Abstract
During reverse transcription of the HIV-1 genome, two strand-transfer events occur. Both events rely on the RNase H cleavage activity of reverse transcriptases (RTs) and template homology. Using a panel of mutants of HIV-1BH10 (group M/subtype B) and HIV-1ESP49 (group O) RTs and in vitro assays, we demonstrate that there is a strong correlation between RT minus-strand transfer efficiency and template-primer binding affinity. The highest strand transfer efficiencies were obtained with HIV-1ESP49 RT mutants containing the substitutions K358R/A359G/S360A, alone or in combination with V148I or T355A/Q357M. These HIV-1ESP49 RT mutants had been previously engineered to increase their DNA polymerase activity at high temperatures. Now, we found that RTs containing RNase H-inactivating mutations (D443N or E478Q) were devoid of strand transfer activity, whereas enzymes containing F61A or L92P had very low strand transfer activity. The strand transfer defect produced by L92P was attributed to a loss of template-primer binding affinity and, more specifically, to the higher dissociation rate constants (koff) shown by RTs bearing this substitution. Although L92P also deleteriously affected the RT's nontemplated nucleotide addition activity, neither nontemplated nucleotide addition activity nor the RT's clamp activities contributed to increased template switching when all tested mutant and WT RTs were considered. Interestingly, our results also revealed an association between efficient strand transfer and the generation of secondary cleavages in the donor RNA, consistent with the creation of invasion sites. Exposure of the elongated DNA at these sites facilitate acceptor (RNA or DNA) binding and promote template switching.
Keywords: reverse transcription, human immunodeficiency virus (HIV), DNA synthesis, retrovirus, ribonuclease, RNase H, strand transfer, template-primer binding
Introduction
The HIV type 1 (HIV-1) genome consists of two positive-sense, single-stranded RNA molecules, which are noncovalently linked near their 5′ ends in a region known as the dimer linkage structure. Upon HIV-1 infection, the viral reverse transcriptase (RT)3 converts the single-stranded viral genome into dsDNA. RTs exhibit DNA polymerase (RNA-dependent and DNA-dependent) and RNase H activities (1, 2). Two strand transfer events are required for reverse transcription of the HIV genome (3). The first occurs during minus-strand DNA synthesis and involves the transfer of the newly synthesized cDNA of the 5′ repeat (R) element of the genome to the complementary 3′ R region to continue reverse transcription. The second strand transfer occurs when the 3′ end of the plus-strand DNA containing the primer-binding site is switched to a complementary sequence in the minus-strand DNA to complete the process (1, 2).
HIV RTs are able to slide, flip orientations, and dissociate from the template during reverse transcription (4, 5). It has been estimated that during retroviral reverse transcription, template switching occurs with frequencies in the range of 3 × 10−4 to 1.4 × 10−3 times per nucleotide, i.e. 3–12 strand transfer events per genome replication (6). High HIV recombination frequencies are a consequence of template switching during reverse transcription and allow the virus to explore a larger sequence space (7). In the HIV-1 genome, around 15–20% of all mutations are associated with recombination (8).
Retroviral recombination does not occur at a uniform frequency along the viral genome. The probability that template switching will occur at a given template position is affected by several factors, including the enzymatic properties of the RT, the availability of nucleotide substrates, and the presence of structured regions in the genomic RNA template (9, 10). In addition, strand transfer and further elongation of the synthesized DNA can be facilitated by the presence of viral and cellular proteins. Thus, it has been shown that the nucleic acid annealing and helix destabilization activities of the viral nucleocapsid protein promote strand transfer (11–15). Besides, cytoplasmic lysates of Jurkat cells were found to increase template switching, although the specific protein responsible for these effects was not identified (16).
The DNA polymerase and RNase H activities of the HIV-1 RT are essential for strand transfer during viral replication (3, 9, 17). In fact, the balance between DNA polymerization and endonucleolytic cleavage of the template RNA seems to have an influence on template switching efficiency. A relatively slow polymerization correlates with higher rates of strand transfer (17), whereas lower RNase H activity associates with decreased template switching efficiency (18, 19). In addition, studies in vitro and ex vivo have demonstrated that RTs devoid of RNase H activity are unable to carry out strand transfer reactions (20, 21). RNase H cleavage of the template promotes strand transfer by removing RNA sequences and making newly single-stranded DNA available for annealing to a second template region or by making cuts in the RNA that contribute to strand transfer by creating an invasion site (22).
Apart from the distinctive catalytic activities of retroviral RTs, these enzymes are able to incorporate nucleotides at the 3′ end of blunt-ended template-primers (23–25). It has been suggested that nontemplated nucleotide addition (also known as tailing activity) may facilitate strand transfer by providing 3′ end overhangs in the DNA, where acceptor RNAs bearing a 5′ complementary sequence could bind (26). RTs can clamp together these complexes and facilitate further extension of the DNA. Based on molecular modeling studies that predicted the participation of HIV-1 RT residues Glu-89, Val-90, Gln-91, Leu-92, Lys-154, and Pro-157 in clamp formation, Herzig et al. (27) showed that the substitution of Pro for Leu-92 rendered an HIV-1 RT without strand transfer activity but retaining wildtype (WT) DNA polymerase and RNase H activities. However, in those studies, the influence of template-primer binding affinity in DNA polymerization was not evaluated.
Several studies have shown that nucleotide analogues can enhance strand transfer by slowing DNA polymerization kinetics (19), and similar effects have been reported for amino acid changes that reduced nucleotide incorporation efficiency, such as V148I or Q151N in HIV-1 RT (18, 28). Template-primer binding kinetics, and specifically the dissociation rate constant (koff), could play a relevant role when DNA synthesis is stalled.
In this work, we have determined the strand transfer efficiencies of previously characterized HIV-1 RTs showing different template-primer binding affinities (29). Among the studied enzymes, we included WT HIV-1BH10 (group M/subtype B) and HIV-1ESP49 (group O) RTs, as well as seven HIV-1ESP49 RT mutants containing the connection subdomain substitutions K358R/A359G/S360A (3M) alone or in combination with F61A, K65R/E478Q, V148I, T355A/Q357M, D443N or E478Q. In previous studies, substitutions at positions 358, 359, and 360 had been introduced in the RT to increase its DNA polymerase activity at high temperatures (29). K358R, A359G, and S360A were also responsible for a large increase in nucleic acid–binding affinity, although the other amino acid changes had different effects on the efficiency of nucleotide incorporation (29). The amino acid substitution L92P was also introduced in different sequence contexts to determine whether the RT's clamp activity had an independent effect on the strand transfer efficiency of those enzymes. Our results show that template switching is largely influenced by template-primer binding affinities, nucleic acid–binding kinetics, and the RT's RNase H activity, but it is mechanistically independent of nontemplated nucleotide addition.
Results
Strand transfer efficiency of HIV-1 RT variants
The R region in the HIV-1 genome is a robust hot spot for recombination (30, 31). Previous studies showed that the upper part of the trans-activating response element (TAR) hairpin structure (Fig. 1) in the 5′ R donor is critical for efficient strand transfer (32). We determined RT strand transfer efficiencies with oligonucleotides representing sequences found in the TAR element. For this purpose, a 20-nt synthetic DNA was labeled at its 5′ end with 32P and annealed to an RNA donor of 33 nucleotides (R33B) containing part of the TAR sequence. After preincubating the RNA/DNA hybrid with the corresponding RT, reactions were initiated by adding a mixture of all four dNTPs and an acceptor oligonucleotide of 35 bases, containing a 16-nt overlap with the sequence of R33B. Full-length 49-nt products are detected when strand transfer occurs, whereas in the absence of template switching, the primer is elongated to the 5′ end of the donor RNA template, rendering a product of 30 nucleotides.
Figure 1.
Secondary structure of the 5′ TAR element of HIV-1BH10 and synthetic oligonucleotides used in strand transfer assays. The 5′ and 3′ ends of donor and acceptor molecules are indicated in the TAR structure. The bottom panel shows a standard primer–donor–acceptor complex with expected donor extension (DE) and strand transfer products (TP) (added nucleotides are shown in blue). The R33B/20A hybrid was also used in template-primer binding affinity and RNase H cleavage assays and for the determination of dissociation rate constants.
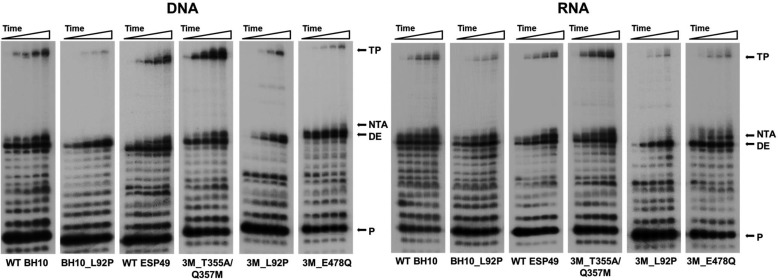
The results of representative strand transfer reactions are shown in Fig. 2. WT BH10 and ESP49 RTs produced significant amounts of transfer product, although the RT with the highest template switching activity was HIV-1ESP49 mutant RT 3M_T355A/Q357M. The amount of transfer product generated with this enzyme was about 2-fold higher than the one obtained with WT ESP49 RT. In contrast, RTs devoid of RNase H activity, such as the mutant 3M_E478Q had almost undetectable strand transfer activity. Similar results were obtained when L92P was introduced in different sequence contexts (i.e. mutant BH10_L92P and 3M_L92P RTs). As shown in Fig. 2, the amounts of strand transfer reaction products were smaller with RNA acceptors than with DNA acceptors. However, the nature of the acceptor had a minor effect on the relative strand transfer efficiencies of the studied RTs. Interestingly, DNA polymerization experiments carried out with dsDNA hybrids D33B/20A in the presence of all four dNTPs and the DNA acceptor 35D provided additional evidence that demonstrated that RNA cleavage was required for template switching (Fig. S1).
