Abstract
Generation of single‐stranded DNA (ssDNA) is required for the template strand formation during DNA replication. Replication Protein A (RPA) is an ssDNA‐binding protein essential for protecting ssDNA at replication forks in eukaryotic cells. While significant progress has been made in characterizing the role of the RPA–ssDNA complex, how RPA is loaded at replication forks remains poorly explored. Here, we show that the Saccharomyces cerevisiae protein regulator of Ty1 transposition 105 (Rtt105) binds RPA and helps load it at replication forks. Cells lacking Rtt105 exhibit a dramatic reduction in RPA loading at replication forks, compromised DNA synthesis under replication stress, and increased genome instability. Mechanistically, we show that Rtt105 mediates the RPA–importin interaction and also promotes RPA binding to ssDNA directly in vitro, but is not present in the final RPA–ssDNA complex. Single‐molecule studies reveal that Rtt105 affects the binding mode of RPA to ssDNA. These results support a model in which Rtt105 functions as an RPA chaperone that escorts RPA to the nucleus and facilitates its loading onto ssDNA at replication forks.
Keywords: replication fork, replication stress, RPA chaperone, Rtt105
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; DNA Replication, Repair & Recombination
Introduction
DNA replication is tightly regulated at multiple levels to ensure that the genome is both accurately and completely duplicated during each cell cycle (Bell & Dutta, 2002; Burgers & Kunkel, 2017). Faults in this regulation can lead to replication errors, and consequently, genome instability. Understanding carcinogenesis and a variety of other diseases (Aguilera & García‐Muse, 2013; Jackson et al, 2014) therefore requires a detailed understanding of how replication is mediated.
The generation of single‐stranded DNA (ssDNA) templates is necessary for DNA replication, but ssDNA is susceptible to secondary structure formation and digestion by nucleases. Therefore, ssDNA exposed during DNA replication must be protected and stabilized, a function performed by replication protein A (RPA; Chase & Williams, 1986). RPA is an evolutionarily conserved protein complex present in all eukaryotes and regulates both DNA replication initiation and elongation (Wobbe et al, 1987; Fairman & Stillman, 1988; Wold & Kelly, 1988; Brill & Stillman, 1989; Wold, 1997). RPA is also important during DNA damage repair and recombination, and RPA‐coated ssDNA plays a role in the activation of the DNA replication checkpoint pathway and the nucleosome assembly pathway (Zou & Elledge, 2003; Maréchal & Zou, 2015; Liu et al, 2017; Zhang et al, 2017). RPA consists of the three related subunits Rfa1, Rfa2, and Rfa3 in Saccharomyces cerevisiae, with apparent masses of approximately 70, 30, and 14 kDa, respectively (Brill & Stillman, 1991). Each RFA gene is essential in budding yeast (Brill & Stillman, 1991), and all three subunits are required for the formation of the functional RPA complex. In vitro, RPA binds to ssDNA via six oligonucleotide binding (OB)‐fold domains with affinities of up to ~ 10−7–10−10 M and a defined 5′→3′ polarity (Bochkarev et al, 1997; Bochkareva et al, 2001, 2002; Fanning et al, 2006; Fan & Pavletich, 2012; Brosey et al, 2013). This high binding affinity has led to the assumption that RPA functions passively and directly binds to exposed ssDNA, including that at the replication fork. However, in vitro binding affinity measurements are not expected to represent all aspects of regulation in living cells. Moreover, previous studies have shown that the binding affinity of RPA for short oligonucleotides is dependent on ssDNA length, and RPA has multiple modes of binding ssDNA determined by the length of ssDNA that it contacts and the number of OB‐fold domains involved (Kim et al, 1994; Bastin‐Shanower & Brill, 2001; Bochkareva et al, 2001; Kolpashchikov et al, 2001; Fanning et al, 2006). These results raise the possibility that other proteins can alter the binding mode of RPA and thereby regulate its function in replication and other DNA metabolic pathways.
The regulator of Ty1 transposition (RTT) genes were first identified in S. cerevisiae in a screen for deficiency in Ty1 transposon transposition (Scholes et al, 2001). 13 of the 21 RTT genes identified in this screen were found to be previously characterized, with confirmed functions in DNA damage response (Game & Mortimer, 1974; Ajimura et al, 1993; Watt et al, 1996; Fortin & Symington, 2002). The other 8 RTT genes were initially uncharacterized (Scholes et al, 2001), but several studies have since shown that some of these 8 genes function in DNA‐processing pathways. For example, Rtt101 is a member of the cullin family of ubiquitin ligases and is required for promoting replication fork progression (Michel et al, 2003; Luke et al, 2006; Zaidi et al, 2008), and functions in the regulation of DNA replication‐coupled nucleosome assembly via the ubiquitination of H3 (Han et al, 2013). Rtt108, also named MMS1, interacts with Rtt101, and the Rtt101MMS1 complex is the budding yeast counterpart of the mammalian CUL4DDB1 ubiquitin ligase (Zaidi et al, 2008). Rtt109 is the acetyltransferase for histone H3 lysine 56 and is critical for genome integrity in part through its role in DNA replication‐coupled nucleosome assembly (Driscoll et al, 2007; Pursell et al, 2007; Fillingham et al, 2008; Li et al, 2008; Burgess et al, 2010). Rtt106 is a histone chaperone that recognizes acetylation on lysine 56 of H3 and plays an important role in transcriptional silencing (Huang et al, 2005, 2007; Li et al, 2008; Su et al, 2012; Zunder et al, 2012). Rtt110, also named Elg1, forms an alternative RFC‐like complex (Ben‐Aroya et al, 2003; Kanellis et al, 2003) that unloads the PCNA clamp during DNA replication (Kubota et al, 2013; Gazy et al, 2015). Therefore, the functions of most of the RTT genes have been elucidated with the exception of RTT105 (YER104W).
A functional dissection of protein complexes based on genetic interactions clustered RTT105 among DNA replication factors, as it shares a similar genetic interaction profile with RFA1 and RFA2 (Collins et al, 2007). However, the biochemical function of Rtt105 remains largely unknown. Here, we show that the Rtt105 protein interacts directly with RPA. We find that cells lacking Rtt105 exhibit defects in the association of RPA with ssDNA at DNA replication forks, and reduced DNA synthesis under replication stress. Mechanistically, we show that Rtt105 promotes both nuclear import of RPA and RPA binding to ssDNA. In vitro single‐molecule and EMSA studies reveal that Rtt105 alters the interaction mode of RPA with ssDNA. We propose a model whereby Rtt105 is analogous to histone chaperones, which mediate histone import and promote nucleosome formation using histones and double‐stranded DNA (dsDNA): Rtt105 is an RPA chaperone that both escorts RPA during nuclear import and promotes the formation of RPA–ssDNA at replication forks.
Results
Rtt105 interacts directly with RPA
We purified Rtt105 from yeast cells using a TAP tag and identified co‐purified proteins using mass spectrometry. There were two groups of top hits among the co‐purified proteins: the three subunits of RPA, and Kap95 (YLR347C), a karyopherin‐beta protein essential for the nuclear import of many proteins, including RPA (Belanger et al, 2011; Fig 1A and Table EV1). To characterize the Rtt105–RPA interaction, we performed reciprocal immunoprecipitation (IP) of TAP‐tagged Rfa1 and detected Rtt105 in the Rfa1‐associated complex (Fig EV1A). Moreover, the RPA–Rtt105 interaction peaked during S phase and diminished at G2/M phase (Fig 1B). In vitro pull‐down assays revealed that purified recombinant maltose‐binding protein‐tagged Rtt105 (MBP‐Rtt105) pulled down recombinant RPA in a concentration‐dependent manner, whereas MBP alone did not bind RPA (Fig 1C). These results establish that Rtt105 binds directly to RPA.
Figure 1. Rtt105 binds RPA in vivo and in vitro .
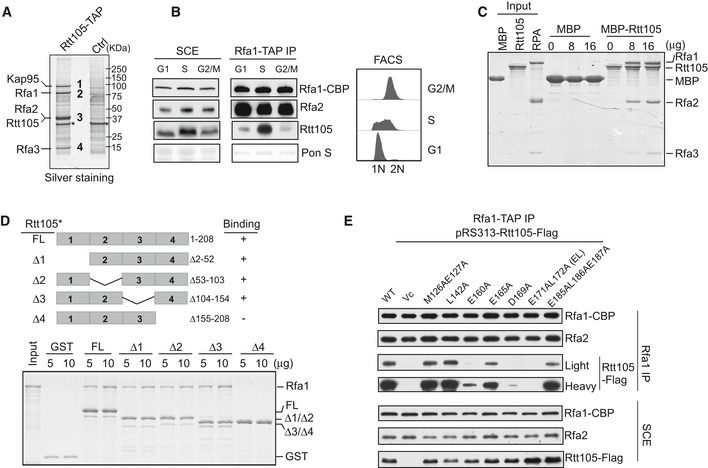
- Identification of Rtt105‐TAP‐associated proteins. Purified proteins were resolved on a 4–12% gel, revealed by silver staining, and identified by mass spectrometry (Table EV1). Asterisk (*) indicates non‐specific band also in control.
- Cells containing Rfa1‐TAP were synchronized at G1, S, and G2/M phases and used for the TAP purification. Rfa1‐TAP‐associated protein complexes were analyzed by Western blot using CBP, Rfa2, and Rtt105 antibodies (left panel). DNA content was monitored by flow cytometry (right panel).
- MBP‐Rtt105 pulled down recombinant RPA, resolved on SDS–PAGE gels and visualized by Coomassie Brilliant Blue (CBB) staining. MBP was used as a negative control.
- GST‐tagged full‐length (FL) and Rtt105 deletion mutants (schematic of the Rtt105 truncations is shown in the upper panel) were purified and used to pull down Rfa1 protein. GST proteins were used as a negative control. Isolated protein complexes were resolved on 15% SDS–PAGE gels and visualized by CBB staining.
- Rfa1‐TAP purification was performed using Rfa1‐TAP rtt105Δ strains expressing WT and indicated Rtt105 mutant forms. pRS313 serves as vector control (Vc).
Source data are available online for this figure.
Figure EV1. Mapping key amino acids for the Rtt105–RPA interaction.
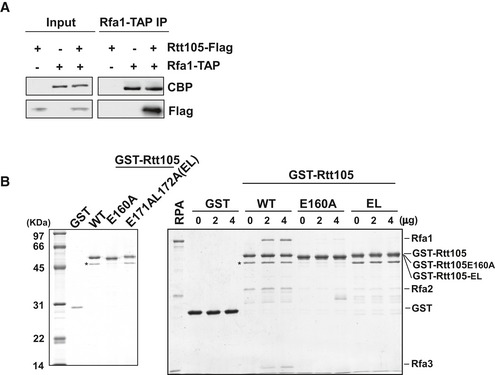
- Identification of RPA–Rtt105 interaction by tandem affinity purification (TAP). TAP‐tagged Rfa1 was purified from yeast cells with or without Flag‐tagged Rtt105 (Rtt105‐Flag), and the co‐purified proteins were resolved on SDS–PAGE and analyzed by Western blotting using antibodies against calmodulin binding peptide (CBP) and Flag. A no‐TAP tag strain was used as a negative control.
- The Rtt105 E160A and E171AL172A (EL) mutations attenuate the RPA–Rtt105 interaction. Left panel: purified recombinant GST‐tagged Rtt105 WT and mutant proteins analyzed by SDS–PAGE with CBB staining. Asterisk (*) indicates non‐specific band; right panel: GST pull‐down assay was performed to determine the interaction of RPA with Rtt105 mutants. The bead‐bound proteins were resolved by SDS–PAGE gels and subjected to silver staining.
