Abstract
The deletion of the metabolizing Glutathione S-transferase Mu 1 (GSTM1) gene has been associated with multiple cancers, metabolic and autoimmune disorders, as well as drug response. It is unusually common, with allele frequency reaching up to 75% in some human populations. Such high allele frequency of a derived allele with apparent impact on an otherwise conserved gene is a rare phenomenon. To investigate the evolutionary history of this locus, we analyzed 310 genomes using population genetics tools. Our analysis revealed a surprising lack of linkage disequilibrium between the deletion and the flanking single nucleotide variants in this locus. Tests that measure extended homozygosity and rapid change in allele frequency revealed signatures of an incomplete sweep in the locus. Using empirical approaches, we identified the Tanuki haplogroup, which carries the GSTM1 deletion and is found in approximately 70% of East Asian chromosomes. This haplogroup has rapidly increased in frequency in East Asian populations, contributing to a high population differentiation among continental human groups. We showed that extended homozygosity and population differentiation for this haplogroup is incompatible with simulated neutral expectations in East Asian populations. In parallel, we found that the Tanuki haplogroup is significantly associated with the expression levels of other GSTM genes. Collectively, our results suggest that standing variation in this locus has likely undergone an incomplete sweep in East Asia with regulatory impact on multiple GSTM genes. Our study provides the necessary framework for further studies to elucidate the evolutionary reasons that maintain disease-susceptibility variants in the GSTM1 locus.
Keywords: copy number variants, structural variation, cellular detoxification, natural selection, soft sweep
Here, we describe the evolutionary forces that shape the variation in the GSTM1 gene, which has been shown to be an important metabolism gene and associated with bladder cancer, autoimmune disorders and response to different drugs. Our results reveal a genetic type common in East Asian populations that may have important evolutionary and biomedical implications.
Thousands of structural variants (SVs, i.e., deletions, duplications, inversions and translocations of large segments of DNA) comprise a major part of genetic variation among humans (Redon et al. 2006; Sudmant et al. 2015b). Several studies have shown that common SVs can have important functional effects, contributing to both normal and pathogenic phenotypic variation (Zhang et al. 2009; Weischenfeldt et al. 2013). In addition, there were multiple recent studies showing adaptive phenotypes that are underlain by SVs in humans (Perry et al. 2007; Girirajan et al. 2011). However, there is still a major gap in our understanding of the functional and evolutionary impact of SVs due to three interrelated factors. First, multiple mutational mechanisms lead to SV formation, affecting the size and sequence content of the resulting variants, as well as the rate of their formation (Hastings et al. 2009). Second, most of the functionally relevant SVs are in complex, segmental duplication regions (Feuk et al. 2006; Marques-Bonet et al. 2009). This genomic context complicates discovery and genotyping of SVs themselves and also leads to higher than usual false negative and false positive single nucleotide variation calls in those regions. Third, the segmental duplication regions also lead to frequent non-allelic homologous recombination events, leading to the formation of new SVs. Linkage disequilibrium-based imputation is essential for many evolutionary genetic analyses (Crisci et al. 2013) and genome-wide association studies (Visscher et al. 2012). Recurrent SVs and sequence exchange between homologous sequences along with recombination break the linkage disequilibrium in segmental duplication-rich loci (Usher et al. 2015; Boettger et al. 2016). As a result, the majority of SVs cannot be imputed accurately using “tag” single nucleotide polymorphisms (Wellcome Trust Case Control Consortium et al. 2010) and the standard, haplotype-based neutrality tests cannot be directly conducted.
To overcome these issues, recent studies have focused on resolving complex haplotype architectures harboring SVs in a locus-specific manner. For example, the reassessment of SVs in the haptoglobin locus revealed recurrent exonic deletions that are associated with blood cholesterol levels (Boettger et al. 2016). Similarly, the reconstruction of the haplotype-level variation in the GYPA and GYPB (Glycophorin A and Glycophorin B) locus has revealed that a specific SV leads to resistance to malaria in African populations (Leffler et al. 2017). To highlight some of the negative results, careful reconstruction of the haplotype-level variation in salivary Amylase (Usher et al. 2015) showed that previous associations in locus with obesity may be false. Similarly, a haplotype level analysis of salivary MUC7 gene showed that previous association of this locus to asthma susceptibility was likely spurious but found surprising evidence of archaic hominin introgression in Africa affecting the structural variation in this locus (Xu et al. 2017b). Overall, with the availability of population-level genome variation datasets, it is possible to scrutinize the evolutionary and functional impact of SVs residing in complex regions of the genome.
Within this context, we focused on resolving the mechanisms and evolutionary forces that lead to the common haplotypes that carry the deletion of the glutathione S-transferase mu 1 (GSTM1) gene in humans (Figure 1A). GSTM1 is a member of the GST gene family, which code for cellular detoxifying enzymes and studied intensively within cancer biology and pharmacogenomics context (Parl 2005). However, these enzymes are functionally relevant to human evolution as well. It has already been shown that they contribute to cellular detoxification of naturally existing products (Hayes et al. 2005; McIlwain et al. 2006). For example, GST enzymes metabolize sulforaphane, which is an abundant product in cruciferous vegetables with unknown metabolic effects (Hayes et al. 2005). Similarly, they metabolize prostaglandin, an important naturally existing metabolite and a target of aspirin (Ricciotti and FitzGerald 2011). Prostaglandin affects multiple evolutionarily relevant processes, including constriction of the blood vessels and platelet aggregation. As such, it is plausible that variation affecting GST gene family may have adaptive effects. In fact, such adaptive evolution in humans has been documented for variation affecting other metabolizing genes. These examples include but not limited to the deletion of the UDP Glucuronosyltransferase Family 2 Member B17 (UGT2B17) (Xue et al. 2008), and single nucleotide variation affecting other metabolizing genes, such as N-acetyltransferase 2 (NAT2) (Mortensen et al. 2011), alcohol dehydrogenase (Sheeley and McAllister 2008), and Glucose-6-phosphate dehydrogenase (G6PD) (Ruwende et al. 1995). Therefore, it is not a stretch to imagine that the members of the metabolizing functions of GST gene family are adaptively relevant. Within that context, the deletion of GSTM1 and other variants in this locus may have a broader effect on the neighboring GSTM gene family, members of which have similar metabolizing functions (Hayes et al. 2005).
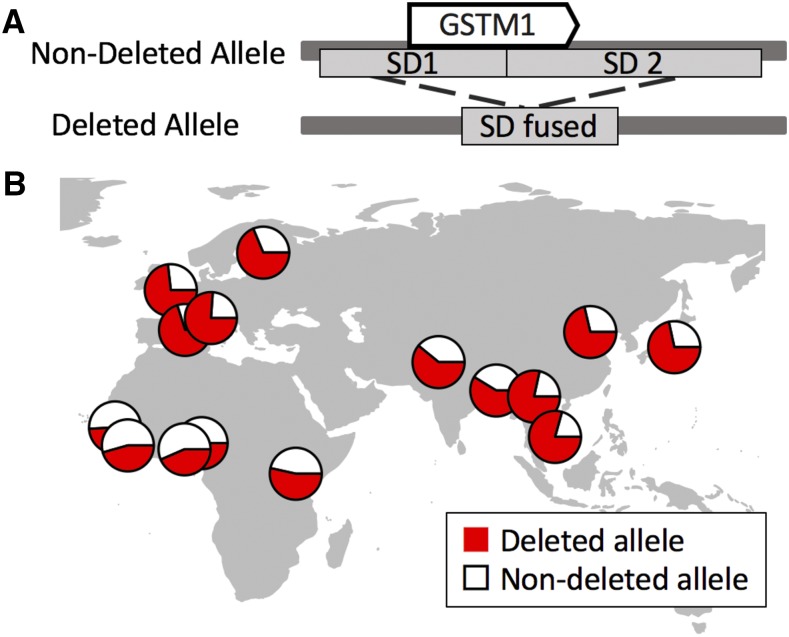
Figure 1.
The formation of the GSTM1 deleted allele and its geographical distribution. Two highly-similar segmental duplications fuse each other and form the deleted allele (upper). The geographical distribution of the GSTM1 deletion allele (red) is shown in the lower figure. The frequency data are from the 1000 Genomes Project phase 3 database (Sudmant et al. 2015a).
In addition to its functional relevance, the GSTM1 gene deletion is one of the most common whole gene deletions observed among human populations (Figure 1B) (Garte et al. 2001; Gaspar et al. 2002; Saadat 2007; Buchard et al. 2007; Fujihara et al. 2009; Piacentini et al. 2011). A whole gene deletion leads to the lack of the corresponding protein and thus it is thought to be deleterious in most cases. Therefore, it is rare for whole gene deletions to be at high frequencies (Conrad et al. 2010; Sudmant et al. 2015b). Nevertheless, the average global frequency of Eurasians with homozygous GSTM1 deletion is higher than 50% (Sudmant et al. 2015a), which puts it among the most frequent whole gene deletions observed in Eurasians according to 1000 Genomes Project database, the other being deletions affecting the LCE3BC (Late Cornified Envelope 3B and C) and the UGT2B17 (uridine diphosphoglucuronosyltransferase), both of which have been shown to be evolving under non-neutral conditions (Xue et al. 2008; Pajic et al. 2016).