Figure 2.
Strand transfer activity of WT and mutant HIV-1 RTs. Assays were carried out with template-primers containing a 5′-32P-radiolabeled DNA primer (20A), an RNA donor (R33B), and DNA or RNA acceptors containing 3′ end sequences that overlap with the 5′ nucleotide sequence of R33B. Reactions were carried out in the presence of RT, donor–primer (R33B/20A), and acceptor (35C or 35D) at concentrations of 6–7.5 nm RTs (active-site concentrations), 30 nm, and 60 nm, respectively. Aliquots were removed at 1, 5, 15, 30, and 60 min. TP, transfer product; DE, donor extension product; NTA, nontemplated nucleotide addition product;, and P, un-extended primer.
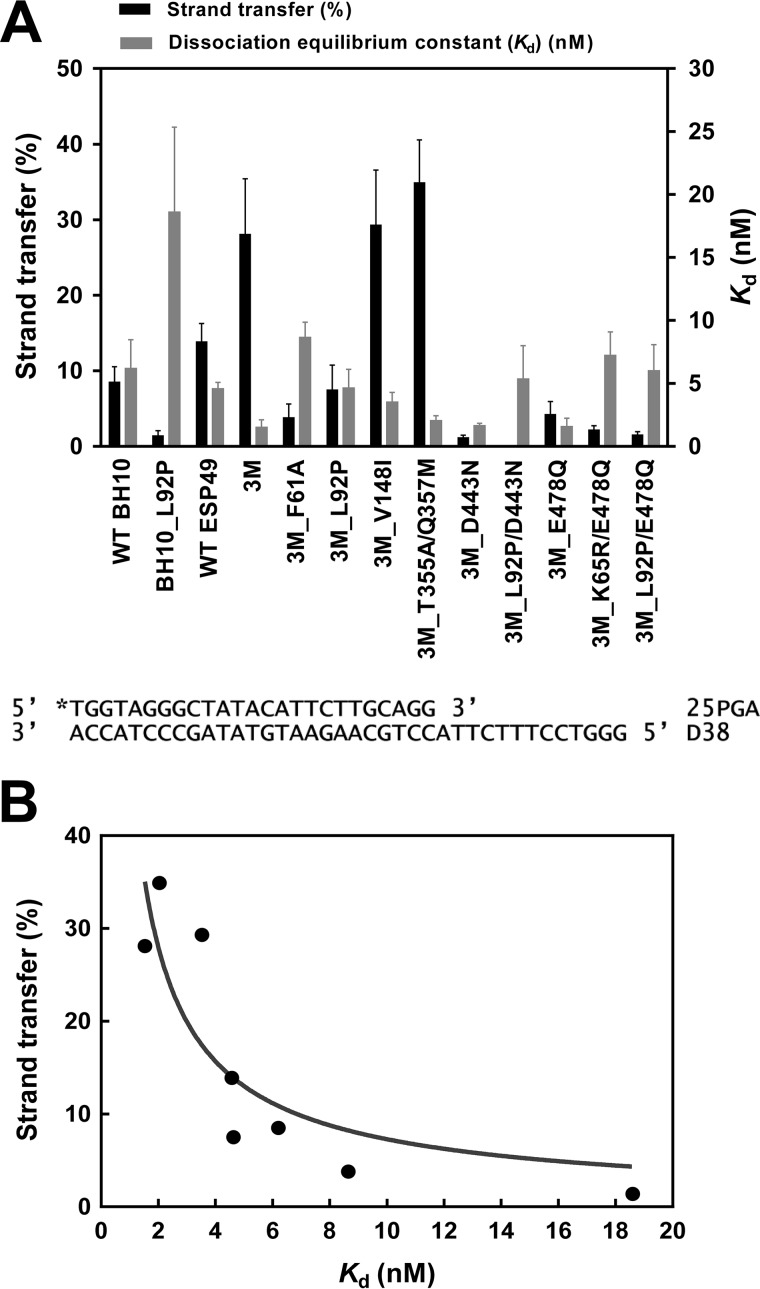
In our assays, the highest strand transfer efficiencies were observed with mutants 3M, 3M_V148I, and 3M_T355A/Q357M both with DNA and RNA acceptors. The differences observed between these mutants and all other tested RTs were statistically significant (p < 0.05, two-tailed t test). In contrast, RTs carrying the amino acid substitution L92P showed very low strand transfer activity (Fig. 3A). These effects were observed not only when L92P was introduced in WT BH10 RT but also when added to ESP49 RT variants containing the three connection subdomain substitutions (i.e. K358R, A359G, and S360A), with or without RNase H-inactivating mutations. We found statistically significant differences between the strand transfer efficiencies of WT BH10 and BH10_L92P, 3M and 3M_L92P, and 3M_D443N and 3M_L92P/D443N RTs (p < 0.05, two-tailed t test). In addition, our results also showed that in the context of the 3M RT sequence, the substitution of Ala for Phe-61 impaired its strand transfer activity.
Figure 3.
Strand transfer efficiencies and dissociation equilibrium constants (Kd) of WT and mutant HIV-1 RTs. A, black bars represent strand transfer efficiencies (average ± S.D.) of all RTs, obtained after incubating the reaction during 60 min using DNA acceptor (35D). Gray bars indicate the mean Kd values ± S.D. for all RTs. Kd values (obtained with template-primer D38/25PGA) were calculated by fitting the data to the quadratic form of the binding equation as described under “Experimental procedures.” Represented values were obtained from at least three independent experiments. B, correlation between the strand transfer efficiencies and binding affinities of all tested enzymes. RTs containing RNase H–inactivating mutations (i.e. all those having D443N or E478Q) were devoid of strand transfer activity and therefore were excluded from the analysis. The correlation was found to be significant (Pearson r test, p < 0.05).
In all cases, the introduction of L92P produced a 3–4-fold increase in the dissociation equilibrium constant (Kd) for DNA/DNA template-primers, suggesting that the strand transfer defect was a consequence of impaired nucleic acid binding. Interestingly, when RTs lacking RNase H activity were excluded from the analysis, we found that strand transfer efficiencies were inversely correlated with template-primer binding affinities (Kd values) obtained with DNA/DNA hybrids (Pearson's correlation coefficient, r = −0.73; p < 0.05) (Fig. 3B).
Contribution of dissociation rate constants (koff) to the strand transfer efficiency of HIV-1 RTs
The dissociation equilibrium constant (Kd) equals koff/kon, where koff and kon correspond to the dissociation and association rate constants, respectively. Dissociation rate constants (koff) for mutant and WT RTs were obtained with the RNA/DNA complex R33B/20A used in strand transfer assays. Most of the RTs had koff values within the range of 0.0051 and 0.0288 s−1 (Table 1). The dissociation rate constants obtained with mutants BH10_L92P and 3M_L92P were rather high and consistent with their lower template-primer binding affinity and minimal strand transfer efficiency. In different sequence contexts (e.g. WT BH10, 3M, etc.), the introduction of L92P produced a 2.5–4.8-fold increase of the koff, explaining in part the deleterious effects of this amino acid substitution in template switching. In agreement with these observations, we found that the Kd values obtained with template-primer R33B/20A for WT BH10 and mutant BH10_L92P RTs were 4.09 ± 0.60 and 12.44 ± 1.96 nm, respectively. The 3.0-fold decrease in binding affinity observed in these assays was consistent with the effects of L92P in the dissociation rate constant. However, unlike the case of Kd values, we found that there was no significant correlation between koff and strand transfer efficiencies of the tested RTs (p = 0.287, Pearson r test).
Table 1.
Dissociation rate constants (koff) of WT and mutant HIV-1 RTs
Reported constants were obtained with the RNA/DNA template-primer R33B/20A and represent the average ± S.D. of at least three independent experiments. Numbers in parentheses represent the fold-increase of the koff relative to the value obtained with WT BH10 RT.