To identify the Rfa1 binding region on Rtt105, we deleted sections of Rtt105 in 50‐amino acid intervals, yielding four truncated forms of Rtt105: Δ2–52, Δ53–103, Δ104–154, and Δ155–208 (Fig 1D). We found that deletion of the C‐terminal 50 amino acids (Δ155–208 = Rtt105Δ4) results in a loss of binding to Rfa1 in vitro (Fig 1D). Mutations of several conserved residues at the Rtt105 C‐terminus, including E160A, D169A, and E171L172AA, reduce the interaction of Rtt105 with RPA both in vivo and in vitro, with the E171L172AA (EL) mutant exhibiting a more pronounced effect (Figs 1E and EV1B). These results indicate that the C‐terminus of Rtt105 mediates the RPA–Rtt105 interaction.
Rtt105 is important for RPA binding at replication forks
We noticed that in the yeast genome database, RTT105 deletion cells are listed as inviable. However, using standard methods we successfully generated haploid rtt105Δ mutant cells in a W303 background (Appendix Fig S1A and B). To confirm the viability of rtt105Δ cells, we also constructed the null mutant cells in several additional genetic backgrounds, namely S288C, DBY747, and BY4742. rtt105Δ mutant cells in these backgrounds are also viable (Appendix Fig S1A and B). Moreover, rtt105Δ cells in all these strain backgrounds are sensitive to various DNA‐damaging agents to a similar degree (Appendix Fig S1A and B). They are also sensitive to DNA‐damaging agents when grown at 16°C (Appendix Fig S2A), a temperature used to perform genome‐wide Rfa1 ChIP‐seq without HU (see below). Furthermore, expression of RTT105 driven by its own promoter in rtt105Δ cells rescues the phenotype of rtt105Δ mutant cells (Appendix Fig S1C). We chose the W303 background, which is our standard yeast background, to perform the remaining experiments.
Because RPA protects ssDNA at replication forks, we next explored whether Rtt105 mediates RPA behavior during replication (Wold, 1997). We used chromatin immunoprecipitation (ChIP) to determine whether deletion of RTT105 affects RPA binding at replication forks under two conditions: hydroxyurea (HU)‐stalled forks and active forks without HU treatment (Fig 2A). First, G1‐phase cells were released into early S phase in the presence of 0.2 M HU for 45 min. HU arrests cells at early S phase and has no apparent effect on initiation of DNA replication from early replication origins. HU treatment also activates the DNA replication checkpoint, which in turn inhibits initiation of DNA replication from late replication origins. Antibodies against Rfa1 or Rfa2 or TAP‐specific antibodies (IgG beads) against Rfa1‐TAP were used to perform ChIP assays (Figs 2A and EV2A). Quantitative PCR (qPCR) analysis of ChIP DNA revealed that Rfa1 and Rfa2 bound at early replication origins (ARS305 and ARS607), but not at distal sites (ARS607 + 14 kb and ARS305 + 12 kb) that remain unreplicated during early S phase in the presence of HU (Liu et al, 2017; Fig EV2B and C). Deletion of RTT105 reduced the association of both assayed RPA subunits with replicating DNA (Fig EV2B and C). Exogenous expression of full‐length Rtt105 rescued the chromatin‐binding defects of Rfa1 in rtt105∆ cells, whereas expression of an Rtt105 mutant lacking its C‐terminus or harboring the E171L172AA mutation (EL) failed to do so (Fig EV2D). We also analyzed the impact of the rtt105∆ mutation on the association of Rfa1 across the genome using ChIP‐seq. Rfa1 ChIP‐seq peaks co‐localized with almost all early replication origins (Figs 2B and EV2E). A calculation of the average Rfa1 ChIP‐seq read density across all fired origins revealed that the binding of Rfa1 at early replication origins was reduced in rtt105∆ cells compared to WT cells at HU‐stalled forks (Fig 2C). These results indicate that Rtt105 is required for RPA binding to HU‐stalled replication forks genome‐wide.
Figure 2. RTT105 deletion leads to a reduction in RPA binding at replicating regions genome‐wide.
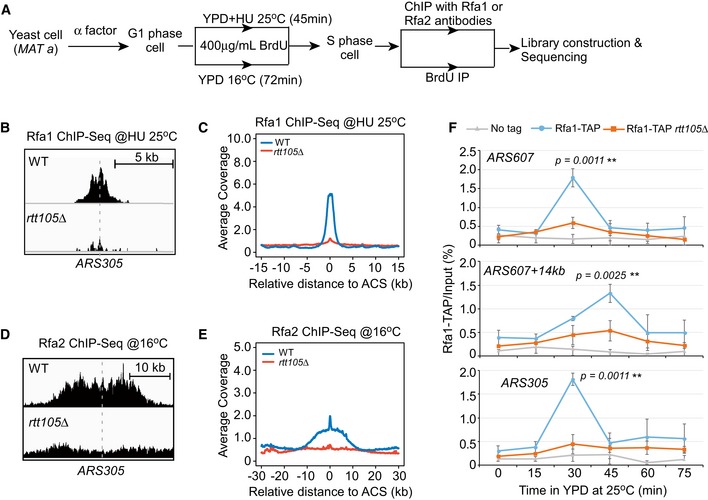
- Schematic for Rfa1 and Rfa2 ChIP‐seq (this figure) and BrdU IP‐seq (Fig 6). Yeast cells were arrested with α‐factor at G1 phase and then released into fresh YPD medium containing BrdU to label newly synthesized DNA as well as to allow cell to enter S phase at two conditions: (i) in medium with 0.2 M HU for 45 min at standard growth temperature (25°C), and (ii) in medium without HU for 72 min at low temperature (16°C) to slow down S‐phase progression. Cells were then collected for Rfa1/Rfa2 ChIP or BrdU IP. The resulted DNAs were detected by sequencing.
- Snapshots of Rfa1 ChIP‐seq peak at ARS305 from WT and rtt105∆ cells released into HU medium.
- The average Rfa1 ChIP‐seq read density from cells released into HU medium around ACS sites. ACS, ARS consensus sequence.
- Snapshots of Rfa2 ChIP‐seq peaks at ARS305 from cells released at 16°C for 72 min.
- The average Rfa2 ChIP‐seq read density surrounding ACS sites from cells released at 16°C for 72 min.
- Rfa1 occupancy analysis of cells progression through S phase. G1‐arrested cells were released into fresh YPD medium to allow cell progression through S phase at standard growth temperature (25°C). Equal amounts of cells were collected at each time point. Rfa1 ChIP was performed using IgG beads for TAP tag. ARS305 and ARS607, early fired replication origins; ARS607 + 14 kb, a corresponding 14‐kb away region of ARS607. The mean and standard deviation (SD) of three biological replicates are shown. The single‐tailed nonparametric Wilcoxon test was performed as described in Materials and Methods (**0.001 ≤ P‐value < 0.01).
Figure EV2. Rtt105 is important for the association of RPA with replication forks.
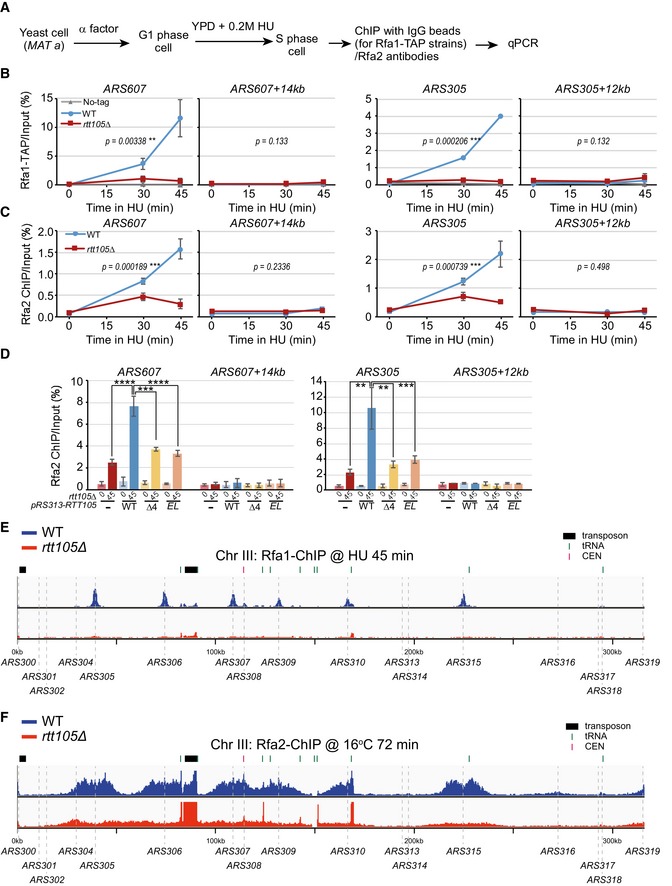
-
AAn outline for ChIP assays for Rfa1 and Rfa2 subunits. G1 yeast cells were released into fresh YPD medium containing 0.2 M HU to arrest cells at early S phase. Equal amounts of cells were collected prior to G1 (0 min) or at different time points following release. Rfa1 and Rfa2 ChIP was performed using IgG beads for TAP‐tagged strains, or anti‐Rfa2 antibody.
-
B, CRPA binding was dramatically decreased at HU‐stalled replication forks upon deletion of RTT105. ChIP DNA and input DNA were analyzed by quantitative real‐time PCR using primers against the replication origins ARS607 and ARS305 and their corresponding distal regions (ARS607 + 14 kb and ARS305 + 12 kb), and the percentage of ChIP DNA over the input DNA was calculated. The mean and standard error (SE) of at least two biological replicates are shown, with P‐values derived from two‐way analysis of variance (ANOVA; **0.001 ≤ P‐value < 0.01, ***P‐value ≤ 0.001).
-
DThe C‐terminus of Rtt105 is important for RPA binding at replication forks. pRS313‐based plasmids expressing Flag‐tagged WT Rtt105 and mutants (Δ4 and EL) were transformed into the rtt105Δ strain. ChIP was performed using Rfa2 antibodies. Vc: pRS313 vector; WT: pRS313‐Rtt105‐Flag; Δ4: pRS313‐Rtt105 (Δ155–208)‐Flag; EL: pRS313‐Rtt105E171AL172A‐Flag. The mean and standard error (SE) of two biological replicates are shown. Statistical significance was evaluated based on Student's t‐tests (**0.001 ≤ P‐value < 0.01, ***0.0001 ≤ P‐value < 0.001; ****P‐value < 0.0001).
-
ESnapshot of Rfa1 ChIP‐seq at chromosome III under HU synchronized cells is shown. Yeast cells were synchronized at G1 phase and then released into fresh medium containing 0.2 M HU and 400 μg/ml BrdU for 45 min at 25°C. Equal amounts of cells were collected just prior to G1 (0 min) or at 45 min following release into early S phase in the presence of HU. Rfa1 ChIP DNA was processed for sequencing. The sequencing reads were mapped to the yeast reference genome.
-
FSnapshot of Rfa2 ChIP‐seq at chromosome III at S phase without HU treatment is shown. G1‐arrested yeast cells were released into S phase at low temperature (16°C) without HU. Yeast cells were collected for Rfa2 ChIP and were further processed for sequencing. The sequencing reads were mapped to the yeast reference genome.