Despite the functional and potential evolutionary importance of the GSTM gene family, a clear understanding of the mechanism through which the GSTM1 deletion and other variations in this locus actually affect phenotype remains elusive. We believe that understanding the underlying evolutionary forces that shape the variation in this locus will give us a framework to better target the functionally relevant haplotypes. Such an evolutionary analysis has been difficult since the GSTM1 gene resides in a complex genomic region with two segmental duplications (SDs) flanking both sides of the gene (Figure 1A). This particular genomic architecture predisposes GSTM1 to non-allelic homologous recombination events, which can lead to formation of new SVs. In fact, the polymorphic deletion observed in humans was almost certainly a result of such a mechanism (Xu et al. 1998). Moreover, we recently reported that recurrent deletions of the GSTM1 gene have happened independently in the human and chimpanzee lineages and that multiple rearrangements have defined genetic variation in this locus in primates (Saitou et al. 2018). These complicate the phylogenetic and broader evolutionary analysis immensely. This complication may also explain the paucity of evolutionary analysis in the GSTM1 locus, which is otherwise the target of more than 500 publications in the last 20 years, most of which are single-locus association studies implicating variation in this locus in susceptibility to multiple cancers (Parl 2005).
Collectively, the functional relevance of the GSTM1 deletion, its mechanistic complexity, and its high allele frequency in human populations make it suitable as a model for studying the evolution of metabolizing gene families. Therefore, in this paper, we analyzed hundreds of genomes to resolve the haplotypic variation that defines the GSTM1 deletion.
Materials and Methods
Materials and Methods
Study Populations:
We primarily used data from 3 populations, YRI (Yoruba in Ibadan), CEU (Utah residents with Northern and Western European ancestry), and CHB (Han Chinese in Beijing) available from the 1000 Genome Project (1000 Genomes Project Consortium et al. 2012; Sudmant et al. 2015b). For the phylogenetic analysis, we also analyzed haplotypes of the chimpanzee reference genome (The Chimpanzee Sequencing Consortium 2005), the Neanderthal genome (Prüfer et al. 2014) and Denisovan genome (Reich et al. 2010).
Confirmation of the genotyping and phasing in our dataset:
To validate the accuracy of the genotyping of the GSTM1 deletion in 1KGP dataset, we used digital droplet polymerase chain reaction experiments to genotype the GSTM1 deletion in 17 humans from 1KGP (samples Available through Coriell Institute for Medical Research) (forward primer: TCGAGGGTGCCATTACATTC; reverse ACTTCTGTCCTGGGTCATTC; probe: /56-FAM/TAGGAGCAG /ZEN/GCAGGTGATGTGAAC /3IABkFQ/). We followed standard protocol provided by Bio-Rad EIF2C1 probe assay (Table S1).
To investigate the accuracy of the 1KGP genotyping of single nucleotide variants, we investigated 74 CHB individuals and 6 sites in the target regions (chr1:110211681-110223007 and chr1:110246810-110255596) reported both in 1KGP and HapMap (this is all the SNPs genotyped in HapMap in the region). Of all the 444 sites, 9 were “NN” in hapmap and 3 showed different results in the two databases. Overall, we found approximately %99.3 concordance between the two datasets.
To validate the accuracy of the phasing in 1KGP project data that we used in this study, we compared haplotypes on 50kb on each side of the deletion in our dataset with phased HapMap genomes. Specifically, we were able to find 322 heterozygous sites in 30 phased HapMap CEU genomes. 320 (∼99.4%) of these are concordant with the 1KGP dataset. We also investigated Sanger sequencing data from seven deleted haplotypes for which we generated long-range PCR amplicons for an earlier study (Saitou et al. 2018). Since, the samples that were used for Sanger sequencing were not present in 1000 Genomes database, we asked whether common polymorphic sites in 1000 Genomes are also polymorphic in the sequenced haplotypes. Here, we defined common by 30% or higher allele frequency. For the 15 common single nucleotide variants that were reported in the 1000 Genomes project that overlap with the region (chr1:110216300-110225620 and chr1:110244324-110246143, the deletion was excluded), we found 14 variants among the 7 haplotypes (one example is represented in Figure S1). The only one that was not polymorphic among the seven haplotype dataset was rs61799140. This SNP has been detected by more than a dozen previous studies based on the dbSNP (https://www.ncbi.nlm.nih.gov/projects/SNP/snp_ref.cgi?rs=61799140). As such, we concluded that the absence of polymorphism in this location among the sequenced dataset is likely because it was not represented among the seven haplotypes, rather than it being a false-positive genotype in the 1000 Genomes dataset. Overall, we are confident that 1000 Genomes adequately captures the genetic variation in this locus and that the variant calling which our further analysis based on was accurate.
Linkage disequilibrium and haplotype analyses:
Vcftools (Danecek et al. 2011) was used to calculate R2 values between the GSTM1 deletion and flanking single nucleotide variations in Han Chinese in Bejing, China (CHB), Utah residents with Northern and Western European ancestry (CEU) and Yoruba in Ibadan, Nigeria (YRI) populations in 1KGP phase 3 datasets (Figure 2A, Figure S2). The Ensembl genome browser (http://asia.ensembl.org/index.html) was used to calculate and visualize linkage disequilibrium (LD) between single nucleotide variations and that (or LD) between the deletion and single nucleotide variations. Based on the R2 values, we used two target regions for the statistical neutrality tests. Two target regions were selected for the following analyses; target1, upstream the deletion (chr1:110211681-110223007 in the hg19) and target2, downstream the deletion (chr1:110246810-110255596 in the hg19), which showed relatively high R2 with the GSTM1 deletion in CHB (Figure 2A). We constructed a phylogenetic tree of the combination of these two regions by VCFtoTree (Xu et al. 2017a) and visualized it in Dendroscope (Huson et al. 2007). For the clustering and the visualization of haplotypes and its cluster analysis, we used the pipeline described in Xu et al., (2017b). The clustering was conducted based only on the haplotypes themselves with neither a priori input of haplogroups nor the deletion status (Figure 3).
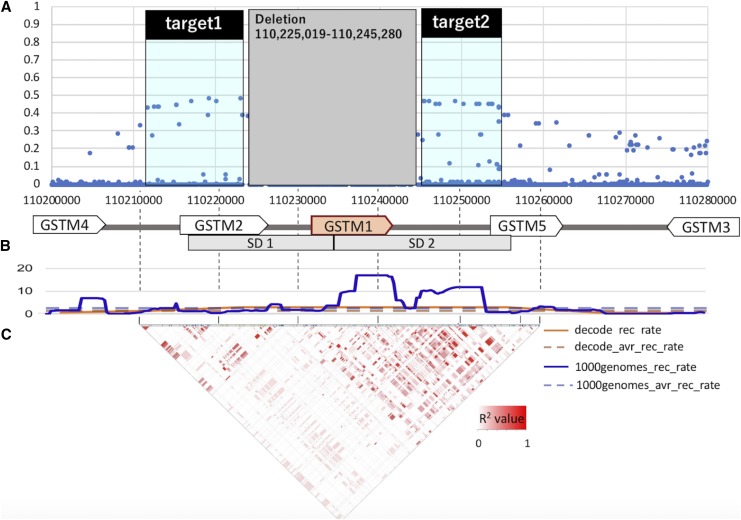
Figure 2.
Linkage disequilibrium in the GSTM1 region in CHB. (A) R2 value between the GSTM1 deletion and flanking single nucleotide variants in CHB. Each dot represent each single nucleotide variants. X-axis indicates chromosomal location and Y-axis indicates the R2 value between the GSTM1 deletion and flanking single nucleotide variants. Target regions were chr1:110211681-110223007 (target1, upstream the deletion) and chr1:110246810-110255596 (target2, downstream the deletion). (B) Recombination rate of the GSTM1 flanking region from DeCode (orange) and 1000genomes (blue). The chromosomal average of recombination rate from decode (orange) and 1000genomes (blue) were also plotted in the graph by broken lines. (C) Linkage disequilibrium (LD, R2 value) between the single nucleotide variants around the GSTM1 gene in CHB. Color coded in a continuous R2 values between each single nucleotide variants from 0 (white) to 1 (red).
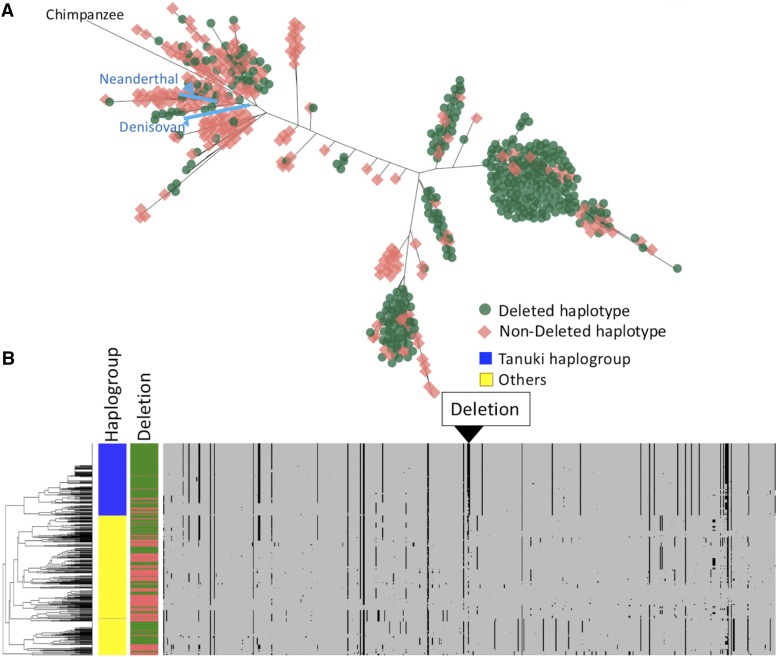
Figure 3.
