Abstract
Rationale: DSP (desmoplakin), the most abundant component of desmosomes, which maintain the mechanical integrity of epithelium, is a genome-wide association study–identified genetic risk locus in human idiopathic pulmonary fibrosis (IPF). Subjects with IPF express a significantly higher level of DSP than control subjects.
Objectives: Determine potential mechanisms by which DSP is regulated in lung fibrosis.
Methods: Matrigel-coated soft and stiff polyacrylamide gels were made to simulate the stiffness of normal and fibrotic lungs. Quantitative chromatin immunoprecipitation and electrophoretic mobility shift assay were used to evaluate transcription factor binding to the DSP promoter. Targeted DNA methylation was achieved by CRISPR (clustered regularly interspaced short palindromic repeats)/dCas9 (deactivated CRISPR-associated protein-9 nuclease)–mediated Dnmt3A (DNA methyltransferase 3A) expression under the guidance of sequence-specific single guide RNAs.
Measurements and Main Results: Stiff matrix promotes DSP gene expression in both human and rodent lung epithelial cells as compared with soft matrix. A conserved region in the proximal DSP promoter is hypermethylated under soft matrix conditions and becomes hypomethylated/demethylated under stiff matrix conditions. Demethylation of this conserved DSP promoter region is associated with transactivation of transcription factor EGR1 (early growth response protein 1), resulting in EGR1-dependent DSP overexpression. Targeted DNA methylation by CRISPR/dCas9/Dnmt3A–mediated epigenome editing blocks EGR1 binding to the DSP promoter and inhibits stiff matrix–induced DSP overexpression.
Conclusions: DSP is a matrix stiffness–regulated mechanosensitive gene. CRISPR/dCas9-Dnmt3A–mediated epigenome editing reverses DSP overexpression by reestablishment of the epigenetic control of DSP under the mechanically homeostatic environment. It provides a useful tool for investigations of the functional role of DSP in the pathogenesis of lung fibrosis.
Keywords: matrix stiffness, desmoplakin, CRISPR/dCas9, epigenome editing, lung fibrosis
At a Glance Commentary
Scientific Knowledge on the Subject
DSP (desmoplakin) gene variants have been associated with idiopathic pulmonary fibrosis in multiple genome-wide association studies. The functional role of DSP in lung fibrosis remains largely unknown.
What This Study Adds to the Field
We demonstrated that matrix stiffness, a mechanical factor relevant to lung fibrosis, regulates DSP gene expression by an epigenetic mechanism involving alteration of DNA methylation in the DSP promoter. Targeted DNA methylation by CRISPR (clustered regularly interspaced short palindromic repeats)/dCas9 (deactivated CRISPR-associated protein-9 nuclease)–mediated epigenome editing effectively reverses stiff matrix–induced DSP overexpression. Our work provides novel mechanistic insights into the regulation of DSP by mechanical signals from the extracellular matrix and demonstrates that CRISPR/dCas9–mediated epigenome editing is a useful tool for further investigations of the functional role of DSP in lung fibrogenesis.
Idiopathic pulmonary fibrosis (IPF) is the most common and severe idiopathic interstitial pneumonia, with a median survival of 2 to 3 years after diagnosis (1). The precise etiology of IPF remains elusive. Both genetic susceptibility and environmental toxins are likely associated with the risk of this disease (2, 3). Evidence for the genetic etiology of IPF includes identification of genetic risk loci and specific risk alleles associated with the disease (4–7).
Genome-wide association studies (GWASs) have identified the DSP (desmoplakin) gene, located at chromosome 6p24, as a significant locus associated with IPF (5, 7–9). Protein encoded by the DSP gene is the most abundant component of desmosomes, a type of intercellular junction responsible for maintaining the structural integrity and mechanical stability of the epithelium. DSP links desmosomal plaques to the intermediate filament network at the intracellular side of desmosomes. Patients with IPF express significantly higher levels of DSP in lung tissues than healthy human control subjects, and expression of DSP differs most significantly by genotype at rs2076295, located in intron 5 of the gene (TT > GT > GG) (5, 7). The mechanisms by which DSP is upregulated in IPF are currently unknown. Understanding the regulation of the DSP gene in lung fibrosis will shed light on further investigations of the functional role of DSP in the pathogenesis of IPF.
Stiffening of the extracellular matrix (ECM) is a prominent feature of human IPF and experimental lung fibrosis in animals (10, 11). Previous studies have shown that stiff matrix promotes fibrogenic phenotypes of lung fibroblasts, including proliferation, myofibroblast differentiation, resistance to apoptosis, and invasion into the basement membrane (BM) (11–15). Unlike fibroblasts, which sense interstitial ECM stiffness through direct contacts, lung epithelial cells are separated from the interstitial ECM by an amorphous, sheet-like, and thin BM that is compliant in nature (16). Currently, there is no direct evidence whether or not lung epithelial cells are capable of sensing stiffened interstitial lung ECM or whether the BM itself stiffens in lung fibrosis. Transmission electron microscopic studies have shown that type II alveolar epithelial cells (AEC II) develop the cytoplasmic processes that penetrate the BM and extended into the lung interstitium (17). AEC II may use their foot processes to directly sense the stiffness of interstitial lung ECM. It has been reported that cells can sense a rigid base that is 10 to 20 μm beneath them (18, 19). The threshold thickness for how deeply cells sense an underlying “hidden” matrix has been defined to be 3.4 μm (18). The alveolar BM has an average thickness of less than 0.1 μm (17), which is an order of magnitude thinner than the threshold thickness that cells can sense. This suggests that alveolar epithelial cells are capable of sensing the rigidity of stiffened fibrotic lung ECM despite the presence of the alveolar BM.
CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein-9 nuclease) is a newly developed, state-of-the-art gene-editing technology. The genome-wide, site-specific gene editing is achieved by single guide RNA (sgRNA)-mediated DNA targeting followed by Cas9 endonuclease–mediated DNA cleavage (20). CRISPR/Cas9 technology is highly versatile and can be repurposed for a variety of new applications. For example, mutations made in the catalytic domain of Cas9 create a deactivated version of Cas9 (dCas9), which preserves the ability of Cas9 to bind DNA at the site defined by sgRNA but no longer cut DNA. The modified version of CRISPR/dCas9 can be used as a DNA-binding platform for generating various chimeric versions of dCas9 fused with transcriptional activators/repressors for targeted gene activation/silencing or with epigenetic modifiers for targeted epigenome editing (21–23).