To rule out the possibility that replication stress caused by HU synchronization affects RPA binding to replication forks, we examined the effect of rtt105Δ on RPA binding at replication forks throughout the cell cycle. To capture the S‐phase association of Rfa1 with replication forks genome‐wide, G1‐phase cells were released into S phase at low temperature (16°C) for 72 min (Aparicio et al, 1997; Yu et al, 2014; Fig 2A). The Rfa2 ChIP‐seq results show that Rfa2 binding to active forks is reduced in rtt105∆ cells (Figs 2D and E, and EV2F). This was confirmed by ChIP‐qPCR when yeast cells were released into S phase at 25°C: Rfa1 binding at all of the tested chromatin regions during S phase was reduced substantially in rtt105Δ cells (Fig 2F). It is unlikely that this reduction in Rfa1 binding was due to the slight S‐phase delay seen in rtt105Δ cells (Appendix Fig S2B). Moreover, the total levels of Rfa1 and Rfa2 protein expression were not reduced in rtt105Δ cells (Appendix Fig S2C). Interestingly, the association of Cdc45 and Mcm6—two components of the active replicative helicase CMG (Bell & Dutta, 2002)—with active replication forks during progression through S phase at 16°C is not affected in rtt105∆ mutant cells (Appendix Fig S3). Taken together, these results support the idea that Rtt105 has a direct role in regulating RPA binding at both HU‐stalled and active DNA replication forks.
Rtt105 mediates RPA nuclear import
To understand how Rtt105 mediates RPA binding to DNA replication forks, we first asked whether Rtt105 is required for RPA nuclear import, given that it associates with Kap95 (importin β), a factor known to be involved in RPA nuclear transport (Belanger et al, 2011). In WT cells, Rfa1 localizes mostly to the nucleus. In contrast, the Rfa1 signal in rtt105∆ cells is diffuse and present in both the cytoplasm and the nucleus (Fig 3A). Exogenous expression of full‐length Rtt105 rescued the localization defects of Rfa1 in rtt105∆ cells, whereas expression of an Rtt105 mutant lacking its C‐terminus failed to do so (Appendix Fig S4A). Finally, we observed that deletion of RTT105 abolishes the interaction between RPA and Kap95 (Appendix Fig S4B). These results indicate that Rtt105 regulates RPA nuclear import at least in part through mediating the association of RPA with Kap95.
Figure 3. Rtt105 has roles in both RPA nuclear import and loading RPA to replicating DNA .
-
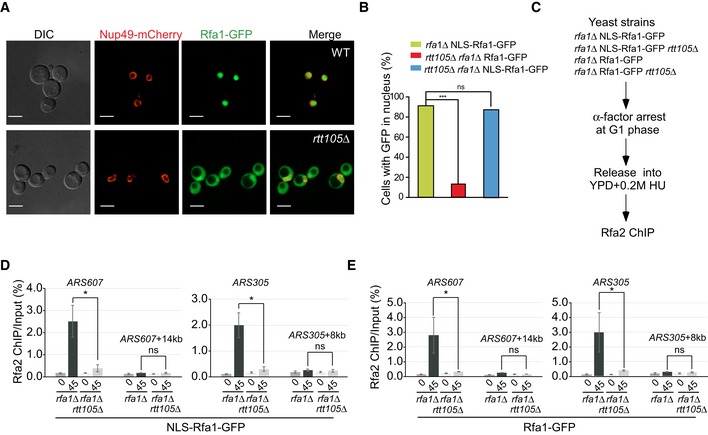
AThe nuclear localization of Rfa1 proteins is altered in rtt105Δ mutant cells. Rfa1 tagged with GFP at its C‐terminus was used to analyze the localization of Rfa1‐GFP fusion protein (green), and the nuclear envelop is visualized with the Nup49‐mCherry fusion protein (red) using fluorescence microscopy. DIC: differential interference contrast. Scale bar: 5 μm.
-
BNLS‐RFA1‐GFP rescues the nuclear localization of Rfa1 in rtt105Δ mutant cells. The wild‐type RFA1 gene was fused with an SV40 large T‐antigen nuclear localization sequence (NLS) at its 5′‐end and a GFP gene at its 3′‐end to obtain a construct, driven by the RFA1 promoter to express NLS‐RFA1‐GFP fusion protein. The engineered construct was then transformed into WT or rtt105Δ mutant yeast cells to replace its endogenous RFA1 expression. The resulting yeast cells were then visualized under microscope, and GFP signals enriched in the nuclei were scored. RFA1‐GFP lacking the NLS sequence was transformed into rtt105Δ rfa1Δ mutant cells as a control. DAPI staining indicates nuclear DNA. Statistical significance was evaluated based on Student's t‐tests (***P‐value < 0.001).
-
CThe experimental scheme for the Rfa2 ChIP in the indicated yeast strains.
-
D, EAn increase in nuclear localization of Rfa1 could not rescue the RPA binding defects at the replication regions in rtt105∆ cells. The percentage of Rfa2 ChIP DNA over the total input DNA was calculated. The mean and standard error (SE) of three biological replicates are shown. Statistical significance was evaluated based on Student's t‐tests (*0.01 ≤ P‐value < 0.05).
DNA replication, and therefore the association of RPA with ssDNA at replication forks, occurs in the nucleus. Therefore, the mis‐localization of RPA to the cytoplasm in rtt105∆ mutant cells should contribute to the reduced association of RPA with DNA replication forks. To test whether this is the sole function of Rtt105, we expressed a fusion protein made up of a prototypical nuclear localization signal (NLS) coding for the peptide PKKKRKV, the full‐length Rfa1 sequence, and a C‐terminal GFP tag (NLS‐Rfa1‐GFP) as the only copy of Rfa1 in cells (Fig 3B). In rtt105∆ cells, the NLS‐Rfa1‐GFP fusion protein co‐localized with DAPI in the nucleus in both WT and rtt105∆ cells (Fig 3B and Appendix Fig S4C), indicating that NLS‐Rfa1‐GFP can bypass the requirement for Rtt105 to achieve nuclear import. Surprisingly, Rfa2 ChIP‐qPCR revealed that the binding of Rfa2 at replication origins remains defective in rtt105∆ cells, even in the presence of NLS‐Rfa1‐GFP (Fig 3C–E). This result suggests that in addition to its role in RPA nuclear import, Rtt105 has some additional role in the regulation of RPA binding to DNA replication forks. Consistent with this interpretation, we observed that rtt105∆ cells are sensitive to DNA‐damaging agents, and expression of NLS‐RFA1‐GFP in rtt105∆ cells failed to rescue the sensitivity to DNA damage of rtt105∆ (Appendix Fig S4D). Moreover, the Rfa1–Rtt105 interaction occurs predominantly in the nucleus. In contrast, the Kap95–Rtt105 interaction is more pronounced at the nuclear periphery (Appendix Fig S5). Taken together, these results demonstrate that Rtt105, in addition to its role in mediating RPA nuclear import, must have an additional function in the nucleus that promotes the association of RPA with DNA replication forks.
Rtt105 facilitates RPA binding to ssDNA, but is not present in the final RPA–ssDNA complex
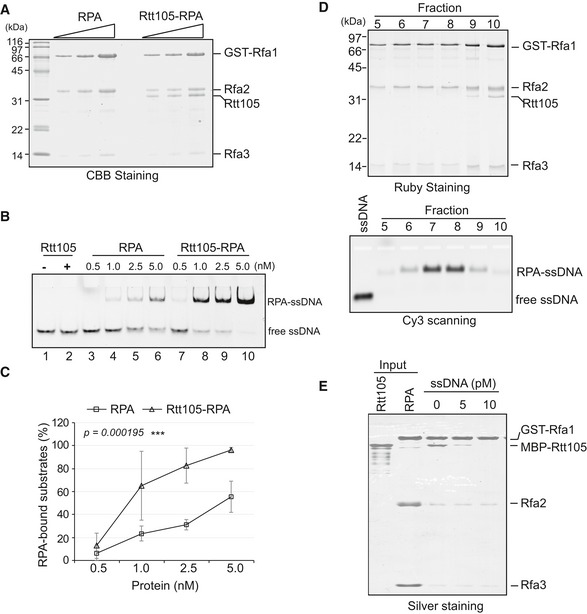
To understand how Rtt105 impacts the binding of RPA to ssDNA at replication forks, we asked whether Rtt105 directly affects RPA–ssDNA binding because no additional replisome components were identified in the Rtt105 co‐purified protein complex (Table EV1). We purified Rtt105 and a stable complex consisting of Rtt105, Rfa1, Rfa2, and Rfa3 (Fig 4A). An electrophoretic mobility shift assay (EMSA; Kim et al, 1994) was first used to determine the ssDNA binding affinity of Rtt105, RPA, and Rtt105–RPA. We incubated increasing amounts of each protein (complex) with a fixed amount of Cy3‐labeled 30‐nt oligonucleotide and resolved the bound complex from the free DNA using non‐denaturing gel electrophoresis. The relative amounts of free and bound DNA were then detected using chemical fluorescence imaging. We found that Rtt105 does not bind ssDNA or dsDNA (Fig EV3A–C). However, the binding efficiency of the Rtt105–RPA complex for ssDNA increased compared to that of RPA alone (Fig 4B and C). In contrast, the Rtt105‐EL mutant protein that cannot bind RPA failed to increase the RPA binding efficiency (Fig EV3D). These results demonstrate that Rtt105 promotes the formation of the RPA–ssDNA complex in vitro and this ability depends on the Rtt105–RPA interaction.
Figure 4. Rtt105 enhances the binding efficiency of RPA to ssDNA in vitro .
-
APurified RPA and Rtt105–RPA complex were resolved on 15% SDS–PAGE gels and analyzed by CBB staining.
-
B, CEMSA was performed to analyze ssDNA binding ability of RPA or Rtt105–RPA complex. Cy3‐labeled 30‐nt ssDNA was used as the substrate, and reaction mixtures were resolved on native PAGE gels. Quantitation of the RPA‐bound ssDNA is shown in (C). The mean values ± SD from three independent experiments are plotted, with P‐values derived from two‐way analysis of variance (ANOVA; ***P‐value < 0.001).
-
DRecombinant Rtt105–RPA complex was mixed with a 30‐nt ssDNA in a 2:1 ratio, and the reaction products were subjected to gel filtration chromatography. Equal volumes of Fractions 5–10 were resolved on SDS–PAGE for detecting proteins by Ruby staining (upper panel) or were resolved by native agarose gel for detection of Cy3‐ssDNA signal (lower panel).
-
EGST‐RPA pull down Rtt105 with increased amount of 30‐nt ssDNA, and the bound proteins were resolved on SDS–PAGE and visualized by silver staining.
Source data are available online for this figure.
Figure EV3. Rtt105 does not possess DNA binding ability, but promotes RPA binding to ssDNA .
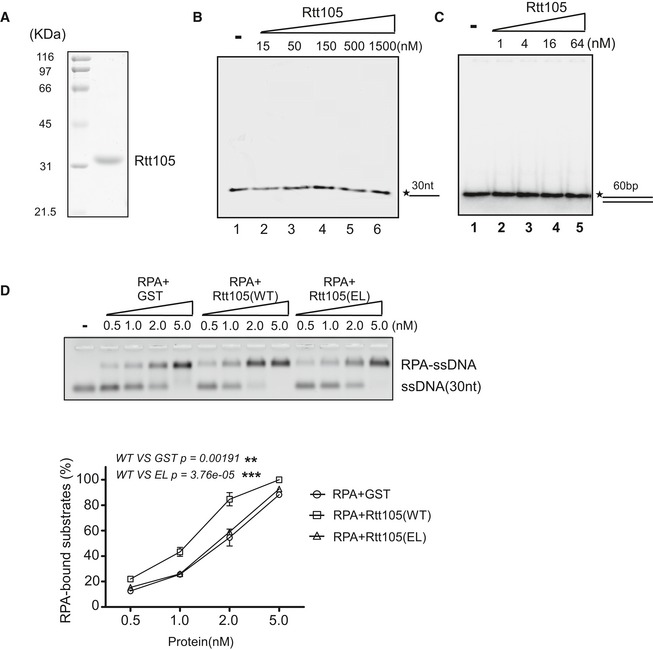
- Recombinant His‐tagged Rtt105 used in EMSAs. Purified protein was resolved on 10% SDS–PAGE and stained with CBB.