Haplotypes of the combined target1 and target2 regions. (A) The phylogenetic relationship of the downstream target 2 region of CEU, YRI, CHB, Neanderthal, Denisovan and Chimpanzee constructed by the maximum-likelihood method. The GSTM1-deleted haplotypes were marked by green and the GSTM1-non-deleted haplotypes were marked by red manually. (B) Clustered haplotypes of the upstream target 1 region and downstream target 2 regions of CHB and CEU. The clustering was conducted based only on the haplotypes themselves with neither a priori input of haplogroups nor the deletion status. The GSTM1-deleted haplotypes were marked by green and the GSTM1-non-deleted haplotypes were marked by red. The Tanuki haplogroup, which showed high population differentiation, was marked by blue and other haplotypes were marked by yellow.
Tests of natural selection on the Target region:
We first constructed a null distribution by calculating the FST (Weir and Cockerham 1984), Tajima’s D (Tajima 1989), π (Nei and Li 1979) and iHS (Voight et al. 2006) values for 1000 randomly chosen regions on the same chromosome where the GSTM1 is located (chromosome 1) and also match our target region by size (∼9kb). Then, we calculated these statistics observed in the downstream regions of the GSTM1 deletion (chr1:110246810-110255596, downstream the deletion, also represented in Figure 2) and compare the results with those from the random regions. We also replicated this with size-matched regions that overlap with all 618 segmental duplications on chromosome 1 to match the genomic features in the GSTM1 region (Figure S3).
We used genomic data of CEU, CHB and YRI populations in 1KGP. Tajima’s D value, iHS and π were calculated for 3kb intervals. XP-CLR were calculated for 2kb intervals. All these measurements were acquired through the 1000 Genomes Selection Browser (Pybus et al. 2014).
To visualize the geographic distribution of the Tanuki haplogroup, which was linked with the deletion in East Asia, we used data from 15 populations in the 1000 Genome Project for haplotype distribution analysis: BEB, CDX, CHB, ESN, FIN, GBR, GWD, IBS, JPT, KHV, LWK, MSL, PJL, TSI, and YRI, which have not experienced recent population admixture or migration. We used the “rworldmap” package (South 2011) to visualize the global distribution of the haplotypes.
Simulations for FST and π values:
For the simulation analyses, we used ms (Hudson 2002) to conduct 1000 simulations matching the size (8,787bp) and the observed Watterson Estimator (θ=9.14) which was calculated by DnaSP (Librado and Rozas 2009) of the downstream target2 region (Figure 2). For each simulation, we generated 216 and 206 haplotypic sequences for YRI and CHB, respectively. We chose these populations and the number of sequences to match the empirical data from the 1000 Genome Project. To accurately model the effect of the demography in our simulations, we used the published parameters for East Asian and African populations (Schaffner et al. 2005) and we implemented the recombination rates of this region (ρ=5) reported by the HapMap Consortium (International HapMap Consortium et al. 2007). Once the sequences are generated, we focused on constructing expected distributions for population differentiation (FST) and nucleotide diversity (π).
The command line for ms simulator was ./ms 422 1000 -t 91.4 -I 2 216 206 -m 1 2 3.2 -n 2 0.077 -en 0.0005 1 0.25 -en 0.001 2 0.077 -ej 0.00875 2 1 -en 0.0425 1 0.125 -r 5 8787.
We calculated FST values between YRI and CHB populations for each single nucleotide variant that was generated in the simulations. We then plotted the values with the matching allele frequencies (0.69-0.70, the range of standard deviation of the observed allele frequency of the variations in Tanuki haplogroup) for each of these variants. Next, we considered the distribution of the simulated π values. To simulate this, we first generated 1000 simulated datasets as described above. We then focused on the segregating sites with alternative allele frequencies at 0.69-0.70, the range of standard deviation of the observed allele frequency of the variations in Tanuki haplogroup. Based on this, we calculated π for each of these haplogroups which carry the alternative allele for each single nucleotide variation in the simulated CHB population (Figure. S4).
We observed that the correlation fit between the simulated and empirical FST values are high (Figure 5A), but notably less than the correlation between the simulated and empirical π values (Figure 5B). We believe that this is due to the fact that we used demographic parameters for two populations (East Asian and African populations (Schaffner et al. 2005) for the FST simulations, while assumed parameters for only the East Asian populations for the π calculations.
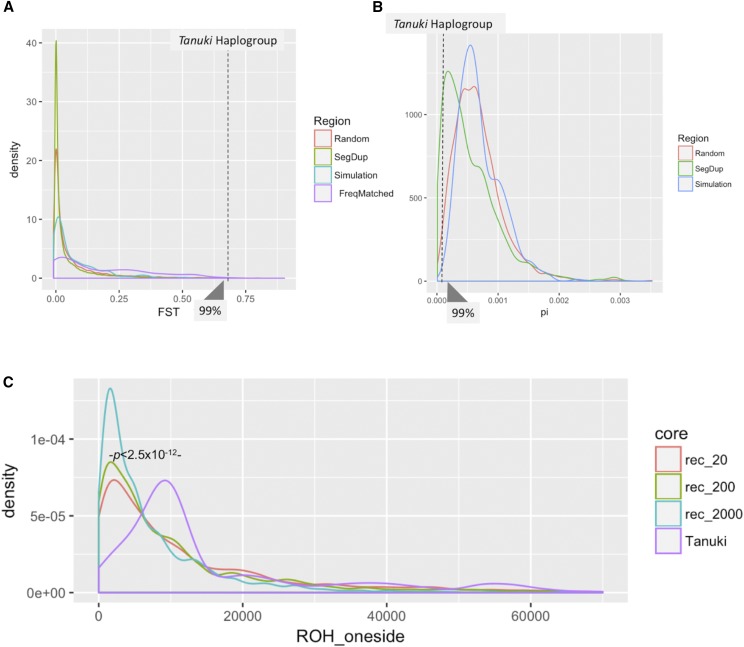
Figure 5.
(A) The average values of FST for Tanuki SNPs (dotted line), the 1000 randomly selected 9kb regions (red), 9kb regions on chromosome 1 that overlap with segmental duplications (green), simulated data under neutrality (blue), and frequency-matched (0.69 - 0.70) single nucleotide variants on chromosome 1 (purple). We found that none of 603 frequency-matched simulated FST is higher than the observed FST for the Tanuki haplogroup. (B) The π value for Tanuki haplotypes (dotted line), the 1000 randomly selected 9kb regions (red), 9kb regions on chromosome 1 that overlap with segmental duplications (green), simulated data under neutrality (blue). The results showed that π observed in the Tanuki haplogroup is lowest among 389 frequency-matched simulated values. (C) The Run of Homozygosity of the Tanuki SNP (purple) and simulated frequency-matched SNPs with 3 different recombination rates, 20 (red), 200 (green), 2000 (blue). We found that the ROH values observed for the Tanuki single nucleotide variants in the 1KGP East Asian populations is significantly higher than what is expected under neutrality based on frequency matched simulation results with r = 200, the most realistic parameter from the observation (Fig. 5C, P < 2.5x10−12, Wilcoxon-Rank-Sum Test).
The Run of Homozygosity analysis:
We used the demographic model of (Schaffner et al. 2005) with theta = 0.00375 per site and Rec rate = 0.001 per site and 4Nem = 9 (Ne = effective population size and m= migration rate) from the empirical observation. We ran more than 7000 simulations per each condition and obtained 70-120 core sites with the frequency of 0.69+-0.031 (Standard deviation calculated from Binomial Distribution) in East Asia and 0 in Africa under neutrality. We calculated both side of ROH for the 70-120 core sites from the simulation. We also calculated one side (downstream) of ROH of the 238 Tanuki homozygous individuals in East Asian populations for the Tanuki SNP as there was a recombination hotspot in the upstream of Tanuki region (Figure. 2B). We used three recombination rates (20, 200, 2000 recombinations per the 200kb region) to see the impact of recombination rates on ROH. r = 200 was the most realistic parameter from the observation.
The command line for ms simulator was ./ms 422 1000 -t 750 -r 200 200000 -I 2 206 216 9 -en 0.0005 2 0.25 -en 0.001 1 0.077 -en 0.0425 2 0.125
To test for potential sampling bias (e.g., consanguinity) in our dataset, we conducted bootstrap resampling of individual phased chromosomes. There are 238 individuals in East Asian populations (CHB, JPT, CHS, KHV, CDX) that are homozygous for Tanuki haplotype. To construct a distribution to investigate whether there is a sampling bias, we randomly resampled from 694 phased Tanuki haplotypes to generate 500 datasets each with 238 homozygous constructs. This allowed us to test whether the observed value of ROH deviates from randomly resampled distribution. We observed no such deviation, suggesting no sampling bias in our analysis (Figure S5).
Age estimation of Tanuki haplogroup and selection on Tanuki haplogroup:
We conducted several age estimates in this paper. First, we used the simple formula detailed (15) in (Kimura and Ohta 1973) and estimated the expected time for the Tanuki haplogroup to reach the current frequency in East Asia and Europe under neutrality.
Second, we used the average sequence differences of the Tanuki haplogroup from a non-Tanuki haplotype (Figure S6, S7) to calculate a coalescent date for the divergence of these two haplogroups. Specifically, based on a more recent (5 million years (Varki and Altheide 2005)) and older (∼12 million years (Moorjani et al. 2016)) divergent estimates between humans and chimpanzees, we estimated the coalescent of Tanuki haplogroup (0.001048 distance from the root haplotype) to be between 300 to 720 thousand years before present.
Third, we calculated the starting date of the putative selection on Tanuki haplogroup based on the run-of-homozygosity in this locus with method detailed by Racimo et al. (2015). Briefly, at a past time when r*L*t (r = recombination rate, L = the length of run of homozygosity, t = the time when the selection on the variant) reaches 1, the ROH shows rapid breaks because of recombination. So, we can assume that the t at that time is the start point of the selection. We used r= (1.450.05)× 10−8/generation (Narasimhan et al. 2017). In the paper, they noted this value and confidence interval from their two estimates for the mutation rate (1.51 and 1.41 × 10−8 per bp per generation (Narasimhan et al. 2017), u = 3 × 10−8/site/generation, and observed averaged ROHaver of the Tanuki Haplogroup (∼16kb).