In this study, we investigated the potential effects of matrix stiffness on the regulation of DSP expression in lung epithelial cells. We developed CRISPR/dCas9–mediated epigenome editing to reverse stiff matrix–induced DSP overexpression by epigenetic reprogramming of DSP.
Some of the results of these studies have been previously reported in the form of an abstract (24).
Methods
Statistical Analysis
Statistical analysis was conducted using GraphPad Prism 7.02 software. Values of P <0.05 were considered significant. More detailed information is provided in the online supplement.
Additional detail on the methods is provided in the online supplement.
Results
Stiff Matrix Promotes DSP/Dsp mRNA and Protein Expression in Human, Mouse, and Rat Lung Epithelial Cells
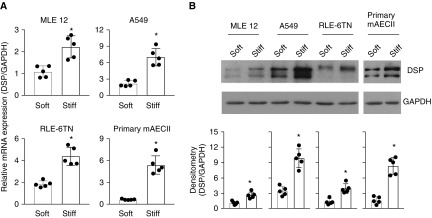
To determine the potential effect of matrix stiffness on DSP expression, we cultured human alveolar epithelial adenocarcinoma cells (A549), primary AEC II isolated from wild-type C57BL6 mice, transformed mouse lung epithelial cells, and transformed rat AEC II on soft (1 kPa) and stiff (20 kPa) polyacrylamide hydrogel matrix substrates, the stiffness grades of which were within the range of normal and fibrotic lungs, respectively (10, 11). We observed that lung epithelial cells cultured on stiff matrix expressed significantly higher levels of DSP/Dsp mRNA and protein than cells cultured on soft matrix, regardless of the origin (human vs. rodent) and state (normal/primary vs. transformed/cancerous) of the cells (Figures 1A and 1B). Stiffness-regulated DSP expression was confirmed in a newly developed, fibroblast-derived ECM system (25) (see Figure E1 in the online supplement). These data suggest that DSP, a GWAS-identified risk allele of IPF, is a matrix stiffness–regulated mechanosensitive gene. Our findings provide a potential mechanism for DSP overexpression in IPF through matrix stiffening.
Figure 1.
Stiff matrix upregulates DSP (desmoplakin) expression in lung epithelial cells. Human and rodent lung epithelial cells were cultured on soft and stiff polyacrylamide gels for 48 hours. (A) Relative mRNA levels of DSP were determined by quantitative PCR (qPCR). (B) Protein levels of DSP were determined by immunoblot. GAPDH was used as reference/loading control. Results are mean ± SD of five independent experiments, each qPCR reaction performed in triplicate. *P < 0.05. A549 = human alveolar epithelial adenocarcinoma cells; mAECII = mouse type II alveolar epithelial cells; MLE = mouse lung epithelial cells; RLE-6TN = transformed rat type II alveolar epithelial cells.
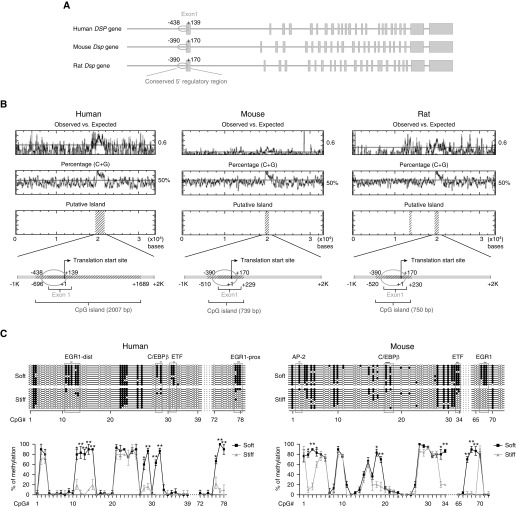
Stiff Matrix Promotes DNA Demethylation in a Conserved 5′ Regulatory Region in Human and Mouse DSP/Dsp Genes
Matrix stiffness regulates both human and rodent DSP/Dsp expression (Figure 1), suggesting a common mechanism involved in mechanoactivation of DSP/Dsp genes. Analysis of DNA sequences revealed a poor overall DNA homology (<3%) in a 20-kb 5′ regulatory region upstream of the translation start site of human and rodent DSP/Dsp genes. However, a high level of DNA homology (78%) was found in a 560-bp region immediately upstream of the translation start site that includes untranslated and a portion of translated exon 1 region (Figure 2A). Furthermore, this 560-bp conserved region is enriched by methylation-prone CpG dinucleotides, and it is the only CpG-rich fragment in the region of the 20-kb 5′ regulatory sequences, exon 1 and intron 1 (except that rat Dsp gene contains an additional small CpG-rich sequence upstream of the 560-bp conserved region) (Figure 2B). Sodium bisulfite sequencing demonstrated that a group of CpG islands in the conserved 560-bp region of human and mouse DSP/Dsp were steadily methylated under soft matrix conditions, but hypomethylated/demethylated under stiff matrix conditions (Figure 2C). The results suggest that matrix stiffness regulates DNA methylation status in the conserved 5′ regulatory region of DSP/Dsp genes. Analysis for the potential binding elements of transcription factors revealed that stiffness-sensitive CpG islands are associated with the binding sites of transcription factor EGR1 (early growth response protein 1), C/EBPβ (CCAAT/enhancer-binding protein β), ETF (EGFR [epidermal growth factor receptor]-specific transcription factor), and AP-2 (activator protein-2) in the conserved 5′ regulatory region (Figure 2C).
Figure 2.