| RTs | koff |
|---|---|
| s−1 | |
| WT BH10 | 0.0141 ± 0.0010 |
| BH10_L92P | 0.0684 ± 0.0130 (4.8) |
| WT ESP49 | 0.0240 ± 0.0034 (1.7) |
| 3M | 0.0203 ± 0.0021 (1.4) |
| 3M_L92P | 0.0913 ± 0.0100 (6.5) |
| 3M_D443N | 0.0051 ± 0.0005 (0.4) |
| 3M_L92P/D443N | 0.0127 ± 0.0014 (0.9) |
| 3M_E478Q | 0.0054 ± 0.0006 (0.4) |
| 3M_L92P/E478Q | 0.0160 ± 0.0018 (1.1) |
| 3M_F61A | 0.0212 ± 0.0020 (1.5) |
| 3M_K65R/E478Q | 0.0057 ± 0.0004 (0.4) |
| 3M_V148I | 0.0288 ± 0.0053 (2.0) |
| 3M_T355A/Q357M | 0.0241 ± 0.0020 (1.7) |
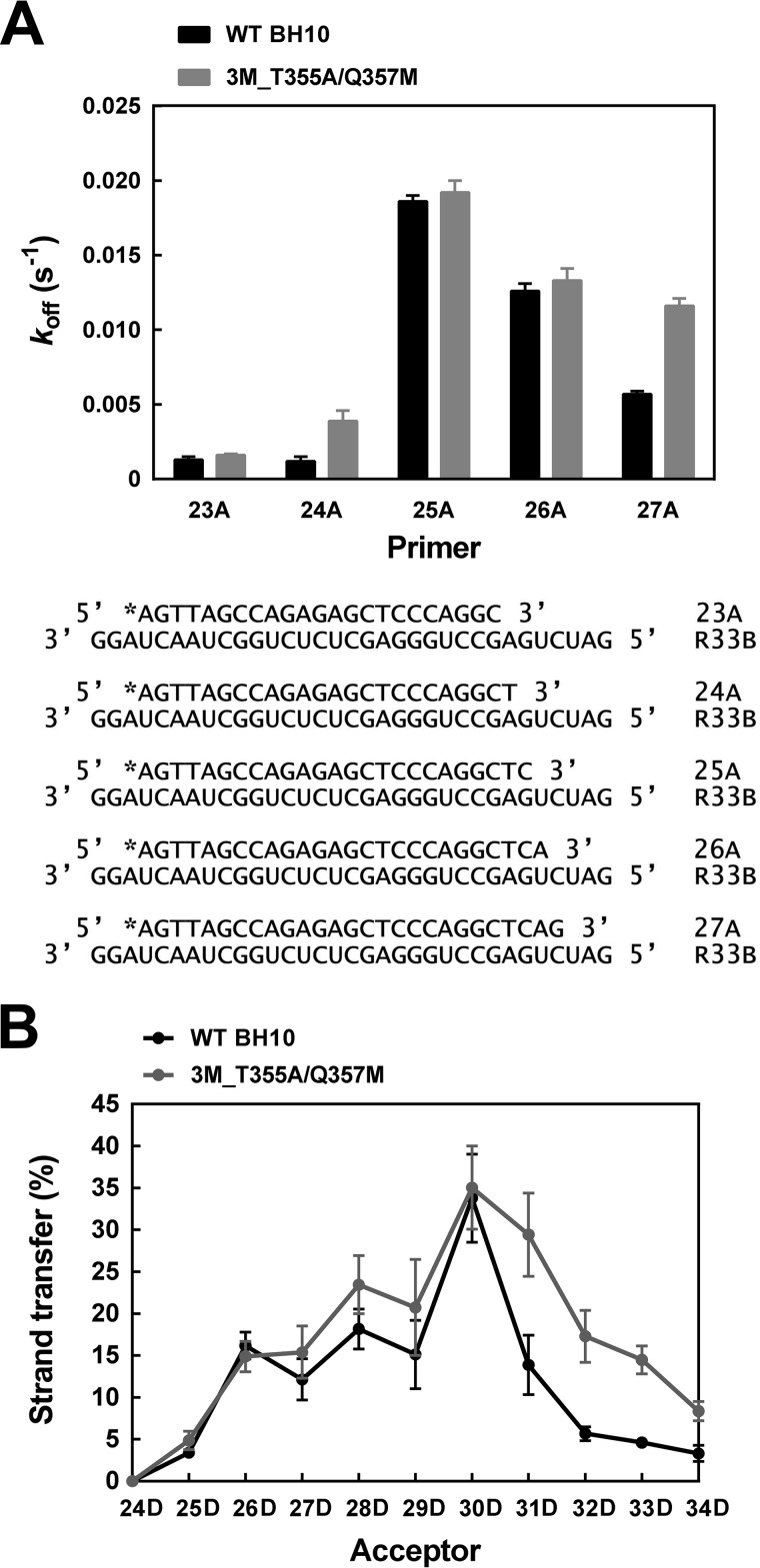
In an attempt to determine the contribution of the koff to template switching efficiency, we obtained dissociation rate constants for template-primers representing potential intermediates of the strand transfer reaction. Donor R33B and acceptors 35C and 35D share an overlapping sequence of 16 nucleotides. Therefore, in the strand transfer assays carried out with those oligonucleotides and the 20A primer, there were 10 positions available for template switching (Fig. 1). We obtained koff values for WT BH10 and mutant 3M_T355A/Q357M RTs and template-primers composed of R33B annealed to primers of 23–27 nucleotides, mimicking extended versions of the 20A primer. As shown in Fig. 4A, the highest koff values were obtained with R33B/25A. A large increase of the dissociation rate constant was observed with this hybrid compared with R33B/24A whose primer was one-nucleotide shorter. These results were consistent with those obtained in strand transfer assays carried out with a series of donor/acceptor duplexes containing overlapping sequences of different lengths. The highest strand transfer efficiencies were obtained with acceptor 30D whose overlapping sequence is complementary to that of primer 25A (Fig. 4B). In agreement with the results of koff determinations, WT BH10 and mutant 3M_T355A/Q357M RTs had similar strand transfer activity profiles, showing maximum efficiencies with the R33B donor and the 30D acceptor.
Figure 4.
Dissociation rate constants (koff) and strand transfer efficiencies of WT BH10 and 3M_T355A/Q357M RTs with primers and acceptors of different lengths. A, dissociation rate constants (koff) determined with RNA/DNA hybrids containing primers of 23–27 nucleotides complexed with the RNA template donor R33B. Reactions were carried out at 37 °C using template-primers R33B/23A, R33B/24A, R33B/25A, R33B/26A, and R33B/27A (10 nm) and an RT concentration of 30 nm. B, effect of the overlapping sequence size on the strand transfer efficiency of WT BH10 and 3M_T355A/Q357M RTs. Strand transfer reactions were carried out with the [32P]DNA/RNA primer–donor complex 20A/R33B, in the presence of DNA acceptors of variable lengths. The used acceptors differ from each other in the number of overlapping nucleotides with the 5′ end of the donor R33B (see Fig. 1 for details). Reported strand transfer efficiencies were obtained after incubating the sample for 60 min and were the averages of three independent determinations.
Effects of RNase H cleavage on strand transfer efficiency
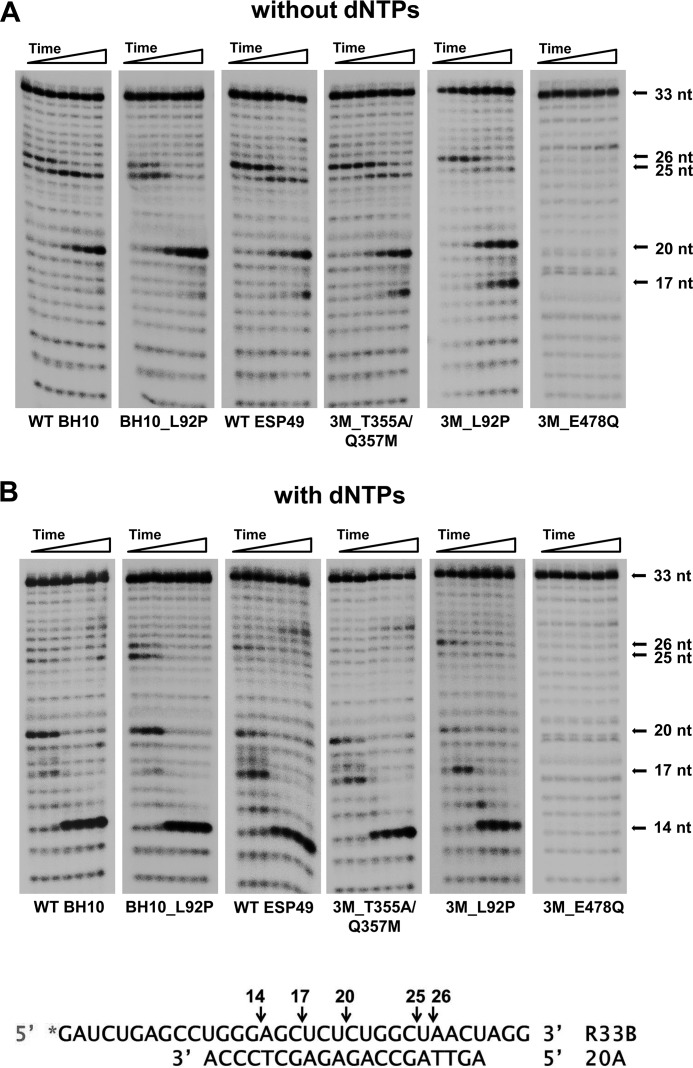
The donor–primer complex R33B/20A was used as substrate to study RNase H cleavage specificity and kinetics with RTs displaying different levels of strand transfer efficiency. As expected, the RNA template remained uncleaved when incubated in the presence of an RT bearing the RNase H active-site substitution E478Q (Fig. 5). However, all other RTs produced similar cleavage patterns when tested with or without dNTPs. The RNase H hydrolysis rate was slightly higher for the HIV-1ESP49 RTs than for the HIV-1BH10 enzymes. Analysis of the cleavage patterns revealed that the most pronounced differences were associated with the amount of 17-nt intermediate generated in reactions carried out in the presence of dNTPs (Fig. 5B). The amino acid substitution L92P had a relatively minor effect on RNase H-mediated cleavage, although we found that 3M_L92P RT produced a larger proportion of secondary cleavage products than WT ESP49 and 3M_T355A/Q357M RTs, in reactions carried out without dNTPs (Fig. 5A). These differences were more pronounced at the 5- and 15-min time points, where the proportion of 17-nt versus 20-nt intermediates was higher for the 3M_L92P RT than for the WT ESP49 enzyme. An increase in RNase H activity was also observed with mutant BH10_L92P RT compared with the WT BH10 enzyme (Fig. 5A). In this case, the largest differences were observed at the 15-min time point, where we detected 42.1 and 30.5% of degraded substrate with BH10_L92P RT and WT BH10 RT, respectively.
Figure 5.