It is important to note some caveats here. First, the coalescence dates depend on the local recombination and mutation rates, and even our best estimates may not be accurate with our current knowledge. Second, given that this locus has likely evolved under adaptive forces, it would be more appropriate to incorporate dating approaches that consider selection. However, we have little insight into the timing and the strength of the selection, and thus we focused on dating approaches that do not depend on selection coefficient as a parameter.
Data availability
Table S1 contains genotyping of the GSTM1 copy number by ddPCR of 17 individuals in YRI population in the 1KGP. Supplemental materials are available at Figshare: Saitou, Marie; Gokcumen, Omer; Satta, Yoko (2018): G3-Supp0626.pdf.; https://doi.org/10.25387/g3.6384782.
Results
Imperfect linkage disequilibrium Between the GSTM1 deletion and flanking single nucleotide variants in the human lineage
To understand the evolutionary mechanisms which maintain the GSTM1 deletion common in humans, we first attempted to resolve the haplotype structure of the locus. We calculated the R2 value between the GSTM1 deletion and flanking single nucleotide variants in Han Chinese in Bejing, China (CHB), Utah residents with Northern and Western European ancestry (CEU) and Yoruba in Ibadan, Nigeria (YRI) populations in 1KGP phase 3 datasets (Figure 2A, Figure S1). Remarkably, we found almost no linkage disequilibrium between the GSTM1 deletion flanking single nucleotide variants in CEU and YRI (Figure S1), and observable, but imperfect linkage disequilibrium in CHB only extending to >2.5kb on each side of the deletion (R2=∼0.45,Figure 2A). One way to explain this low level of linkage disequilibrium is by invoking a recombination hotspot in the region. However, we found that the reported recombination in both deCODE (Kong et al. 2010) and 1000 genomes phase 3 datasets for the region (1000 Genomes Project Consortium et al. 2010) is high, but not exceptionally high to be categorized as a hotspot (3-17 cM/Mb, (Mc Vean et al. 2004)) (Figure 2B).
One mechanism that can explain the imperfect linkage disequilibrium between the deletion and the neighboring variants can be recurrent formation of deletions. In other words, if two or more independent deletion events have occurred in different haplotypic backgrounds, then we expect (i) different deletion breakpoints, and (ii) LD to be intact between the variants on each side of the deletion as described by Boettger et al. (Boettger et al. 2016). In a previous study, we conducted an analysis of the breakpoints of this deletion, but, due to repetitive nature of this locus, could not identify the exact breakpoints of this deletion (Saitou et al. 2018). In parallel, we found that there is relatively weak, but observable linkage disequilibrium (R2=∼0.5) between variants on each side of the deletion in CHB (Figure 2C). Overall, our results cannot rule out recurrent formation of the GSTM1 deletion in the human lineage.
To further understand the haplotypic variation in this locus, we constructed a phylogenetic tree using sequence variation data in the “target” region flanking the deletion for 620 phased haplotypes from YRI, CEU, CHB populations, as well as chimpanzee, Denisovan, Neanderthal genomes (Figure 3A). As expected from the low levels of linkage disequilibrium between single nucleotide variants and the deletion, we observed no clear separation between haplotypes with and without the deletion. Instead, we found multiple branches that are predominantly populated with deleted haplotypes and others with non-deleted haplotypes without notable population structuring. These observations are concordant with the recent study documenting the genetic variation in this locus in a Russian population (Khrunin et al. 2016). It is important to note here, however, that Neanderthal and Denisovan haplotypes, as well as the branching point for the chimpanzee reference haplotype, all cluster with the predominantly non-deleted haplotypes. This suggests that the deletion is a derived variant that likely evolved after the human-Neanderthal divergence (i.e., in the last 1 Million years).
Despite these general insights, the observable lack of linkage disequilibrium between the GSTM1 deletion and the neighboring variants reduces the power of disease-association studies (e.g., GWAS) that rely on imputation of the GSTM1 deletion genotype using nearby single nucleotide variants. Even considering multiple single nucleotide variants within a single population, we were not able to impute the deletion accurately in any of the study populations (YRI, CHB, and CEU) (Figure 3B). This is concordant with the previous finding reported the difficulty to predict the copy number of the GSTM1 gene by the flanking haplotypes in CEU (Khrunin et al. 2016). As such, direct genotyping, rather than imputation, may be more robust approach to study GSTM1 deletion.
A potential signature of an incomplete sweep in East Asian populations in the GSTM1 locus
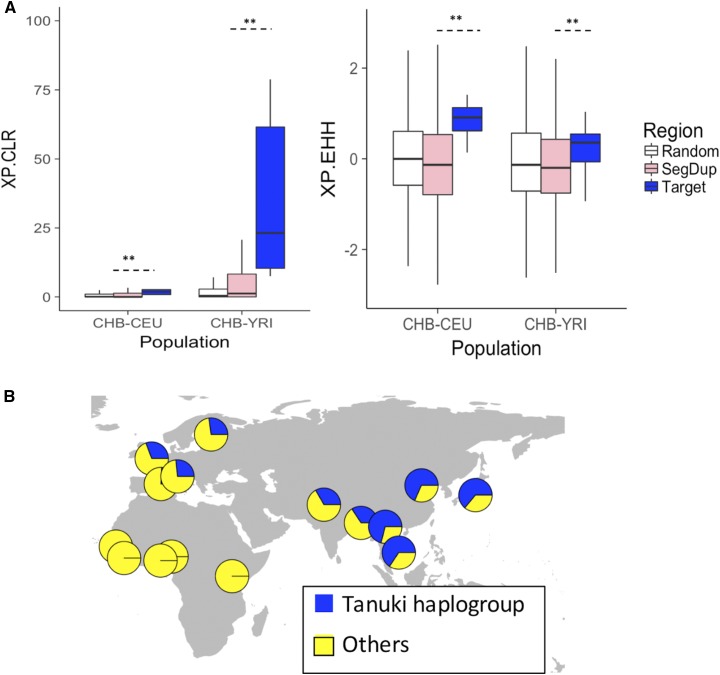
To reveal any potential signatures of adaptive evolution affecting variation in this locus, including the GSTM1 deletion, we conducted statistical neutrality tests using single nucleotide variation data from the sequences flanking the GSTM1 deletion. Specifically, we first constructed a null empirical distribution by calculating the FST (Weir and Cockerham 1984), Tajima’s D (Tajima 1989), π (Nei and Li 1979), iHS (Voight et al. 2006), XP-EHH and XP-CLR values for 1000 randomly chosen regions on the same chromosome where the GSTM1 is located (chromosome 1) and also match our target region by size (∼9kb). Then, we calculated these statistics observed in the downstream regions of the GSTM1 deletion and compared the results with those from the random regions (Figure 4A, Figure. S2). We also replicated this with randomly selected, size-matched regions that overlap with all of the 618 segmental duplications on chromosome 1 to match the genomic features in the GSTM1 region (Figure. S2). We found that both XP-CLR and XP-EHH tests showed significant differences as compared to neutrality when CHB population is involved (p values < 0.01, Wilcoxon rank sum test with continuity correction). These tests measures the change in allele frequency in one population has occurred more quickly than expected by drift alone and the difference in extended homozygosity between two populations (Figure 4A, Table 1). XP-EHH (Sabeti et al. 2007) is cross-population extended haplotype homozygosity and XP-CLR (Chen et al. 2010) is multi-locus allele frequency differentiation between two populations. Collectively, the unusually high XP-CLR and XP-EHH values in CHB population suggest an incomplete sweep has shaped the distribution of Tanuki haplogroup in this population.
Figure 4.
(A) Neutrality tests on the target region (chr1:110246810-110255596, downstream the deletion, also represented in Figure. 2.), the 1000 randomly selected 9kb regions, and all the segmental duplications on chromosome one, which contains the GSTM1 in CEU, CHB and YRI populations in 1KGP. XP-CLR and XP-EHH were calculated for 2kb intervals. The stars show significant differences (P < 0.01, Wilcoxon Rank Sum Test). (B) The geographical distribution of the Tanuki haplogroup calculated from the 1KGP phase3 data.
Table 1. XP-EHH and XP-CLR between CHB-YRI, CEU-YRI and CHB-CEU. Significant values were shaded.
| XP-CLR | |||
|---|---|---|---|
| XP-EHH | YRI | CEU | CHB |
| YRI | 3.18 (P = 0.244) | 35.6 (p=0.000707) | |
| CEU | −0.338 (P = 0.995) | 3.54 (p=0.00543) | |
| CHB | 0.232 (p=5.32e-0.7) | 0.902 (p=2.20e-16) | |
To investigate the haplotypic background of this putative sweep, we focused on the single nucleotide variants that showed the highest population differentiation (Figure 4A). We first showed that they are in high linkage disequilibrium with each other (R2 > 0.84), indicating that they represent a single haplotype group in East Asian populations (Figure S8, Table S2, called Tanuki haplogroup from hereon) in the target2 region (Figure 2A). We first investigated the global distribution of the haplogroup and found that it represents the major allele (>70% allele frequency) in East Asian populations, but it is virtually absent in sub-Saharan African populations (Figure 4B, Figure S9).