Stiff matrix promotes DNA demethylation in a conserved 5′ regulatory region in human and mouse DSP/Dsp (desmoplakin) genes. (A) Schematic diagram of human, mouse, and rat DSP/Dsp genes. The conserved 560-bp DNA sequences (78% homology) are indicated by ovals. Columns represent DSP/Dsp exons. (B) Analysis of CpG dinucleotides in a 30+-kb region, including the 20-kb 5′ regulatory region, exon 1, and intron 1. CpG-rich regions are indicated by areas marked with diagonals. (C) DNA methylation profiling analysis for the 560-bp conserved region in human alveolar epithelial adenocarcinoma cells and mouse primary type II alveolar epithelial cells cultured on soft and stiff matrix. Ten clones were sequenced for each condition. Solid circles indicate methylated CpG islands. Open circles indicate unmethylated CpG islands. Results are mean ± SD of three independent experiments. *P < 0.05; **P < 0.01. AP-2 = activator protein-2; C/EBPβ = CCAAT/enhancer-binding protein β; dist = distal; EGR1 = early growth response protein 1; ETF = EGFR [epidermal growth factor receptor]-specific transcription factor; prox = proximal.
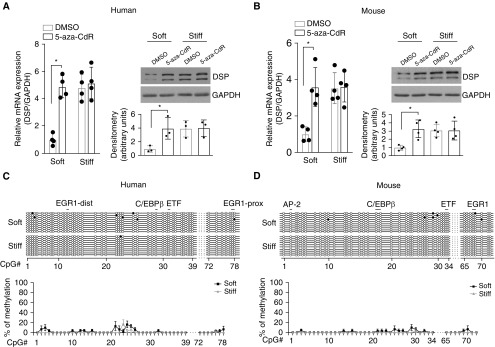
Inhibition of DNA Methyltransferase Activity Promotes Human and Mouse DSP/Dsp Expression on Soft Matrix
To assess whether matrix stiffness–dependent DSP/Dsp expression is regulated by DNA methylation, we cultured human A549 and primary mouse AEC II on soft and stiff matrix substrates in the presence or absence of 5-aza-2′-deoxycytidine, a potent DNA methyltransferase inhibitor. Inhibition of DNA methyltransferase activity increased levels of DSP/Dsp mRNA and protein in human and mouse lung epithelial cells cultured on soft matrix, whereas no significant changes in DSP/Dsp expression were observed in cells cultured on stiff matrix (Figures 3A and 3B). Sodium bisulfite sequencing confirmed DNA demethylation in the conserved 560-bp region after 5-aza-2′-deoxycytidine treatment in human and mouse lung epithelial cells cultured on both soft and stiff matrix substrates (Figures 3C and 3D). Together, these data suggest that matrix stiffness regulates DSP expression by a mechanism involved in alterations of DNA methylation.
Figure 3.
Inhibition of DNA methyltransferase promotes DSP (desmoplakin)/Dsp expression on soft matrix. (A) Human (A549) and (B) mouse (primary mouse type II alveolar epithelial cells) lung epithelial cells were cultured on soft and stiff matrix in the presence or absence of 5 mM 5-aza-2′-deoxycytidine (5-aza-CdR) for 48 hours. Levels of DSP/Dsp mRNA and protein were determined by quantitative PCR (qPCR) and immunoblot analysis. GAPDH/Gapdh were used as reference/loading controls. DMSO is a solvent of 5-aza-CdR. (C and D) DNA methylation profiles in the conserved 560-bp regulatory region were determined by sodium bisulfite sequencing. Ten randomly picked DNA clones were sequenced for each condition. Solid circles indicate methylated CpG islands. Open circles indicate unmethylated CpG islands. Results are mean ± SD of three or four independent experiments, each qPCR reaction performed in triplicate. *P < 0.05. AP-2 = activator protein-2; C/EBPβ = CCAAT/enhancer-binding protein β; dist = distal; EGR1 = early growth response protein 1; ETF = EGFR [epidermal growth factor receptor]-specific transcription factor; prox = proximal.
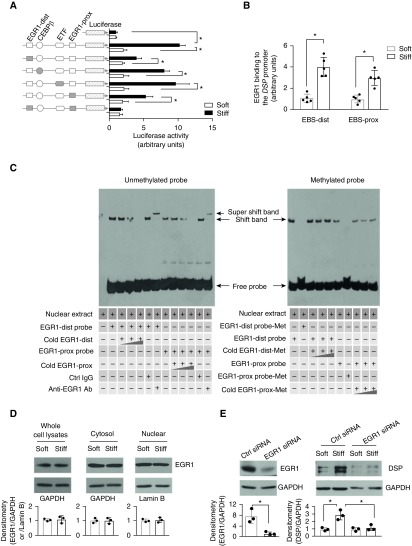
Stiff Matrix–induced DSP Overexpression Is Associated with EGR1-mediated Transactivation of the DSP Promoter
We cloned the 560-bp DNA fragment of the conserved, CpG-rich regulatory region in human DSP and made deletion mutations to the binding sequences of both proximal and distal EGR1, C/EBPβ, and ETF. Luciferase-based promoter reporter assay demonstrated that this conserved regulatory region had the promoter activity, and the promoter activity was regulated by matrix stiffness (Figure 4A). Deletions of the proximal, the distal, or both EGR1 binding elements inhibited stiff matrix–induced promoter activation, whereas deletions of the C/EBPβ- and the ETF-binding elements did not (Figure 4A). These findings suggest that stiff matrix activates the DSP promoter and that activation requires the binding of transcription factor EGR1 to the DSP promoter. Quantitative chromatin immunoprecipitation assay showed that nuclear EGR1 bindings to the proximal and the distal EGR1 binding sites (EBSs) were increased on stiff matrix as compared with soft matrix (Figure 4B), suggesting that transactivation of transcription factor EGR1 may be involved in stiff matrix–induced DSP promoter activation.
Figure 4.
EGR1 (early growth response protein 1) transactivation is associated with stiff matrix–induced DSP (desmoplakin) overexpression. (A) Schematic depicts the wild-type (open shapes) and mutated (solid shapes) 560-bp regulatory sequences. Promoter activity was determined by a luciferase-based assay. (B) The bindings of EGR1 to the distal and proximal EGR1 binding sites (EBS) under soft versus stiff matrix conditions were measured by quantitative chromatin immunoprecipitation. (C) The bindings of nuclear EGR1 from human alveolar epithelial adenocarcinoma cells with unmethylated or methylated DSP promoter probes were determined by electrophoretic mobility shift assay. (D) Levels of EGR1 in the whole-cell lysates, cytoplasmic fraction, and nuclear fraction were determined by immunoblot. GAPDH and lamin B were used as loading controls for the cytoplasmic and nuclear proteins, respectively. (E) Knockdown of EGR1 by siRNA was confirmed by immunoblot. Effects of EGR1 knockdown on stiff matrix–induced DSP expression were determined by immunoblot. Results are mean ± SD of at least three or five independent experiments. *P < 0.05. Ab = antibody; C/EBPβ = CCAAT/enhancer-binding protein β; Ctrl = control; dist = distal; ETF = EGFR [epidermal growth factor receptor]-specific transcription factor; Met = methylated; prox = proximal.