- Rtt105 does not bind ssDNA. 30‐nt ssDNA was 5′‐labeled with [γ‐32P] ATP. Rtt105 and DNA were incubated at room temperature (RT) for 20 min. The reaction products were resolved on 4% native PAGE on ice. The asterisk denotes the γ‐32P label.
- Rtt105 does not bind dsDNA. A 60‐bp dsDNA substrate was 5′‐labeled with [γ‐32P] ATP. Rtt105 and DNA were incubated at room temperature (RT), and the reaction products were resolved on 4% native PAGE on ice. The asterisk denotes the γ‐32P label.
- The Rtt105‐EL mutant protein is deficient in promoting RPA–ssDNA binding. Recombinant GST‐tagged WT and mutant were used in EMSAs. After incubation with RPA on ice for more than 2 h in titrations, Cy3‐labeled 30‐nt ssDNA was added as the substrate. The reaction mixtures were resolved on 2% native agarose gels, and the Cy3 signals were detected. At least three independent experiments were performed. The mean values ± SD from three independent experiments are shown. P‐values were derived from two‐way analysis of variance (ANOVA; **0.001 ≤ P‐value < 0.01, ***P‐value ≤ 0.001).
We noticed that the position of the DNA‐protein complex on the native gels is similar in reactions with or without Rtt105, suggesting that Rtt105 is not a stable component of the RPA–ssDNA complex. To test this idea, we mixed the purified Rtt105–RPA complex with ssDNA in a 2:1 ratio and separated the reaction products using gel filtration column chromatography: Rtt105 was not in the RPA–ssDNA complex (Fig 4D, compare Fractions 9 and 10 to Fractions 5–8). Consistent with this idea, we found that with an increased amount of 30‐nt ssDNA, the fraction of Rtt105 that was co‐purified with GST‐RPA decreased in the GST‐RPA pull‐down assays (Fig 4E). Together, these results suggest that while Rtt105 promotes the binding of RPA to ssDNA, Rtt105 itself does not remain associated with the final product.
Rtt105 alters the ssDNA binding mode of RPA
To better understand how Rtt105 promotes RPA binding to ssDNA in vitro, we performed a series of EMSAs using three different lengths of oligodeoxythymidine (oligo(dT), 17, 23, and 30 nt) and compared the effect of Rtt105 on ssDNA binding (Appendix Fig S6). We chose to use these short oligo(dT)s because each short oligo allows one RPA trimer to bind, thereby avoiding the complication of cooperative binding from a second RPA complex. Moreover, it has been shown using these short oligos that yeast RPA engages ssDNA via at least two binding modes: with three OB‐fold domains of Rfa1 contacting 12–23 nt of ssDNA, and with three OB‐fold domains of Rfa1 and one OB‐fold domain of Rfa2 contacting 23‐ to 27‐nt ssDNA (Kim et al, 1994; Bastin‐Shanower & Brill, 2001). Consistent with published results (Bastin‐Shanower & Brill, 2001), we observed that the binding constant for RPA in the absence of Rtt105 ranges from 0.11 × 108 M−1 for oligo(dT)17 to approximately 0.34 × 108 M−1 for oligo(dT)30. Remarkably, Rtt105 promotes the binding of RPA to all of these different lengths of ssDNA substrates (Appendix Fig S6). Importantly, when normalized against RPA binding to the corresponding length of oligo(dT) in the absence of Rtt105, we observed that Rtt105 stimulated RPA binding to (dT)23 and (dT)30 to a similar degree (14.9‐fold and 15.4‐fold). In contrast, the stimulatory effect for the oligo(dT)17 was much smaller (3.1‐fold; Appendix Fig S6). These results suggest that Rtt105 changes the ssDNA binding mode of RPA, likely by facilitating RPA to adopt an extended conformation (see Discussion).
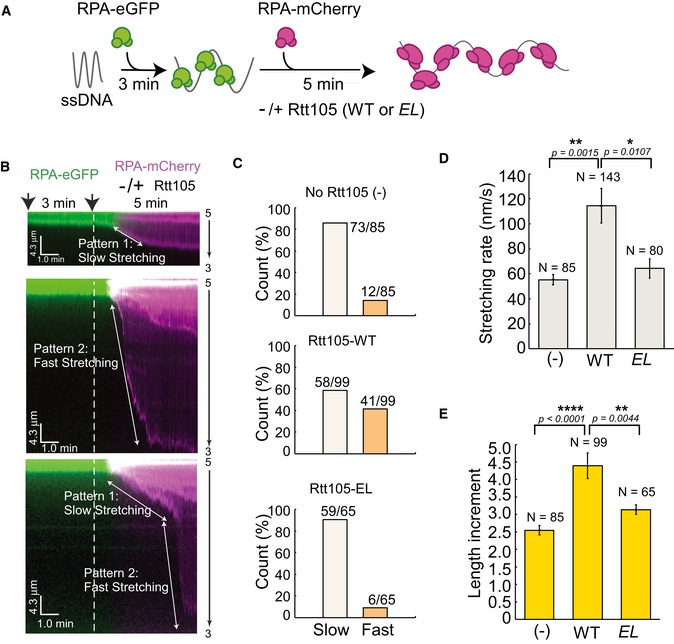
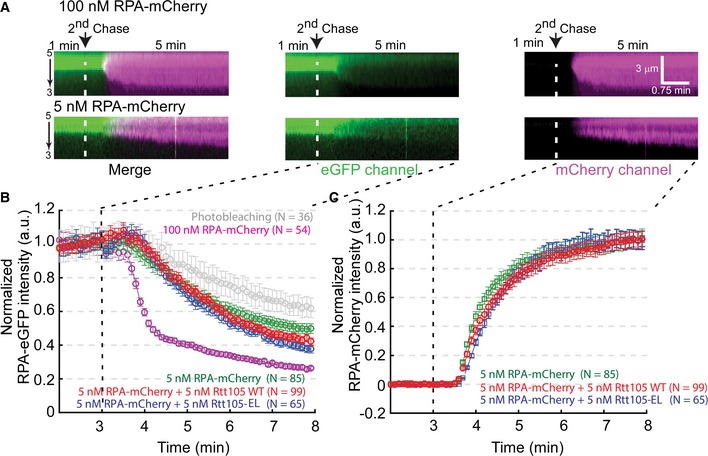
To gain additional mechanistic insight on how Rtt105 affects the RPA–ssDNA interaction, we used an ssDNA curtain assay to visualize the behavior of RPA binding to individual ssDNA molecules in real time (Gibb et al, 2014). ssDNA has a relatively short contour length, and therefore cannot be maintained in an extended conformation necessary for microscopic observation. Therefore, the ssDNA curtain assay relies on bound fluorescent RPA to visualize ssDNA. ssDNA‐RPA‐eGFP complexes were generated inside a microfluidic flow cell by flowing 1 nM RPA‐eGFP in buffer onto stationary ssDNA for 3 min. During this incubation, RPA‐eGFP binds and therefore extends ssDNA. 5 nM RPA‐mCherry protein pre‐mixed with or without 5 nM Rtt105‐WT or the Rtt105‐EL mutant was then injected into the flow cell to allow for the exchange of RPA‐eGFP for RPA‐mCherry (Fig 5A). The RPA‐eGFP signal decreased at the expense of the RPA‐mCherry signal, which increased as RPA molecules exchanged (Fig EV4A). When Rtt105 was included, both the dissociation rate of RPA‐eGFP and the binding rate of RPA‐mCherry remained unaffected (Fig EV4B and C). However, based on the stretching rate defined by the change in the ssDNA length upon binding by RPA‐mCherry in unit time, two distinct patterns were observed in the presence of Rtt105 (Fig 5B and C, Appendix Fig S7, and Movie EV1): (i) pattern 1 (58/99, 58.6%), slow stretching; and (ii) pattern 2 (41/99, 41.4%), fast stretching. We also observed some individual DNA molecules with mixed patterns, a slow‐stretching pattern first, followed by a fast‐stretching pattern (Fig 5B, lower panel). Without Rtt105, pattern 1 was dominant (73/85, 85.6%). This was also the case with the Rtt105‐EL mutant: pattern 1 (59/65, 90.8%; Fig 5C, lower panel; Movie EV2). Moreover, on average, the stretching process was relatively fast upon the addition of the Rtt105‐RPA‐mCherry complex (Fig 5D). Finally, Rtt105 increased ssDNA length upon binding with RPA‐mCherry. Without Rtt105, comparing to the ssDNA molecule length after the first chase with 1 nM RPA‐eGFP in the flow, the ssDNA length increased by ~ 2.6‐fold (2.55 ± 0.13) when chased with 5 nM RPA‐mCherry only. In the presence of Rtt105, the ssDNA molecule length increased by ~ 4.4‐fold (4.39 ± 0.36; Fig 5E). These faster and more extensive stretching events depended on the Rtt105–RPA interaction, as the Rtt105‐EL mutant had no apparent effect on either the distribution pattern of RPA or stretching rate (Fig 5D and E). Taken together, these results provide additional evidence supporting the idea that Rtt105 affects the mode of RPA binding to ssDNA in vitro.
Figure 5. Rtt105 affects the binding mode of RPA to ssDNA .
-
ASchematic for ssDNA curtain assay. Briefly, ssDNA curtains were formed by flowing 1 nM RPA‐eGFP for 3 min. Then, 5 nM RPA‐mCherry pre‐mixed with or without 5 nM Rtt105 wild‐type (WT) or E171L172AA (EL) mutant proteins was injected into the flow cell. Data were acquired with a TIRF microscope (Nikon, Inverted Microscope Eclipse Ti‐E). Lasers were shuttered at 2‐s intervals, and the exposure time was 100 ms. Each kymogram with two channels, in which green and red represent eGFP and mCherry signal, respectively.
-
B, CKymograms showing most ssDNA molecules two distinct stretching patterns of RPA–ssDNA molecules. Pattern 1, slow stretching; pattern 2, fast stretching. The counts of the two patterns in each reaction are plotted in (C). Note that a fraction of individual ssDNA shows both patterns.
-
DRtt105 increases the rate of ssDNA stretching upon RPA binding. Stretching rate was defined as the length change of ssDNA upon binding to RPA‐mCherry in unit time as shown in (B) (white arrow). All error bars represent SD for each data set. Statistical significance was evaluated based on Student's t‐tests (*0.01 ≤ P‐value < 0.05; **0.001 ≤ P‐value < 0.01).
-
ERtt105 increases the extent of ssDNA stretching upon RPA binding. Length increment = DNA length at 8 min/DNA length at 3 min. All error bars represent SD for each data set. Statistical significance was evaluated based on Student's t‐tests (**0.001 ≤ P‐value < 0.01; ****P‐value < 0.0001).
Figure EV4. RPA‐eGFP can be rapidly replaced on ssDNA by RPA‐mCherry.
- The exchange of RPA‐eGFP with free RPA‐mCherry is dependent on the concentration of RPA‐mCherry. Each kymogram shows a single RPA–ssDNA complex at an indicated experimental condition. Each kymogram with two channels was also split into two single‐color kymograms, in which the green color represents eGFP signal and the red color represents mCherry signal.
- Rtt105 does not significantly affect the disassociation rate of RPA‐eGFP from ssDNA, as indicated by a 5 nM RPA‐mCherry chase. Quantitation of the green color signal intensity versus time for each experimental condition of the second chase. Each curve represents the normalized average calculated from the indicated numbers of ssDNA molecules. Error bars represent the standard deviation for each data set.
- Rtt105 increased the RPA‐mCherry binding to ssDNA. Quantitation of the mCherry signal intensity versus time for each indicated the second chase condition. Each curve represents the normalized average calculated from the indicated numbers of ssDNA molecules. Error bars represent the standard deviation for each data set.