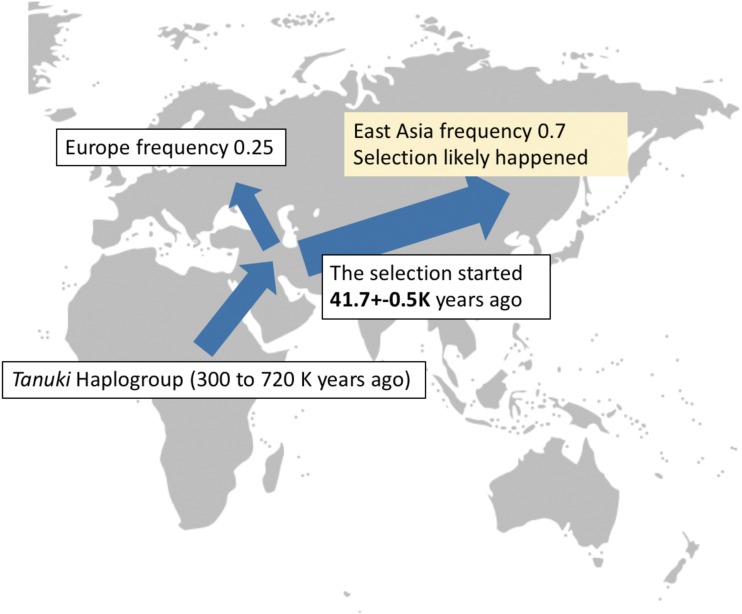
To determine the age of this haplogroup, we constructed a phylogenetic tree (Figure S6) of all Tanuki haplotypes in CEU and CHB populations (no Tanuki haplotype was found among YRI genomes), along with a non-Tanuki haplotype and the chimpanzee reference genome haplotype as outgroups. We surmised that Tanuki haplotype diverged from other human haplotypes at least 300 thousand years ago (∼300K to 720K YBP). Even considering the relatively large deviation of this estimate, this date is far earlier than the out-of-Africa migrations. As such, we conclude that this haplogroup has originated in Africa, but has increased in frequency in Eurasia. It is plausible that some Tanuki haplotypes still remain in African populations, but at a low allele frequency.
To further understand the haplotypic background of the significantly high XP-CLR and XP-EHH values in Asian populations as compared to European and African populations, we constructed a network of Tanuki haplotypes (Figure S7). We found that a vast majority of Tanuki haplotypes in CHB population has identical sequences, suggesting a rapid and recent increase in the allele frequency in the East Asian populations. This observation explains the unusually high XP-CLR and XP-EHH values observed for CHB population and further support the notion that a incomplete sweep has shaped the distribution of Tanuki haplogroup in this population.
Allele frequency of the Tanuki haplogroup is unusual in East Asia, but not in Europe
The high XP-CLR and XP-EHH values along with very high frequency of Tanuki haplogroup in CHB potentially point to the effect of an local sweep. The alternative, null hypothesis would be that the observed difference in allele frequency is due to the effect of drift alone. To further distinguish between these two scenarios, we conducted multiple empirical and simulation based analyses. First, we investigated whether the allele frequency of Tanuki haplogroup in East Asian and European populations can be explained by drift alone. To do this, we first assumed that the putative target for selection is an allele that reached to approximately 70% allele frequency in East Asia, but remain at 25% allele frequency in Europe (Figure 4B). Then, we used the simple formula detailed (15) in (Kimura and Ohta 1973) and estimated that it would take ∼580 and ∼160 thousand years under neutrality for that single haplotype to increase in allele frequency from 1/2Ne in ancestral African population to 70% and 25% observed in Asia and Europe, respectively. This results suggest that the allele frequency of Tanuki haplotypes in Europe can be explained by neutrality alone, but not in East Asia.
To further interrogate whether the high allele frequency of Tanuki haplotypes can be explained by genetic drift alone, we conducted simulation-based analyses of the genetic variation we observed (for detailed conditions for simulations, see Methods). Specifically, we used ms (Hudson 2002) to generate 1000 simulated datasets comprising sequences matching the size (8,787bp) of the target2 region of GSTM1 (Figure 2). For these simulations, we used demographic parameters previously laid out for CHB and YRI populations (Schaffner et al. 2005) to accurately model the effect of drift on nucleotide diversity and allele frequency.
Once the sequences were generated, we calculated FST values for each single nucleotide variant that was generated in the simulations between two simulated populations (YRI and CHB). To verify the accuracy of our simulations, we first compared our simulated sequences to empirical distribution of FST between YRI and CHB populations for randomly selected single nucleotide variants across the genome (Figure 5A). We then plotted the FST values with the matching allele frequencies (0.69-0.70, that represent the deviation of the observed allele frequency of the variations in Tanuki in Asian populations) for each of these variants observed in the simulation. We found that none of 603 frequency-matched simulated FST is higher than the observed FST for the Tanuki haplogroup (>99th percentile, Figure 5A). Overall, these results suggest that the unusually high allele frequency of Tanuki haplotypes in CHB population is unlikely to be explained by neutral forces alone.
Haplotype homozygosity provides further evidence for a selective sweep
In parallel to the allele frequency based analyses described above, we conducted tests based on haplotype similarity. We first followed the reasoning outlined by Kim and Satta (Kim and Satta 2008). Briefly, if a recent sweep indeed increased the allele frequency of a particular existing haplotype group, we expect that the nucleotide diversity would decrease among the haplotypes that were sweeped, but not the others. Indeed, we found that the nucleotide diversity of haplotypes that belongs to the Tanuki haplogroup, which make up >70% of haplotypes in East Asians, is approximately six times lower (π=0.00012) in East Asians as compared to the nucleotide diversity observed in haplotypes that do not belong to the Tanuki haplogroup in the same population (π=0.00076).
To explore this issue, we have considered the π values for Tanuki and nonTanuki haplotypes across a larger genomic region surrounding the GSTM1, including sequences that are not overlapping segmental duplications in multiple East Asian populations (CHB, CHS, JPT and KHV) (Figure. S10). These results also corroborates our initial finding that Tanuki haplotypes show unusually low π as compared to simulated expectations (Figure. 5B).
To quantify this observation, we used simulated sequences (as described above) to construct an expected, neutral distribution of nucleotide diversity (π) values. Specifically, we wanted to test whether π calculated for the haplotypes that belong to the Tanuki haplogroup is lower than expected distribution under neutrality. To simulate this, we first generated 1000 simulated datasets as described above. From the simulated dataset, we chose haplogroups within the simulated with alternative allele frequencies at 0.69-0.70 to match that of Tanuki haplogroup in frequency (please see Figure S4). Then, we plotted the π values for these simulated haplogroups. The results showed that π observed in the Tanuki haplogroup is lowest among 389 frequency-matched simulated values (<1st percentile, Figure 5B).
To test whether the low nucleotide diversity observed in Tanuki haplogroup is also translated into extended homozygosity, a hallmark of a selective sweep, we simulated expected Runs of Homozygosity (ROH) assuming demographic parameters outlined in (Schaffner et al. 2005) and different recombination rates (see methods for details). We found that the ROH values observed for the Tanuki single nucleotide variants in the 1KGP East Asian populations is significantly higher than what is expected under neutrality based on frequency matched simulation results with r = 200, the most realistic parameter from the observation (Figure 5C, P < 2.5x10−12, Wilcoxon-Rank-Sum Test). The averaged ROH of the Tanuki haplogroup was 16Kb.
This calculation also allowed us to calculate a potential date for the sweep using the approach outlined elsewhere (Racimo et al. 2015) (see methods). Briefly, this method uses the length of ROH to estimate the age of the beginning of a sweep under given recombination and mutation rates. Using this method, we estimated this date to be 41.70.5K years ago when the selection on Tanuki Haplogroup began (Figure 6). Combined, the observed values of population differentiation and haplotype diversity are inconsistent with the neutral evolution of this locus and are parsimonious with our hypothesis that the Tanuki haplogroup has increased its frequency under adaptive evolution in the East Asian populations.
Figure 6.
The evolutionary history of the Tanuki haplogroup. Based on our analyses, we argue that the Tanuki haplogroup originated before Out of Africa migrations (∼300K years before present) and spread out to Eurasia. We further argue that in Europe, the frequency of Tanuki haplogroup increased approximately to 0.25 under neutrality. In Asia, the selection on the Tanuki haplogroup started 41.7K years ago and pushed frequency of the Tanuki haplogroup to 0.7 under selection.
Potential functional impact of Tanuki haplogroup
By carefully resolving the different haplotype groups in the GSTM1 locus, we were able to detect a putatively adaptive haplogroup (Tanuki haplogroup). The exact underlying evolutionary reason and the functional impact of the increase in frequency of the Tanuki haplogroup remain excellent venues for future studies. One obvious question is whether the GSTM1 deletion is the actual target of positive selection. Indeed, we discovered that Tanuki haplogroup is significantly, albeit imperfectly, linked with the GSTM1 deletion in the CHB population (R2=∼0.47) (Table S2). It is important to note that despite the low linkage disequilibrium, ∼91% of the Tanuki haplotypes harbor the GSTM1 deletion. We reasoned that if the GSTM1 deletion was the target for the selective sweep that we observed, then other haplotypes carrying the GSTM1 deletion also show evidence for selection. However, we did not observe any unusual increase in allele frequency of other haplotypes that carry the deletion. As such, it is highly plausible, as drastic an event as a whole gene deletion is, GSTM1 deletion may not be the main target of the selective sweep that we observed.