Next, we evaluated whether DNA methylation regulates the binding of nuclear EGR1 to the DSP promoter. Incubation of nuclear extracts of A549 with biotin-labeled, unmethylated DSP promoter probes containing the proximal or distal EBS showed DNA–protein complex formation (Figure 4C, left panel), whereas incubation with biotin-labeled, methylated probes did not (Figure 4C, right panel). The formation of complexes was dose-dependently inhibited by addition of unlabeled (“cold”), unmethylated probes, demonstrating that the DNA–protein complexes were EGR1-binding sequence specific. The presence of EGR1 in the DNA–protein complexes was further demonstrated by addition of EGR1-specific antibody (IgG served as control), which caused a supershift of DNA–protein complexes, owing to the formation of DNA–protein–antibody triple complexes (Figure 4C, left panel). In contrast, addition of competitive methylated DNA probes did not inhibit the formation of DNA–protein complexes (unmethylated DSP probes and nuclear EGR1) (Figure 4C, right panel). Together, these data suggest that the state of DNA methylation at the EGR binding sites regulates nuclear EGR1 binding to the DSP promoter. Methylated EBSs inhibit, whereas demethylated EBSs promote, the EGR1 binding to the DSP promoter.
Additional studies showed that matrix stiffness did not regulate EGR1 expression or its cytoplasm/nuclear distribution (Figure 4D). Knockdown of EGR1 expression inhibited stiff matrix–induced DSP overexpression (Figure 4E). Collectively, these data suggest that stiff matrix–induced DSP overexpression requires EGR1-dependent activation of the DSP promoter, and that DNA methylation state at the EGR1 binding sites regulates the ability of EGR1 binding to and activation of the DSP promoter.
CRISPR/dCas9-Dnmt3A–mediated Targeted DNA Methylation at the EGR1 Binding Sites Reverses Stiff Matrix–induced DSP Expression
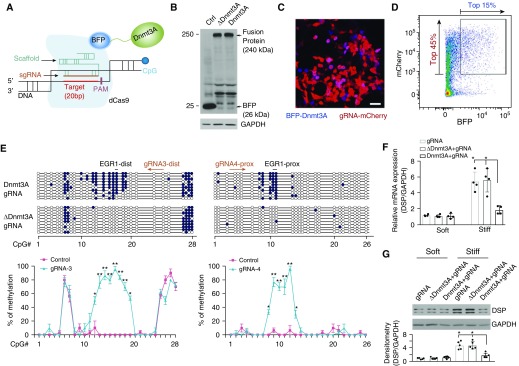
Next, we used state-of-the-art CRISPR/dCas9 technology for targeted DNA methylation at the EGR1 binding sites to determine whether stiff matrix–induced DSP overexpression occurs through an alteration of DNA methylation state in the DSP promoter. We coexpressed dCas9 and the catalytic domain of Dnmt3A, a de novo DNA methyltransferase (26, 27), as a fusion protein and designed specific sgRNAs for spatially controlled Dnmt3A expression at the EGR1 binding sites (Figure 5A). A pilot study, which aimed to optimize sgRNAs for targeted DNA methylation, indicated that gRNA4-prox and gRNA3-dist guided efficient DNA methylation at the proximal and distal EBS, respectively (Figure E2). To facilitate DNA transfection, we generated lentiviral vectors that encoded dCas9-BFP-Dnmt3A complementary DNA (cDNA) (Figure E3), the control dCas9-BFP-∆Dnmt3A (inactive Dnmt3A) cDNA (Figure E4), mU6-sgRNA-mCherry cDNA, and the control mU6-scrambled sgRNA-mCherry cDNA. Lentivirus-mediated expression of dCas9-BFP-Dnmt3A/∆Dnmt3A fusion proteins and sgRNA-mCherry in A549 cells was confirmed by immunoblot (Figure 5B). Confocal fluorescent microscopy showed that most of infected A549 cells expressed gRNAs, as indicated by mCherry-positive signals, but to a lesser extent expressed dCas9-BFP-Dnmt3A fusion protein (blue signals) (Figure 5C). This indicates that only a portion of infected cells had dual expression of gRNAs-mCherry (red) and dCas9-BFP-Dnmt3A (blue). To purify the cells with dual expression at higher levels, we performed fluorescence-activated cell sorting and selected a group of cells with top 45% red color and top 15% blue color, which together represented top 10.8% of cells with dual expression of gRNAs and dCas9-Dnmt3A at higher levels (Figure 5D). We used a similar approach to obtain the control cells that expressed scrambled gRNAs and dCas9-∆Dnmt3A (data not shown). Cells expressing both dCas9-Dnmt3A and sgRNAs showed a significant increase in DNA methylation at the proximal and distal EBS regions as compared with cells expressing inactive dCas9-∆Dnmt3A/sgRNAs (Figure 5E). gRNA4-prox induced DNA methylation in the proximal EBS region, which spanned 36 nucleotides (nt), with the methylation efficiency in CpG islands ranging from 30.0 to 96.7%. gRNA3-dist induced DNA methylation in the distal EBS region that spanned 50 nt, with the methylation efficiency ranging from 50.0% to 96.7%. (Figures 5E and E2). No significant increases in DNA methylation were observed in the control cells expressing dCas9-∆Dnmt3A/sgRNAs (Figure 5E), dCas9-Dnmt3A/scrambled sgRNA, and dCas9-∆Dnmt3A/scrambled sgRNAs (data not shown). Together, these data demonstrated that targeted DNA methylation by sgRNA-mediated sequence-specific binding and dCas9-Dnmt3A methyltransferase activity induces highly efficient DNA methylation at the EGR1 binding sites in the DSP promoter.
Figure 5.