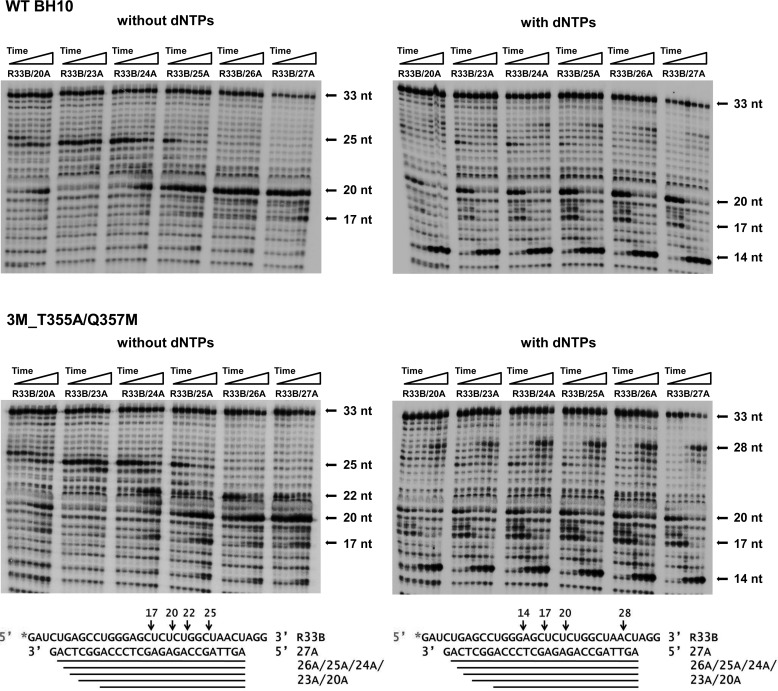
RNase H activity of WT and mutant HIV-1 RTs. Cleavage of a [32P]RNA/DNA substrate (R33B/20A) (30 nm) was carried out at 37 °C in the presence of the corresponding RT at 50 nm (active enzyme concentration). Aliquots were removed at 0.25, 0.5, 1, 5, 15, 30, and 60 min. A and B show RNase H cleavage patterns obtained in reactions carried out in the absence or in the presence of dNTPs, respectively. Arrows indicate the position of the uncleaved RNA substrate (33 nucleotides) and the hydrolysis products of 14, 17, 20, 25, and 26 nucleotides. The locations of cleavage sites on the RNA template are indicated below.
The results obtained with the R33B/20A hybrid suggested that secondary cleavages could affect strand transfer efficiency. In addition, cleavage patterns obtained with different RTs showed a preferential hydrolysis of the RNA template at specific positions, rendering products of 14, 17, 20, 25, and 26 nucleotides. This preference was also observed with template-primers that contained R33B annealed to elongated primers of 23–27 nucleotides. In the absence of dNTPs, we identified two major cleavage sites rendering products of 20 and 25 nucleotides (Fig. 6). The intensity of bands corresponding to those cleavage products varied depending on the template-primer used in the assays. Although the 25-nt RNA predominated in assays carried out with hybrids R33B/23A and in a lesser extent with R33B/24A, shorter products of 20 nucleotides were generated with template-primers R33B/25A, R33B/26A, and R33B/27A with both tested RTs. Observed cleavages were consistent with a distance of 14–16 nucleotides between the DNA polymerase and RNase H active sites (Fig. 6).
Figure 6.
Effect of the primer length on the polymerization-dependent RNase H cleavage patterns obtained with WT BH10 and 3M_T355A/Q357M RTs. Cleavage of 32P-labeled R33B by the RNase H activity of both RTs was monitored with hybrids containing primers of 20 and 23–27 nucleotides. Reactions were carried out in the presence of 30 nm template-primer and 50 nm RT, and aliquots were withdrawn at 0.25, 0.5, 1, 5, 15, 30, and 60 min. Both enzymes were assayed in the absence and in the presence of dNTPs. In polymerization-dependent RNase H activity assays, the concentration of each nucleotide was 500 μm.
Polymerase-dependent RNase H hydrolysis was observed while synthesizing fully-elongated primers. Thus, in the presence of dNTPs and after 5-min incubations, WT BH10 and 3M_T355A/Q357M RTs produced relatively large amounts of 14-nt RNA products with all tested hybrids. Besides, both enzymes rendered similar cleavage patterns, although hydrolysis was somewhat more efficient with the 3M_T355A/Q357M RT. In assays carried out with complexes R33B/25A, R33B/26A, and in a lesser extent with R33B/27A, we observed the generation of intermediate oligonucleotide products of 17–20 nucleotides (Fig. 6), consistent with the creation of invasion sites that could facilitate template switching. The cleavage sites involved in the generation of RNAs of 17–20 nucleotides occur at the UCUCU sequence, adjacent to the loop found in the TAR structure (Fig. 1). The early appearance of those cleavage products is consistent with the dissociation rate constants obtained with both RTs, because RNA degradation could facilitate the dissociation of template-primer and RT during the polymerization reaction. The koff values obtained for WT BH10 and 3M_T355A/Q357M RT and template-primers R33B/25A and R33B/26A were in the range of 0.0126 and 0.0192 s−1 (Fig. 4), indicating that ∼50% of the template-primer was dissociated from the enzyme after 50–80 s of incubation. These values are consistent with the significant cleavage observed at the UCUCU sequence with both enzymes after a 1-min incubation (Fig. 6).
Nontemplated nucleotide addition and strand transfer are mechanistically independent
Several studies have suggested that nontemplated base addition could facilitate template switching by creating 3′ overhangs where complementary 5′ template ends could bind (23, 25, 26). We found significant tailing activity levels (i.e. overextension of the 3′ end of the primer) with RTs displaying higher levels of strand transfer activity.
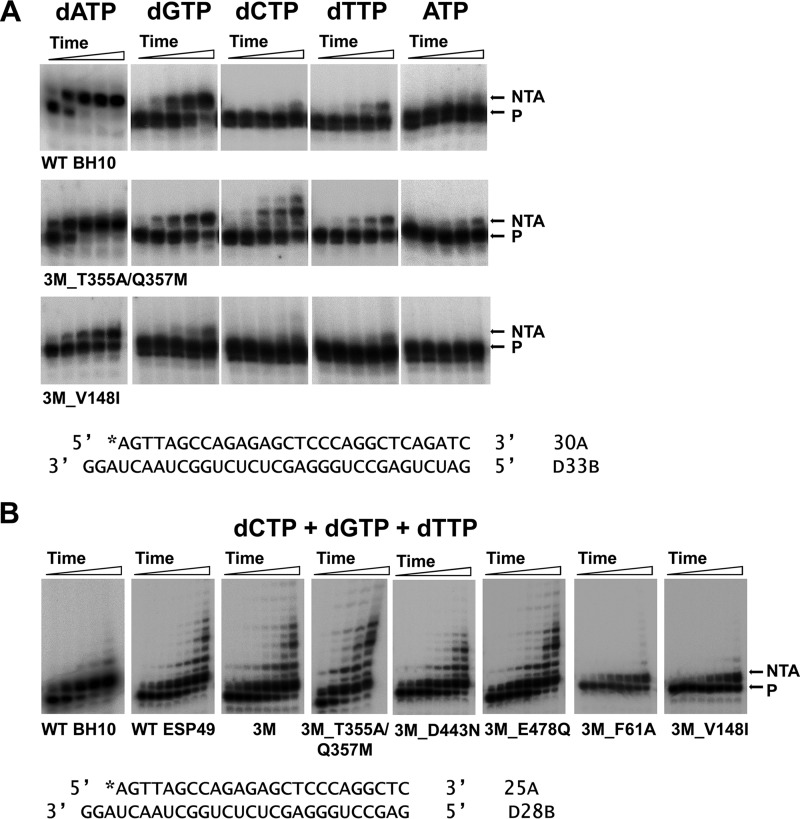
The nontemplated nucleotide addition activity of all studied HIV-1 RTs was assessed with the blunt-ended template-primer D33B/30A. In agreement with previous reports (23, 24), all RTs showed a marked preference for incorporating dATP at 3′ end of the primer (Table 2). In contrast, dTTP and ATP were poor substrates of the reaction. Interestingly, WT BH10 and ESP49 RTs, as well as mutants 3M, 3M_T355A/Q357M, 3M_E478Q, and to a lesser extent 3M_D443N showed the highest levels of nontemplated nucleotide addition activity (kobs values for the incorporation of dATP were in the range of 0.13 to 0.23 min−1) (Table 2). In contrast, 3M derivatives carrying mutations such as F61A, K65R, L92P, or V148I showed very low levels of nontemplated nucleotide addition. These amino acid substitutions map in the vicinity of the polymerase active site and, with the exception of K65R, do not interact with incoming dNTP (Protein Data Bank code 1RTD). Interestingly, the nontemplated nucleotide addition activity of mutant 3M_V148I was low (Fig. 7A) despite showing the highest strand transfer efficiencies, similar to those of mutants 3M and 3M_T355A/Q357M (Fig. 3A). Nontemplated dATP incorporation rates for 3M and 3M_T355A/Q357M RTs were >15 times higher than for mutant 3M_V148I RT (Table 2).
Table 2.
Rates of nontemplated nucleotide addition of WT and mutant HIV-1 RTs using blunt-ended DNA/DNA substrates
Reactions were carried out at 37 °C with DNA/DNA hybrid D33B/30A (30 nm), using a RT concentration of 60 nm. The dNTP concentration in the reaction was 500 μm. Reported incorporation rates (koff) represent the average ± S.D. of at least three independent experiments.
| RTs |
kobs |
||||
|---|---|---|---|---|---|
| dATP | dGTP | dCTP | dTTP | ATP | |
| min−1 | |||||
| WT BH10 | 0.227 ± 0.008 | (1.81 ± 0.06) × 10−2 | (8.50 ± 3.00) × 10−4 | (3.74 ± 0.11) × 10−3 | <5 × 10−5 |
| BH10_L92P | (5.00 ± 1.98) × 10−4 | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 |
| WT ESP49 | 0.216 ± 0.011 | (6.11 ± 0.38) × 10−3 | (1.18 ± 0.18) × 10−3 | (1.43 ± 0.14) × 10−3 | (4.30 ± 0.58) × 10−4 |
| 3M | 0.198 ± 0.006 | (1.75 ± 0.02) × 10−2 | (3.98 ± 0.35) × 10−3 | (3.69 ± 0.17) × 10−3 | (4.40 ± 0.60) × 10−4 |
| 3M_T355A/Q357M | 0.214 ± 0.011 | (1.86 ± 0.08) × 10−2 | (1.60 ± 0.05) × 10−2 | (5.04 ± 0.15) × 10−3 | (1.31 ± 0.02) × 10−3 |
| 3M_D443N | 0.130 ± 0.008 | (9.02 ± 0.30) × 10−3 | (8.49 ± 0.25) × 10−3 | (2.67 ± 0.10) × 10−3 | (1.00 ± 0.08) × 10−3 |
| 3M_E478Q | 0.203 ± 0.018 | (1.25 ± 0.03) × 10−2 | (1.05 ± 0.02) × 10−2 | (4.01 ± 0.12) × 10−3 | (1.82 ± 0.06) × 10−3 |
| 3M_F61A | (3.33 ± 0.09) × 10−2 | (5.11 ± 0.18) × 10−3 | (1.20 ± 0.31) × 10−4 | (1.74 ± 0.14) × 10−3 | <5 × 10−5 |
| 3M_K65R/E478Q | (1.18 ± 0.03) × 10−2 | (9.40 ± 1.10) × 10−4 | <5 × 10−5 | (9.50 ± 2.40) × 10−5 | <5 × 10−5 |
| 3M_L92P | (3.22 ± 0.26) × 10−3 | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 |
| 3M_L92P/D443N | (1.02 ± 0.12) × 10−3 | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 |
| 3M_L92P/E478Q | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 | <5 × 10−5 |
| 3M_V148I | (1.30 ± 0.04) × 10−2 | (2.08 ± 0.07) × 10−3 | (4.70 ± 0.63) × 10−4 | (1.09 ± 0.22) × 10−3 | <5 × 10−5 |
Figure 7.