To further follow up this thread, we asked whether the GSTM1 gene has no fitness consequences and hence accumulating loss of function mutations, such as the deletion. Under this scenario, the GSTM1 deletion just swept along with the remainder of the Tanuki haplogroup with scant adaptive consequences. Contradicting this possibility, it was reported that the GSTM1 gene sequence is conserved among great apes, as well as between humans and archaic humans. Specifically, we found only 4 nonsynomous variants between human and chimpanzee GSTM1, and found none among human, Neanderthal and Denisovan GSTM1 genes (Table S3). In addition, only two commonly observed nonsynonymous mutations were reported for GSTM1 gene in human population, both of which have little effect on the GSTM1 function (Moyer et al. 2007; Tatewaki et al. 2009). ExAC database allowed us to be more specific (Lek et al. 2016). The expected number of loss-of-function variants at the GSTM1 locus is 8.4 but observed number was 2 in the Exome analysis in 60,706 humans. In addition, the frequency of these loss-of-function variants are extremely low (<0.00003133). It is important to note that the deletion of the entire gene in approximately half of human genomes is not currently reported in ExAC database. However, our point here is that if GSTM1 has no fitness effect in humans, we expect to find relatively high frequencies of loss of function variants in addition to the deletion. Instead, virtually all the nondeleted GSTM1 haplotypes carry an intact, open-reading frame, further supporting the notion that both deleted and nondeleted haplotypes are maintained in the population.
These results suggest that it is likely that the GSTM1 deletion has fitness effects, but it is independent of the selective sweep we observed for the Tanuki haplogroup. To explain these observations, we hypothesize that Tanuki haplogroup affect the function of multiple genes in the GSTM gene family and not just GSTM1. Consequently, the putative adaptive phenotype is a result of combination of these effects. Such a perspective has precedent where other studies described likely adaptive haplotypes that have effects on multiple metabolism gene families, such as IRX (Claussnitzer et al. 2015) and FAD (Fumagalli et al. 2015).
To test whether Tanuki haplogroup indeed affect other neighboring genes, we investigated functional variants that are linked with this haplogroup. Specifically, we identified variants with unusually high PHRED scores (>90th percentile) (Kircher et al. 2014), GWAS variants (MacArthur et al. 2017), and nonsynomous variants (Figure S11). We found no nonsynomous or GWAS variants that is linked with the Tanuki haplogroup. Nevertheless, we found one variant that has a high PHrED score (rs61799140). It is thus plausible that Tanuki haplogroup may have regulatory effect on nearby genes. To further test this, we used gene expression data from Gtex portal (Lonsdale et al. 2013; Wheeler et al. 2016). Our results showed that this haplogroup is associated with significant decreases in expression of the GSTM5 gene in most tissues, especially in the brain, but increases the expression of GSTM3 in skeletal muscle (Figure S12). As such, one potential argument would be that the selected effect is not the functioning of a single gene, but the overall regulatory impact of the Tanuki haplogroup on the GSTM locus.
Discussion
Working With complex genomic structural variation loci
Our results provide a case study for the evolutionary impact of common haplotypic variation in a complex locus, involving both functional single nucleotide and copy number variants. One particular challenge in this locus that we verified is the general lack of linkage disequilibrium between common, neighboring variants. Specifically, we found a lack of strong linkage disequilibrium between the GSTM1 deletion and neighboring variants. This finding corroborates our previous work at a deeper evolutionary depth, where we found evidence for multiple gene conversion events differentiating human and chimpanzee GSTM locus (Saitou et al. 2018). As such, most genome-wide association studies would not have effectively interrogated the potential biomedical impact of the GSTM1 deletion because they have only investigated the single nucleotide variants, which do not adequately tag the deletion variant. In fact, when we consider the best case scenario for such studies and focus on East Asia, where the linkage disequilibrium is relatively high (R2 = 0.4), statistical power for a single nucleotide variant based genome-wide association study would still be low. For example, based on Hong and Park (2012)’s calculations, the statistical power to detect an association would be only 26.5% when assuming a relatively standard experimental setup (e.g., the odds ratio of the phenotype 1.3, 1000 cases and 1000 controls, etc.).
This is not an exceptional case since the majority of deletions reside in similar haplotypic architectures (Sudmant et al. 2015b), complicating both evolutionary analysis, as well as disease association studies that depend on imputation to interrogate structural variants. Almost one-third of the deletions reported in 1000 genomes have R2 < 0.6 with neighboring variants (Sudmant et al. 2015b). As such genome-wide association studies will lose significant amount of power when assessing the effect of such SVs on tested trait if they are using imputation-based genotyping. Consequently, it is important to conduct direct genotyping of the deletion for such analyses or carefully resolve the haplotype structure within the locus to accurately assess the biomedical impact of these variants. Such locus-specific analyses indeed led to important connections between several structural variants and human diseases (Rothman et al. 2010; Usher et al. 2015; Boettger et al. 2016).
Evolution of the GSTM1 locus
Here, we report an incomplete sweep that increase the allele frequency increase of n existing haplogroup (Tanuki) in East Asian populations, putatively affecting the function of multiple GSTM genes (as summarized in Figure 6). Recent studies have shown the importance of sweeps on standing variation (Schrider and Kern 2017), rather than hard sweeps on novel variants (Hernandez et al. 2011), to be the dominating type of positive selection acting on human genome.
These insights fit well with our observations for the Tanuki haplogroup as well as with the emerging, broader picture where genes involved in metabolism have been shown to evolve under complex adaptive forces. For example, the GSTM1 deletion and multiple NAT (N-acetyltransferase) variants were often considered together as the leading candidates for explaining the genetic basis of bladder cancer susceptibility (García-Closas et al. 2005). NAT2 has been reported to be evolving under balancing selection (Mortensen et al. 2011). In addition, variations in other xenobiotic metabolism genes, such as members of the SLC (solute carrier) gene family, have also been shown to be evolving adaptively (Sabeti et al. 2007). Some of these variations involve gene deletions. For example, the deletion of UGT2B17 (uridine diphosphoglucuronosyl-transferase) has been shown to have increased in frequency in East Asian populations under adaptive forces (Xue et al. 2008). Overall, functional variation affecting metabolizing genes may be maintained adaptively in the human populations due to varying environmental pressures.
Implications to evolutionary medicine
As mentioned above, the variation in the GSTM locus has been the subject of more than 500 studies within the context of multiple diseases (Parl 2005). However, given that majority of these studies are correlations, they lack mechanistic or evolutionary insights as to why the genetic variation in this locus is relevant. The emerging picture is that of a gene family (GSTM1-5) that broadly metabolizes multiple carcinogenic substances, for example, 4-nitroquinoline-1-oxide (NQO) (Hayes et al. 2005). On top of that functional layer, it has been shown that this gene family is riddled with common genetic variation, including the unusually common deletion of the GSTM1. These genetic variants, as expected, were associated with multiple cancers (Parl 2005). The mechanistic explanation would be that the reduced the GSTM function leads to higher susceptibility to carcinogenic substance exposure (Hayes et al. 2005).
However, there seems to be some functional redundancy among the GSTM gene family members. As a consequence, the association between single variants to traits may not completely capture the biomedical impact of the variation in this locus as a whole. For example, the GSTM2 may compensate some of the lost function due to the GSTM1 deletion (Bhattacharjee et al. 2013). As such, the overall functional impact of a variant depends on its genomic context. Moreover, a recent pathway-level analysis revealed that glutathione conjugation pathway, for which the GSTM genes are central, is a significant player in determining breast cancer susceptibility (Wang et al. 2017). The implication being that rather than single variants, the combined effect of multiple variants affecting the function of the genes in this pathway eventually contributes to the overall disease susceptibility. This is not a new concept (Jin et al. 2014). Such insights into the collective but varied impact of variation in a given locus have been discussed in evolutionary context in multiple species (Lin et al. 2015; Salojärvi et al. 2017). We are now in a position to quantitatively address this issue, especially using genealogical approaches as we described for the GSTM1 here.
From an evolutionary medicine point of view, it is important to highlight two interrelated features of the GSTM locus. First, even when a single haplotype affects susceptibility to disease, this may be due to pleiotropic effects. Indeed, we described multiple putative functional effects of the Tanuki haplogroup in East Asian populations, including the loss of function due to the GSTM1 deletion and the decrease of the GSTM5 expression. This result exemplifies the benefits of a haplotype-level understanding of the genetic variation. We argue based on our results and those of others (Liu et al. 2005; Claussnitzer et al. 2015) that the co-occurrence of multiple functionally and biomedically relevant variants in particular haplotypes should be treated as the norm, rather than the exception. As exemplified in this study, functional analysis of haplotypes that are under selection may provide crucial targets for future mechanistic studies.
Second, our results add to the growing list of population-specific haplotypes that may contribute to disease susceptibility, further underlying the importance of conducting genetic epidemiology studies in ancestrally diverse populations (Rosenberg et al. 2010; Wojcik et al. 2017). This is especially pertinent to the GSTM locus, given that environmental toxins and carcinogens, which may vary from one population to the other, are the primary target for this gene family. Such loci are also targets for local selection in humans, may be best exemplified by the recent study on Arsenic adaptation in Argentinian Andes population (Schlebusch et al. 2015).
Conclusion
In this study, we report a particular haplogroup (Tanuki haplogroup) carrying the deletion allele that has likely evolved under non-neutral conditions and reached a high allele frequency in East Asian populations. This haplogroup has a broad regulatory effect on the metabolizing GSTM gene family. Overall, our study adds to the emerging notion that complex loci involving and structural variants may contribute to adaptive and biomedically relevant phenotypic variation (Boettger et al. 2016; Sekar et al. 2016).
Acknowledgments
This study is supported by MS’s fund from Astellas Foundation for Research on Metabolic Disorders, Graduate Program for Leaders in Life Innovation, The University of Tokyo Life Innovation Leading Graduate School from MEXT, Japan and Grant-in-Aid for Japan Society for the Promotion of Science (JSPS) Fellows Grant Number 264456. This work is also supported by OG’s start-up funds from University at Buffalo Research Foundation.