Targeted DNA methylation by CRISPR (clustered regularly interspaced short palindromic repeats)/dCas9 (deactivated CRISPR-associated protein-9 nuclease)–Dnmt3A (DNA methyltransferase 3A)–mediated epigenome editing reverses mechanoinductive DSP (desmoplakin) expression. (A) Schematic depiction of targeted DNA methylation. (B) Lentivirus-mediated expression of BFP (blue fluorescent protein), dCas9-BFP-Dnmt3A, and dCas9-BFP-∆Dnmt3A fusion proteins were determined by immunoblot. (C) Confocal fluorescent microscopy showed expression of guide RNA (gRNA)-mCherry (red) and dCas9-BFP-Dnmt3A (blue) in infected human alveolar epithelial adenocarcinoma (A549) cells. Scale bar, 20 μm. (D) Purification of A549 cells with dual expression of gRNA-mCherry (red) and dCas9-BFP-Dnmt3A (blue) by fluorescence-activated cell sorting. The boxed area indicates 10.8% cells with dual expression at relatively higher levels (top 45% red and top 15% blue). (E) DNA methylation profiling analysis for cells with dual expression of gRNA and dCas9-BFP-Dnmt3A/∆Dnmt3A. Ten clones were sequenced for each condition. Solid circles indicate methylated CpG islands. Open circles indicate unmethylated CpG islands. (F) Relative mRNA levels of DSP were determined by quantitative PCR. (G) Protein levels of DSP were determined by immunoblot. GAPDH was used as reference/loading control. Results are mean ± SD of three to five independent experiments, each quantitative PCR reaction performed in triplicate. *P < 0.05; **P < 0.01. Ctrl = control; dist = distal; EGR1 = early growth response protein 1; PAM = protospacer adjacent motif; prox = proximal; sgRNA = single guide RNA.
To determine the effects of targeted DSP promoter methylation on matrix stiffness–regulated DSP expression, we cultured A549 cells expressing dCas9-Dnmt3A/sgRNAs and the control cells expressing dCas9-∆Dnmt3A/sgRNAs on soft and stiff matrix. Stiff matrix–induced DSP overexpression was blocked in cells expressing Dnmt3A/sgRNAs and preserved in the control cells expressing dCas9-∆Dnmt3A/sgRNAs (Figures 5F and 5G). Collectively, these findings demonstrated that targeted DNA methylation by CRISPR/dCas9-Dnmt3A–mediated epigenome editing reverses mechanoinductive DSP expression.
DSP Intron 5 Harbors an Enhancer Element at the 3′ End
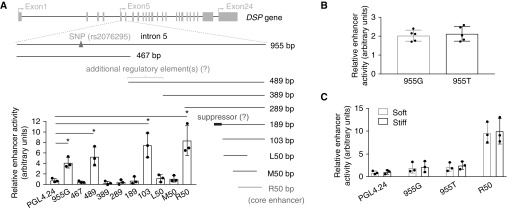
The IPF-associated genetic variant rs2076295 is located in intron 5 of the DSP gene (Figure 6A) and is associated with differential expression of DSP in human lungs (5). We investigated the potential mechanisms by which rs2076295 SNP regulates DSP gene expression. Previous studies demonstrated that a vast majority of disease-associated DNA polymorphisms are mapped within DNA enhancers (28). Therefore, we first determined whether DSP intron 5 has the enhancer activity. We cloned a 955-bp full-length intron 5 of human DSP gene. DNA sequencing confirmed that the genotype of rs2076295 we cloned was guanine (G), which represents the minor variant allele in humans. Enhancer activity assays demonstrated that human DSP intron 5 (955G) indeed contained enhancer activity (Figure 6A). To determine the core enhancer region in DSP intron 5, we generated a series of deletion mutants and found that the core enhancer element was located in a 50-bp region at the 3′ terminus in intron 5 (Figure 6A). In addition, the 3′ terminal 489-bp and 103-bp fragments showed the enhancer activity, whereas the 389-bp, 289-bp, and 189-bp fragments did not (Figure 6A). These findings suggest that an enhancer suppressor may exist upstream of the 103-bp fragment, likely located in an 86-bp region at the 5′ terminus of the 189-bp fragment. Interestingly, the 489-bp fragment containing the presumed suppressor displayed the enhancer activity, although at a relatively lower level than the more 3′ terminal fragments (103-bp and 50-bp fragments). We speculate that there might be additional regulatory elements in the 5′ terminus of the 489-bp fragment (Figure 6A). To determine whether the minor and major alleles of rs2076295 differ in their enhancer activity, we substituted guanine (G) (minor allele) to thymine (T) (major allele) in intron 5. The enhancer activity did not significantly differ in between DSP intron 5 with the minor allele and with the major allele (Figure 6B). Furthermore, we did not find that matrix stiffness regulates the enhancer activity of the full-length intron 5 (either with the major or the minor variant allele) or the core enhancer (R50) region at the 3′ terminus (Figure 6C). Collectively, we identified that human DSP intron 5 harbors a DNA enhancer element at the 3′ end. Neither the genetic variants of rs2076295 nor matrix stiffness regulates the enhancer activity of DSP intron 5.
Figure 6.
Human DSP (desmoplakin) intron 5 contains DNA enhancer activity. (A) Schematic depictions of human DSP gene and wild-type full-length and deletion mutations of intron 5. The SNP at rs2076295, the core enhancer region, and the presumed enhancer suppressor and additional regulatory elements are indicated. DNA enhancer activity assays were performed as described in Methods. (B) DNA enhancer activity assays for DSP intron 5 with the major (955T) and minor (955G) variant allele. (C) Effects of matrix stiffness on the enhancer activity of DSP intron 5 with 955T, intron 5 with 955G, and the core enhancer region (R50); PGL4.24, empty vector. Results are mean ± SD of three or five independent experiments, each performed in triplicate. *P < 0.05.