Nontemplated nucleotide addition activity of WT and mutant HIV-1 RTs. Reactions were carried out with blunt-ended DNA/DNA hybrids D33B/30A (A) or D28B/25A (B). Primers 30A and 25A were labeled with 32P at their 5′ ends. In these assays, RTs were supplied at 60 nm, whereas the template-primer concentration was 30 nm. A, nontemplated nucleotide addition activity of WT BH10, 3M_T355A/Q357M, and 3M_V148I RTs in reactions carried out in the presence of a single nucleotide (at 500 μm final concentration). Aliquots were withdrawn at 1, 5, 15, 30, and 60 min. B, nontemplated nucleotide addition activity of WT and mutant HIV-1 RTs determined in the presence of a mixture of dCTP, dGTP, and dTTP (at 500 μm each). Aliquots were withdrawn at 0.5, 1, 2.5, 5, 10, 15, 30, and 60 min, except for WT BH10 and 3M_T355A/Q357M RTs, where time points were obtained after incubating the reaction during 1, 5, 15, 30, and 60 min.
Several RTs were able to incorporate significant amounts of dGTP and dCTP on blunt-ended template-primer. In the case of dCTP, we observed some tailing activity (i.e. incorporation of two or more nucleotides), which was significantly enhanced in the presence of mixtures of dGTP, dCTP, and dTTP (Fig. 7B). This tailing activity was most pronounced with WT ESP49 RT and with mutant RTs 3M, 3M_T355A/Q357M, 3M_E478Q, and 3M_D443N (Fig. S2). In contrast, 3M_F61A and 3M_V148I RTs were devoid of tailing activity. Nucleotide incorporation kinetics using different combinations of nucleotides confirmed the reduced ability of 3M_V148I RT to incorporate nucleotides at the 3′ ends of blunt-ended template-primers (Table S1), supporting the notion that unlike template-primer binding kinetics the nontemplated nucleotide addition has a very small impact on template switching activity.
Discussion
One of the hallmarks of retroviral replication is their high frequency of recombination. There are two obligatory strand transfer events during reverse transcription, but the presence of breaks in the packaged viral RNA facilitate template switching and the generation of recombinant genomes (9, 33). According to the dynamic copy-choice model, at steady state, template switching is determined by the relative contributions of DNA polymerization and RNA degradation during reverse transcription (17). However, several factors such as the structure of the viral RNA, the intrinsic properties of the RT, and viral and cellular proteins that interact with the reverse transcription complex may affect this balance (34). Our results demonstrate that strand transfer efficiencies correlate with template-primer binding affinity and reveal that nucleic acid binding kinetics may be affected by secondary RNase H cleavages that could facilitate the creation of invasion sites favoring template switching.
In this study, the effect of template-primer affinity on the efficiency of the strand transfer reaction has been demonstrated with a panel of HIV-1 RTs of biotechnological interest engineered to increase their DNA polymerization activity at high temperatures (29). The 3M and 3M_T355A/Q357M mutants are HIV-1 group O RTs with amino acid substitutions in the connection subdomain (Fig. 8) that conferred about 2–3-fold increased affinity for a model template-primer (29). These enzymes were also the ones showing the highest strand transfer efficiencies in our assays. In a recent report, authors claimed that in HIV-1 RT, the amino acid substitution L92P abolished template switching without affecting the enzyme's DNA polymerase and RNase H activities (27). However, our results demonstrate that the defect of this mutant in strand transfer is a consequence of the loss of template-primer binding affinity (Kd). These effects on the Kd have been demonstrated with RTs having different combinations of amino acid substitutions (Fig. 3). The role of Leu-92 in template-primer binding can be explained by its contribution to the stability of the RT, because this residue locates at the interface between the 66- and 51-kDa subunits of the enzyme (35). In addition, Leu-92 interacts with the DNA template in the ternary structure of HIV-1 RT bound to a DNA/DNA hybrid and an incoming dNTP (Fig. 8).
Figure 8.
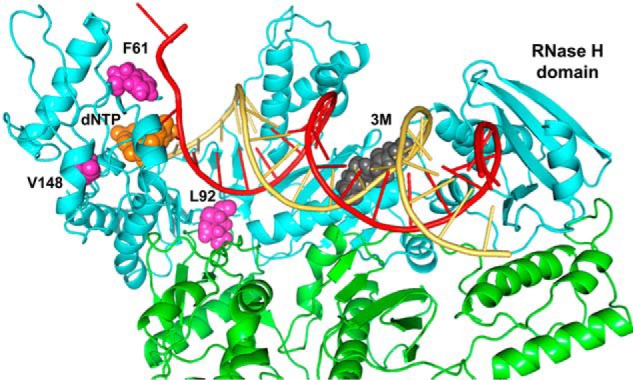
Molecular model of HIV-1ESP49 RT in a ternary complex with dsDNA and dTTP, showing the location of substituted amino acids in the studied mutants. The model was built by using the structure of WT HIV-1 RT (HXB2) strain bound to dsDNA and dTTP as template (29). RT subunits are represented by cyan and green ribbons. The incoming dTTP is shown in orange; the side chains of Phe-61, Leu-92, and Val-148 are shown in purple, and the side chains of connection subdomain residues 358–360 are shown in dark gray. The template and primer strands are depicted in red and yellow, respectively.
It has been shown that HIV-1 RT dissociates during strand transfer (5). L92P promotes the dissociation of the RT from the template-primer. Thus, our studies with different enzymes reveal that L92P produced more than 3-fold increases in the RT/template-primer dissociation rate constant (koff). Previous studies carried out with two mutant HIV-1 RTs bearing sequences from viruses obtained before and after treatment with antiretroviral drugs showed that the RT with the higher association rate (kon) was also characterized by its higher strand transfer activity (36). In contrast, both RTs had similar koff values. Although the contribution of the kon has not been accurately determined in our assays, our data indicate that the koff plays an important role in template switching. In addition, it is still possible that the relative contribution of koff and kon to strand transfer could also depend on the specific template-primer and/or RT analyzed.
It is well established that RNase H activity is required for template switching (20, 21, 37). However, strand transfer efficiencies do not seem to be affected by the RNA cleavage rates. Moreover, RNase H active–site mutations (e.g. D443N or E478Q) abrogated strand transfer activity. Although our studies showed that the HIV-1ESP49 RTs were slightly more active than the HIV-1BH10 RT variants in RNase H cleavage assays, differences detected among those enzymes were small. Interestingly, L92P mutants with impaired strand transfer efficiency and reduced template-primer affinities had a slightly higher RNase H activity when tested with the R33B/20A hybrid (Fig. 5). The different RNase H cleavage patterns observed with WT BH10 and 3M_T355A/Q357M RTs were consistent with the higher activity of the latter enzyme. Besides, we found that both RTs generated multiple cuts at positions 17–20 with primers of 24 or more nucleotides (Fig. 6). These cuts are located at positions adjacent to the hairpin loop in the TAR element used in our assays and may reflect the generation of invasion sites that could facilitate strand transfer. These observations are consistent with the evidence showing that strand transfer requires docking of the nascent DNA into acceptor RNA or DNA followed by strand invasion (38). DNA 3′-directed RNase H cleavages (either primary or secondary) are known to determine strand transfer efficiency (22), probably by creating invasion sites that constitute a rate-limiting step with most tested templates (39).
The results of the RNase H cleavage experiments in the presence of dNTPs (i.e. polymerase-dependent RNase H cleavage) with elongated DNA primers were also consistent with determinations of dissociation rate constants (koff values) using the same hybrids. Higher rates appear to result from partial degradation of the RNA donor and correlate well with strand transfer efficiencies obtained with different acceptors (Fig. 4). In agreement with those observations, higher efficiencies were obtained with the 3M_T355A/Q357M RT, rendering maximum values with the acceptor that had the same nucleotide sequence than the 25A primer.
RTs derived from the HIV-1ESP49 strain, including mutants 3M, 3M_T355A/Q357M, 3M_D443N, and 3M_E478Q, had a strong tailing activity, particularly in the presence of mixtures of dGTP, dTTP, and dCTP. In the presence of dCTP, those enzymes were able to incorporate several nucleotides at the 3′ end of the growing DNA chain, as shown previously for murine leukemia virus RTs (25, 40). Interestingly, WT BH10 RT was a proficient enzyme in nontemplated nucleotide addition experiments but had very low tailing activity. HIV-1 RTs, as well as murine leukemia virus and avian myeloblastosis virus RTs, shared a preference for incorporating A at the 3′ end of blunt template-primers (23, 25, 41).