Rtt105 is important for DNA synthesis under replication stress
As RPA is essential for DNA replication (Brill & Stillman, 1991; Yeeles et al, 2015), we sought to determine whether deletion of RTT105 affects DNA synthesis by monitoring BrdU incorporation during DNA replication under the conditions described in Fig 2A. DNA samples were denatured and subjected to immunoprecipitation (IP) with antibodies against BrdU to enrich for nascent DNA (BrdU IP). The BrdU signal was enriched at replication origins in both HU‐treated (Figs 6A and EV5A) and untreated WT cells (Figs 6C and EV5B). The average BrdU level around the fired replication origins was substantially reduced in rtt105∆ cells compared with WT cells under HU conditions (Figs 6A and B, and EV5A). Measurement of cellular dNTP concentrations revealed that the dNTP levels in rtt105Δ cells were higher than those in WT cells (Appendix Fig S8A), indicating that the defect in DNA replication in cells lacking Rtt105 is not caused by reduced dNTP levels. We also found that the rtt105∆ mutation exaggerated the growth and drug sensitivity of rtt101∆ mutation (Appendix Fig S8B). Rtt101 is a Cul4 E3 ubiquitin ligase required for replication through damaged templates and difficult replicating regions (Luke et al, 2006). These results indicate that Rtt105 is required for efficient DNA synthesis under replication stress.
Figure 6. Rtt105 is important for DNA replication and genome stability maintenance.
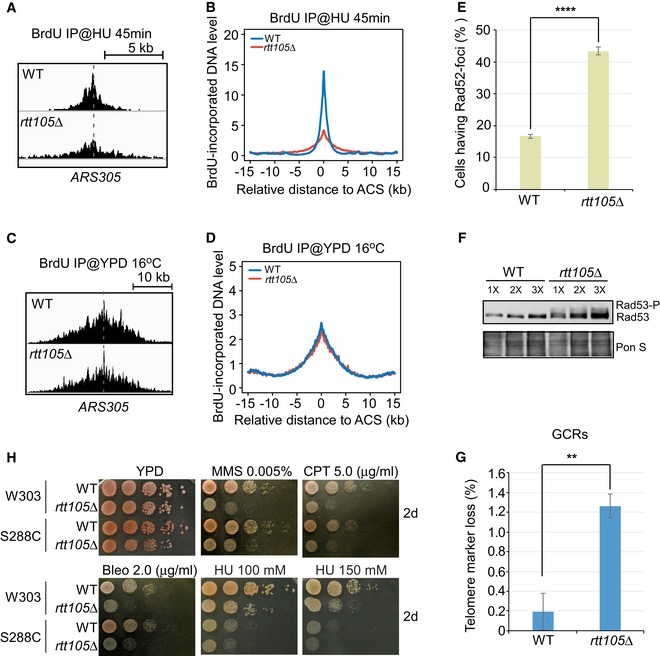
- Snapshots of BrdU IP‐seq peaks at ARS305 from cells released into YPD medium containing HU for 45 min.
- The average BrdU IP‐seq read density around fired ACS sites from cells released into YPD medium containing HU for 45 min.
- Snapshots of BrdU IP‐seq peaks at ARS305 from cells released into YPD medium at 16°C for 72 min.
- The average BrdU IP‐seq read density around ACS sites from cells released into 16°C for 72 min.
- Rad52‐GFP foci were counted in WT and rtt105Δ cells, and the percentage of cells with Rad52‐foci is reported. Error bars indicate one standard deviation from three independent experiments with at least 150 cells counted in each replicate. Statistical significance was evaluated based on Student's t‐tests (****P‐value < 0.0001).
- Rad53 phosphorylation was analyzed in WT and rtt105Δ cells by immunoblotting of protein extracts with anti‐Rad53 antibodies. Ponceau S (Pon S) staining was applied as a loading control.
- Deletion of RTT105 leads to an increased rate of gross chromosomal rearrangements (GCRs). Wild‐type (WT) and rtt105Δ mutant strains integrated with the yWSS439‐5oriΔ yeast artificial chromosomes (YACs). GCRs rates in the evaluation of telomere marker loss were assayed and calculated as previously described (Huang & Koshland, 2003). Data represent the mean (±SD) of three independent experiments. Statistical significance was evaluated based on Student's t‐tests (**0.001 ≤ P‐value < 0.01).
- Ten‐fold serial dilutions of WT or rtt105Δ cells in two different backgrounds were assayed on normal growth media (YPD) and on media containing the indicated DNA‐damaging agents, methyl‐methanesulfonate (MMS), camptothecin (CPT), bleomycin (Bleo), and hydroxyurea (HU).
Figure EV5. Snapshots of BrdU IP‐seq of chromosome III .
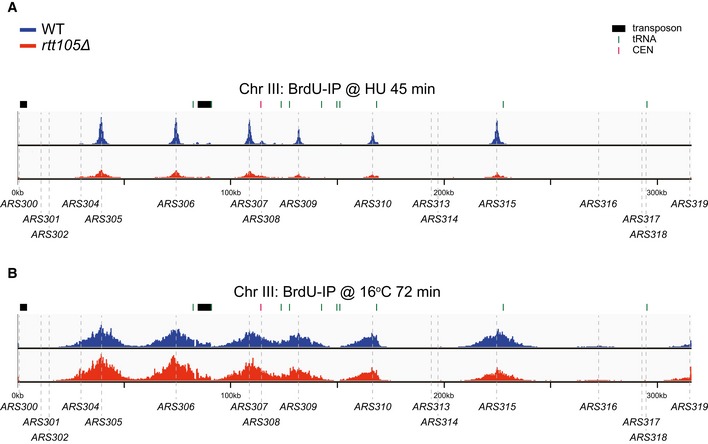
- Snapshot of BrdU IP‐seq at chromosome III for cells under HU treatment is shown.
- Snapshot of BrdU IP‐seq at chromosome III for cells at S phase without HU treatment (16°C, 72 min) is shown. The sequencing reads were mapped to the yeast reference genome.
The average BrdU density at active replication forks (16°C, 72 min) showed only a minor defect on DNA synthesis (Figs 6C and D, and EV5B), suggesting that Rtt105 deficiency does not cause a dramatic global reduction in DNA synthesis under normal conditions. However, because the association of RPA with DNA replication forks is reduced in rtt105∆ mutant cells (Fig 2E), we tested whether Rtt105 is required for normal replication using other sensitive assays. We observed that rtt105∆ cells display a 5‐ to 10‐min delay in progression through S phase (Appendix Fig S2B). Moreover, the colony size of rtt105Δ cells is smaller than that of WT cells (Appendix Fig S9B). Finally, the rtt105∆ mutation exhibits synthetic lethality when combined with temperature‐sensitive mutants of either ORC2 (a subunit of the origin recognition complex) or POL3 (the catalytic subunit of DNA polymerase δ; Appendix Fig S8C). Taken together, these results indicate that the rtt105∆ mutation also likely impacts normal replication function, but its effect is minor. This could be because RPA molecules exceed the amount needed for protecting ssDNA under normal growth conditions (see Discussion).
Rtt105 is important for genome integrity
Cells defective in DNA synthesis in general exhibit genome instability (Zeman & Cimprich, 2013). Therefore, we used multiple approaches to monitor the genome stability of rtt105∆ mutant cells. We observed that Rad52‐GFP foci, which mark spontaneous chromosome breaks during S phase (Lisby et al, 2001, 2003), occurred at a higher frequency in rtt105Δ cells than in WT cells under normal growth conditions (Fig 6E). Consistent with this observation, we also found an increase in Rad53 phosphorylation in rtt105Δ cells compared to WT cells (Fig 6F), indicating checkpoint activation in rtt105Δ cells in the absence of exogenous stress. These results are consistent with the idea that spontaneous chromosome breaks are elevated in cells lacking Rtt105.
To examine the effect of rtt105∆ on chromosome aberration, we measured the rates of gross chromosomal rearrangement (GCR) and found that the GCR rate in rtt105Δ cells is higher than in WT cells (Fig 6G). Moreover, the rtt105∆ cells are highly sensitive to DNA‐damaging agents such as camptothecin (CPT), methyl‐methanesulfonate (MMS), and bleomycin (Bleo), and mildly sensitive to HU (Fig 6H). In addition to increased sensitivity to these exogenous DNA‐damaging agents, rtt105Δ cells were also sensitive to HO‐induced DSBs [Appendix Fig S9A; HO is an endonuclease and yeast cells contain a single defined HO site at the MAT locus (Strathern et al, 1982)]. Finally, the rtt105∆ mutation exhibits synthetic growth defects with mutations in RAD52, RAD51, and YKU70/80 (Appendix Fig S9B and C). Rad52 and Rad51 are essential for homologous recombination (HR), whereas Ku70/80 is essential for non‐homologous end joining (NHEJ). These results indicate that Rtt105 may also contribute to DNA damage repair pathways.
Discussion
Rtt105 was a previously uncharacterized protein. Here, we show that Rtt105 interacts with RPA and regulates RPA nuclear import as well as the mode of RPA binding to ssDNA in vitro. However, Rtt105 does not remain stably associated with the final RPA–ssDNA complex. In cells, we show that Rtt105 is needed for the association of RPA with both active and HU‐stalled DNA replication forks. Interestingly, DNA synthesis in rtt105∆ is dramatically reduced under HU‐induced replication stress, but not under normal growth conditions. Finally, rtt105∆ mutant cells exhibit defects in genome integrity. Taking our results as a whole, we propose that Rtt105 functions as an “RPA chaperone” that both accompanies RPA during nuclear import and promotes RPA–ssDNA complex formation at replication forks (Fig 7).
Figure 7. A proposed model for Rtt105 acting as a chaperone for RPA .
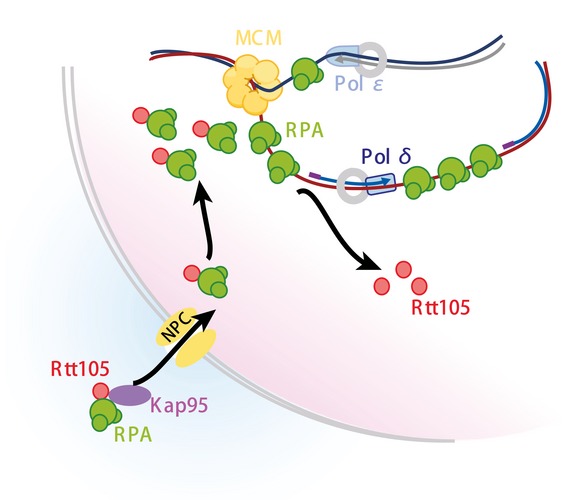
Rtt105 accompanies RPA during nuclear import and promotes RPA–ssDNA complex formation at replication forks. Rtt105 does not remain stably associated with the final RPA–ssDNA complex.