Discussion
The major findings in this study are that DSP, a GWAS-identified genetic risk locus in human subjects with IPF, is a matrix stiffness–regulated mechanosensitive gene. This mechanoinductive DSP expression is preserved in both human and rodent lung epithelial cells. This is the first report that links DSP overexpression to matrix stiffness, a mechanical factor known to be critical for the development/progression of lung fibrosis. Wnt/beta-catenin signaling is activated in epithelial cells in IPF and is linked to epithelial cell injury and hyperplasia (29). However, addition of Wnt3a did not regulate DSP expression in cultured human and rodent lung epithelial cells (Figure E5). In an earlier study, it was found that transforming growth factor-β1 promotes a modest increase in expression of DSP mRNA and protein in cultured airway epithelial cells (30). The mechanisms by which transforming growth factor-β1 regulates DSP expression remain unknown. In this study, we demonstrate that stiff matrix promotes DSP expression through induction of demethylation of a conserved regulatory region in the DSP promoter, which results in transactivation of EGR1 and EGR1-dependent DSP expression. Importantly, we used CRISPR/dCas9–mediated epigenome editing and showed that targeted DNA methylation in the DSP promoter reestablishes epigenetic control of DSP and reverses stiff matrix–induced DSP overexpression.
DNA methylation is known to be essential for cellular adaptation to environmental signals (31). Chemical inhibitors have been used to remodel aberrant epigenetic landscapes (32). Such an approach exerts epigenetic inhibition at the global genome level and is, thus, nonselective. CRISPR/dCas9–mediated epigenome editing uses coexpression of epigenetic modifiers and sgRNA for targeted epigenetic modifications at a specific gene locus (21–23, 33). Because demethylation of the EGR1 binding sequences is critical for stiff matrix–induced DSP overexpression, we reasoned that targeted DNA methylation instructs a repressive epigenetic state mimicking the epigenetic control of DSP under the mechanically homeostatic environment and reverses matrix stiffness–induced DSP overexpression. To optimize sgRNAs for targeted DNA methylation, we designed nine distinct sgRNAs with different orientation and distance to the proximal and distal EGR1 binding sequences. We found that each of these sgRNAs induces strong increases in CpG methylation. The length of methylated DNA sequences ranges from 20 to 62 nt region. The nearest methylation site is 13 nt away from gRNA (gRNA3-dist and gRNA4-prox), and the farthest site is 99 nt away from gRNA (gRNA3-prox). All methylation occurs at the 3′ terminus of the protospacer adjacent motif sequence. Our findings demonstrate that CRISPR/dCas9-Dnmt3A mediates highly efficient DNA methylation. The specificity of DNA methylation depends on the design of sgRNAs. CRISPR/dCas9-Dnmt3A/sgRNAs we developed offer a useful tool for the investigation of the functional role of DSP in lung fibrogenesis. In addition, CRISPR-mediated gene editing has been used to determine the potential regulatory role of GWAS-identified genetic variants in cancer-associated gene expression and function (34).
Many disease-associated SNPs are enriched in enhancers (28, 35). It is believed that SNPs may regulate enhancer activity by alteration of the binding of transcription factors to enhancers, resulting in differential gene expression. In this study, we investigated whether the SNP at rs2076295 locus may be involved in enhancer activity and, as such, regulates differential DSP expression. We discovered that intron 5 of human DSP, in which the SNP of rs2076295 resides, indeed contains DNA enhancer activity. We identified that the core enhancer element is located in a 50-bp region at the 3′ terminus of DSP intron 5. However, the SNP rs2076295 is approximately 700 bp upstream of the core enhancer region. Furthermore, we found no significant difference in the enhancer activity between intron 5 with the major variant allele (T) and with the minor variant allele (G) under both soft and stiff matrix conditions. Thus, our data do not support that the rs2076295 genetic variants regulate DSP expression by alteration of the enhancer activity of intron 5. A major limitation of our enhancer studies was that the findings were made from enhancer reporter-based in vitro studies. Nonetheless, data obtained in the current study could provide useful information for future functional characterization of the enhancer in DSP intron 5 using transgenic strategies (e.g., bacterial artificial chromosome and/or animal transgenesis) in the endogenous context. In addition, it is noteworthy that rs2076295 contains a binding site for transcription factor PU.1, which has been found in many functional enhancers (36). Previous studies also suggest that PU.1 plays an important role in asthmatic airway remodeling, macrophage activation, and alternative splicing of target genes (37, 38). Whether PU.1 is involved in the regulation of DSP remains to be determined.
IPF lungs have increased DSP expression relative to the control lungs, and expression of DSP decreases with age in the control lungs but not in IPF lungs (5, 7). Interestingly, the risk minor allele (G) of rs2076295 is associated with decreased, rather than increased, whole-lung expression of DSP. The current study provides a novel potential mechanism for DSP overexpression in IPF, but it does not explain an association of rs2076295 with decreased DSP expression. Hypothetically, if decreased DSP expression due to the risk allele of rs2076295 is causatively linked to the pathogenesis of IPF, increased DSP expression might be a protective response to IPF. Alternatively, both decreased and increased DSP expression could be detrimental. For instance, a lower-than-baseline level of DSP might render type I alveolar epithelial cells more vulnerable to lung injury, whereas stiff matrix–induced overexpression of DSP in the fibrotic phase might impair the repair of injured lung epithelium. The functional role of aberrant DSP expression in IPF currently remains unknown. It has been shown that ectopic expression of DSP in non–small-cell lung cancer cell line H157 cells inhibits cell proliferation, anchorage-independent growth, migration, and invasion (39). Similarly, we observed that suppression of DSP expression, either by CRISPR/dCas9-Dnmt3A–mediated targeted DSP promoter methylation or by siRNA-mediated gene knockdown, significantly increased A549 cell proliferation (Figures E6B and E6C) and migration (Figures E7A and E7B) on stiff matrix. Furthermore, overexpression of DSP significantly inhibited A549 cell proliferation on soft matrix (Figure E6E) but had no effects on cell proliferation (Figure E6D) and migration on stiff matrix (Figure E7C), presumably because DSP expression was already upregulated on stiff matrix (plastic surfaces). Future studies will test mechanoinductive DSP expression in the regulation of primary lung epithelial cell phenotype and function in the context of lung fibrosis and will elucidate whether DSP overexpression promotes or protects against lung fibrosis.