Overhangs generated by retroviral RTs can be used in TA cloning, GC cloning, cosmid library preparations, and for efficient ligation of adaptor molecules to DNA ends in next-generation sequencing. Furthermore, template-switch and nontemplated C addition at the 5′ of the cDNA are also commonly used for the preparation of libraries using standard methods such as SMART or Peregrine (42–44). Although nucleic acid hybrids formed by the association of short (i.e. one or two nucleotides) complementary 3′ ends of growing DNA chains and 5′ ends of acceptor strands are thermodynamically unstable, RTs can stabilize those complexes and continue the polymerization reaction (26). Although this clamp activity has been proposed as a relevant factor contributing to facilitate template switching, our results provide evidence suggesting a relatively minor impact on strand transfer efficiency.
This conclusion is based on the lack of correlation between the strand transfer efficiencies and tailing activities of the studied mutants. For example, the 3M_V148I RT has low nontemplated nucleotide addition activity but high strand transfer efficiency. The low tailing activity of the mutant was observed in the presence of single nucleotides and with mixtures of two or more dNTPs. Val-148 locates in the vicinity of the nucleotide-binding site but does not interact with the incoming dNTP. Its low activity in nontemplated nucleotide addition assays could be related to an indirect effect on the side chain of the dNTP-binding residue Gln-151 (Fig. 8). The amino acid substitutions L92P and F61A also had negative impacts on the RT's tailing activity, although tested enzymes had low strand transfer activity. Phe-61 (together with Trp-24) acts on template-primer binding and remodeling of the catalytic site (45). Specifically, the F61A substitution has been shown to increase the accuracy of HIV-1 RT, although the mutant showed very low processivity compared with the WT enzyme (46, 47).
In summary, our results demonstrate that template-primer binding affinity and RNase H activity are critical properties of the RT, required for efficient template switching. Secondary cleavages in the donor RNA due to efficient endonucleolytic activity generate invasion sites while promoting RT dissociation from the template-primer. These processes appear to be critical for strand transfer, and amino acid substitutions altering those RT properties, such as L92P or RNase H-inactivating mutations, have a major impact on template switching. In contrast, nontemplated nucleotide addition does not appear to be strongly associated with strand transfer efficiency, and the contribution of the RT's clamp activity to recombination events is expected to be negligible, although under specific conditions this property might be exploited for biotechnological applications.
Experimental procedures
Expression and purification of RTs
The WT group O HIV-1ESP49 (WT ESP49) RT and mutants containing the connection subdomain substitutions K358R/A359G/S360A (3M) alone, or in combination with F61A, K65R/E478Q, V148I, T355A/Q357M, or E478Q were obtained and purified in our laboratory as described previously (29). Expression and purification of WT and mutant HIV-1BH10 (WT BH10) RTs were carried out as described previously (48, 49). In all cases, RT p66 subunits were co-expressed with HIV-1 protease in Escherichia coli XL1 Blue cells to obtain p66/p51 heterodimers that were purified by ion-exchange chromatography followed by immobilized metal-affinity chromatography on Ni2+-nitrilotriacetic acid–agarose (50, 51). All RTs were quantified by active-site titration prior to biochemical studies (52). For normalization purposes, the RT concentrations reported in this work refer to the amount of catalytically active enzyme as determined by active-site titration at saturating template-primer concentrations (53, 54).
Site-directed mutagenesis
Site-directed mutagenesis was carried out by following the QuikChangeTM (Stratagene) standard protocol. Plasmids derived of p66RTB and containing the HIV-1 RT-coding sequence and a poly(CAG) tail encoding a His6 tag at its 3′ end were used as templates. Mutations L92P, D443N, and/or E478Q were introduced in a sequential manner into the coding sequence of the 3M variant of group O HIV-1ESP49 RT (3M_L92P, 3M_D443N, 3M_L92P/D443N, and 3M_L92P/E478Q) or into WT HIV-1BH10 RT (BH10_L92P), by using primers described in Table S2. Mutagenesis was verified by sequencing of the whole RT-coding region.
Nucleotides and template-primers
Stock solutions of dNTPs and rNTPs (at 100 mm) were obtained from GE Healthcare. Synthetic oligonucleotides were obtained from Sigma and Integrated DNA Technologies. HPLC-purified oligonucleotides were labeled at their 5′ termini with 1–2 μCi of [γ-32P]ATP (10 mCi/ml; 3000 Ci/mmol, PerkinElmer Life Sciences) and 5 units of T4 polynucleotide kinase (New England Biolabs) in 70 mm Tris-HCl buffer, pH 7.6, containing 10 mm MgCl2 and 5 mm dithiothreitol (DTT). Template-primer solutions were prepared at 300 nm in different buffers as follows: (i) 100 mm Hepes buffer, pH 7.0, containing 45 mm magnesium acetate and 45 mm NaCl (for strand transfer, RNase H, and nontemplated nucleotide addition assays); (ii) 550 mm Hepes buffer, pH 7.0, containing 150 mm magnesium acetate and 150 mm NaCl (for DNA binding affinity assays); and (iii) 100 mm Hepes buffer, pH 7.0, containing 30 mm magnesium acetate and 30 mm NaCl (for the determination of dissociation rate constants).
Strand transfer assays
The strand transfer activity of the RTs was determined in vitro using three synthetic oligonucleotides: a 5′ 32P-radiolabeled DNA primer (20A), an RNA donor of 33 nucleotides (R33B), and 35-nucleotide RNA or DNA acceptors (35C or 35D, respectively) containing sequences partially homologous to those of R33B. Strand transfer control experiments carried out without RNA involved the use of a hybrid containing a 5′ 32P-radiolabeled DNA (20A), a DNA donor of 33 nucleotides (D33B), and the 35D acceptor. The nucleotide sequences of RNA donors and acceptors correspond to those found in the TAR element of HIV-1HXB2 (GenBankTM accession number K03455) (Fig. 1). In addition, strand transfer efficiency of WT BH10 RT and HIV-1ESP49 RT mutant 3M_T355A/Q357M was measured with DNA acceptor oligonucleotides of different lengths, varying from 24 to 35 nucleotides and containing sequences of 5 to 16 nucleotides homologous to those of the RNA or DNA donor. In these assays, the labeled primer 20A was annealed to the donor RNA or DNA at a molar ratio of donor to primer of 4:1. RTs (12–15 nm) were preincubated 10 min at 37 °C with 60 nm template-primer in 20 mm Hepes buffer, pH 7.0, containing 9 mm magnesium acetate, 9 mm NaCl, 120 mm potassium acetate, 1 mm dithiothreitol (DTT), and 5% PEG 6000. Reactions were started by adding an equal volume (15 μl) of a solution of 150 mm potassium acetate, 1 mm DTT, and 5% PEG 6000, containing all four dNTPs (1 mm each), and the corresponding acceptor RNA or DNA at 120 nm concentration. Strand transfer reactions were incubated for 1, 5, 15, 30, and 60 min and then terminated by adding an equal volume of stop solution (10 mm EDTA in 90% formamide containing 1 mg/ml xylene cyanol FF and 1 mg/ml bromphenol blue). The strand transfer products were visualized on denaturing 20% polyacrylamide-8 m urea gels. Strand transfer efficiencies were determined by phosphorimaging with a BAS1500 scanner (Fuji), using the program Tina version 2.09 (Raytest Isotopenmessgerate Gmbh, Staubenhardt, Germany). Yields were calculated as the amount of fully extended products (up to 49 nucleotides long) relative to the total amount of template-primer used in the assay.
DNA-binding affinity assays
The dissociation equilibrium constants (Kd) for RTs and hybrids R33B/20A (Fig. 1) and D38/25PGA (Fig. 3) were obtained as described previously (29). DNA primers 20A and 25PGA were labeled at their 5′ end with 32P and annealed to RNA (R33B) or DNA (D38) in 550 mm Hepes buffer, pH 7.0, containing 150 mm magnesium acetate and 150 mm NaCl. RTs (4–6 nm) were preincubated during 10 min at 37 °C with increasing concentrations of template-primer complexes (ranging from 0.5 to 60 nm) in 100 mm Hepes buffer, pH 7.0, containing 30 mm magnesium acetate, 30 mm NaCl, 130 mm potassium acetate, 1 mm DTT, and 5% PEG 6000. Reactions were initiated by adding an equal volume (10 μl) of 150 mm potassium acetate, 1 mm DTT, and 5% PEG 6000, containing the corresponding dNTP at 1 mm concentration (dTTP for D38/25PGA and dGTP for R33B/20A). Aliquots of 3–4 μl were removed after 10, 20, 30, and 40 s, and mixed with equal amounts of sample loading buffer (stop solution). Results were analyzed by denaturing PAGE and phosphorimaging as described above. The amounts of RT bound to template-primer at time 0 were extrapolated from the nucleotide incorporation data at 10–40 s, and the data were fit to Equation 1,
| (Eq. 1) |
where [RT·TP], Kd, [RTT], and [TP] represent the productive template-primer complex concentration, the equilibrium dissociation constant for RT-nucleic acid binding, the active enzyme concentration, and the template-primer concentration, respectively (54). Calculations were done using GraphPad Prism version 6.00 for Windows (GraphPad Software, La Jolla, CA).
Determination of dissociation rate constants (koff)
Dissociation rate constants were determined with hybrids containing 5′ 32P-radiolabeled DNA primers and RNA or DNA templates. In these assays, we used seven different RNA/DNA complexes (R33B/20A, R33B/23A, R33B/24A, R33B/25A, R33B/26A, and R33B/27A) and one DNA/DNA complex (D33B/20A) (sequences shown in Figs. 1 and 4). Radioactively labeled primers were annealed to templates at 4:1 molar ratios of template over primer. RTs (60 nm) were preincubated 1 min at 37 °C in the presence of template-primer (20 nm) in 20 mm Hepes buffer, pH 7.0, containing 6 mm magnesium acetate, 6 mm NaCl, 130 mm potassium acetate, 1 mm DTT, and 5% PEG 6000. Then, salmon sperm DNA was added to a concentration of 5.5 mg/ml, and the incubation was continued for 0, 5, 10, 15, 20, 25, 30, 40, 50, 50, 60, 70, 80, 100, 120, and 150 s, before adding the corresponding dNTP (at 50 μm final concentration). Samples were then incubated during 7 s, and reactions were stopped by adding an equal volume of sample loading buffer. Products were analyzed by denaturing PAGE and quantified by phosphorimaging. Dissociation rate constants (koff) for all RTs were determined after fitting the data to a one-phase decay equation ([P] = exp(−koff × t)), using the GraphPad Prism software.
