 76 patient visits (130 body samples and 244 environmental samples) were typed and divided into 11 type groups based on the unique combinations of PFGE pulsotype, SCCmec type, agr type, and presence/absence of PVL. Healthcare-associated MRSA accounted for 54% of strains, and strains considered as community-associated accounted for 33%. A total of 419 typed and confirmed VRE isolates from 80 patient visits (122 body samples and 297 environmental samples) showed a marked heterogeneity and were divided into 44 groups belonging to 5 species. Complete same-visit data with at least 1 strain isolated from the patient and at least 1 strain isolated from environmental sites included in our panels were available for 44 visits in the case of MRSA and 45 in the case of VRE.
For MRSA, 43 times out of 44 (98%) a strain type isolated from the patient exhibited a matching typing group with a strain isolated from 1 or more sites included in our environmental panel. For VRE, a type and species match was present 41 out of 45 times (91%). Figures 2 and 3 provide examples for comparison of multiple within-visit MRSA and VRE isolates obtained from sites included in our panels, and sites excluded from our panels.
Figure 2.
Molecular typing results of methicillin-resistant Staphylococcus aureus isolates from body and environmental sites within a single patient visit. A, Patient 14, facility 4, 14-day follow-up visit. B, Patient 17, facility 6, 14-day follow-up visit. The bed curtain is not part of the panel. All other sites referred to in this figure are part of the panel. Abbreviations: ID, infectious disease; MRSA, methicillin-resistant Staphylococcus aureus; TV, television; PFGE, pulsed-field gel electrophoresis; PVL, Panton-Valentine leukocidin.
Figure 3.
Molecular typing results of vancomycin-resistant Enterococcus (VRE) isolates from body and environmental sites within a single patient visit. A, Patient 50, facility 2, 60-day follow-up visit. B, Patient 4, facility 1, 14-day follow-up visit. Bedside table, bed curtain, and door knob are not part of the panel. All other sites referred to in this figure are part of the panel. Abbreviations: ID, infectious disease; PFGE, pulsed-field gel electrophoresis; TV, television; VREF, vancomycin-resistant Enterococcus faecium; VREFs, vancomycin-resistant Enterococcus faecalis.
DISCUSSION
Patient refusal, lack of resources, limited availability of dedicated trained personnel, and timing coordination issues are among the challenges that prevent many NFs from carrying out much-needed multidrug-resistant organism (MDRO) surveillance screening programs. Our panels would offer a dramatic improvement because it requires considerably fewer resources and allows flexibility and consistency at the same time. Furthermore, since the consent and presence of patients are not required, the predictable logistics of sample collection and laboratory workflow would save considerable personnel time. Also, a wider range of professional and support figures would be qualified to carry out part of the screening workload.
With this study, we provide a proof of concept and a real-life example that carefully selected environmental colonization panels correlate with patient MRSA and VRE colonization. This correlation was consistently verified across multiple facilities and reflects a preponderance of concurrent, same-strain co-colonization, rather than independent events. Our panels can be used to rule out patients unlikely to be colonized and to carry out facility-wide surveillance protocols and potentially could be used directly or with some modifications to monitor the effectiveness of interventions aimed at reducing patient’s colonization burden. In addition to directly benefiting patients, environmental panels can be a valuable tool to gauge the effectiveness of process and policy improvements, increase facility awareness of MRSA and VRE burden, and monitor changes in the MDRO ecology within the facility over time. The evaluation of the best performing sites to include in the panels is an important one. Because a certain degree of interfacility variation is inevitable, it is possible that different NFs may need different panel compositions depending on room and furniture characteristics and positioning, cleaning protocols, and similar variables. Future studies should address this issue in a larger sample of facilities.
In our case, several isolates from sites not included in the panels are unrelated to strains colonizing the patient (Figures 2B and 3B). These findings illustrate the importance of a correct choice of multiple sampling sites for environmental screening, especially in settings with high prevalence of the target organism. The performance of our panels may be affected by overall colonization burden, since it is known that a higher prevalence of the target condition will increase the positive predictive value of a test and decrease its negative predictive value. To this end, it is worth noting that VRE prevalence varies more by geographic location than MRSA prevalence [28].
Our study has strengths and limitations. The very close clonal type match in concordant patient-panel MRSA pairs is a very encouraging result, confirming the potential for this simple, indirect screening method. However, some typing techniques such as PFGE may underestimate the underlying strain heterogeneity in this pathogen [29]. To limit inclusion of unrelated strains in the same group, we performed typing based on 3 independent parameters in addition to restriction profiles. Notably, the better concordance for MRSA (98%) compared to VRE (91%) might be due to a higher prevalence of the latter in our facilities. Overall, these results confirm that when our panels are positive, they provide a direct link to the presence of the specific strain type in the patient, rather than a generally increased likelihood of colonization.