Rtt105 is an RPA chaperone
It was reported that importin protein Kap95 is important for RPA nuclear import (Belanger et al, 2011). We observed that the nuclear localization of Rfa1 is compromised in rtt105∆ cells. Moreover, the interaction between RPA and Kap95 is substantially reduced in rtt105∆ cells, indicating that Rtt105 mediates the RPA–Kap95 interaction and nuclear import. In addition to nuclear import, we provide several lines of evidence indicating that Rtt105 also functions in loading RPA onto ssDNA at replication forks. First, the interaction of Kap95 with Rtt105 occurs mainly at the nuclear periphery, whereas the Rtt105–RPA interaction occurs predominantly within the nucleus. Second, the association of Rfa2 with replication forks remains defective in rtt105∆ cells even after the targeting Rfa1 into the nucleus using a strong nuclear localization signal that bypasses the requirement for Rtt105. Third, we found that Rtt105 increases the binding efficiency of RPA with ssDNA in vitro, likely by mediating a change in the mode of RPA binding to ssDNA. RPA is a heterotrimer of RPA70, RPA32, and RPA14 with six OB‐fold domains, four of which engage with ssDNA during binding. It has been reported that RPA adopts multiple binding modes to interact with ssDNA, in which different numbers of OB‐fold domains contact different lengths of ssDNA (Kim et al, 1994; Kolpashchikov et al, 2001; Bochkareva et al, 2002; Fanning et al, 2006; Fan & Pavletich, 2012; Brosey et al, 2013). Yeast RPA adopts at least two binding modes, with OB‐A, OB‐B, and OB‐C of Rfa1 contacting 12–23 nt of ssDNA, and with these three OB‐fold domains along with OB‐D in Rfa2 contacting 23–27 nt of ssDNA (Bastin‐Shanower & Brill, 2001). We observed that Rtt105 stimulates RPA binding to 17‐nt, 23‐nt, and 30‐nt ssDNA. Moreover, the stimulatory effect is dramatically higher for 23 and 30 nt than for 17 nt. These results suggest that Rtt105 could impact the conformational transition of the RPA complex and that Rtt105 is more important when more OB‐fold domains of RPA are needed to protect a longer ssDNA. Our ssDNA curtain data support this idea: We found that in the presence of Rtt105, RPA's capacity to stretch ssDNA is substantially increased. This aspect of Rtt105 function is analogous to the RPA conformational change induced by force reported before. Using a combination of atomic force microscopy imaging and mechanical manipulation of single ssDNA tethers, it was reported that a mechanical force mediates the switch of the RPA‐bound ssDNA from amorphous aggregation to a much more regular extended conformation (Chen et al, 2015). Thus, in addition to mediating RPA, Rtt105 also functions in assisting RPA to adopt a mode for efficient interaction with ssDNA.
The relationship between Rtt105 and RPA is analogous to the relationship between histone chaperones and histones (Akey & Luger, 2003; Gurard‐Levin et al, 2014; Hammond et al, 2017). Both histones and RPA are major DNA binding proteins in eukaryotic cells, with histones protecting dsDNA and RPA protecting ssDNA (Wold, 1997; Akey & Luger, 2003). Histone chaperones are engaged with histones, from nuclear import to the formation of nucleosomes (Keck & Pemberton, 2012; Hammond et al, 2017). For example, histone chaperone Asf1 forms a complex with histone H3–H4 and importin 4 (Kap123; Campos et al, 2010; Jasencakova et al, 2010), and Nap1 forms a complex with H2A–H2B and Kap114 (importin 9; Straube et al, 2010). In addition to histone import, histone chaperones also promote nucleosome formation. However, these histone chaperones are not present in nucleosomes, the final products that they assist to form (Akey & Luger, 2003). Similarly, while Rtt105 promotes the association of RPA with ssDNA, Rtt105 is not present in the final RPA–ssDNA complex. Therefore, the functions of Rtt105 in regulating RPA nuclear import and ssDNA binding are quite analogous to histone chaperones in regulating histone import and promoting nucleosome assembly, and hence, we propose that Rtt105 functions as an RPA chaperone.
Rtt105 is important for DNA synthesis under replication stress
The association of RPA with chromatin is significantly reduced at HU‐stalled forks as well as active forks under normal growth conditions in cells lacking Rtt105. RPA is essential for DNA replication in eukaryotes (Wobbe et al, 1987; Fairman & Stillman, 1988; Wold & Kelly, 1988; Yeeles et al, 2015). Therefore, we tested whether Rtt105 has a role in DNA replication. Interestingly, we did not detect a global reduction in DNA synthesis in rtt105∆ mutant cells under normal replication conditions using BrdU IP‐seq, suggesting that the reduced association of RPA with active DNA forks is not likely due to reduced DNA replication. One trivial explanation for the apparent discordance between the reduction in RPA binding and apparent normal DNA synthesis in rtt105∆ cells could be a technique issue, where inefficient RPA ChIP exaggerates the degree of RPA reduction at replication forks. Another possibility is that the amount of RPA at DNA replication forks is not reduced in rtt105∆ mutant cells enough to cause a global reduction in DNA synthesis under unstressed conditions. Consistent with this explanation, it is estimated that human RPA is in 6‐ to 10‐fold excess relative to the amount needed to protect ssDNA under normal replication conditions (Toledo et al, 2013, 2017). Alternatively, during normal replication, the amount of ssDNA within unperturbed fork is relatively less and shorter at length, as suggested by direct observation of replication bubble region via electron microscopy (Sogo et al, 2002). Under this circumstance, we reasoned such small ssDNA is moderately stable to allow smooth replisome progression, even dramatically reduced RPA amount when the Rtt105 function is trivialized. On the contrary, exposure to HU may trigger potentially longer ssDNA formation, where an instant requirement for more RPA binding must be fulfilled to maintain fork integrity. Therefore, RPA paucity aggravated by lacking escorted Rtt105 could explain the sudden destabilization of fork that eventually causes replication collapse. This hypothesis would be in consistence with our experimental evidence suggesting Rtt105 is particularly important to uphold DNA replication under replication stress. Moreover, it is possible that another RPA chaperone contributes to the delivery of RPA under normal growth conditions. For example, histone chaperone CAF‐1 is critical for DNA replication‐coupled nucleosome assembly from yeast through humans, and yet yeast cells lacking CAF‐1 exhibit only a minor growth defect because two other histone chaperones, Rtt106 and FACT, compensate (Li et al, 2008; Yang et al, 2016). Furthermore, it is possible that Rad51 could complement the reduced levels of RPA in rtt105∆ mutant cells. Despite the lack of direct evidence regarding functional relationships between RPA and Rad51 specifically during DNA replication, there are several lines of evidence that might support this notion. First, previous experiments suggest that Rad52 can help to load Rad51 onto unperturbed replication fork to complete alkylated DNA replication induced by MMS treatment (González‐Prieto et al, 2013). It is proposed that ssDNA generated during lagging strand synthesis acts as substrate for Rad52‐facilitated Rad51 loading (Sung, 1997; González‐Prieto et al, 2013). Second, double‐strand breaks that arise from replication fork collapse may be repaired by break‐induced replication (BIR) in both human cells and yeast (Llorente et al, 2008; Malkova & Ira, 2013; Costantino et al, 2014; Alexander & Orr‐Weaver, 2016). BIR DNA synthesis can generate long‐lived ssDNA intermediate that can be stabilized and protected by RPA, and overexpression of Rad51 can suppress the BIR defect of rfa1 hypomorphic mutants (Ruff et al, 2016). Nonetheless, while global DNA synthesis is not reduced in rtt105∆ cells, we present several lines of evidence supporting the idea that Rtt105 has a role in normal DNA replication. First, we observed that the colony size of rtt105Δ cells is relatively smaller than that of WT cells and that S‐phase progression is slightly delayed. Second, rtt105Δ cells exhibit a synthetic lethal phenotype with DNA replication mutants including mutations to ORC and DNA polymerase. These results indicate that Rtt105 has a role, although not an essential one, in DNA replication.
Remarkably, we observed that DNA synthesis as detected by BrdU incorporation is dramatically reduced in rtt105∆ cells treated with HU. A reduction in DNA synthesis likely leads to a reduced amount of RPA at HU‐stalled replication forks. Therefore, the reduction in RPA at HU‐stalled replication forks detected in rtt105∆ cells is likely due to the combination of defects in RPA loading and nuclear import mediated by Rtt105 and subsequent reduced DNA synthesis. HU treatment is known to deplete dNTPs required for DNA synthesis and induce DNA replication stress. These results suggest that rtt105∆ mutant cells can tolerate reduced association of RPA with ssDNA under normal growth conditions, but experience difficulty under replication stress caused by HU. DNA replication stress can be caused by a variety of internal events and external agents that impede normal replication progression, including collisions between the DNA replication and gene transcription machineries, DNA‐damaging agents, and oncogene activation in precancerous lesions in human cells, for example (Aguilera & García‐Muse, 2013; Zeman & Cimprich, 2013). It is likely that more ssDNA is generated under replication stress than under unstressed conditions, which may explain why Rtt105 plays a greater role in DNA synthesis under stress conditions. Consistent with this interpretation, we observed that Rtt105 genetically interacts with Rtt101, an E3 ubiquitin ligase that is required for replicating through damaged regions and natural pause sites (Luke et al, 2006), which induce replication stress. Therefore, we suggest that Rtt105 is needed more for the regulation of the association of RPA with ssDNA during DNA replication stress.
Is Rtt105 involved in other aspects DNA metabolism?
In addition to DNA replication, RPA is also important for DNA repair and for activation of the DNA replication checkpoint kinase, Mec1 (ATR in human cells; Maréchal & Zou, 2015). The phenotypes of rtt105∆ cells reported here suggest that Rtt105 may also contribute to these processes. For example, rtt105Δ cells are sensitive to DNA‐damaging agents and the HO‐induced double‐strand break and displayed increased spontaneous chromosome breaks and abnormal chromosome rearrangements. Moreover, we found that rtt105Δ cells exhibit a synthetic sick phenotype with mutations at key genes involved in HR and NHEJ. These results indicate that Rtt105's role in these repair processes may be complicated, and further in‐depth analysis is needed in the future.
In addition to RPA, we also detected Rim1 co‐purifying with Rtt105 (Table EV1). Rim1 is an ssDNA‐binding protein important for mitochondrial DNA replication (Van Dyck et al, 1992). Therefore, it would be interesting to determine whether Rtt105 also has a role in regulating Rim1 localization and ssDNA binding in the mitochondria.
The protein sequence of Rtt105 is highly conserved within the Saccharomyces clade. While Rtt105 has a sequence orthologue in Schizosaccharomyces pombe, it does not appear to have sequence orthologues in higher eukaryotes. It is noteworthy that Rtt106 and Rtt109 do not have sequence orthologues in higher eukaryotic cells. However, when the crystal structures of these two proteins were solved, it was apparent that these proteins were, respectively, functional homologs of a histone chaperone (Daxx; Drané et al, 2010; Su et al, 2012) and a histone acetyltransferase enzyme (p300/CBP; Tang et al, 2008). We suggest that a similar situation exists with Rtt105. In Xenopus, an RPA binding protein, XRIPα, was identified in complex with importin β. XRIPα is required for RPA's nuclear import, and its orthologues are present in higher eukaryotic cells from Drosophila to human cells, but not in yeast (Jullien et al, 1999). It would be interesting to test whether XRIPα is the functional homolog of Rtt105 in higher eukaryotic cells.
Materials and Methods
Yeast strains, plasmids, and culture
All yeast strains used in this study were derived from the W303 (leu2‐3, 112 ura3‐1 his3‐11, trp1‐1, ade2‐1 can 1‐100) except where indicated. All genotypes (Appendix Table S1), plasmid constructions, and oligonucleotides (Appendix Table S2) are listed in Appendix Supplementary Methods. Standard culture media and genetic techniques were used to generate all the yeast strains.
Yeast spot assays
To analyze phenotypes of yeast strain, serial dilutions (10‐fold) of fresh cultures concentrated to an OD600 of 0.6 were spotted onto YPD plates containing different concentration of drugs. Plates were incubated at 30°C for the time noted in each figure legend. Each spot assay was performed with three replicates with one representative result shown.