RNase H activity assays
To determine RNase H activity of RTs, an RNA oligonucleotide (R33B) was labeled at its 5′ end with 32P, purified with a Quick SpinTM column (Roche Applied Science), and then annealed to DNA primers of different lengths, depending on the assay (20A, 23A, 24A, 25A, 26A, and 27A) (Figs. 5 and 6). The RNA/DNA molar ratio was 4:1. RNase H cleavage reactions were carried out in 10 mm Hepes buffer, pH 7.0, containing 4.5 mm magnesium acetate, 4.5 mm NaCl, 130 mm potassium acetate, 1 mm DTT, and 5% PEG 6000. In these assays, the template-primer concentration was 30 nm, whereas RTs were supplied at 50 nm. RNase H activity assays were carried out in the presence of 60 nm acceptor DNA (35D) with or without dNTPs (500 μm each). Aliquots were taken after 0.25-, 0.5-, 1-, 5-, 15-, 30-, and 60-min incubations and mixed with equal volumes of the sample loading buffer. Products were analyzed by denaturing PAGE and quantified by phosphorimaging.
Nontemplated nucleotide addition
Nontemplated nucleotide addition activity was measured with blunt-ended DNA/DNA heteropolymeric template-primers (D33B/30A and D28B/25A) (Fig. 7). Primers were labeled at their 5′ end with 32P and annealed to the template DNA in 100 mm Hepes buffer, pH 7.0, containing 45 mm magnesium acetate and 45 mm NaCl. A 4-fold excess of template over primer was used in the annealing reaction. RTs (120 nm) were preincubated during 10 min at 37 °C with the labeled template-primers (60 nm), in 20 mm Hepes buffer, pH 7.0, containing 9 mm magnesium acetate, 9 mm NaCl, 120 mm potassium acetate, 1 mm DTT, and 5% PEG 6000. Nucleotide incorporation reactions were initiated by adding an equal volume (10 μl) of 150 mm potassium acetate, 1 mm DTT, and 5% PEG 6000, containing the corresponding dNTP (at 1 mm concentration) or mixtures of 2–4 dNTPs (1 mm each). Aliquots were removed after 0.5, 1, 2.5, 5, 10, 15, 30, and 60 min, and reactions were terminated by adding an equal volume of sample loading buffer. Products were analyzed by denaturing PAGE and phosphorimaging. The efficiency of nontemplated nucleotide addition was determined as the amount of dNTP incorporated to the blunt-ended DNA complex relative to the template-primer used in the reaction. The nucleotide incorporation rate (kobs) for dATP was determined by fitting the data to a one-phase exponential association equation ([P] = A·(1 − exp(−kobs × t))), using the GraphPad Prism software. Nucleotide incorporation reactions using dGTP, dCTP, dTTP, and ATP were too slow to be calculated using the exponential equation, and in those cases, the data were fit using a linear regression model.
Author contributions
J. L. data curation; J. L. and T. M. formal analysis; J. L., T. M., and L. M.-A. investigation; J. L. methodology; J. L., T. M., and L. M.-A. writing-review and editing; L. M.-A. conceptualization; L. M.-A. supervision; L. M.-A. funding acquisition; L. M.-A. writing-original draft; L. M.-A. project administration.
Supplementary Material
This work was supported in part by Spanish Ministry of Economy and Competitiveness Grant BIO2016-76716-R (AEI/FEDER, UE) and an institutional grant of the Fundación Ramón Areces. HIV-1ESP49 3M and 3M_E478Q RTs and methods for their use are the subject of patents and patent applications that have been licensed by Consejo Superior de Investigaciones Científicas to Sygnis AG/Expedeon Ltd. T. M. and L. M.-A. receive royalty payments for the sale of relevant enzymes and kits (SunScriptTM RT).
This article contains Figs. S1–S2 and Tables S1–S2.
- RT
- reverse transcriptase
- R
- repeat
- TAR
- trans-activating response element
- nt
- nucleotide.
References
- 1. Hu W. S., and Hughes S. H. (2012) HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2, a006882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Menéndez-Arias L., Sebastián-Martín A., and Álvarez M. (2017) Viral reverse transcriptases. Virus Res. 234, 153–176 10.1016/j.virusres.2016.12.019 [DOI] [PubMed] [Google Scholar]
- 3. Basu V. P., Song M., Gao L., Rigby S. T., Hanson M. N., and Bambara R. A. (2008) Strand transfer events during HIV-1 reverse transcription. Virus Res. 134, 19–38 10.1016/j.virusres.2007.12.017 [DOI] [PubMed] [Google Scholar]
- 4. Liu S., Abbondanzieri E. A., Rausch J. W., Le Grice S. F., and Zhuang X. (2008) Slide into action: dynamic shuttling of HIV reverse transcriptase on nucleic acid substrates. Science 322, 1092–1097 10.1126/science.1163108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muchiri J. M., Rigby S. T., Nguyen L. A., Kim B., and Bambara R. A. (2011) HIV-1 reverse transcriptase dissociates during strand transfer. J. Mol. Biol. 412, 354–364 10.1016/j.jmb.2011.07.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Onafuwa-Nuga A., and Telesnitsky A. (2009) The remarkable frequency of human immunodeficiency virus type 1 genetic recombination. Microbiol. Mol. Biol. Rev. 73, 451–480 10.1128/MMBR.00012-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vuilleumier S., and Bonhoeffer S. (2015) Contribution of recombination to the evolutionary history of HIV. Curr. Opin. HIV AIDS 10, 84–89 10.1097/COH.0000000000000137 [DOI] [PubMed] [Google Scholar]
- 8. Schlub T. E., Grimm A. J., Smyth R. P., Cromer D., Chopra A., Mallal S., Venturi V., Waugh C., Mak J., and Davenport M. P. (2014) Fifteen to twenty percent of HIV substitution mutations are associated with recombination. J. Virol. 88, 3837–3849 10.1128/JVI.03136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delviks-Frankenberry K., Galli A., Nikolaitchik O., Mens H., Pathak V. K., and Hu W.-S. (2011) Mechanisms and factors that influence high frequency retroviral recombination. Viruses 3, 1650–1680 10.3390/v3091650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Simon-Loriere E., Rossolillo P., and Negroni M. (2011) RNA structures, genomic organization and selection of recombinant HIV. RNA Biol. 8, 280–286 10.4161/rna.8.2.15193 [DOI] [PubMed] [Google Scholar]
- 11. Cameron C. E., Ghosh M., Le Grice S. F., and Benkovic S. J. (1997) Mutations in HIV reverse transcriptase which alter RNase H activity and decrease strand transfer efficiency are suppressed by HIV nucleocapsid protein. Proc. Natl. Acad. Sci. U.S.A. 94, 6700–6705 10.1073/pnas.94.13.6700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derebail S. S., Heath M. J., and DeStefano J. J. (2003) Evidence for the differential effects of nucleocapsid protein on strand transfer in various regions of the HIV genome. J. Biol. Chem. 278, 15702–15712 10.1074/jbc.M211701200 [DOI] [PubMed] [Google Scholar]
- 13. Levin J. G., Mitra M., Mascarenhas A., and Musier-Forsyth K. (2010) Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 7, 754–774 10.4161/rna.7.6.14115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Piekna-Przybylska D., and Bambara R. A. (2011) Requirements for efficient minus strand strong-stop DNA transfer in human immunodeficiency virus 1. RNA Biol. 8, 230–236 10.4161/rna.8.2.14802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Darlix J. L., Godet J., Ivanyi-Nagy R., Fossé P., Mauffret O., and Mély Y. (2011) Flexible nature and specific functions of the HIV-1 nucleocapsid protein. J. Mol. Biol. 410, 565–581 10.1016/j.jmb.2011.03.037 [DOI] [PubMed] [Google Scholar]
- 16. Warrilow D., Warren K., and Harrich D. (2010) Strand transfer and elongation of HIV-1 reverse transcription is facilitated by cell factors in vitro. PLoS ONE 5, e13229 10.1371/journal.pone.0013229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hwang C. K., Svarovskaia E. S., and Pathak V. K. (2001) Dynamic copy choice: steady state between murine leukemia virus polymerase and polymerase-dependent RNase H activity determines frequency of in vivo template switching. Proc. Natl. Acad. Sci. U.S.A. 98, 12209–12214 10.1073/pnas.221289898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Svarovskaia E. S., Delviks K. A., Hwang C. K., and Pathak V. K. (2000) Structural determinants of murine leukemia virus reverse transcriptase that affect the frequency of template switching. J. Virol. 74, 7171–7178 10.1128/JVI.74.15.7171-7178.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nikolenko G. N., Svarovskaia E. S., Delviks K. A., and Pathak V. K. (2004) Antiretroviral drug resistance mutations in human immunodeficiency virus type 1 reverse transcriptase increase template switching frequency. J. Virol. 78, 8761–8770 10.1128/JVI.78.16.8761-8770.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tanese N., Telesnitsky A., and Goff S. P. (1991) Abortive reverse transcription by mutants of Moloney murine leukemia virus deficient in the reverse transcriptase-associated RNase H function. J. Virol. 65, 4387–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. DeStefano J. J., Mallaber L. M., Rodriguez-Rodriguez L., Fay P. J., and Bambara R. A. (1992) Requirements for strand transfer between internal regions of heteropolymeric templates by human immunodeficiency virus reverse transcriptase. J. Virol. 66, 6370–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Purohit V., Balakrishnan M., Kim B., and Bambara R. A. (2005) Evidence that HIV-1 reverse transcriptase employs the DNA 3′ end-directed primary/secondary RNase H cleavage mechanism during synthesis and strand transfer. J. Biol. Chem. 280, 40534–40543 10.1074/jbc.M507839200 [DOI] [PubMed] [Google Scholar]