Because each of our environmental sites showed different degrees of positive and negative concordance in relation to patient colonization, our panel choice required striking a balance between those 2 desirable characteristics. For example, our current MRSA panel exhibited a poor positive concordance in validation facilities vs a very good negative concordance. Nevertheless, if needed, it is possible to choose a specific panel that focuses more on sensitivity or on specificity. We believe our observations may help in guiding informed choices toward such goals.
Most of our patients were housed in double rooms. However, most objects used in our panels were located close to each patient’s bed (eg, side table) or attached to it (eg, bed railing, nurse call button, bed controls). Also, each wheelchair was typically used by a specific patient. The toilet seat, on the contrary, is a site that study patients shared with other patients. Furthermore, any object can be touched by healthcare workers and visitors. Although our study does not focus on tracking transmission, it is important to verify the correlation within patients’ strains and environmental panel strains. Our molecular typing results attest to a clear relationship between patients and their immediate environment.
These environmental panels will likely not provide perfect accuracy in identifying colonized patients. The NPV of our panels may not be high enough for the test to be used in isolation if the goal is to diagnose the presence of impending and potentially lethal conditions, such as cancer or myocardial ischemia, just to name a few. However, the use of these panels is an exciting step for estimating the burden of MRSA and VRE within a facility and may identify high-risk patients who could serve as a reservoir for transmission. In turn, screening and, if needed, decolonization of high-risk patients may reduce the burden of infections and their sequelae in NFs and potentially in the community once the patients are discharged. Future interventional studies should investigate how use of environmental panels for colonization surveillance improves patient outcomes and cost-effectiveness of such an approach.
Notably, we were able to identify patient colonization characteristics, making them more likely to fall in the false-negative category. Indeed, the proportion of false negatives varied according to the burden of patient colonization expressed in terms of number of colonized body sites (Figure 1). For example, among 341 visits in which the patient was colonized with VRE, 136 yielded no VRE on the environmental panel. The majority of those (131) were colonized at 1 or 2 body sites, while only 5 were colonized at 3 or more sites. For MRSA, among the 154 visits in which the patient was colonized, 60 visits yielded a negative panel. In 49 of such instances, the patient was colonized at 1 or 2 body sites and only in 11 at 3 or more sites. Patients with a higher colonization burden may shed more heavily in the environment, thus explaining our observations. Further investigation into the relationships among body site colonization burden, environmental positivity, and transmission within a facility are needed.
Cleaning practices did not change during our study in any of the 6 facilities we followed. However, it is expected that they could change once facilities use the panels to monitor environmental colonization in real time. Therefore, consideration must be given to the interpretation of environmental panel results and the effectiveness of this strategy for detecting MRSA- and VRE-colonized patients if cleaning practices are altered in response to the results. Future investigations should align sampling times with environmental cleaning. Although only 6 facilities within a single metropolitan area were included in the study, they nevertheless represent diverse populations in terms of ethnicity, socioeconomic status, facility governance style and star rating, and referral hospital. We anticipate that our simple algorithm can allow selection of useful environmental panels in other NFs and long-term care settings, and that it can be scaled up.
In conclusion, targeted environmental screening could be a useful surveillance strategy to define the burden of MRSA and VRE in NFs and monitor efforts to reduce spread of resistance within facilities.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments. We thank the patients and their families who participated in this study.
Disclaimer. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention (CDC).
Financial support. This work was supported by the National Institutes of Health (RO1 AG041780 and K24 AG050685 to L. M.), the University of Michigan Pepper Center (MICHR PESC F039569 to M. C.), and the CDC (BAA 200-2016-91954 to L. M. and P. K. P.).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Harris-Kojetin L, Sengupta M, Park-Lee E, et al. Long-term care providers and services users in the United States: data from the National Study of Long-Term Care Providers, 2013–2014. Vital Health Stat 3 2016; x–xii: 1–105. [PubMed] [Google Scholar]