Chromatin immunoprecipitation (ChIP), BrdU immunoprecipitation (BrdU IP), and ssDNA library preparation for sequencing (ChIP‐seq and BrdU IP‐seq)
ChIP‐seq and BrdU IP‐seq were performed according to procedures previously described with minor modifications (Yu et al, 2014). All yeast cells were arrested at G1 phase with α‐factor and then either released into fresh YPD media containing 200 mM hydroxyurea (Sigma) for 45 min at 25°C to capture HU‐stalled replication forks or released into fresh YPD media for 72 min at 16°C to capture active replication forks. For BrdU IP, 400 mg/l BrdU (Sigma) was added to YPD medium. Cells were cross‐linked with 1% formaldehyde (Sigma) at 25°C for 20 min and quenched with 0.125 M glycine for 5 min. The DNA was sheared to an average size of 200 bp using a Bioruptor (Diagenode, Inc.). The Rfa1/Rfa2 protein‐bound DNA was isolated as ChIP assay described in Appendix Supplementary Methods (due to limited amount of Rfa1 antibodies, we performed Rfa2 ChIP in the experiment at 16°C for 72 min). After binding with Protein G Sepharose (GE Healthcare) and following extensive washing, chromatin was reverse‐cross‐linked in eluting buffer (10 mM Tris–HCl, pH 8.0, 10 mM EDTA, 1% SDS, 150 mM NaCl, and 5 mM DTT) overnight at 65°C. BrdU IP was performed using denatured DNA and BrdU antibodies (GE Healthcare). The input DNA and ChIP DNA were then subjected to library construction following procedures as previously described (Yu et al, 2014), and the samples were then sequenced with the Illumina HiSeq 2500 system. The data were mapped to the S. cerevisiae reference genome (sacCer3) using Bowtie2 software. More than 1 million reads were successfully mapped for each sample. Reads coverage was calculated with a step size of 10 bp and normalized to total reads. Signal‐enriched regions were called by MACS2 software. The average coverage around ACS was plotted to show the distribution of protein during DNA replication. The ACS information is from previous reports (Eaton et al, 2010; Liu et al, 2017). Information regarding transposons, rDNA, telomere, and centromere was abandoned for a normal DNA replication pattern analysis.
Purification of Rtt105–RPA complex
Recombinant Rtt105 and RPA protein purification and in vitro pull‐down assay are described in Appendix Supplementary Methods. To purify Rtt105–RPA complex, two plasmids (p11td‐scRPA, gift from Dr. Marc Wold, University of Iowa; and pQlinkN‐Rtt105, this study) were separately transformed into Escherichia coli strain BL21 (DE3). Inoculation and induction of RPA complex followed the procedures above. No tagged Rtt105 was induced by adding 1 mM IPTG at 37°C for 5 h in 1 l culture. After French press and clarified by centrifugation at 40,000 g for 1 h, equal volumes of both lysates were mixed for 1–2 h at 4°C on shaker before chromatography steps. Subsequent purification of Rtt105–RPA complex was adapted from a previous RPA purification method (Binz et al, 2006) using both Affi‐Blue Gel (Bio‐Rad) and Hydroxyapatite (Bio‐Rad). Rtt105–RPA complex was eluted with HI buffer containing 80 mM potassium phosphate. Proteins from these purification steps were further purified through Mono‐Q (GE Healthcare) and Superdex 200 (Increase 10/300; GE Healthcare) chromatography. The resulted protein complex was dialyzed in 1× TBS with 10% glycerol and 1 mM DTT overnight, and concentrated by Ultra Centrifugal Filter Unit (Millipore).
Electrophoresis mobility shift assay (EMSA)
DNA substrates used in this study were labeled with either [γ‐32P] using T4 polynucleotide kinase (NEB) or Cy3 (Invitrogen) at 5′‐end. 5 nM Cy3‐labeled DNA substrate or 1 nM γ‐32P‐labeled DNA substrate was incubated with indicated amounts of proteins at room temperature in 1× binding buffer (25 mM Tris, pH 7.5, 200 mM NaCl, 5 mM MgCl2, 1 mM DTT, 5% glycerol, and 0.05% Triton X‐100) for 20 min. The reaction mixture (20 μl in total) was then loaded with 4 μl 6× loading dye and resolved in either a 4% native acrylamide/Bis gel or 2% agarose (Biowest) gel in cold 0.5× TBE buffer (44.5 mM Tris, 44.5 mM boric acid, and 0.5 mM EDTA, pH 8.3). Native acrylamide/Bis gels were dried on Whatman DE3 paper in a Model 583 Gel Dryer (Bio‐Rad). Signals were detected on a Typhoon FLA 9500 imager (GE Healthcare), and band intensities were quantified by ImageJ (NIH). The mean values ± SD from three independent experiments were plotted, with P‐values derived from two‐way analysis of variance (ANOVA). At least three independent biological repeats were performed, and similar results were obtained each time.
Single‐stranded DNA (ssDNA) curtain assay
Single‐tethered ssDNA curtain assay was performed as previously described with minor modifications (Gibb et al, 2014). ssDNA‐RPA‐eGFP complex was firstly chased with 1 nM RPA‐eGFP in buffer A (40 mM Tris–HCl (pH 7.5), 2 mM MgCl2, 150 mM NaCl, 1 mM DTT, and 0.2 mg/ml BSA) for 3 min and then secondly chased with m nM RPA‐mCherry and n nM wild‐type Rtt105 (WT) or Rtt105E171L172AA mutant (EL) proteins. We defined five different experimental conditions: (1) Case 1: m = 0, and n = 0 (a photo bleaching test); (2) Case 2: m = 100, and n = 0; (3) Case 3: m = 5, and n = 0; (4) Case 4: m = 5, and n = 5; (5) Case 5: m = 5, and n = 5. All experimental data were acquired with a custom‐built prism‐type total internal reflection fluorescence (TIRF) microscope (Nikon, Inverted Microscope Eclipse Ti‐E). The microscope was mounted with OBIS 488‐nm and 561‐nm LS 100‐mW lasers. The real laser powers before the prism were measured: (i) 488 nm, 9.9 mW; and (ii) 561 nm, 16.0 mW.
The stretching rate of ssDNA bound by RPA‐mCherry (nm/s) defined by the ssDNA length change in unit time was measured as shown in the kymographs in Fig 3D (white arrows), and then, they were plotted in histograph (Appendix Fig S2). Single‐Gaussian function was used to fit Case 3 of 5 nM RPA‐mCherry and Case 5 of 5 nM RPA‐mCherry + 5 nM Rtt105‐m6, and double‐Gaussian functions were for Case 4 of 5 nM RPA‐mCherry + 5 nM Rtt105‐WT. The cutoff of 80 nm/s was determined by plotting a dash line between two Gaussian peaks in the data of Case 4. Slow‐stretching event was defined as the rate was < 80 nm/s, and the fast‐stretching event was that the rate was more than 80 nm/s. The error bars for all rate distributions represent 70% confidence intervals obtained through bootstrap analysis.
Accession numbers
The high‐throughput sequencing data sets have been deposited at the NCBI GEO database with the accession identifiers GSE87356 and GSE101536.
Author contributions
SL, ZX, and JX carried out the majority of experiments. ZX, CY, HG, PZ, and XW performed the ChIP‐seq analysis. DX guided the initial tests of EMSAs. LZ and ZQ performed the ssDNA curtain assay. AC and SS measured the cellular dNTP levels. LL and DL carried out some of site mutagenesis mapping. SW and YS guided florescence experiments. SZ, JL, and XC guided the DNA damage sensitivity assay. SL, JF, and QL wrote the manuscript. KC and JH commented on the manuscript. QL and JF supervised the overall research project.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 6
Acknowledgements
We thank Marc Wold for providing the yeast RPA expression constructs, Steven Brill for providing yeast Rfa1 and Rfa2 antibodies, Oscar Aparcio for the BrdU incorporation strains and plasmids, Chao Tang and Xiaojing Yang for providing GFP and mCherry plasmids, and Wenfeng Qian for providing various genetic background yeast strains. We thank Yun Zhang for assistance with the Illumina sequencing. We thank Hongxia Lv and Xiaochen Li for assistance with live‐cell imaging. We thank Zhiguo Zhang, Lilin Du, and Hengyao Niu for critical reading of the manuscript. We thank Huiqiang Lou and Lee Zou for early discussion. This work was supported by grants from NSFC [31725015 (QL) and 31671332 (JF)]. Work in AC's laboratory was funded by the Swedish Research Council, the Swedish Cancer Society, and the Knut and Alice Wallenberg Foundation.
The EMBO Journal (2018) 37: e99154
Contributor Information
Jianxun Feng, Email: fengjx@pku.edu.cn.
Qing Li, Email: li.qing@pku.edu.cn.
References
- Aguilera A, García‐Muse T (2013) Causes of genome instability. Annu Rev Genet 47: 1–32 [DOI] [PubMed] [Google Scholar]
- Ajimura M, Leem SH, Ogawa H (1993) Identification of new genes required for meiotic recombination in Saccharomyces cerevisiae . Genetics 133: 51–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey CW, Luger K (2003) Histone chaperones and nucleosome assembly. Curr Opin Struct Biol 13: 6–14 [DOI] [PubMed] [Google Scholar]
- Alexander JL, Orr‐Weaver TL (2016) Replication fork instability and the consequences of fork collisions from rereplication. Genes Dev 30: 2241–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Weinstein DM, Bell SP (1997) Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell 91: 59–69 [DOI] [PubMed] [Google Scholar]
- Bastin‐Shanower SA, Brill SJ (2001) Functional analysis of the four DNA binding domains of Replication Protein A: the role of RPA2 in ssDNA binding. J Biol Chem 276: 36446–36453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belanger KD, Griffith AL, Baker HL, Hansen JN, Kovacs LAS, Seconi JS, Strine AC (2011) The karyopherin Kap95 and the C‐termini of Rfa1, Rfa2, and Rfa3 are necessary for efficient nuclear import of functional RPA complex proteins in Saccharomyces cerevisiae . DNA Cell Biol 30: 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Dutta A (2002) DNA replication in eukaryotic cells. Annu Rev Biochem 71: 333–374 [DOI] [PubMed] [Google Scholar]
- Ben‐Aroya S, Koren A, Liefshitz B, Steinlauf R, Kupiec M (2003) ELG1, a yeast gene required for genome stability, forms a complex related to replication factor C. Proc Natl Acad Sci USA 100: 9906–9911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz SK, Dickson AM, Haring SJ, Wold MS (2006) Functional assays for replication protein A (RPA). Methods Enzymol 409: 11–38 [DOI] [PubMed] [Google Scholar]
- Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L (1997) Structure of the single‐stranded‐DNA‐binding domain of replication protein A bound to DNA. Nature 385: 176–181 [DOI] [PubMed] [Google Scholar]
- Bochkareva E, Belegu V, Korolev S, Bochkarev A (2001) Structure of the major single‐stranded DNA‐binding domain of replication protein A suggests a dynamic mechanism for DNA binding. EMBO J 20: 612–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochkareva E, Korolev S, Lees‐Miller SP, Bochkarev A (2002) Structure of the RPA trimerization core and its role in the multistep DNA‐binding mechanism of RPA. EMBO J 21: 1855–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brill SJ, Stillman B (1989) Yeast replication factor‐A functions in the unwinding of the SV40 origin of DNA replication. Nature 342: 92–95 [DOI] [PubMed] [Google Scholar]
- Brill SJ, Stillman B (1991) Replication factor‐A from Saccharomyces cerevisiae is encoded by three essential genes coordinately expressed at S phase. Genes Dev 5: 1589–1600 [DOI] [PubMed] [Google Scholar]
- Brosey CA, Yan C, Tsutakawa SE, Heller WT, Rambo RP, Tainer JA, Ivanov I, Chazin WJ (2013) A new structural framework for integrating replication protein A into DNA processing machinery. Nucleic Acids Res 41: 2313–2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PMJ, Kunkel TA (2017) Eukaryotic DNA replication fork. Annu Rev Biochem 86: 417–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess RJ, Zhou H, Han J, Zhang Z (2010) A role for Gcn5 in replication‐coupled nucleosome assembly. Mol Cell 37: 469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos EI, Fillingham J, Li G, Zheng H, Voigt P, Kuo W‐HW, Seepany H, Gao Z, Day LA, Greenblatt JF, Reinberg D (2010) The program for processing newly synthesized histones H3.1 and H4. Nat Struct Mol Biol 17: 1343–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase JW, Williams KR (1986) Single‐stranded DNA binding proteins required for DNA replication. Annu Rev Biochem 55: 103–136 [DOI] [PubMed] [Google Scholar]