- 23. Golinelli M.-P., and Hughes S. H. (2002) Nontemplate nucleotide addition by HIV-1 reverse transcriptase. Biochemistry 41, 5894–5906 10.1021/bi0160415 [DOI] [PubMed] [Google Scholar]
- 24. Golinelli M.-P., and Hughes S. H. (2002) Nontemplated base addition by HIV-1 RT can induce nonspecific strand transfer in vitro. Virology 294, 122–134 10.1006/viro.2001.1322 [DOI] [PubMed] [Google Scholar]
- 25. Ohtsubo Y., Nagata Y., and Tsuda M. (2017) Efficient N-tailing of blunt DNA ends by Moloney murine leukemia virus reverse transcriptase. Sci. Rep. 7, 41769 10.1038/srep41769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oz-Gleenberg I., Herschhorn A., and Hizi A. (2011) Reverse transcriptases can clamp together nucleic acid strands with two complementary bases at their 3′ termini for initiating DNA synthesis. Nucleic Acids Res. 39, 1042–1053 10.1093/nar/gkq786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herzig E., Voronin N., Kucherenko N., and Hizi A. (2015) A novel Leu-92 mutant of HIV-1 reverse transcriptase with a selective deficiency in strand transfer causes a loss of viral replication. J. Virol. 89, 8119–8129 10.1128/JVI.00809-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Operario D. J., Balakrishnan M., Bambara R. A., and Kim B. (2006) Reduced dNTP interaction of human immunodeficiency virus type 1 reverse transcriptase promotes strand transfer. J. Biol. Chem. 281, 32113–32121 10.1074/jbc.M604665200 [DOI] [PubMed] [Google Scholar]
- 29. Matamoros T., Barrioluengo V., Abia D., and Menéndez-Arias L. (2013) Major groove binding track residues of the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase enhance cDNA synthesis at high temperatures. Biochemistry 52, 9318–9328 10.1021/bi401390x [DOI] [PubMed] [Google Scholar]
- 30. Moumen A., Polomack L., Roques B., Buc H., and Negroni M. (2001) The HIV-1 repeated sequence R is a robust hotspot for copy-choice recombination. Nucleic Acids Res. 29, 3814–3821 10.1093/nar/29.18.3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vo M. N., Barany G., Rouzina I., and Musier-Forsyth K. (2006) Mechanistic studies of mini-TAR RNA/DNA annealing in the absence and presence of HIV-1 nucleocapsid protein. J. Mol. Biol. 363, 244–261 10.1016/j.jmb.2006.08.039 [DOI] [PubMed] [Google Scholar]
- 32. Berkhout B., Vastenhouw N. L., Klasens B. I., and Huthoff H. (2001) Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA 7, 1097–1114 10.1017/S1355838201002035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Coffin J. M. (1979) Structure, replication, and recombination of retrovirus genomes: some unifying hypotheses. J. Gen. Virol. 42, 1–26 10.1099/0022-1317-42-1-1 [DOI] [PubMed] [Google Scholar]
- 34. Negroni M., and Buc H. (2000) Copy-choice recombination by reverse transcriptases: reshuffling of genetic markers mediated by RNA chaperones. Proc. Natl. Acad. Sci. U.S.A. 97, 6385–6390 10.1073/pnas.120520497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Menéndez-Arias L., Abraha A., Quiñones-Mateu M. E., Mas A., Camarasa M. J., and Arts E. J. (2001) Functional characterization of chimeric reverse transcriptases with polypeptide subunits of highly divergent HIV-1 group M and O strains. J. Biol. Chem. 276, 27470–27479 10.1074/jbc.M104342200 [DOI] [PubMed] [Google Scholar]
- 36. Nguyen L. A., Daddacha W., Rigby S., Bambara R. A., and Kim B. (2012) Altered strand transfer activity of a multi-drug-resistant human immunodeficiency virus type 1 reverse transcriptase mutant with a dipeptide fingers domain insertion. J. Mol. Biol. 415, 248–262 10.1016/j.jmb.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brincat J. L., Pfeiffer J. K., and Telesnitsky A. (2002) RNase H activity is required for high-frequency repeat deletion during Moloney murine leukemia virus replication. J. Virol. 76, 88–95 10.1128/JVI.76.1.88-95.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moumen A., Polomack L., Unge T., Véron M., Buc H., and Negroni M. (2003) Evidence for a mechanism of recombination during reverse transcription dependent on the structure of the acceptor RNA. J. Biol. Chem. 278, 15973–15982 10.1074/jbc.M212306200 [DOI] [PubMed] [Google Scholar]
- 39. Hanson M. N., Balakrishnan M., Roques B. P., and Bambara R. A. (2006) Evidence that creation of invasion sites determines the rate of strand transfer mediated by HIV-1 reverse transcriptase. J. Mol. Biol. 363, 878–890 10.1016/j.jmb.2006.08.068 [DOI] [PubMed] [Google Scholar]
- 40. Zhu Y. Y., Machleder E. M., Chenchik A., Li R., and Siebert P. D. (2001) Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. BioTechniques 30, 892–897 [DOI] [PubMed] [Google Scholar]
- 41. Clark J. M. (1988) Novel non-templated nucleotide addition reactions catalyzed by procaryotic and eucaryotic DNA polymerases. Nucleic Acids Res. 16, 9677–9686 10.1093/nar/16.20.9677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Levin J. Z., Yassour M., Adiconis X., Nusbaum C., Thompson D. A., Friedman N., Gnirke A., and Regev A. (2010) Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715 10.1038/nmeth.1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Langevin S. A., Bent Z. W., Solberg O. D., Curtis D. J., Lane P. D., Williams K. P., Schoeniger J. S., Sinha A., Lane T. W., and Branda S. S. (2013) Peregrine: a rapid and unbiased method to produce strand-specific RNA-Seq libraries from small quantities of starting material. RNA Biol. 10, 502–515 10.4161/rna.24284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Head S. R., Komori H. K., LaMere S. A., Whisenant T., Van Nieuwerburgh F., Salomon D. R., and Ordoukhanian P. (2014) Library construction for next-generation sequencing: overviews and challenges. BioTechniques 56, 61–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Agopian A., Depollier J., Lionne C., and Divita G. (2007) p66 Trp-24 and Phe-61 are essential for accurate association of HIV-1 reverse transcriptase with primer/template. J. Mol. Biol. 373, 127–140 10.1016/j.jmb.2007.07.044 [DOI] [PubMed] [Google Scholar]
- 46. Fisher T. S., and Prasad V. R. (2002) Substitutions of Phe-61 located in the vicinity of template 5′-overhang influence polymerase fidelity and nucleoside analog sensitivity of HIV-1 reverse transcriptase. J. Biol. Chem. 277, 22345–22352 10.1074/jbc.M200282200 [DOI] [PubMed] [Google Scholar]
- 47. Fisher T. S., Darden T., and Prasad V. R. (2003) Substitutions at Phe-61 in the β3-β4 hairpin of HIV-1 reverse transcriptase reveal a role for the fingers subdomain in strand displacement DNA synthesis. J. Mol. Biol. 325, 443–459 10.1016/S0022-2836(02)01225-1 [DOI] [PubMed] [Google Scholar]
- 48. Matamoros T., Deval J., Guerreiro C., Mulard L., Canard B., and Menéndez-Arias L. (2005) Suppression of multidrug-resistant HIV-1 reverse transcriptase primer unblocking activity by α-phosphate-modified thymidine analogues. J. Mol. Biol. 349, 451–463 10.1016/j.jmb.2005.04.004 [DOI] [PubMed] [Google Scholar]
- 49. Betancor G., Puertas M. C., Nevot M., Garriga C., Martínez M. A., Martinez-Picado J., and Menéndez-Arias L. (2010) Mechanisms involved in the selection of HIV-1 reverse transcriptase thumb subdomain polymorphisms associated with nucleoside analogue therapy failure. Antimicrob. Agents Chemother. 54, 4799–4811 10.1128/AAC.00716-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Boretto J., Longhi S., Navarro J. M., Selmi B., Sire J., and Canard B. (2001) An integrated system to study multiply substituted human immunodeficiency virus type 1 reverse transcriptase. Anal. Biochem. 292, 139–147 10.1006/abio.2001.5045 [DOI] [PubMed] [Google Scholar]
- 51. Alvarez M., Matamoros T., and Menéndez-Arias L. (2009) Increased thermostability and fidelity of DNA synthesis of wild-type and mutant HIV-1 group O reverse transcriptases. J. Mol. Biol. 392, 872–884 10.1016/j.jmb.2009.07.081 [DOI] [PubMed] [Google Scholar]
- 52. Kati W. M., Johnson K. A., Jerva L. F., and Anderson K. S. (1992) Mechanism and fidelity of HIV reverse transcriptase. J. Biol. Chem. 267, 25988–25997 [PubMed] [Google Scholar]
- 53. Reardon J. E. (1992) Human immunodeficiency virus reverse transcriptase: steady-state and pre-steady-state kinetics of nucleotide incorporation. Biochemistry 31, 4473–4479 10.1021/bi00133a013 [DOI] [PubMed] [Google Scholar]
- 54. Menéndez-Arias L. (1998) Studies on the effects of truncating α-helix E′ of p66 human immunodeficiency virus type 1 reverse transcriptase on template-primer binding and fidelity of DNA synthesis. Biochemistry 37, 16636–16644 10.1021/bi981830g [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.