Genome editing provides an exciting opportunity for treatment of severe forms of human disease (40, 41). The efficient epigenome editing for reversing mechanoinductive DSP expression holds promise, both as a tool for the functional study and as a potential novel approach for fibrosis therapy. However, CRISPR/Cas9 gene editing can generate unwanted off-target effects that may confound research experiments and also have potential implications for therapeutic uses. Various strategies have been developed to substantially reduce genome-wide off-target effects of the commonly used Cas9 nuclease (SpCas9). For example, hyper-accurate Cas9 variant (HypaCas9) has demonstrated high genome-wide specificity without compromising on-target efficiency in human cells (42). New CRISPR methods and reagents along with whole-genome sequencing will be considered in the following functional studies.
In summary, DSP as a major desmosomal protein plays a critical role in the maintenance of epithelial barrier function and the regulation of normal wound healing (43, 44). DSP has been shown to regulate proliferation, migration, and differentiation of cancer cells (45–48). We speculate that aberrant DSP expression in IPF may not only represent a robust and persistent epithelial response to chronic/repetitive lung injury but also actively participate in aberrant lung repair and/or the restoration of lung epithelial function. Our work provides novel mechanistic insights into regulation of DSP overproduction by mechanical signals from the stiffened matrix. We have developed CRISPR/dCas9–mediated epigenome editing for efficient reversion of matrix stiffness–dependent DSP expression. Future studies will investigate functional significance of DSP in the pathogenesis of lung fibrosis and determine whether the DSP risk allele alters disease progression and survival in human IPF.
Footnotes
Supported by NIH grants R01HL124076 (Y.Z.), R01EY027924 (Y.Z.), and P01HL114470 (V.J.T.); American Heart Association Grant-in-Aid 14GRNT2018023 (Y.Z.) and Postdoctoral Fellowship 17POST33660976 (J.Q.); and University of Alabama at Birmingham School of Medicine AMC21 R01 Award (Y.Z.).
Author Contributions: J.Q. and Y.Z. designed the study; J.Q., L.Z., Z.Z., and S.L. performed experiments; J.Q., L.Z., and Y.Z. analyzed data; P.C., M.L.L., and V.J.T. provided experimental materials and/or participated in discussion; and J.Q. and Y.Z. wrote the manuscript.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201711-2242OC on June 20, 2018
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.American Thoracic Society; European Respiratory Society. American Thoracic Society/European Respiratory Society International Multidisciplinary Consensus Classification of the Idiopathic Interstitial Pneumonias. This joint statement of the American Thoracic Society (ATS), and the European Respiratory Society (ERS) was adopted by the ATS board of directors, June 2001 and by the ERS Executive Committee, June 2001. Am J Respir Crit Care Med. 2002;165:277–304. doi: 10.1164/ajrccm.165.2.ats01. [DOI] [PubMed] [Google Scholar]
- 2.Garantziotis S, Schwartz DA. Host-environment interactions in pulmonary fibrosis. Semin Respir Crit Care Med. 2006;27:574–580. doi: 10.1055/s-2006-957329. [DOI] [PubMed] [Google Scholar]
- 3.Taskar VS, Coultas DB. Is idiopathic pulmonary fibrosis an environmental disease? Proc Am Thorac Soc. 2006;3:293–298. doi: 10.1513/pats.200512-131TK. [DOI] [PubMed] [Google Scholar]
- 4.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, et al. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mushiroda T, Wattanapokayakit S, Takahashi A, Nukiwa T, Kudoh S, Ogura T, et al. Pirfenidone Clinical Study Group. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. J Med Genet. 2008;45:654–656. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]
- 7.Mathai SK, Pedersen BS, Smith K, Russell P, Schwarz MI, Brown KK, et al. Desmoplakin variants are associated with idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2016;193:1151–1160. doi: 10.1164/rccm.201509-1863OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobbs BD, de Jong K, Lamontagne M, Bossé Y, Shrine N, Artigas MS, et al. COPDGene Investigators; ECLIPSE Investigators; LifeLines Investigators; SPIROMICS Research Group; International COPD Genetics Network Investigators; UK BiLEVE Investigators; International COPD Genetics Consortium. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nat Genet. 2017;49:426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allen RJ, Porte J, Braybrooke R, Flores C, Fingerlin TE, Oldham JM, et al. Genetic variants associated with susceptibility to idiopathic pulmonary fibrosis in people of European ancestry: a genome-wide association study. Lancet Respir Med. 2017;5:869–880. doi: 10.1016/S2213-2600(17)30387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booth AJ, Hadley R, Cornett AM, Dreffs AA, Matthes SA, Tsui JL, et al. Acellular normal and fibrotic human lung matrices as a culture system for in vitro investigation. Am J Respir Crit Care Med. 2012;186:866–876. doi: 10.1164/rccm.201204-0754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Mih JD, Shea BS, Kho AT, Sharif AS, Tager AM, et al. Feedback amplification of fibrosis through matrix stiffening and COX-2 suppression. J Cell Biol. 2010;190:693–706. doi: 10.1083/jcb.201004082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, Yang N, Fiore VF, Barker TH, Sun Y, Morris SW, et al. Matrix stiffness-induced myofibroblast differentiation is mediated by intrinsic mechanotransduction. Am J Respir Cell Mol Biol. 2012;47:340–348. doi: 10.1165/rcmb.2012-0050OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y, Huang X, Hecker L, Kurundkar D, Kurundkar A, Liu H, et al. Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J Clin Invest. 2013;123:1096–1108. doi: 10.1172/JCI66700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rahaman SO, Grove LM, Paruchuri S, Southern BD, Abraham S, Niese KA, et al. TRPV4 mediates myofibroblast differentiation and pulmonary fibrosis in mice. J Clin Invest. 2014;124:5225–5238. doi: 10.1172/JCI75331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen H, Qu J, Huang X, Kurundkar A, Zhu L, Yang N, et al. Mechanosensing by the α6-integrin confers an invasive fibroblast phenotype and mediates lung fibrosis. Nat Commun. 