- Chen J, Le S, Basu A, Chazin WJ, Yan J (2015) Mechanochemical regulations of RPA's binding to ssDNA. Sci Rep 5: 9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M (2007) Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446: 806–810 [DOI] [PubMed] [Google Scholar]
- Costantino L, Sotiriou SK, Rantala JK, Magin S, Mladenov E, Helleday T, Haber JE, Iliakis G, Kallioniemi OP, Halazonetis TD (2014) Break‐induced replication repair of damaged forks induces genomic duplications in human cells. Science 343: 88–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drané P, Ouararhni K, Depaux A, Shuaib M, Hamiche A (2010) The death‐associated protein DAXX is a novel histone chaperone involved in the replication‐independent deposition of H3.3. Genes Dev 24: 1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll R, Hudson A, Jackson SP (2007) Yeast Rtt109 promotes genome stability by acetylating histone H3 on lysine 56. Science 315: 649–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM (2010) Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairman MP, Stillman B (1988) Cellular factors required for multiple stages of SV40 DNA replication in vitro . EMBO J 7: 1211–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Pavletich NP (2012) Structure and conformational change of a replication protein A heterotrimer bound to ssDNA. Genes Dev 26: 2337–2347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res 34: 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fillingham J, Recht J, Silva AC, Suter B, Emili A, Stagljar I, Krogan NJ, Allis CD, Keogh M‐C, Greenblatt JF (2008) Chaperone control of the activity and specificity of the histone H3 acetyltransferase Rtt109. Mol Cell Biol 28: 4342–4353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin GS, Symington LS (2002) Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51–DNA complexes. EMBO J 21: 3160–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC, Mortimer RK (1974) A genetic study of X‐ray sensitive mutants in yeast. Mutat Res 24: 281–292 [DOI] [PubMed] [Google Scholar]
- Gazy I, Liefshitz B, Parnas O, Kupiec M (2015) Elg1, a central player in genome stability. Mutat Res Rev Mutat Res 763: 267–279 [DOI] [PubMed] [Google Scholar]
- Gibb B, Ye LF, Gergoudis SC, Kwon Y, Niu H, Sung P, Greene EC (2014) Concentration‐dependent exchange of Replication Protein A on single‐stranded DNA revealed by single‐molecule imaging. PLoS One 9: e87922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González‐Prieto R, Muñoz‐Cabello AM, Cabello‐Lobato MJ, Prado F (2013) Rad51 replication fork recruitment is required for DNA damage tolerance. EMBO J 32: 1307–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurard‐Levin ZA, Quivy J‐P, Almouzni G (2014) Histone chaperones: assisting histone traffic and nucleosome dynamics. Annu Rev Biochem 83: 487–517 [DOI] [PubMed] [Google Scholar]
- Hammond CM, Strømme CB, Huang H, Patel DJ, Groth A (2017) Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol 18: 141–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Zhang H, Zhang H, Wang Z, Zhou H, Zhang Z (2013) A Cul4 E3 ubiquitin ligase regulates histone hand‐off during nucleosome assembly. Cell 155: 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Koshland D (2003) Chromosome integrity in Saccharomyces cerevisiae: the interplay of DNA replication initiation factors, elongation factors, and origins. Genes Dev 17: 1741–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou H, Katzmann D, Hochstrasser M, Atanasova E, Zhang Z (2005) Rtt106p is a histone chaperone involved in heterochromatin‐mediated silencing. Proc Natl Acad Sci USA 102: 13410–13415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Zhou H, Tarara J, Zhang Z (2007) A novel role for histone chaperones CAF‐1 and Rtt106p in heterochromatin silencing. EMBO J 26: 2274–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AP, Laskey RA, Coleman N (2014) Replication proteins and human disease. Cold Spring Harb Perspect Biol 6: a013060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasencakova Z, Scharf AND, Ask K, Corpet A, Imhof A, Almouzni G, Groth A (2010) Replication stress interferes with histone recycling and predeposition marking of new histones. Mol Cell 37: 736–743 [DOI] [PubMed] [Google Scholar]
- Jullien D, Görlich D, Laemmli UK, Adachi Y (1999) Nuclear import of RPA in Xenopus egg extracts requires a novel protein XRIPalpha but not importin alpha. EMBO J 18: 4348–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellis P, Agyei R, Durocher D (2003) Elg1 forms an alternative PCNA‐interacting RFC complex required to maintain genome stability. Curr Biol 13: 1583–1595 [DOI] [PubMed] [Google Scholar]
- Keck KM, Pemberton LF (2012) Histone chaperones link histone nuclear import and chromatin assembly. Biochem Biophys Acta 1819: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim C, Paulus BF, Wold MS (1994) Interactions of human replication protein A with oligonucleotides. Biochemistry 33: 14197–14206 [DOI] [PubMed] [Google Scholar]
- Kolpashchikov DM, Khodyreva SN, Khlimankov DY, Wold MS, Favre A, Lavrik OI (2001) Polarity of human replication protein A binding to DNA. Nucleic Acids Res 29: 373–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota T, Nishimura K, Kanemaki MT, Donaldson AD (2013) The Elg1 replication factor C‐like complex functions in PCNA unloading during DNA replication. Mol Cell 50: 273–280 [DOI] [PubMed] [Google Scholar]
- Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z (2008) Acetylation of histone H3 lysine 56 regulates replication‐coupled nucleosome assembly. Cell 134: 244–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R (2003) Colocalization of multiple DNA double‐strand breaks at a single Rad52 repair centre. Nat Cell Biol 5: 572–577 [DOI] [PubMed] [Google Scholar]
- Liu S, Xu Z, Leng H, Zheng P, Yang J, Chen K, Feng J, Li Q (2017) RPA binds histone H3‐H4 and functions in DNA replication–coupled nucleosome assembly. Science 355: 415–420 [DOI] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS (2008) Break‐induced replication: what is it and what is it for? Cell Cycle 7: 859–864 [DOI] [PubMed] [Google Scholar]
- Luke B, Versini G, Jaquenoud M, Zaidi IW, Kurz T, Pintard L, Pasero P, Peter M (2006) The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol 16: 786–792 [DOI] [PubMed] [Google Scholar]
- Malkova A, Ira G (2013) Break‐induced replication: functions and molecular mechanism. Curr Opin Genet Dev 23: 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maréchal A, Zou L (2015) RPA‐coated single‐stranded DNA as a platform for post‐translational modifications in the DNA damage response. Cell Res 25: 9–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel JJ, McCarville JF, Xiong Y (2003) A role for Saccharomyces cerevisiae Cul8 ubiquitin ligase in proper anaphase progression. J Biol Chem 278: 22828–22837 [DOI] [PubMed] [Google Scholar]
- Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA (2007) Yeast DNA polymerase epsilon participates in leading‐strand DNA replication. Science 317: 127–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruff P, Donnianni RA, Glancy E, Oh J, Symington LS (2016) RPA stabilization of single‐stranded DNA is critical for break‐induced replication. Cell Rep 17: 3359–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ (2001) Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics 159: 1449–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602 [DOI] [PubMed] [Google Scholar]
- Strathern JN, Klar AJS, Hicks JB, Abraham JA, Ivy JM, Nasmyth KA, McGill C (1982) Homothallic switching of yeast mating type cassettes is initiated by a double‐stranded cut in the MAT locus. Cell 31: 183–192 [DOI] [PubMed] [Google Scholar]
- Straube K, Blackwell JS, Pemberton LF (2010) Nap1 and Chz1 have separate Htz1‐nuclear import and assembly functions. Traffic 11: 185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D, Hu Q, Li Q, Thompson JR, Cui G, Fazly A, Davies BA, Botuyan MV, Zhang Z, Mer G (2012) Structural basis for recognition of H3K56‐acetylated histone H3‐H4 by the chaperone Rtt106. Nature 483: 104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung P (1997) Function of Yeast Rad52 protein as a mediator between Replication Protein A and the Rad51 recombinase. J Biol Chem 272: 28194–28197 [DOI] [PubMed] [Google Scholar]
- Tang Y, Holbert MA, Wurtele H, Meeth K, Rocha W, Gharib M, Jiang E, Thibault P, Verrault A, Cole PA, Marmorstein R (2008) Fungal Rtt109 histone acetyltransferase is an unexpected structural homolog of metazoan p300/CBP. Nat Struct Mol Biol 15: 738–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo LI, Altmeyer M, Rask M‐B, Lukas C, Larsen Dorthe H, Povlsen Lou K, Bekker‐Jensen S, Mailand N, Bartek J, Lukas J (2013) ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155: 1088–1103 [DOI] [PubMed] [Google Scholar]
- Toledo L, Neelsen KJ, Lukas J (2017) Replication catastrophe: when a checkpoint fails because of exhaustion. Mol Cell 66: 735–749 [DOI] [PubMed] [Google Scholar]
- Van Dyck E, Foury F, Stillman B, Brill SJ (1992) A single‐stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J 11: 3421–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ (1996) SGS1, a homologue of the Bloom's and Werner's syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae . Genetics 144: 935–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wobbe CR, Weissbach L, Borowiec JA, Dean FB, Murakami Y, Bullock P, Hurwitz J (1987) Replication of simian virus 40 origin‐containing DNA in vitro with purified proteins. Proc Natl Acad Sci USA 84: 1834–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS, Kelly T (1988) Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc Natl Acad Sci USA 85: 2523–2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold MS (1997) Replication Protein A: a heterotrimeric, single‐stranded DNA binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem 66: 61–92 [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang X, Feng J, Leng H, Li S, Xiao J, Liu S, Xu Z, Xu J, Li D, Wang Z, Wang J, Li Q (2016) The histone chaperone FACT contributes to DNA replication‐coupled nucleosome assembly. Cell Rep 14: 1128–1141 [DOI] [PubMed] [Google Scholar]
- Yeeles JTP, Deegan TD, Janska A, Early A, Diffley JFX (2015) Regulated eukaryotic DNA replication origin firing with purified proteins. Nature 519: 431–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C, Gan H, Han J, Zhou Z‐X, Jia S, Chabes A, Farrugia G, Ordog T, Zhang Z (2014) Strand‐specific analysis shows protein binding at replication forks and PCNA unloading from lagging strands when forks stall. Mol Cell 56: 551–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi IW, Rabut G, Poveda A, Scheel H, Malmström J, Ulrich H, Hofmann K, Pasero P, Peter M, Luke B (2008) Rtt101 and Mms1 in budding yeast form a CUL4DDB1‐like ubiquitin ligase that promotes replication through damaged DNA. EMBO Rep 9: 1034–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman MK, Cimprich KA (2013) Causes and consequences of replication stress. Nat Cell Biol 16: 2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Gan H, Wang Z, Lee J‐H, Zhou H, Ordog T, Wold MS, Ljungman M, Zhang Z (2017) RPA interacts with HIRA and regulates H3.3 deposition at gene regulatory elements in mammalian cells. Mol Cell 65: 272–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Elledge SJ (2003) Sensing DNA damage through ATRIP recognition of RPA‐ssDNA complexes. Science 300: 1542–1548 [DOI] [PubMed] [Google Scholar]
- Zunder RM, Antczak AJ, Berger JM, Rine J (2012) Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing. Proc Natl Acad Sci USA 109: E144–E153 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Table EV1
Movie EV1
Movie EV2
Review Process File
Source Data for Figure 1
Source Data for Figure 4
Source Data for Figure 6