- 2. Mody L, Krein SL, Saint S, et al. A targeted infection prevention intervention in nursing home residents with indwelling devices: a randomized clinical trial. JAMA Intern Med 2015; 175:714–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murphy CR, Quan V, Kim D, et al. Nursing home characteristics associated with methicillin-resistant Staphylococcus aureus (MRSA) burden and transmission. BMC Infect Dis 2012; 12:269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ludden C, Cormican M, Vellinga A, Johnson JR, Austin B, Morris D. Colonisation with ESBL-producing and carbapenemase-producing Enterobacteriaceae, vancomycin-resistant enterococci, and meticillin-resistant Staphylococcus aureus in a long-term care facility over one year. BMC Infect Dis 2015; 15:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Benenson S, Cohen MJ, Block C, Stern S, Weiss Y, Moses AE; JIRMI Group Vancomycin-resistant enterococci in long-term care facilities. Infect Control Hosp Epidemiol 2009; 30:786–9. [DOI] [PubMed] [Google Scholar]
- 6. Sexton T, Clarke P, O’Neill E, Dillane T, Humphreys H. Environmental reservoirs of methicillin-resistant Staphylococcus aureus in isolation rooms: correlation with patient isolates and implications for hospital hygiene. J Hosp Infect 2006; 62:187–94. [DOI] [PubMed] [Google Scholar]
- 7. Boyce JM, Potter-Bynoe G, Chenevert C, King T. Environmental contamination due to methicillin-resistant Staphylococcus aureus: possible infection control implications. Infect Control Hosp Epidemiol 1997; 18:622–7. [PubMed] [Google Scholar]
- 8. Senn L, Clerc O, Zanetti G, et al. The stealthy superbug: the role of asymptomatic enteric carriage in maintaining a long-term hospital outbreak of ST228 methicillin-resistant Staphylococcus aureus. MBio 2016; 7:e02039–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dancer SJ. Importance of the environment in methicillin-resistant Staphylococcus aureus acquisition: the case for hospital cleaning. Lancet Infect Dis 2008; 8:101–13. [DOI] [PubMed] [Google Scholar]
- 10. Ostrowsky BE, Trick WE, Sohn AH, et al. Control of vancomycin-resistant enterococcus in health care facilities in a region. N Engl J Med 2001; 344:1427–33. [DOI] [PubMed] [Google Scholar]
- 11. Mody L, Kauffman CA, Donabedian S, Zervos M, Bradley SF. Epidemiology of Staphylococcus aureus colonization in nursing home residents. Clin Infect Dis 2008; 46:1368–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McKinnell JA, Huang SS, Eells SJ, Cui E, Miller LG. Quantifying the impact of extranasal testing of body sites for methicillin-resistant Staphylococcus aureus colonization at the time of hospital or intensive care unit admission. Infect Control Hosp Epidemiol 2013; 34:161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gibson KE, McNamara SE, Cassone M, Perri MB, Zervos M, Mody L; Targeted Infection Prevention Study Team, Ann Arbor, Michigan Methicillin-resistant Staphylococcus aureus: site of acquisition and strain variation in high-risk nursing home residents with indwelling devices. Infect Control Hosp Epidemiol 2014; 35:1458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meurman O, Routamaa M, Peltonen R. Screening for methicillin-resistant Staphylococcus aureus: which anatomical sites to culture?J Hosp Infect 2005; 61:351–3. [DOI] [PubMed] [Google Scholar]
- 15. Boyce JM, Havill NL, Otter JA, Adams NM. Widespread environmental contamination associated with patients with diarrhea and methicillin-resistant Staphylococcus aureus colonization of the gastrointestinal tract. Infect Control Hosp Epidemiol 2007; 28:1142–7. [DOI] [PubMed] [Google Scholar]
- 16. Saeb A, Mody L, Gibson K. How are nursing homes cleaned? Results of a survey of 6 nursing homes in Southeast Michigan. Am J Infect Control 2017; 45:e119–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, Tenover FC. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol 2003; 41:5113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Donabedian SM, Perri MB, Abdujamilova N, et al. Characterization of vancomycin-resistant Enterococcus faecium isolated from swine in three Michigan counties. J Clin Microbiol 2010; 48:4156–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tenover FC, McDougal LK, Goering RV, et al. Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J Clin Microbiol 2006; 44:108–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lina G, Piémont Y, Godail-Gamot F, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis 1999; 29:1128–32. [DOI] [PubMed] [Google Scholar]
- 21. Okuma K, Iwakawa K, Turnidge JD, et al. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J Clin Microbiol 2002; 40:4289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kondo Y, Ito T, Ma XX, et al. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother 2007; 51:264–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol 2005; 43:5026–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Witte W, Werner G, Cuny C. Subtyping of MRSA isolates belonging to a widely disseminated clonal group by polymorphism of the dru sequences in mec-associated DNA. Int J Med Microbiol 2001; 291:57–62. [DOI] [PubMed] [Google Scholar]
- 25. Strommenger B, Cuny C, Werner G, Witte W. Obvious lack of association between dynamics of epidemic methicillin-resistant Staphylococcus aureus in central Europe and agr specificity groups. Eur J Clin Microbiol Infect Dis 2004; 23:15–9. [DOI] [PubMed] [Google Scholar]
- 26. McGee S. Simplifying likelihood ratios. J Gen Intern Med 2002; 17:646–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rabe-Hesketh S, Skrondal A. Classical latent variable models for medical research. Stat Methods Med Res 2008; 17:5–32. [DOI] [PubMed] [Google Scholar]
- 28. Montoya A, Cassone M, Mody L. Infections in nursing homes: epidemiology and prevention programs. Clin Geriatr Med 2016; 32:585–607. [DOI] [PubMed] [Google Scholar]
- 29. Enright MC, Robinson DA, Randle G, Feil EJ, Grundmann H, Spratt BG. The evolutionary history of methicillin-resistant Staphylococcus aureus (MRSA). Proc Natl Acad Sci U S A 2002; 99:7687–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.