We would like to thank Skyler Resendez for careful reading of the earlier versions of this manuscript. We would like to thank Takafumi Ishida, Jun Ohashi, Naoko Fujito and Mehmet Somel for discussions with regards to population genetic analyses. This study constitutes a part of the doctoral thesis of Marie Saitou submitted to The University of Tokyo, Japan.
Footnotes
Supplemental material available at Figshare: https://doi.org/10.25387/g3.6384782.
Communicating editor: S. Tishkoff
Figures: Saitou, Marie; Gokcumen, Omer; Satta, Yoko (2018): G3-Supp0626.pdf. figshare. Dataset.
Literature Cited
- 1000 Genomes Project Consortium, Abecasis G. R., Altshuler D., Auton A., Brooks L. D., et al. , 2010. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. 10.1038/nature09534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Abecasis G. R., Auton A., Brooks L. D., et al. , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee P., Paul S., Banerjee M., Patra D., Banerjee P., et al. , 2013. Functional compensation of glutathione S-transferase M1 (GSTM1) null by another GST superfamily member, GSTM2. Sci. Rep. 3: 2704 10.1038/srep02704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettger L. M., Salem R. M., Handsaker R. E., Peloso G. M., Kathiresan S., et al. , 2016. Recurring exon deletions in the HP (haptoglobin) gene contribute to lower blood cholesterol levels. Nat. Genet. 48: 359–366. 10.1038/ng.3510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchard A., Sanchez J. J., Dalhoff K., Morling N., 2007. Multiplex PCR detection of GSTM1, GSTT1, and GSTP1 gene variants: simultaneously detecting GSTM1 and GSTT1 gene copy number and the allelic status of the GSTP1 Ile105Val genetic variant. J. Mol. Diagn. 9: 612–617. 10.2353/jmoldx.2007.070030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Patterson N., Reich D., 2010. Population differentiation as a test for selective sweeps. Genome Res. 20: 393–402. 10.1101/gr.100545.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M., Dankel S. N., Kim K.-H., Quon G., Meuleman W., et al. , 2015. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 373: 895–907. 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad D. F., Pinto D., Redon R., Feuk L., Gokcumen O., et al. , 2010. Origins and functional impact of copy number variation in the human genome. Nature 464: 704–712. 10.1038/nature08516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisci J. L., Poh Y.-P., Mahajan S., Jensen J. D., 2013. The impact of equilibrium assumptions on tests of selection. Front. Genet. 4: 235 10.3389/fgene.2013.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. 10.1093/bioinformatics/btr330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciotti E., FitzGerald G. A., 2011. Prostaglandins and Inflammation. Arterioscler. Thromb. Vasc. Biol. 31: 986–1000. 10.1161/ATVBAHA.110.207449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L., Carson A. R., Scherer S. W., 2006. Structural variation in the human genome. Nat. Rev. Genet. 7: 85–97. 10.1038/nrg1767 [DOI] [PubMed] [Google Scholar]
- Fujihara J., Yasuda T., Iida R., Takatsuka H., Fujii Y., et al. , 2009. Cytochrome P450 1A1, glutathione S-transferases M1 and T1 polymorphisms in Ovambos and Mongolians. Leg. Med. (Tokyo) 11: S408–S410. 10.1016/j.legalmed.2009.01.073 [DOI] [PubMed] [Google Scholar]
- Fumagalli M., Moltke I., Grarup N., Racimo F., Bjerregaard P., et al. , 2015. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349: 1343–1347. 10.1126/science.aab2319 [DOI] [PubMed] [Google Scholar]
- García-Closas M., Malats N., Silverman D., Dosemeci M., Kogevinas M., et al. , 2005. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet 366: 649–659. 10.1016/S0140-6736(05)67137-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garte S., Gaspari L., Alexandrie A.-K., Ambrosone C., Autrup H., et al. , 2001. Metabolic Gene Polymorphism Frequencies in Control Populations. Cancer Epidemiol. Biomarkers Prev. 10: 1239–1248. [PubMed] [Google Scholar]
- Gaspar P., Hutz M. H., Salzano F. M., Hill K., Hurtado M., et al. , 2002. Polymorphisms of CYP1a1, CYP2e1, GSTM1, GSTT1, and TP53 genes in Amerindians. Am. J. Phys. Anthropol. 119: 249–256. 10.1002/ajpa.10128 [DOI] [PubMed] [Google Scholar]
- Girirajan S., Campbell C. D., Eichler E. E., 2011. Human copy number variation and complex genetic disease. Annu. Rev. Genet. 45: 203–226. 10.1146/annurev-genet-102209-163544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastings P. J., Lupski J. R., Rosenberg S. M., Ira G., 2009. Mechanisms of change in gene copy number. Nat. Rev. Genet. 10: 551–564. 10.1038/nrg2593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes J. D., Flanagan J. U., Jowsey I. R., 2005. Glutathione transferases. Annu. Rev. Pharmacol. Toxicol. 45: 51–88. 10.1146/annurev.pharmtox.45.120403.095857 [DOI] [PubMed] [Google Scholar]
- Hernandez R. D., Kelley J. L., Elyashiv E., Melton S. C., Auton A., et al. , 2011. Classic selective sweeps were rare in recent human evolution. Science 331: 920–924. 10.1126/science.1198878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E. P., Park J. W., 2012. Sample size and statistical power calculation in genetic association studies. Genomics Inform. 10: 117–122. 10.5808/GI.2012.10.2.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. 10.1093/bioinformatics/18.2.337 [DOI] [PubMed] [Google Scholar]
- Huson D. H., Richter D. C., Rausch C., Dezulian T., Franz M., et al. , 2007. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics 8: 460 10.1186/1471-2105-8-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium, Frazer K. A., Ballinger D. G., Cox D. R., Hinds D. A., et al. , 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449: 851–861. 10.1038/nature06258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L., Zuo X.-Y., Su W.-Y., Zhao X.-L., Yuan M.-Q., et al. , 2014. Pathway-based analysis tools for complex diseases: a review. Genomics Proteomics Bioinformatics 12: 210–220. 10.1016/j.gpb.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khrunin A. V., Filippova I. N., Aliev A. M., Tupitsina T. V., Slominsky P. A., et al. , 2016. GSTM1 copy number variation in the context of single nucleotide polymorphisms in the human GSTM cluster. Mol. Cytogenet. 9: 30 10.1186/s13039-016-0241-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. L., Satta Y., 2008. Population genetic analysis of the N-acylsphingosine amidohydrolase gene associated with mental activity in humans. Genetics 178: 1505–1515. 10.1534/genetics.107.083691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Ohta T., 1973. The age of a neutral mutant persisting in a finite population. Genetics 75: 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M., Witten D. M., Jain P., O’Roak B. J., Cooper G. M., et al. , 2014. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 46: 310–315. 10.1038/ng.2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A., Thorleifsson G., Gudbjartsson D. F., Masson G., Sigurdsson A., et al. , 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature 467: 1099–1103. 10.1038/nature09525 [DOI] [PubMed] [Google Scholar]
- Leffler E. M., Band G., Busby G. B. J., Kivinen K., Le Q. S., et al. , 2017. Resistance to malaria through structural variation of red blood cell invasion receptors. Science 356 10.1126/science.aam6393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M., Karczewski K. J., Minikel E. V., Samocha K. E., Banks E., et al. , 2016. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536: 285–291. 10.1038/nature19057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P., Rozas J., 2009. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. 10.1093/bioinformatics/btp187 [DOI] [PubMed] [Google Scholar]
- Lin Y. L., Pavlidis P., Karakoc E., Ajay J., Gokcumen O., 2015. The evolution and functional impact of human deletion variants shared with archaic hominin genomes. Mol. Biol. Evol. 32: 1008–1019. 10.1093/molbev/msu405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.-Y., Zhang Y.-Y., Lu Y., Long J.-R., Shen H., et al. , 2005. A survey of haplotype variants at several disease candidate genes: the importance of rare variants for complex diseases. J. Med. Genet. 42: 221–227. 10.1136/jmg.2004.024752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonsdale J., Thomas J., Salvatore M., Phillips R., Lo E., et al. , 2013. The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J., Bowler E., Cerezo M., Gil L., Hall P., et al. , 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 45: D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques-Bonet T., Kidd J. M., Ventura M., Graves T. A., Cheng Z., et al. , 2009. A burst of segmental duplications in the genome of the African great ape ancestor. Nature 457: 877–881. 10.1038/nature07744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlwain C. C., Townsend D. M., Tew K. D., 2006. Glutathione S-transferase polymorphisms: cancer incidence and therapy. Oncogene 25: 1639–1648. 10.1038/sj.onc.1209373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium, Abecasis G. R., Auton A., Brooks L. D., DePristo M. A., et al. , 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491: 56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mc Vean G. A. T., Myers S. R., Hunt S., Deloukas P., Bentley D. R., et al. , 2004. The Fine-Scale Structureof Recombination Rate Variationin the Human Genome. Science 304: 581–584. [DOI] [PubMed] [Google Scholar]
- Moorjani P., Amorim C. E. G., Arndt P. F., Przeworski M., 2016. Variation in the molecular clock of primates. Proc. Natl. Acad. Sci. USA 113: 10607–10612. 10.1073/pnas.1600374113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen H. M., Froment A., Lema G., Bodo J.-M., Ibrahim M., et al. , 2011. Characterization of genetic variation and natural selection at the arylamine N-acetyltransferase genes in global human populations. Pharmacogenomics 12: 1545–1558. 10.2217/pgs.11.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer A. M., Salavaggione O. E., Hebbring S. J., Moon I., Hildebrandt M. T., et al. , 2007. Glutathione S-transferase T1 and M1: gene sequence variation and functional genomics. Clin. Cancer Res. 13: 7207–7216. 10.1158/1078-0432.CCR-07-0635 [DOI] [PubMed] [Google Scholar]