2016;7:12564. doi: 10.1038/ncomms12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang X, Scott HA, Monickaraj F, Xu J, Ardekani S, Nitta CF, et al. Basement membrane stiffening promotes retinal endothelial activation associated with diabetes. FASEB J. 2016;30:601–611. doi: 10.1096/fj.15-277962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vaccaro CA, Brody JS. Structural features of alveolar wall basement membrane in the adult rat lung. J Cell Biol. 1981;91:427–437. doi: 10.1083/jcb.91.2.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buxboim A, Rajagopal K, Brown AE, Discher DE. How deeply cells feel: methods for thin gels. J Phys Condens Matter. 2010;22:194116. doi: 10.1088/0953-8984/22/19/194116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 20.Doudna JA, Charpentier E. Genome editing: the new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. doi: 10.1038/nbt.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, et al. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247, e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qu J, Chen H, Liu R-M, Thannickal VJ, Zhou Y. Reversing stiff matrix-induced desmoplakin (DSP) expression by CRISPR/dCas9-mediated epigenome editing [abstract] Washington DC: Presented at the American Thoracic Society International Conference. May 19–24, 2017. [Google Scholar]
- 25.Subbiah R, Hwang MP, Du P, Suhaeri M, Hwang JH, Hong JH, et al. Tunable crosslinked cell-derived extracellular matrix guides cell fate. Macromol Biosci. 2016;16:1723–1734. doi: 10.1002/mabi.201600280. [DOI] [PubMed] [Google Scholar]
- 26.Feng J, Chang H, Li E, Fan G. Dynamic expression of de novo DNA methyltransferases Dnmt3a and Dnmt3b in the central nervous system. J Neurosci Res. 2005;79:734–746. doi: 10.1002/jnr.20404. [DOI] [PubMed] [Google Scholar]
- 27.Gowher H, Jeltsch A. Molecular enzymology of the catalytic domains of the Dnmt3a and Dnmt3b DNA methyltransferases. J Biol Chem. 2002;277:20409–20414. doi: 10.1074/jbc.M202148200. [DOI] [PubMed] [Google Scholar]
- 28.Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Königshoff M, Balsara N, Pfaff EM, Kramer M, Chrobak I, Seeger W, et al. Functional Wnt signaling is increased in idiopathic pulmonary fibrosis. PLoS One. 2008;3:e2142. doi: 10.1371/journal.pone.0002142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida M, Romberger DJ, Illig MG, Takizawa H, Sacco O, Spurzem JR, et al. Transforming growth factor-beta stimulates the expression of desmosomal proteins in bronchial epithelial cells. Am J Respir Cell Mol Biol. 1992;6:439–445. doi: 10.1165/ajrcmb/6.4.439. [DOI] [PubMed] [Google Scholar]
- 31.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 32.Rius M, Lyko F. Epigenetic cancer therapy: rationales, targets and drugs. Oncogene. 2012;31:4257–4265. doi: 10.1038/onc.2011.601. [DOI] [PubMed] [Google Scholar]
- 33.Vojta A, Dobrinić P, Tadić V, Bočkor L, Korać P, Julg B, et al. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu S, Liu Y, Zhang Q, Wu J, Liang J, Yu S, et al. Systematic identification of regulatory variants associated with cancer risk. Genome Biol. 2017;18:194. doi: 10.1186/s13059-017-1322-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gosselin D, Link VM, Romanoski CE, Fonseca GJ, Eichenfield DZ, Spann NJ, et al. Environment drives selection and function of enhancers controlling tissue-specific macrophage identities. Cell. 2014;159:1327–1340. doi: 10.1016/j.cell.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guillouf C, Gallais I, Moreau-Gachelin F. Spi-1/PU.1 oncoprotein affects splicing decisions in a promoter binding-dependent manner. J Biol Chem. 2006;281:19145–19155. doi: 10.1074/jbc.M512049200. [DOI] [PubMed] [Google Scholar]
- 38.Qian F, Deng J, Lee YG, Zhu J, Karpurapu M, Chung S, et al. The transcription factor PU.1 promotes alternative macrophage polarization and asthmatic airway inflammation. J Mol Cell Biol. 2015;7:557–567. doi: 10.1093/jmcb/mjv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L, Chen Y, Cui T, Knösel T, Zhang Q, Albring KF, et al. Desmoplakin acts as a tumor suppressor by inhibition of the Wnt/β-catenin signaling pathway in human lung cancer. Carcinogenesis. 2012;33:1863–1870. doi: 10.1093/carcin/bgs226. [DOI] [PubMed] [Google Scholar]
- 40.Gao X, Tao Y, Lamas V, Huang M, Yeh WH, Pan B, et al. Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature. 2018;553:217–221. doi: 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao HK, Hatanaka F, Araoka T, Reddy P, Wu MZ, Sui Y, et al. In vivo target gene activation via CRISPR/Cas9-mediated trans-epigenetic modulation. Cell. 2017;171:1495–1507, e15. doi: 10.1016/j.cell.2017.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, et al. Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature. 2017;550:407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAleer MA, Pohler E, Smith FJ, Wilson NJ, Cole C, MacGowan S, et al. Severe dermatitis, multiple allergies, and metabolic wasting syndrome caused by a novel mutation in the N-terminal plakin domain of desmoplakin. J Allergy Clin Immunol. 2015;136:1268–1276. doi: 10.1016/j.jaci.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ou J, Lowes C, Collinson JM. Cytoskeletal and cell adhesion defects in wounded and Pax6+/- corneal epithelia. Invest Ophthalmol Vis Sci. 2010;51:1415–1423. doi: 10.1167/iovs.09-4023. [DOI] [PubMed] [Google Scholar]
- 45.Asimaki A, Saffitz JE. Remodeling of cell-cell junctions in arrhythmogenic cardiomyopathy. Cell Commun Adhes. 2014;21:13–23. doi: 10.3109/15419061.2013.876016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rampazzo A, Nava A, Malacrida S, Beffagna G, Bauce B, Rossi V, et al. Mutation in human desmoplakin domain binding to plakoglobin causes a dominant form of arrhythmogenic right ventricular cardiomyopathy. Am J Hum Genet. 2002;71:1200–1206. doi: 10.1086/344208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Uzumcu A, Norgett EE, Dindar A, Uyguner O, Nisli K, Kayserili H, et al. Loss of desmoplakin isoform I causes early onset cardiomyopathy and heart failure in a Naxos-like syndrome. J Med Genet. 2006;43:e5. doi: 10.1136/jmg.2005.032904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huber O, Petersen I. 150th Anniversary series: desmosomes and the hallmarks of cancer. Cell Commun Adhes. 2015;22:15–28. doi: 10.3109/15419061.2015.1039642. [DOI] [PubMed] [Google Scholar]