- Narasimhan V. M., Rahbari R., Scally A., Wuster A., Mason D., et al. , 2017. Estimating the human mutation rate from autozygous segments reveals population differences in human mutational processes. Nat. Commun. 8: 303 10.1038/s41467-017-00323-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M., Li W. H., 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76: 5269–5273. 10.1073/pnas.76.10.5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajic P., Lin Y.-L., Xu D., Gokcumen O., 2016. The psoriasis associated deletion of late cornified envelope genes and has been maintained under balancing selection since Human Denisovan divergence. BMC Evol. Biol. 16: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parl F. F., 2005. Glutathione S-transferase genotypes and cancer risk. Cancer Lett. 221: 123–129. 10.1016/j.canlet.2004.06.016 [DOI] [PubMed] [Google Scholar]
- Perry G. H., Dominy N. J., Claw K. G., Lee A. S., Fiegler H., et al. , 2007. Diet and the evolution of human amylase gene copy number variation. Nat. Genet. 39: 1256–1260. 10.1038/ng2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini S., Polimanti R., Porreca F., Martínez-Labarga C., De Stefano G. F., et al. , 2011. GSTT1 and GSTM1 gene polymorphisms in European and African populations. Mol. Biol. Rep. 38: 1225–1230. 10.1007/s11033-010-0221-0 [DOI] [PubMed] [Google Scholar]
- Prüfer K., Racimo F., Patterson N., Jay F., Sankararaman S., et al. , 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505: 43–49. 10.1038/nature12886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pybus M., Dall’Olio G. M., Luisi P., Uzkudun M., Carreño-Torres A., et al. , 2014. 1000 Genomes Selection Browser 1.0: a genome browser dedicated to signatures of natural selection in modern humans. Nucleic Acids Res. 42: D903–D909. 10.1093/nar/gkt1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F., Sankararaman S., Nielsen R., Huerta-Sánchez E., 2015. Evidence for archaic adaptive introgression in humans. Nat. Rev. Genet. 16: 359–371. 10.1038/nrg3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., Ishikawa S., Fitch K. R., Feuk L., Perry G. H., et al. , 2006. Global variation in copy number in the human genome. Nature 444: 444–454. 10.1038/nature05329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Green R. E., Kircher M., Krause J., Patterson N., et al. , 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468: 1053–1060. 10.1038/nature09710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N. A., Huang L., Jewett E. M., Szpiech Z. A., Jankovic I., et al. , 2010. Genome-wide association studies in diverse populations. Nat. Rev. Genet. 11: 356–366. 10.1038/nrg2760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman N., Garcia-Closas M., Chatterjee N., Malats N., Wu X., et al. , 2010. A multi-stage genome-wide association study of bladder cancer identifies multiple susceptibility loci. Nat. Genet. 42: 978–984. 10.1038/ng.687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwende C., Khoo S. C., Snow R. W., Yates S. N., Kwiatkowski D., et al. , 1995. Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature 376: 246–249. 10.1038/376246a0 [DOI] [PubMed] [Google Scholar]
- Saadat M., 2007. GSTM1 null genotype associated with age-standardized cancer mortality rate in 45 countries from five continents: an ecologic study. Int. J. Cancer Res. 3: 74–91. 10.3923/ijcr.2007.74.91 [DOI] [Google Scholar]
- Sabeti P. C., Varilly P., Fry B., Lohmueller J., Hostetter E., et al. , 2007. Genome-wide detection and characterization of positive selection in human populations. Nature 449: 913–918. 10.1038/nature06250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M., Satta Y., Gokcumen O., Ishida T., 2018. Complex evolution of the GSTM gene family involves sharing of GSTM1 deletion polymorphism in humans and chimpanzees. BMC Genomics 19: 293 10.1186/s12864-018-4676-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salojärvi J., Smolander O.-P., Nieminen K., Rajaraman S., Safronov O., et al. , 2017. Genome sequencing and population genomic analyses provide insights into the adaptive landscape of silver birch. Nat. Genet. 49: 904–912. 10.1038/ng.3862 [DOI] [PubMed] [Google Scholar]
- Schaffner S. F., Foo C., Gabriel S., Reich D., Daly M. J., et al. , 2005. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 15: 1576–1583. 10.1101/gr.3709305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch C. M., Gattepaille L. M., Engström K., Vahter M., Jakobsson M., et al. , 2015. Human Adaptation to Arsenic-Rich Environments. Mol. Biol. Evol. 32: 1544–1555. 10.1093/molbev/msv046 [DOI] [PubMed] [Google Scholar]
- Schrider D. R., Kern A. D., 2017. Soft Sweeps Are the Dominant Mode of Adaptation in the Human Genome. Mol. Biol. Evol. 34: 1863–1877. 10.1093/molbev/msx154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A., Bialas A. R., de Rivera H., Davis A., Hammond T. R., et al. , 2016. Schizophrenia risk from complex variation of complement component 4. Nature 530: 177–183. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeley S. L., McAllister B. F., 2008. Patterns of natural selection at the Alcohol dehydrogenase gene of Drosophila americana. Fly (Austin) 2: 243–246. 10.4161/fly.6875 [DOI] [PubMed] [Google Scholar]
- South A., 2011. rworldmap: A New R package for Mapping Global Data. R J. 3: 35–43. [Google Scholar]
- Sudmant P. H., Mallick S., Nelson B. J., Krumm N., Huddleston J., et al. , 2015a Global diversity, population stratification, and selection of human copy number variation. Science 349: aab3761 10.1126/science.aab3761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudmant P. H., Rausch T., Gardner E. J., Handsaker R. E., Abyzov A., et al. , 2015b An integrated map of structural variation in 2,504 human genomes. Nature 526: 75–81. 10.1038/nature15394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F., 1989. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatewaki N., Maekawa K., Atori N. K., Urose K. K., 2009. Genetic Variations and Haplotype Structures of the Glutathione S-transferase Genes, GSTT1 and GSTM1, in a Japanese Patient Population Naoko. 24: 118–126. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing Consortium , 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437: 69–87. 10.1038/nature04072 [DOI] [PubMed] [Google Scholar]
- Usher C. L., Handsaker R. E., Esko T., Tuke M. A., Weedon M. N., et al. , 2015. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat. Genet. 47: 921–925. 10.1038/ng.3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A., Altheide T. K., 2005. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. 15: 1746–1758. 10.1101/gr.3737405 [DOI] [PubMed] [Google Scholar]
- Visscher P. M., Brown M. A., McCarthy M. I., Yang J., 2012. Five years of GWAS discovery. Am. J. Hum. Genet. 90: 7–24. 10.1016/j.ajhg.2011.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voight, B. F., S. Kudaravalli, X. Wen, and J. K. Pritchard, 2006 A map of recent positive selection in the human genome. PLoS Biol. 4: 0446–0458. [DOI] [PMC free article] [PubMed]
- Wang W., Xu Z. Z., Costanzo M., Boone C., Lange C. A., et al. , 2017. Pathway-based discovery of genetic interactions in breast cancer. PLoS Genet. 13: e1006973 10.1371/journal.pgen.1006973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir B. S., Cockerham C. C., 1984. Estimating F-Statistics for the Analysis of Population Structure. Evolution 38: 1358–1370. 10.1111/j.1558-5646.1984.tb05657.x [DOI] [PubMed] [Google Scholar]
- Weischenfeldt J., Symmons O., Spitz F., Korbel J. O., 2013. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet. 14: 125–138. 10.1038/nrg3373 [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium, Craddock N., Hurles M. E., Cardin N., Pearson R. D., et al. , 2010. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature 464: 713–720. 10.1038/nature08979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler H. E., Shah K. P., Brenner J., Garcia T., Aquino-Michaels K., et al. , 2016. Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues. PLoS Genet. 12: e1006423 10.1371/journal.pgen.1006423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcik G., Graff M., Nishimura K. K., Tao R., Haessler J., et al. , 2017. Genetic Diversity Turns a New PAGE in Our Understanding of Complex Traits. bioRxiv 188094 10.1101/188094 [DOI] [Google Scholar]
- Xue Y., Sun D., Daly A., Yang F., Zhou X., 2008. Adaptive Evolution of UGT2B17 Copy-Number Variation. Am. J. Hum. Genet. 83: 337–346. 10.1016/j.ajhg.2008.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Jaber Y., Pavlidis P., Gokcumen O., 2017a VCFtoTree: a user-friendly tool to construct locus-specific alignments and phylogenies from thousands of anthropologically relevant genome sequences. BMC Bioinformatics 18: 426 10.1186/s12859-017-1844-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Pavlidis P., Taskent R. O., Alachiotis N., Flanagan C., et al. , 2017b Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol. Biol. Evol. 34: 2704–2715. 10.1093/molbev/msx206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, S.-J., Y.-P. Wang, B. Roe, W. R. Pearson, and G. G. Deletion, 1998 Characterization of the Human Class Mu Glutathione S -Transferase Gene Cluster and the GSTM1 Deletion *. 273: 3517–3527. [DOI] [PubMed]
- Zhang F., Gu W., Hurles M. E., Lupski J. R., 2009. Copy number variation in human health, disease, and evolution. Annu. Rev. Genomics Hum. Genet. 10: 451–481. 10.1146/annurev.genom.9.081307.164217 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Table S1 contains genotyping of the GSTM1 copy number by ddPCR of 17 individuals in YRI population in the 1KGP. Supplemental materials are available at Figshare: Saitou, Marie; Gokcumen, Omer; Satta, Yoko (2018): G3-Supp0626.pdf.; https://doi.org/10.25387/g3.6384782.